Abstract
Chronic inflammatory periodontal disease develops in part from the infiltration of a large number of classically activated inflammatory macrophages that release inflammatory cytokines important for disease progression, including inflammasome-dependent interleukin (IL)-1β. Streptococcus gordonii is a normally commensal oral microorganism; while not causative, recent evidence indicates that commensal oral microbes are required for the full development of periodontal disease. We have recently reported that inflammatory macrophages counterintuitively allow for the increased survival of phagocytosed S. gordonii over nonactivated or alternatively activated macrophages. This survival is dependent on increased reactive oxygen species production within the phagosome of the inflammatory macrophages, and resistance by the bacterium and can result in S. gordonii damaging the phagolysosomes. Here, we show that activated macrophages infected with live S. gordonii release more IL-1β than non-activated macrophages infected with either live or dead S. gordonii, and that the survival of oral Streptococci are more dependent on macrophage activation than other Gram positive microbes, both classical pathogens and commensals. We also find that S. gordonii–dependent inflammatory macrophage inflammasome activation requires the cytoplasmic NLRP6. Overall, our results suggest S. gordonii is capable of evading immune destruction, increasing inflammatory mediators, and increasing inflammatory macrophage response, and that this ability is increased under conditions of inflammation. This work reveals additional mechanisms by which normally commensal oral streptococci-macrophage interactions can change, resulting in increased release of mature IL-1β, potentially contributing to an environment that perpetuates inflammation.
Keywords: commensal, inflammasome, macrophages, streptococcus gordonii
Inflammatory activated macrophages interactions with live S. gordonii result in enhanced production of the periodontal disease relevant cytokine IL-1β, via mechanisms involving an NLRP6 inflammasome.
1. Introduction
Macrophages are an important cell type in the oral cavity. They function in the maintenance of mucosal immunity, including tolerance and tissue homeostasis, by physically interacting with, and responding to, oral microbes.1–4 During the development and progression of periodontal disease from gingivitis to periodontitis, the number of inflammatory or classically activated (M1) macrophages in the oral cavity increases, along with the inflammatory immune response that they promote.5–8 These inflammatory macrophages contribute to periodontal disease by further promoting inflammation and alveolar bone resorption.6,7,9–11 The importance of inflammatory macrophages in disease progression has been demonstrated through studies in which depletion of macrophages reduces alveolar bone resorption by modulating the host immune response,9 and recruitment of unactivated or alternatively activated (M0 or M2) macrophages by CCL2 reduces alveolar bone loss in mouse models of periodontitis.12
As part of the inflammatory response, macrophages produce cytokines and chemokines to orchestrate the immune response.13 One such cytokine, interleukin (IL)-1β, is produced in an inflammasome-dependent manner and acts to promote inflammation, stimulate fever, and recruit and activate other immune cells.13,14 Increased release of IL-1β along with enhanced inflammasome component expression occurs in gingivitis and chronic periodontitis7,10 and has emerged as a possible therapeutic target in periodontal disease as well as in other chronic inflammatory diseases.11,15,16
Streptococci are part of the normal oral flora; however, they are also able to colonize extraoral sites and contribute to systemic disease.17–19 Classically, periodontal disease was thought to be driven by specific pathogenic bacteria, mainly those classified as red complex bacteria.20 However, recent evidence indicates periodontal disease is not caused by a single organism, but rather is due to the development of a dysbiotic, or imbalanced from healthy, community of microorganisms that includes some normally commensal organisms along with a minority of a keystone pathogen, such as Porphyromonas gingivalis, capable of driving this dysbiosis.21–24Streptococcus gordonii, an oft-studied model oral streptococcus, can promote periodontal disease by working in conjunction with P. gingivalis, thus acting as an accessory pathogen to enhance the pathogenicity of P. gingivalis.25–27 In addition, S. gordonii can penetrate dental tubules, and is found in patients with apical periodontitis, allowing for P. gingivalis migration.28,29 While such intramicrobiome interactions are becoming better understood, there remains a gap in the mechanistic understanding of how dysbiosis of the oral microbiome alters oral microbial-immune cell interactions, including how normally commensal members such as S. gordonii may contribute to disease.
We have previously shown that S. gordonii is better able to survive within, and damage the phagosomes of, inflammatory-activated macrophages over nonactivated macrophages in a reactive oxygen species–dependent manner.30 Here, we show that live S. gordonii taken up by inflammatory activated macrophages results in increased production of the inflammasome-dependent and periodontal disease–relevant cytokine IL-1β, and that this is mechanistically dependent on the NLRP6 inflammasome.
2. Materials and methods
2.1. Cell culture
RAW264.7 macrophages (ATCC) were grown in RPMI medium (Lonza or Corning) supplemented with 10% fetal bovine serum (FBS) (Corning) and 2 mM L-glutamine (Corning) at 37 °C in 5% CO2. Prior to bacterial killing assays macrophages were stimulated with 20 ng/mL recombinant mouse interferon γ (IFNγ) (GenScript) for 24 h and with 0.1 µg/mL lipopolysaccharide (LPS) (Salmonella enterica serotype Minnesota strain Re595; MilliporeSigma) for 2 h. Prior to cytokine analysis experiments macrophages were stimulated with 20 ng/mL IFNγ only.
Human monocyte-derived macrophages were isolated from blood obtained from healthy donors in accordance with our institutional review board–approved protocol (ID: MODCR00005631) as previously described.30 Briefly, peripheral blood mononuclear cells (PBMCs) were isolated by isosmotic density gradient centrifugation using 1-Step polymorphs (Accurate Chemical). The PBMCs were plated on glass coverslips, allowing only PBMCs to adhere in RPMI medium supplemented with 10% FBS, 2 mM L-glutamine, and 100 U/mL penicillin and 100 μg/mL streptomycin (Corning) at 37 °C and 5% CO2. The adherent monocytes were differentiated for 5 to 7 d with 50 ng/mL recombinant human granulocyte macrophage colony-stimulating factor (GenScript). Differentiated macrophages were activated with 20 ng/mL recombinant human IFNγ (BioLegend) for 48 h.
Tibias and femurs were isolated from C57BL/J6 wild-type (WT) (purchased from the Jackson Laboratory) and NLRP6 KO mice (provided by Dr. Gabriel Núñez from University of Michigan Medical School).31 Progenitor cells were seeded at 1 × 106 cells/mL in RPMI medium supplemented with 10% FBS, 2 mM L-glutamine, and 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C and 5% CO2. Adherent cells were differentiated with 50 ng/mL recombinant mouse granulocyte macrophage colony-stimulating factor (GenScript) for 5 d and activated with 20 ng/mL IFNγ for 24 h.
