Abstract
Differences in pathogenesis and the probability of becoming a chronic carrier depend on the age at which hepatitis B virus (HBV) infection is acquired, ranging from 82% in infants less than 6 months of age to 15 to 30% in older children. HBV genotypes from 22 pediatric patients from two areas that differ in prevalence have been determined. Phylogenetic analysis shows a clear difference between the genotype distribution in Buenos Aires, a low-prevalence area, and that found in Gualeguay, Entre Ríos, a high-prevalence area. While the analysis allocated the sequences in the Buenos Aires group to genotypes A (36%), D (9%), and F (55%), the Gualeguay group presented exclusively genotype A isolates with very low nucleotide divergence, which suggests a strong founder viral population. The high prevalence of genotype F in the Buenos Aires group and its high intragroup heterogeneity agree with the American origin of this genotype.
Hepatitis B virus (HBV) is an etiologic agent of acute and chronic liver disease distributed throughout the world. Approximately 5% of the world population is infected with HBV, and chronically infected patients with active liver disease run a serious risk of developing cirrhosis and hepatocellular carcinoma.
Differences in pathogenesis and the probability of becoming a chronic carrier depend on age. The percentage of individuals who become chronically infected with HBV is related to the age at which the infection is acquired. It ranges from 82% in infants less than 6 months of age (5) to 15 to 30% in older children (3, 5). Therefore, a large reservoir of infection is maintained predominantly by the transmission of HBV to infants.
The serologic and genetic classification of that virus has been established. A genetic classification based on the comparison of complete HBV genomes has defined six genotypes, named A to F (11, 12). It has also been shown that the pattern of clustering obtained with S-gene sequences agrees with the classification based on complete genomes (2, 10, 13). Therefore, accumulated S-gene sequences may be used to assess the worldwide molecular epidemiology of HBV.
In the present study, HBV isolates from children chronically infected were analyzed to determine the molecular epidemiology of HBV in the pediatric population.
Serum samples were collected from children undergoing treatment at the Hospital de Niños Ricardo Gutiérrez, which deals with most of the nation’s pediatric HBV-infected patients. Two groups of patients were studied, and all of them met the following eligibility criteria: positivity for hepatitis B surface antigen, chronic hepatitis indicated by liver biopsy, and no relation to each other. Serum samples of 11 children (average age, 11.4 years) were collected from patients living in Buenos Aires, which has been described as a low-prevalence area. Eleven other serum samples were obtained from children (average age, 12.9 years) living in Gualeguay, a country town of 36,000 inhabitants that is 150 miles from Buenos Aires and is suspected to be a high-prevalence area.
For HBV DNA amplification, a nested PCR was performed to amplify part of the surface gene. Briefly, DNA extracted from 200 μl of serum by the phenol-chloroform method was amplified with primers 163 (5′ CCC AAT ACC ACA TCA TCCA 3′, positions 758 to 740) and 376 (5′ CTC ATC TTC TTC TTG GTT CTT CTG GA 3′, positions 425 to 450) (8) for the first round of amplification. In the second round, primers HBS1 (5′ CAA GGT ATG TTG CCC GTT TG 3′, positions 455 to 474) and HBS2 (5′ AAA GCC CTG CGA ACC ACT GA 3′, positions 713 to 694) were used (15). Sequencing of the PCR product was done with the Femtomol sequencing kit (Promega Corp.) and primers HBS1 and HBS2.
A phylogenetic analysis was carried out by using those sequences. DNA alignments were generated with the Clustal W program. Phylogenetic trees were constructed with PHYLIP, version 3.5c (6). Bootstrap analysis was done by using the programs SEQBOOT (to generate 100 reshuffled sequences), DNA-DIST, KITSCH, and CONSENSE.