WT, NLRP6 knockout, and NLRP3 knockout immortalized mouse bone marrow–derived macrophages (iBMDMs) were generously provided by the lab of Dr. Gabriel Núñez from University of Michigan Medical School. Cells were grown in RMPI medium supplemented with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate (Fisher Scientific), and 50 µg/mL gentamycin (Amresco) but maintained without antibiotics after initial passages. Cells were stimulated with 20 ng/mL mouse rIFNγ (GenScript) for 24 h before experiments to differentiate to an M1-like macrophage or left unstimulated.32,33
Human monocytic THP-1 cells (ATCC), THP-1 ASC-GFP cells (provided by Dr. Emad Alnemri of Thomas Jefferson University), and caspase-4 knockout THP-1 (Invivogen) were maintained in RPMI medium supplemented with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM HEPES (Fisher Scientific), 1.5 g/L sodium bicarbonate, and 0.05 mM 2-mercaptoethanol. Two days prior to an experiment, cells were differentiated to macrophages with 100 nM PMA (Cayman Chemicals) for 24 h then allowed to rest in media without PMA for an additional 24 h.34 Cells were stimulated with 20 ng/mL human IFNγ for 24 h to activate toward an M1-like macrophage or left unstimulated.35 For studies looking specifically at IL-1β production of inflammasome activation, LPS was not used to stimulate cells, as LPS alone can activate inflammasome pathways.36,37
2.2. Bacterial culture
S. gordonii strains DL1, SK12, SK9, and 38 were grown in brain heart infusion (BHI) medium (BD Biosciences) supplemented with 0.5% yeast extract (MP Biomedicals) at 37 °C and 5% CO2. All experiments used mid-log phase bacteria cultures and multiplicity of infection (MOI) was calculated by counting using Petroff-Hausser Counter or based on OD600. For experiments using heat-killed bacteria, S. gordonii was grown to mid-log phase then incubated at 80 °C for 60 min prior to use. Throughout the article, S. gordonii refers to S. gordonii strain DL1, unless otherwise indicated.
Staphylococcus epidermidis clinical isolate 94309-0594 and S. aureus strain UAMS-1 (provided by Dr. Anthony Campagnari of the University at Buffalo). Bacteria were grown in BHI medium at 37 °C. S. mutans 25175 (provided by Dr. Stefan Ruhl of the University at Buffalo) was grown in BHI medium at 37 °C. For all bacterial killing assay experiments, mid-log cultures were used.
2.3. Bacteria killing assay
To determine bacterial survival within macrophages, a modified gentamycin resistance assay was used.30,38,39 Briefly, macrophages were seeded in duplicate on 12-well plates and stimulated overnight with 20 ng/mL IFNγ (human [BioLegend] or mouse [GenScript] as required) and with 0.1 µg/mL LPS (S. minnesota Re 595) for 2 h. Mid-log S. gordonii was sonicated to break up chains and added to macrophages at an MOI of 10:1. Plates were centrifuged to synchronize contact of bacteria with macrophages (125 g for 1 min). Cells were incubated at 37 °C for 30 min, then one set of wells was washed extensively with PBS to remove external bacteria, and macrophages were lysed with sterile H2O and were serial diluted and plated on BHI or Todd-Hewitt plates to determine the initial number of bacteria taken up by the macrophages. For the other set of wells, 150 µg/mL gentamycin was added and incubated for 30 min at 37 °C, after which the media was replace with fresh RPMI and incubated for an additional 1.5 h. Again, macrophages were lysed with sterile H2O and serial diluted and plated on bacterial media plates and incubated at 37 °C and 5% CO2 overnight. After incubation, the number of colony forming units (CFU) taken up and CFU survival 2 h postphagocytosis are determined. The ratio of surviving (2.5 h) bacteria to phagocytosed (0.5 h) bacteria gave us percent survival of bacteria within macrophages.
2.4. Cytokine analysis
Macrophages were seeded on 12 or 24-well plates at 5 × 105 cells/mL. Mid-log S. gordonii were added to macrophages at an MOI = 10:1. Plates were centrifuged to synchronize contact of bacteria with macrophages (125 g for 1 min) and incubated at 37 °C for 6, 12, or 24 h. For experiments with small molecule inhibitors, 20 µM of each inhibitor was added 30 min before addition of bacteria: Ac-LEVD-CHO (caspase-4 inhibitor; Cayman Chemicals), Ac-YVAD-CHO (caspase-1 inhibitor; Cayman Chemicals), and MCC950 (NLRP3 inhibitor; Cayman Chemicals). After incubation, cell supernatants were collected and spun down to remove cell debris and bacteria. Levels of human TNFα and IL-1β were measured by enzyme-linked immunosorbent assay (R&D Systems) according to the manufacturer's instructions. Concentrations (pg/mL) are normalized to amount per 1 × 105 cells.
2.5. ASC speckle
To quantify inflammasome activation in human macrophages we analyzed ASC speckle formation as described.36 Briefly, THP-1 ASC-GFP cells were seeded at 5 × 105 cells/mL on 12-well plates with 18-mm cover glass and differentiated to macrophages and stimulated as described previously . Prior to adding bacteria, 20 µM Z-VAD-FMK (AdooQ Bioscience) was added to prevent cell death, then bacteria were added at an MOI = 100:1. After 1 h, any external bacteria were killed by incubation with 150 µg/mL gentamycin for 30 min, after which the media was replaced to fresh RPMI. Bacteria were incubated for a total of 6 or 12 h, then coverslips were fixed in 4% PFA, permeabilized with 0.1% Triton-X100, and counterstained with 0.5 µg/mL DAPI. For a positive control of ASC-associated inflammasome activation, cells were stimulated with 1 μg/mL E. coli LPS for 4 h then incubated with 5 μM nigericin (Cayman Chemicals) for 30 min. Cells were imaged using a 20 × objective on a Nikon Eclipse TE2000-u instrument equipped with a Spot RT740 Camera with at least 1,000 cells imaged per coverslip for at least 3 independent experiments. Percent speckle-positive was calculated using thresholding and analyzing particles on FIJI,40 in which the ratio of cell number (as determined by DAPI) and speckle number (as determined by high intensity small green fluorescent protein [GFP] positive points) were calculated, giving percent cells with activated ASC positive inflammasomes. For imaging cells with bacteria, S. gordonii was labeled with 25 µg/mL Alexa Fluor 568 carboxylic acid, succinimidyl ester (Invitrogen) in 0.1 M sodium bicarbonate for 5 min.
2.6. Immunoblot
Macrophages were incubated with bacteria (as with cytokine analysis) at an MOI = 10:1 in 0.3% FBS-supplemented RPMI in 6-well plates. After 6 or 24 h incubation, protein lysates were collected by rinsing cells with PBS, then scraping and collecting using extraction buffer (50 mM Tris HCl (pH 7.4), 0.5 mM MgCl2, 150 mM NaCl, 0.2 mM EDTA, 1% Triton-X100, 1 × Pierce Protease Inhibitor [Thermo Fisher Scientific]). Protein isolates were concentrated by precipitating with ice-cold acetone then resuspending in extraction buffer. Protein concentrations were measured and normalized using Bio-Rad Bradford Assay. Antibodies used for detection of protein in the cell lysate and supernatant were human reactive α-IL-1β and α-caspase-1 (Cell Signaling Technology; #43811, proptosis sampler kit) or mouse reactive α-IL-1β and α-caspase-1 (Cell Signaling Technology; #20836, inflammasome sampler kit) and α-Actin (BD; #612656).
2.7. Caspase-1 fluorometric assay
The 1 × 106 macrophages were incubated with mid-log S. gordonii at an MOI = 10:1 for 6 h. Cleaved caspase-1 was detected with YVAD-AFC in cell lysates according to manufacturer instructions (Caspase-1/ICE Fluorometric Assay Kit; BioVision). Fold increase of caspase-1 activity was determined by comparing bacteria infected macrophages with uninfected macrophages.
2.8. Cell death assays
Propidium iodide (Biotium) acquisition was measured by flow cytometry. Lactate dehydrogenase (LDH) cytotoxicity was measured using a CyQUANT LDH Cytotoxicity Assay Kit (Invitrogen) according to the manufacturer’s directions. Percent maximum LDH release was compared with maximum LDH release by addition of lysis buffer to macrophages.
3. Results
3.1. Representative oral streptococcus S. gordonii consistently survives better within inflammatory macrophages
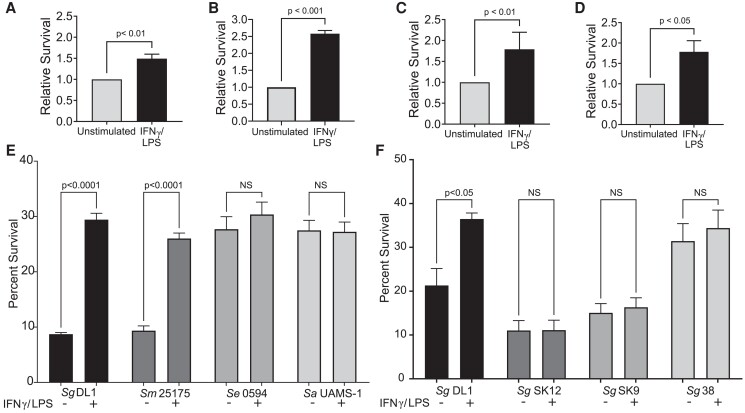
Inflammatory activated macrophages generally have increased microbicidal activity41,42; however, we have shown that S. gordonii has a higher survival rate within M1 activated macrophages over M2 macrophages.30 Further testing found that this increase in survival within activated macrophages was consistent regardless of the macrophage source (Fig. 1A–D); the survival within inflammatory macrophages relative to nonactivated macrophages increased about 2-fold, though the absolute survival rates varied between macrophage source. The intracellular survival rate was analyzed for a variety of traditionally commensal bacteria with pathogenic potential and was found to be unique to the tested viridans streptococci S. gordonii and S. mutans (Fig. 1E). While S. epidermidis and S. aureus had survival similar to S. gordonii DL1 within inflammatory activated macrophages, this survival did not depend on the macrophage activation state. We also tested survival of various S. gordonii strains with varying pathogenicity in an animal model of infective endocarditis. S. gordonii SK12 and SK9 were shown to be less pathogenic in the model, whereas S. gordonii DL1 and 38 were shown to have more pathogenic potential.43 Previously, our lab has shown that this pathogenicity is a good indicator of survivability in phagocytes in which S. gordonii DL1 survives better than SK12. Along these lines, we see that overall S. gordonii DL1 and 38 survive better within macrophages compared with SK12 and SK9 (Fig. 1F). However, the differential survival between activation state of the macrophage was only seen with S. gordonii DL1.
Fig. 1.
Oral viridans streptococci have a unique ability to survive within M1-activated macrophages over unstimulated macrophages. (A–D) Relative survival of S. gordonii DL1 within macrophages from different sources comparing unstimulated and IFNγ/LPS-activated macrophages: (A) RAW264.7 mouse macrophages, (B) WT iBMDMs, (C) THP-1 human monocytic cell line–differentiated macrophages, (D) human primary peripheral blood monocyte–derived macrophages. Shown are mean ± SEM of 3 or more independent experiments. P values calculated by Student's t test. (E) Survival of Gram-positive bacteria within RAW264.7 macrophages. Shown are mean ± SEM of 3 independent experiments. (F) Survival of various S. gordonii strains within RAW264.7 macrophages. S. gordonii SK12 and SK9 are strains known to not cause endocarditis in animal models,43 and SK12 has reduced reactive oxygen resistance30 compared with S. gordonii DL1. S. gordonii DL1 and 38 are known to be more pathogenic in animal models of endocarditis.43P values calculated by ordinary 1-way analysis of variance followed by Sidak's multiple comparisons test. Sa = Staphylococcus aureus; Se = Staphylococcus epidermidis; Sg = Streptococcus gordonii; Sm = Streptococcus mutans.
3.2. IL-1β release is increased in inflammatory activated macrophages infected with live S. gordonii
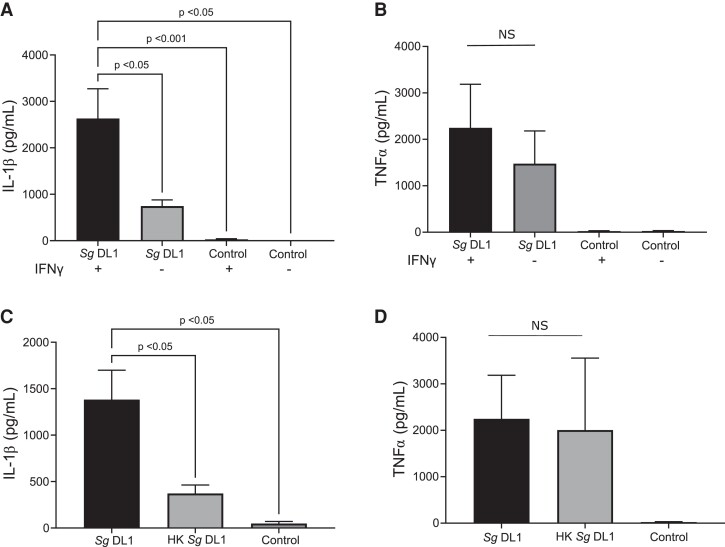
Inflammatory, or M1, macrophages release a myriad of proinflammatory cytokines to recruit other immune cells and elicit an immune response. We next examined 2 proinflammatory cytokines highly prevalent in periodontal disease, IL-1β and tumor necrosis factor α (TNFα),7,8,44,45 to determine if there were inducible changes to the proinflammatory response by human macrophages in response incubation with S. gordonii (Fig. 2). In THP-1–derived macrophages activated to an inflammatory state with IFNγ, IL-1β, but not TNFα, release was significantly increased over nonactivated macrophages when incubated with live S. gordonii (Fig. 2A, B). We also found that macrophages produced significantly less IL-1β when incubated with heat-killed bacteria (Fig. 2C), which are not able to damage the macrophage phagosome.30 This difference was not seen for TNFα release (Fig. 2D), suggesting that the combination of both inflammatory activation of macrophages and live S. gordonii was a prerequisite for maximal IL-1β production and release.
Fig. 2.
Live S. gordonii stimulates significantly increased IL-1β release from IFNγ-activated macrophages. Inflammatory cytokine release measured by enzyme-linked immunosorbent assay (ELISA) from PMA differentiated the THP-1 human monocytic cell line. Cytokines measured were IL-1β (A, C) and TNFα (B, D). (A, B) THP-1 cells were polarized with IFNγ or left unstimulated prior to incubation with S. gordonii DL1 for 6 h (B) or 24 h (A). (C, D) THP-1 cells were polarized with IFNγ and incubated with live or heat-killed (HK) S. gordonii DL1 for 6 h (D) or 24 h (C). In each case, control indicated the THP-1 cells alone. Shown are mean ± SEM of 4 or more independent experiments. P values were calculated by ordinary 1-way analysis of variance followed by Tukey's multiple comparisons test where appropriate. NS indicates that the analysis of variance was not significant with α = 0.05. Concentrations (pg/mL) are per 1 × 105 cells/mL. Sg = Streptococcus gordonii.