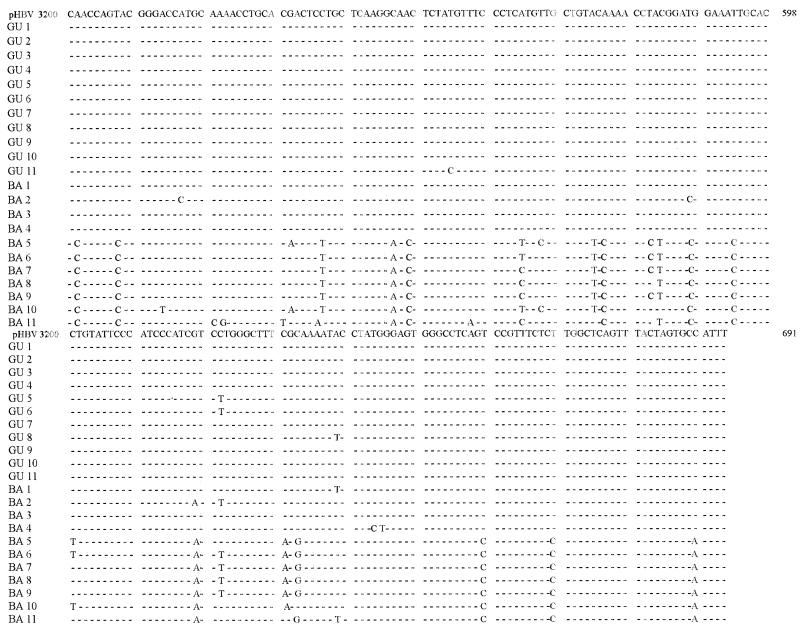
The partial S-gene nucleotide sequences (nucleotides 498 to 691) of the HBV genomes from the 22 children are given in Fig. 1.
FIG. 1.
Nucleotide sequences of the 22 pediatric HBV isolates aligned with that of clone pHBV-3200 (14). The nucleotide sequences correspond to nucleotides 498 to 691 from the EcoRI site. The designations used to indicate geographic origin are as follows: GU, Gualeguay; BA, Buenos Aires.
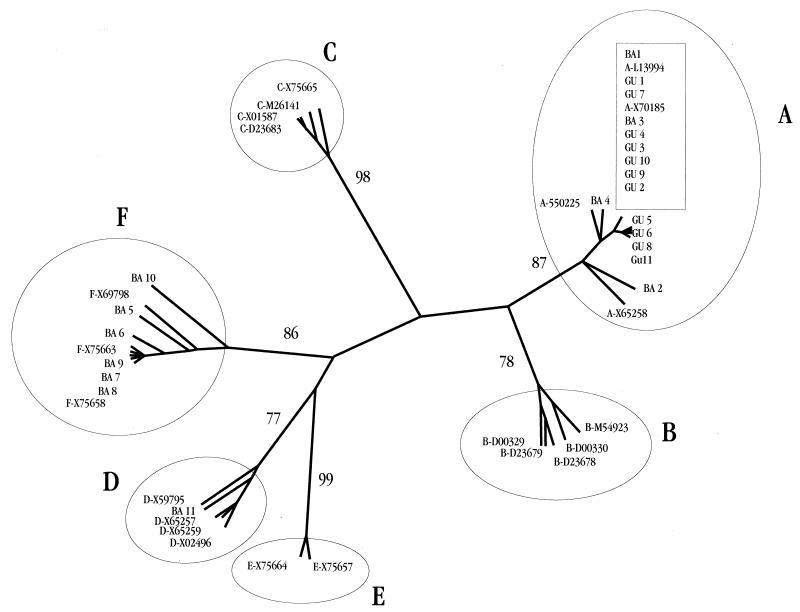
The phylogenetic analysis showed a clear difference between the genotype distribution found in the Buenos Aires group, a low-prevalence area, and that found in the Gualeguay group, a high-prevalence area. While the sequences of the Buenos Aires group were allocated to genotypes A (36%), D (9%), and F (55%), those of the Gualeguay group belonged exclusively to genotype A isolates (Fig. 2).
FIG. 2.
Unrooted tree based on 44 partial S-gene sequences: 22 from the present study and 22 from GenBank. The DNA-DIST and KITSCH programs were used. The bootstrap value for each genotype group is indicated. The different genotypes of GenBank sequences are designated by the letters A to F in front of the accession numbers.