3.3. Live S. gordonii stimulated inflammasome activation in inflammatory activated macrophages
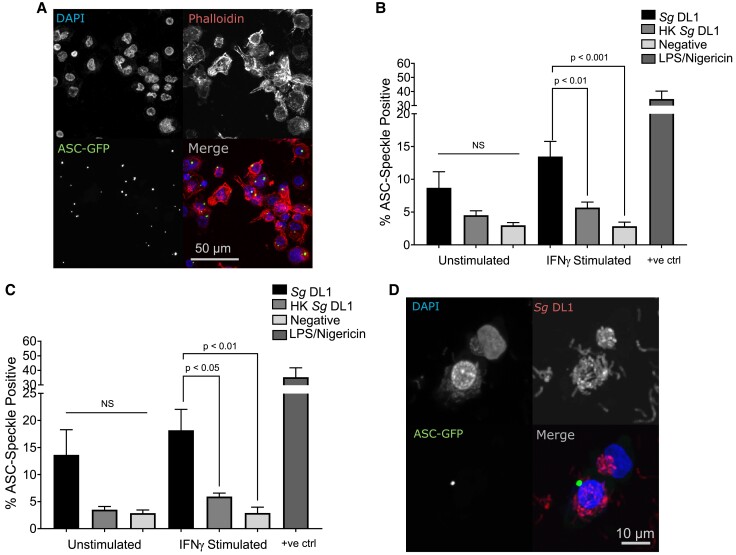
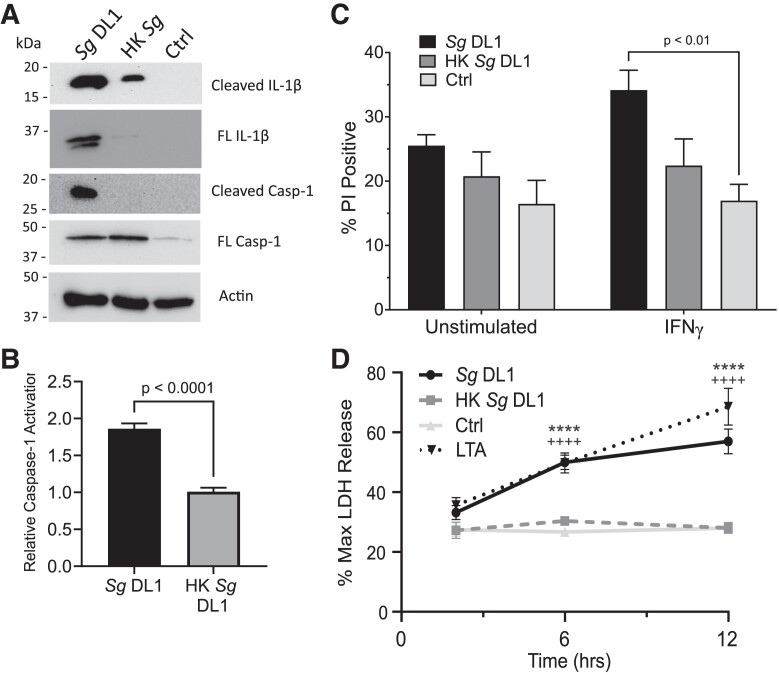
Given that IL-1β processing and release requires activation of the inflammasome complex and caspase-1,46,47 we next tested inflammasome, and ultimately caspase-1, activation in macrophages upon interaction with S. gordonii. Activation of the inflammasome complex itself often requires a cytosolic stimulus, and we have previously shown that S. gordonii is capable of damaging and potentially escaping the phagosome30; therefore, we hypothesized that this allows for activation of the cytoplasmic inflammasome. To examine inflammasome activation, we took advantage of THP-1 macrophages that express ASC, an important scaffold protein in most inflammasomes, conjugated to GFP. When the inflammasome is activated and the large multimeric protein complex is formed, ASC oligomerizes and the diffuse GFP forms a large single speck within the cell (Fig. 3A).36 Quantification of this oligomerization showed that IFNγ-stimulated human macrophages incubated with live S. gordonii had active inflammasomes in significantly more cells than both macrophages alone and macrophages incubated with heat-killed S. gordonii, after both 6-h (Fig. 3B) and 12-h (Fig. 3C) incubations. We confirmed that this inflammasome oligomerization does in fact occur concomitantly in macrophages associated with S. gordonii bacterium and that the THP-1 macrophages do indeed phagocytose the bacteria (Fig. 3D and Supplementary Fig. 1). In addition to inflammasome speck formation, we found increased activation (cleavage) of IL-1β and caspase-1 in the macrophage supernatant upon infection with live S. gordonii (Fig. 4A). Measurement of functional caspase-1 also indicated a significant increase in active caspase-1 when macrophages were infected with live vs heat-killed S. gordonii (Fig. 4B). Overall, these results show that live S. gordonii activates inflammasomes in proinflammatory human macrophages.
Fig. 3.
Live S. gordonii activates inflammasomes in IFNγ-stimulated macrophages. (A, B) Percent ASC speckle–positive cells after incubation (A: 6 h; B: 12 h) with live or heat-killed (HK) S. gordonii in PMA-differentiated THP-1 Asc-GFP cells polarized with IFNγ. Positive control: 4 h 1 µg/mL LPS + 30 min 20 µM nigericin. (C) Representative image of ASC speckle–positive cells. (D) Representative image of ASC speckle–positive cell containing S. gordonii. Shown are mean ± SEM of at least 4 independent experiments in which at least 1,000 cells per condition were analyzed. P values were calculated by ordinary 1-way analysis of variance followed by Tukey's multiple comparisons test. Sg = Streptococcus gordonii.
Fig. 4.
Live S. gordonii activates caspase-1 in macrophages. (A) Representative immunoblot showing caspase-1 and IL-1β cleavage. THP-1 cells were incubated with live or heat-killed (HK) S. gordonii DL1 for 24 h. Actin was used as a loading control. (B) Fold change in caspase-1 activation relative to THP-1 cells without bacteria added measured by YVAD-AFC fluorescence. (C) Propidium iodide (PI) acquisition by THP-1 cells after 24 h incubation. THP-1 cells were polarized with IFNγ or left unstimulated prior to incubation with live or HK S. gordonii. (D) LDH cytotoxicity after 2, 6, and 12 h incubation with live or HK S. gordonii with IFNγ-stimulated THP-1 cells. Transfected LTA was used as a positive control for pyroptosis. Percent release was determined compared with maximum LDH release by cells incubated with lysis buffer. Shown are mean ± SEM of 3 independent experiments. P value was calculated by unpaired t test (B) or ordinary 2-way analysis of variance followed by Dunnett's multiple comparisons in which each group was compared with control (+ indicates control vs LTA, * indicates control vs S. gordonii DL1) (C). Ctrl = control; Sg = Streptococcus gordonii.
3.4. Cell death upon inflammasome activation
Inflammasome-dependent caspase-1 activation leads to cleavage of gasdermin D, which can form pores in the cell membrane. Extended periods of gasdermin D pore formation can result in ion flux, membrane destabilization, and eventual cell death (pyroptosis).48–50 We measured cell cytotoxicity as an indicator of pyroptosis using propidium iodide influx and LDH release (Fig. 4C, D). There was a significant increase in propidium iodide–positive cells when IFNγ THP-1 cells were incubated with live S. gordonii over THP-1 cells alone (Fig. 4C). Additionally, there was a significant increase in LDH release from THP-1 cells incubated with live S. gordonii over HK S. gordonii or cells alone (Fig. 4D).
3.5. Mechanism of inflammasome activation in macrophages by S. gordonii
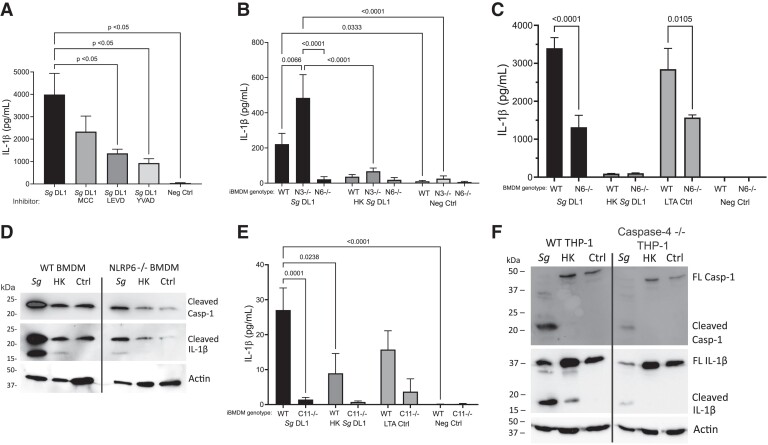
There are many ways that inflammasomes can be activated, including via sensing ionic or enzymatic changes within the cell cytoplasm as well as through direct interaction with and activation by bacterial components. For example, NLRP3 can be activated by cathepsin B release from the phagolysosome upon damage.51 Previous work from our lab has shown that S. gordonii can cause phagolysosomal damage,30 so we initially wanted to determine if NLRP3 was required for inflammasome-dependent IL-1β release. Looking at THP-1 macrophages treated with an NLRP3-specific small molecule inhibitor (MCC950) we did not see significant changes in IL-1β release when the activated macrophages were incubated with S. gordonii (Fig. 5A).
Fig. 5.
Determining inflammasome pathway activation by S. gordonii in macrophages. (A) IL-β release from PMA-differentiated, IFNγ-activated THP-1 macrophages with indicated inflammasome inhibitor added during incubation with S. gordonii for 6 h. MCC950 (MCC) indicates NLRP3 inhibitor, Ac-LEVD-CHO (LEVD) indicate scaspase-4/5 inhibitor, and Ac-YVAD-CHO (YVAD) indicates caspase-1 inhibitor. (B) IL-1β release as determined by enzyme-linked immunosorbent assay (ELISA) from IFNγ-stimulated immortalized BMDMs derived from WT, NLRP3 knockout, or NLRP6 knockout mice incubated with like or heat-killed (HK) S. gordonii for 24 h. (C) IL-1β release as determined by ELISA from BMDM derived from WT or NLRP6 knockout mice incubated with live or HK S. gordonii for 24 h. Transfected LTA (28 μg) was used as a positive control for NLRP6 activation.31 (D) Representative immunoblot showing caspase-1 and IL-1β cleavage. WT or NLRP6 knockout primary BMDMs were incubated with live or HK S. gordonii for 24 h. Actin was used as a loading control. (E) IL-1β release from caspase-11 knockout or WT iBMDMs incubated with live or HK S. gordonii for 24 h. (F) Representative immunoblot showing caspase-1 and IL-1β cleavage. WT or caspase-4 knockout THP-1 cells were incubated with live or HK S. gordonii for 6 h. Again, actin was used as a loading control. Shown are mean± SEM of 3 independent experiments. P values were calculated by 1-way analysis of variance followed by Dunnett's multiple comparisons (A, C, E) or 2-way analysis of variance followed by Sidak's multiple comparisons test (B). Ctrl = control; Sg = Streptococcus gordonii.
Recently, it has been found that the lesser described NLRP6 can be directly activated by Gram-positive bacteria cell wall component lipoteichoic acid (LTA) and that this NLRP6 inflammasome involves activity of both caspase-1 and caspase-4/5 (caspase-11 in mice).31 Therefore, we next examined if NLRP6 activation is the mechanism through which S. gordonii activates macrophage inflammasomes. When caspase-4 was inhibited in human THP-1 cells with a small molecule inhibitor (Ac-LEVD-CHO), we saw a significant decrease in IL-1β release (Fig. 5A). There was also a significant decrease in IL-1β production when caspase-1 was inhibited (Ac-YVAD-CHO), confirming that IL-1β release is dependent on caspase-1 activation. LTA and nigericin were used as controls for NLRP6 and NLRP3 inflammasome activation, respectively, confirming the effectiveness of the small molecule inhibitors (Supplementary Fig. 2A, B).
To verify the inhibitor results, we also tested NLRP3, NLRP6, and capase-11 knockout mouse macrophages and capase-4 knockout THP-1 cells. Consistent with the inhibitor results, NLRP3 knockout iBMDMs did not show a decrease in IL-1β production upon incubation with S. gordonii, while NLRP6 iBMDMs did (Fig. 5B). In BMDMs isolated directly from NLRP6 knockout mice, we also saw a significant decrease in IL-1β release as compared with WT BMDMs (Fig. 5C). Transfected LTA was used as a control for NLRP6 inflammasome activation, confirming the knockout. We also tested for activation of caspase-1 and IL-1β cleavage and found reductions of both in NLRP6 knockout BMDMs as compared with WT BMDMs (Fig. 5D).
Further confirmation of this phenotype was done through examining IL-1β release in mouse caspase-11 knockout iBMDMs, in which we saw a significant decrease in IL-1β production by caspase-11 knockout cells incubated with live S. gordonii compared with WT iBMDMs (Fig. 5E). We additionally looked at the human caspase-4 knockout THP-1 cells and saw a decrease in caspase-1 and IL-1β cleavage by Western blot in the knockout cells compared with WT THP-1 s (Fig. 5F). Together, these data indicate NLRP6 and caspase-4 (or mouse caspase-11) are important for S. gordonii–dependent active IL-1β production and caspase-1 cleavage.
4. Discussion
The multiple pathways of inflammasome activation exist for immune protection; however, many bacteria have mechanisms to either promote or inhibit inflammasome activation and pyroptosis for their benefit.52 For example, many of the bacteria associated with periodontal disease are inflammophilic, feeding off nutrients provided from the inflammatory immune response and tissue breakdown.53 The well-studied periodontal pathogen P. gingivalis is capable of inhibiting some aspects of macrophage inflammation such as iNOS-mediated killing and Toll-like receptor signaling while promoting destructive inflammation.54–57 Recent research has also come to appreciate traditionally commensal bacteria as important players in periodontal disease,23,24,58,59 including oral streptococci and the representative species S. gordonii.26,27,60,61 Here, we show that when macrophages interact with S. gordonii under conditions of existing inflammation, which allows the bacterium to increase its ability to evade immune destruction, the macrophages have increased NLRP6-dependent inflammasome activation and increase the release of the inflammatory cytokine IL-1β.