When series of paired sequences of Gualeguay samples were compared, they differed by only two nucleotides, at most (average nucleotide divergence of 0.36%). In contrast, genotype A genomes from Buenos Aires presented an average intragroup heterogeneity of 1.84%. The average divergence values of both groups were significantly different (P < 0.0001, t test).
The presence of only genotype A, the high similarity among all of the S-gene sequences, and the absence of genotype F suggest the existence of a strong founder viral population in Gualeguay.
Serologic data indicate that one or more family members (including five mothers) were HBV infected in 82% of the cases. Although we cannot confirm vertical transmission because of the absence of prenatal serologic data on the mothers, that route cannot be ruled out. None of the 11 children had received a blood transfusion or blood derivatives, and they had not undergone any surgery. Therefore, household or even vertical transmission is associated with 82% of those cases. The route of transmission for the others is not clear. That region is now suspected of having a greater prevalence of HBV infection, and a further epidemiological study is in progress.
The distribution of HBV genotypes from a pediatric population in Buenos Aires agrees with previous results from adult patients. In a previous report, we showed the occurrence of genotypes A, D, B, and mainly F in chronically infected adults from Buenos Aires, Argentina (13).
The presence of genotypes A and D reflects the immigrant origins of the Buenos Aires population. Buenos Aires is a cosmopolitan city which has received immigration from the Mediterranean area, where genotype D predominates. Thus, we should expect genotype D rather than A to be prevalent. Magnius and Norder (9) suggested that genotype D may have replaced genotype A in the Mediterranean area, including North Africa. If this replacement occurred after the spread of HBV strains to America, it would explain the important presence of genotype A in Argentina. Therefore, genotype D might have been introduced into Argentina by the more recent immigration from the Mediterranean area.
We have also shown that even in the important virus reservoir of the chronically infected infants from Buenos Aires, genotype F predominates, with a high average nucleotide divergence value of 2.29% (range, 0 to 4.81%).
The genetic relationship of our genotype F strains to the other American published sequences belonging to that genotype was studied. Twenty-two isolates from Central America, five from Argentina, one from Colombia, and one from Brazil were included in the phylogenetic analysis with partial S-gene sequence information (1, 13). The same three different clusters formerly described by Arauz-Ruiz et al. on the basis of the complete S-gene sequences (1) were found, even when two different programs (KITSCH and UPGMA) were used to construct the phylogenetic tree (Fig. 3). While almost all Central American strains belong to cluster I, most of the Argentinian samples (pediatric as well as adult samples) are included in cluster II, together with the Brazilian and Colombian strains. The average intergroup nucleotide divergence between the two clusters (2.72%) is an extremely significantly different (P < 0.0001, t test) from the average intragroup divergences (0.86 and 1.44% for clusters I and II, respectively). These results suggest the existence of those two subgroups. None of the Argentinian isolates belong to the third cluster, which is the most divergent subgroup, F. Consequently, Central and South American isolates seem to fit into different clusters, which indicate different replication and survival of each genetic subgroup in those geographic areas.
FIG. 3.
Cladogram based on 36 partial S-gene sequences which belong to genotype F: 11 from Argentina, 22 from Central America, 1 from Colombia, 1 from Brazil, and 1 from France. The other three sequences, from a woodchuck hepatitis virus strain (WHV), were chosen as the outgroup. Two different methods (KITSCH and UPGMA) were used independently on a distance matrix calculated by DNA-DIST and yielded the same pattern of clustering in both cases. The designations used to indicate geographic origin are as follows: CA, Central America; SA, South America; Arg, Argentina; Br, Brazil, Co, Colombia.
Moreover, the coexistence of highly divergent genomes suggests a long natural history for genotype F in its host, as has been indicated for other viruses, such as simian immunodeficiency virus and human T-cell leukemia/lymphoma virus type I (4, 7). Like the phylogenetic analysis performed by Norder et al. (11), the above results imply an ancient origin of genotype F.