Previous research has described the involvement and importance of general inflammation, including the inflammasome and IL-1β, in periodontal disease.8,10,11,62–64 Inflammasomes are activated by recognition of pathogen-associated molecular patterns and damage-associated molecular patterns within the cytosol by cytosolic pattern recognition receptors.52 A multitude of mechanisms can activate a large variety of cytosolic pattern recognition receptors; the well-studied NLR NLRP3 can be activated by several stimuli, including sterile and pathogen associated molecules, as well as damage to phagosomes.49–51,65,66 We have previously shown that S. gordonii can damage phagolysosomes,30 so we initially hypothesized that S. gordonii may activate an NLRP3 inflammasome through this pathway. In fact, we found that the NLRP3 inflammasome is not the primary inflammasome pathway activated by S. gordonii but instead is dependent on NLRP6 and caspase-4 (mouse caspase-11) (Fig. 5). Interestingly, the NLRP3 knockout cells showed a significant increase in IL-1β release compared with WT that was not seen with the small molecule inhibitors. The reasoning for this discrepancy is unclear. The NLRP6 inflammasome has recently been described to sense LTA of Gram-positive bacteria,31 suggesting that bacterial products released from the damaged phagosome into the cytosol may be the primary activator in this case.
NLRP6 is important for activation of both IL-1β and IL-18 in macrophages and epithelial cells,31,67 but it can also be important for increased bacterial pathogenicity in infection.31,68,69 Conflicting information exists on the importance of NLRP6 in the exacerbation of colitis, another major disease associated with microbial dysbiosis, with recent research showing no change in gut microbiome and a gender-based susceptibility to induced colitis between WT and Nlrp6−/− mice.70,71 However, research is coming to appreciate that there may be an important role NLRP6 could play in the oral cavity as NLRP6 is increased, along with other inflammasome components, in periodontal disease.72 NLRP6-activated caspase-1 can lead to the release of IL-1β and IL-18 in gingival fibroblasts,72 though NLRP6 may also inhibit inflammatory responses in human periodontal ligament cells associated with apical periodontitis.73 NLRP6 is known to be activated by LTA from Gram-positive bacterial cell walls,31 and LTA from S. mutans, the major etiological agent of dental caries, can activate NLRP6 in human dental pulp cells.74 However, many of the bacteria traditionally associated with periodontal disease, including P. gingivalis, are Gram-negative inflammophilic bacteria.75–77 These traditional periodontal pathogens, however, generally make up a minority of the periodontal disease microbiome, with much of the disease-associated dysbiotic microbiome consisting of accessory pathogens, many of which are Gram positive.78,79 Our results suggest that upon streptococci interaction with inflammatory macrophages, an increase of IL-1β production is produced via NLRP6 that in turn could be beneficial to the periodontal-associated bacteria through feeding into the positive feedback loop of disease development.80 The results presented here thus suggest that, in addition to the role that S. gordonii has in promoting the attachment and growth of periodontal pathogens,81 it may have an active role in driving chronic inflammation in the oral environment once inflammation is initiated. Important future studies will need to examine the in vivo requirements of NLRP6 in the development of periodontal disease, as well as the potential changing interactions of oral streptococci with other immune cells known to be important in periodontal disease, including neutrophils.
Supplementary Material
Acknowledgments
The authors thank Drs. Núñez, Alnemri, Campagnari, and Ruhl for providing reagents, as noted in the Materials and Methods. The graphical abstract was created with BioRender.com. Microscopy was performed in the Optical Imaging and Analysis Facility (UB Dental Sciences).
Contributor Information
Sarah Metcalfe, Department of Oral Biology, School of Dental Medicine, University at Buffalo, 3435 Main street, Buffalo, NY 14214, United States.
Michelle Panasiewicz, Department of Oral Biology, School of Dental Medicine, University at Buffalo, 3435 Main street, Buffalo, NY 14214, United States.
Jason G Kay, Department of Oral Biology, School of Dental Medicine, University at Buffalo, 3435 Main street, Buffalo, NY 14214, United States.
Author contributions
J.G.K. and S.M conceptualized the study; S.M. and M.P. performed the experimental investigations; S.M, M.P., and J.G.K. analyzed the data; S.M. and J.G.K. wrote the original manuscript draft; all authors contributed to review, editing, and revision of the manuscript.
Supplementary material
Supplementary materials are available at Journal of Leukocyte Biology online.
Funding
This work was funded in part by National Institutes of Health grants R03DE025062 (J.G.K), R01DE028307 (J.G.K), F31DE029400 (S.M.), and T32DE02526.
References
- 1. Dixon DR, Reife RA, Cebra JJ, Darveau RP. Commensal bacteria influence innate status within gingival tissues: a pilot study. J Periodontol. 2004:75(11):1486–1492. 10.1902/jop.2004.75.11.1486 [DOI] [PubMed] [Google Scholar]
- 2. Moutsopoulos NM, Konkel JE. Tissue-Specific immunity at the oral mucosal barrier. Trends Immunol. 2018:39(4):276–287. 10.1016/j.it.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dutzan N, Konkel JE, Greenwell-Wild T, Moutsopoulos NM. Characterization of the human immune cell network at the gingival barrier. Mucosal Immunol. 2016:9(5):1163–1172. 10.1038/mi.2015.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Merry R, Belfield L, McArdle P, McLennan A, Crean S, Foey A. Oral health and pathology: a macrophage account. Br J Oral Maxillofac Surg. 2012:50(1):2–7. 10.1016/j.bjoms.2010.10.020 [DOI] [PubMed] [Google Scholar]
- 5. Gonzalez OA, Novak MJ, Kirakodu S, Stromberg A, Nagarajan R, Huang CB, Chen KC, Orraca L, Martinez-Gonzalez J, Ebersole JL. Differential gene expression profiles reflecting macrophage polarization in aging and periodontitis gingival tissues. Immunol Invest. 2015:44(7):643–664. 10.3109/08820139.2015.1070269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Topoll HH, Zwadlo G, Lnage DE, Sorg C. Phenotypic dynamics of macrophage subpopulations during human experimental gingivitis. J Periodontal Res. 1989:24(2):106–112. 10.1111/j.1600-0765.1989.tb00864.x [DOI] [PubMed] [Google Scholar]
- 7. Yang J, Zhu Y, Duan D, Wang P, Xin Y, Bai L, Liu Y, Xu Y. Enhanced activity of macrophage M1/M2 phenotypes in periodontitis. Arch Oral Biol. 2018:96:234–242. 10.1016/j.archoralbio.2017.03.006 [DOI] [PubMed] [Google Scholar]
- 8. Yu T, Zhao L, Huang X, Ma C, Wang Y, Zhang J, Xuan D. Enhanced activity of the macrophage M1/M2 phenotypes and phenotypic switch to M1 in periodontal infection. J Periodontol. 2016:87(9):1092–1102. 10.1902/jop.2016.160081 [DOI] [PubMed] [Google Scholar]
- 9. Lam RS, O'Brien-Simpson NM, Lenzo JC, Holden JA, Brammar GC, Walsh KA, McNaughtan JE, Rowler DK, Van Rooijen N, Reynolds EC. Macrophage depletion abates Porphyromonas gingivalis-induced alveolar bone resorption in mice. J Immunol. 2014:193(5):2349–2362. 10.4049/jimmunol.1400853 [DOI] [PubMed] [Google Scholar]
- 10. Kinane DF, Adonogianaki E, Moughal NA. Bioassay of interleukin 1 (IL-1) in human gingival crevicular fluid during experimental gingivitis. Archs Oral Biol. 1992:37(2):153–156. 10.1016/0003-9969(92)90011-V [DOI] [PubMed] [Google Scholar]
- 11. Hasturk H, Kantarci A, Goguet-Surmenian E, Blackwood A, Andry C, Serhan CN, Van Dyke TE. Resolvin E1 regulates inflammation at the cellular and tissue level and restores tissue homeostasis in vivo. J Immunol. 2007:179(10):7021–7029. 10.4049/jimmunol.179.10.7021 [DOI] [PubMed] [Google Scholar]
- 12. Zhuang Z, Yoshizawa-Smith S, Glowacki A, Maltos K, Pacheco C, Shehabeldin M, Mulkeen M, Myers N, Chong R, Verdelis K, et al. Induction of M2 macrophages prevents bone loss in murine periodontitis models. J Dent Res. 2018:98(2):200–208. 10.1177/0022034518805984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arango Duque G, Descoteaux A. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol. 2014:5(11):491. 10.3389/fimmu.2014.00491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delaleu N, Bickel M. Interleukin-1b and interleukin-18: regulation and activity in local inflammation. Periodontol 2000. 2004:35(1):42–52. 10.1111/j.0906-6713.2004.003569.x [DOI] [PubMed] [Google Scholar]
- 15. Van Dyke TE. The management of inflammation in periodontal disease. J Periodontol. 2008:79(8s):1601–1608. 10.1902/jop.2008.080173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011:117(14):3720–3732. 10.1182/blood-2010-07-273417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Han YW, Wang X. Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J Dent Res. 2013:92(6):485–491. 10.1177/0022034513487559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Douglas CW, Heath J, Hampton KK, Preston FE. Identity of viridans streptococci isolated from cases of infective endocarditis. J Med Microbiol. 1993:39(3):179–182. 10.1099/00222615-39-3-179 [DOI] [PubMed] [Google Scholar]
- 19. Yombi J, Belkhir L, Jonckheere S, Wilmes D, Cornu O, Vandercam B, Rodriguez-Villalobos H. Streptococcus gordonii septic arthritis: two cases and review of literature. BMC Infect Dis. 2012:12(1):215. 10.1186/1471-2334-12-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998:25(2):134–144. 10.1111/j.1600-051X.1998.tb02419.x [DOI] [PubMed] [Google Scholar]
- 21. Hajishengallis G, Lamont RJ. Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 2016:24(6):477–489. 10.1016/j.tim.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011:10(5):497–506. 10.1016/j.chom.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ebersole JL, Dawson D 3rd, Emecen-Huja P, Nagarajan R, Howard K, Grady ME, Thompson K, Peyyala R, Al-Attar A, Lethbridge K, et al. The periodontal war: microbes and immunity. Periodontol 2000. 2017:75(1):52–115. 10.1111/prd.12222 [DOI] [PubMed] [Google Scholar]
- 24. Irie K, Novince CM, Darveau RP. Impact of the oral commensal Flora on alveolar bone homeostasis. J Dent Res. 2014:93(8):801–806. 10.1177/0022034514540173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuboniwa M, Tribble GD, James CE, Kilic AO, Tao L, Herzberg MC, Shizukuishi S, Lamont RJ. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol Microbiol. 2006:60(1):121–139. 10.1111/j.1365-2958.2006.05099.x [DOI] [PubMed] [Google Scholar]
- 26. Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, Demuth DR. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology. 2002:148(6):1627–1636. 10.1099/00221287-148-6-1627 [DOI] [PubMed] [Google Scholar]
- 27. Daep CA, Novak EA, Lamont RJ, Demuth DR. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011:79(1):67–74. 10.1128/IAI.00361-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park OJ, Kim J, Kim HY, Kwon Y, Yun CH, Han SH. Streptococcus gordonii induces bone resorption by increasing osteoclast differentiation and reducing osteoblast differentiation. Microb Pathog. 2018:126:218–223. 10.1016/j.micpath.2018.11.005 [DOI] [PubMed] [Google Scholar]
- 29. Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011:7(3):e1002012. 10.1371/journal.ppat.1002012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Croft AJ, Metcalfe S, Honma K, Kay JG. Macrophage polarization alters postphagocytosis survivability of the commensal Streptococcus gordonii. Infect Immun. 2018:86(3):e00858-17. 10.1128/IAI.00858-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hara H, Seregin SS, Yang D, Fukase K, Chamaillard M, Alnemri ES, Inohara N, Chen GY, Nunez G. The NLRP6 inflammasome recognizes lipoteichoic acid and regulates gram-positive pathogen infection. Cell. 2018:175(6):1651–1664.e14. 10.1016/j.cell.2018.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ying W, Cheruku PS, Bazer FW, Safe SH, Zhou B. Investigation of macrophage polarization using bone marrow derived macrophages. J Vis Exp. 2013:(76):50323. 10.3791/50323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paulina Achita AB. Immortalized murine macrophage cell line as a model for macrophage polarization into classically activated M(IFNγ+LPS) or alternatively activated M(IL-4) macrophages. J Clin Cell Immunol. 2015:6(2):4172. 10.4172/2155-9899.1000318 [DOI] [Google Scholar]
- 34. Lund ME, To J, O'Brien BA, Donnelly S. The choice of phorbol 12-myristate 13-acetate differentiation protocol influences the response of THP-1 macrophages to a pro-inflammatory stimulus. J Immunol Methods. 2016:430:64–70. 10.1016/j.jim.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 35. Genin M, Clement F, Fattaccioli A, Raes M, Michiels C. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer. 2015:15(1):577. 10.1186/s12885-015-1546-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fernandes-Alnemri T, Wu J, Yu JW, Datta P, Miller B, Jankowski W, Rosenberg S, Zhang J, Alnemri ES. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007:14(9):1590–1604. 10.1038/sj.cdd.4402194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013:341(6151):1250–1253. 10.1126/science.1240988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vaudaux P, Waldvogel FA. Gentamicin antibacterial activity in the presence of human polymorphonuclear leukocytes. Antimicrob Agents Chemother. 1979:16(6):743–749. 10.1128/AAC.16.6.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Honma K, Mishima E, Inagaki S, Sharma A. The OxyR homologue in Tannerella forsythia regulates expression of oxidative stress responses and biofilm formation. Microbiology. 2009:155(6):1912–1922. 10.1099/mic.0.027920-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012:9(7):676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Atri C, Guerfali FZ, Laouini D. Role of human macrophage polarization in inflammation during infectious diseases. Int J Mol Sci. 2018:19(6):1801. 10.3390/ijms19061801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Price JV, Vance RE. The macrophage paradox. Immunity. 2014:41(5):685–693. 10.1016/j.immuni.2014.10.015 [DOI] [PubMed] [Google Scholar]