Finally, we observe that a highly populated and cosmopolitan city like Buenos Aires, has a heterogeneous distribution of HBV genotypes, where American genotype F prevails. In contrast, the combination of a low human population density with more emigration than immigration and a strong founder viral population produces the high prevalence of a unique HBV genotype. In Gualeguay, it is genotype A.
These results suggest that similar characteristics in the environment of another small city in Argentina could bring about the same epidemiological predicament, i.e., the existence of a unique HBV population which belongs to genotype A or F or another genotype.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the sequences reported here are AFO 43559 to AFO 43580.
Acknowledgments
This work has been supported by the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and the Secretaría de Ciencia y Técnica de la Universidad de Buenos Aires (FA132).
REFERENCES
- 1.Arauz-Ruiz P, Norder H, Visoná K A, Magnius L O. Molecular epidemiology of hepatitis B virus in Central America reflected in the genetic variability of the small S gene. J Infect Dis. 1997;176:851–858. doi: 10.1086/516507. [DOI] [PubMed] [Google Scholar]
- 2.Arauz-Ruiz P, Norder H, Visoná K A, Magnius L O. Genotype F prevails in HBV infected patients of Hispanic origin in Central America and may carry the precore stop mutants. J Med Virol. 1997;51:305–312. [PubMed] [Google Scholar]
- 3.Beasly R P, Hwang L Y, Linn C C, Leu M L, Stevens C E, Szmuness W, Chen K P. Incidence of hepatitis B infections in preschool children in Taiwan. J Infect Dis. 1982;146:198–204. doi: 10.1093/infdis/146.2.198. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, Telfer P, Gettie A, Reed P, Zhang L, Ho D, Marx P A. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J Virol. 1997;70:3617–3627. doi: 10.1128/jvi.70.6.3617-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coursaget P, Yvonnet B, Chotard J, Vincelot P, Sarr M, Diouf C, Chiron J P, Diop-Mar I. Age and sex related study of hepatitis B virus chronic carrier state in infants from an endemic area (Senegal) J Med Virol. 1987;22:1–5. doi: 10.1002/jmv.1890220102. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein J. PHYLIP: phylogeny inference package (version 3.5c). Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 7.Gessain A, Gallo R C, Franchini G. Low degree of human T-cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J Virol. 1992;66:2288–2295. doi: 10.1128/jvi.66.4.2288-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkins A E, Gilson R J C, Gibert N, Wreghitt J J G, Ahlers-de Boer Y, Tedder R S, Alexander G J M. Hepatitis B surface mutations associated with infection after liver transplantation. J Hepatol. 1996;24:8–14. doi: 10.1016/s0168-8278(96)80179-6. [DOI] [PubMed] [Google Scholar]
- 9.Magnius L O, Norder H. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology. 1995;38:24–34. doi: 10.1159/000150411. [DOI] [PubMed] [Google Scholar]
- 10.Norder H, Hammas B, Lofdahl S, Couroucé A M, Magnius L O. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J Gen Virol. 1992;73:1201–1208. doi: 10.1099/0022-1317-73-5-1201. [DOI] [PubMed] [Google Scholar]
- 11.Norder H, Couroucé A M, Magnius L O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo R I, Imai M, Miyakawa Y, Mayuml M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 13.Telenta P F S, Palacios Poggio G, López J L, González J, Lemberg A, Campos R H. Increased prevalence of genotype F hepatitis B virus isolates in Buenos Aires, Argentina. J Clin Microbiol. 1997;35:1873–1875. doi: 10.1128/jcm.35.7.1873-1875.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong S, Li Y, Vikvitski L, Trepo C. Active hepatitis B replication in the presence of anti-HBe is associated with viral variants containing an inactive pre-C region. Virology. 1990;176:596–603. doi: 10.1016/0042-6822(90)90030-u. [DOI] [PubMed] [Google Scholar]
- 15.Yokosuka O, Omata M, Hosoda K, Tada M, Ehata T, Ohto M. Detection and direct sequencing of hepatitis B virus genome by DNA amplification method. Gastroenterology. 1991;100:175–181. doi: 10.1016/0016-5085(91)90598-f. [DOI] [PubMed] [Google Scholar]