- 43. Young Lee S, Cisar JO, Bryant JL, Eckhaus MA, Sandberg AL. Resistance of Streptococcus gordonii to polymorphonuclear leukocyte killing is a potential virulence determinant of infective endocarditis. Infect Immun. 2006:74(6):3148–3155. 10.1128/IAI.00087-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gorska R, Gregorek H, Kowalski J, Laskus-Perendyk A, Syczewska M, Madalinski K. Relationship between clinical parameters and cytokine profiles in inflamed gingival tissue and serum samples from patients with chronic periodontitis. J Clin Periodontol. 2003:30(12):1046–1052. 10.1046/j.0303-6979.2003.00425.x [DOI] [PubMed] [Google Scholar]
- 45. Zhou LN, Bi CS, Gao LN, An Y, Chen F, Chen FM. Macrophage polarization in human gingival tissue in response to periodontal disease. Oral Dis. 2019:25(1):265–273. 10.1111/odi.12983 [DOI] [PubMed] [Google Scholar]
- 46. Dinarello CA. Interleukin-1 beta, interleukin-18, and the interleukin-1 beta converting enzyme. Ann N Y Acad Sci. 1998:856(1):1–11. 10.1111/j.1749-6632.1998.tb08307.x [DOI] [PubMed] [Google Scholar]
- 47. Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992:356(6372):768–774. 10.1038/356768a0 [DOI] [PubMed] [Google Scholar]
- 48. Ruhl S, Shkarina K, Demarco B, Heilig R, Santos JC, Broz P. ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science. 2018:362(6417):956–960. doi: 10.1126/science.aar7607 [DOI] [PubMed] [Google Scholar]
- 49. Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009:7(2):99–109. 10.1038/nrmicro2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vande Walle L, Lamkanfi M. Pyroptosis. Curr Biol. 2016:26(13):R568–R572. 10.1016/j.cub.2016.02.019 [DOI] [PubMed] [Google Scholar]
- 51. Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016:16(7):407–420. 10.1038/nri.2016.58 [DOI] [PubMed] [Google Scholar]
- 52. Ashida H, Mimuro H, Ogawa M, Kobayashi T, Sanada T, Kim M, Sasakawa C. Cell death and infection: a double-edged sword for host and pathogen survival. J Cell Biol. 2011:195(6):931–942. 10.1083/jcb.201108081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hajishengallis G. The inflammophilic character of the periodontitis-associated microbiota. Mol Oral Microbiol. 2014:29(6):248–257. 10.1111/omi.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Papadopoulos G, Weinberg EO, Massari P, Gibson FC, Wetzler LM, Morgan EF, Genco CA. Macrophage-Specific TLR2 signaling mediates pathogen-induced TNF-dependent inflammatory oral bone loss. J Immunol. 2013:190(3):1148–1157. 10.4049/jimmunol.1202511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Slocum C, Coats SR, Hua N, Kramer C, Papadopoulos G, Weinberg EO, Gudino CV, Hamilton JA, Darveau RP, Genco CA. Distinct lipid A moieties contribute to pathogen-induced site-specific vascular inflammation. PLoS Pathog. 2014:10(7):e1004215. 10.1371/journal.ppat.1004215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang M, Krauss JL, Domon H, Hosur KB, Liang S, Magotti P, Triantafilou M, Triantafilou K, Lambris JD, Hajishengallis G. Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal. 2010:3(109):ra11. 10.1126/scisignal.2000697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hajishengallis G, Lamont RJ. Polymicrobial communities in periodontal disease: their quasi-organismal nature and dialogue with the host. Periodontol 2000. 2021:86(1):210–230. 10.1111/prd.12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Irie K, Tomofuji T, Ekuni D, Morita M, Shimazaki Y, Darveau RP. Impact of oral commensal Bacteria on degradation of periodontal connective tissue in mice. J Periodontol. 2015:86(7):899–905. 10.1902/jop.2015.150006 [DOI] [PubMed] [Google Scholar]
- 59. Devine DA, Marsh PD, Meade J. Modulation of host responses by oral commensal bacteria. J Oral Microbiol. 2015:7(1):26941. 10.3402/jom.v7.26941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mahmoud MY, Steinbach-Rankins JM, Demuth DR. Functional assessment of peptide-modified PLGA nanoparticles against oral biofilms in a murine model of periodontitis. J Control Release. 2019:297:3–13. 10.1016/j.jconrel.2019.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hajishengallis G, Lambris JD. Microbial manipulation of receptor crosstalk in innate immunity. Nat Rev Immunol. 2011:11(3):187–200. 10.1038/nri2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Page RC. The role of inflammatory mediators in the pathogenesis of periodontal disease. J Periodontal Res. 1991:26(3):230–242. 10.1111/j.1600-0765.1991.tb01649.x [DOI] [PubMed] [Google Scholar]
- 63. Shibata K. Historical aspects of studies on roles of the inflammasome in the pathogenesis of periodontal diseases. Mol Oral Microbiol. 2018:33(3):203–211. 10.1111/omi.12217 [DOI] [PubMed] [Google Scholar]
- 64. Jandinski JJ, Stashenko P, Feder LS, Leung CC, Peros WJ, Rynar JE, Deasy MJ. Localization of interleukin-1 beta in human periodontal tissue. J Periodontol. 1991:62(1):36–43. 10.1902/jop.1991.62.1.36 [DOI] [PubMed] [Google Scholar]
- 65. Bortoluci KR, Medzhitov R. Control of infection by pyroptosis and autophagy: role of TLR and NLR. Cell Mol Life Sci. 2010:67(10):1643–1651. 10.1007/s00018-010-0335-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dinarello CA. The C3a receptor, caspase-1, and release of IL-1b. Blood. 2013:122(20):3394–3395. 10.1182/blood-2013-08-518282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Levy M, Thaiss A, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, et al. Microbiota-Modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015:163(6):1428–1443. 10.1016/j.cell.2015.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Anand PK, Malireddi RK, Lukens JR, Vogel P, Bertin J, Lamkanfi M, Kanneganti TD. NLRP6 Negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012:488(7411):389–393. 10.1038/nature11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ghimire L, Paudel S, Jin L, Baral P, Cai S, Jeyaseelan S. NLRP6 Negatively regulates pulmonary host defense in gram-positive bacterial infection through modulating neutrophil recruitment and function. PLoS Pathog. 2018:14(9):e1007308. 10.1371/journal.ppat.1007308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth CS, Gordon JI, et al. NLRP6 Inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011:145(5):745–757. 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lemire P, Robertson SJ, Maughan H, Tattoli I, Streutker CJ, Platnich JM, Muruve DA, Philpott DJ, Girardin SE. The NLR protein NLRP6 does not impact gut Microbiota composition. Cell Rep. 2017:21(13):3653–3661. 10.1016/j.celrep.2017.12.026 [DOI] [PubMed] [Google Scholar]
- 72. Liu W, Liu J, Wang W, Wang Y, Ouyang X. NLRP6 Induces pyroptosis by activation of caspase-1 in gingival fibroblasts. J Dent Res. 2018:97(12):1391–1398. 10.1177/0022034518775036 [DOI] [PubMed] [Google Scholar]
- 73. Lu WL, Zhang L, Song DZ, Yi XW, Xu WZ, Ye L, Huang DM. NLRP6 Suppresses the inflammatory response of human periodontal ligament cells by inhibiting NF-kappaB and ERK signal pathways. Int Endod J. 2019:52(7):999–1009. 10.1111/iej.13091 [DOI] [PubMed] [Google Scholar]
- 74. Tian XX, Li R, Liu C, Liu F, Yang LJ, Wang SP, Wang CL. NLRP6-caspase 4 inflammasome activation in response to cariogenic bacterial lipoteichoic acid in human dental pulp inflammation. Int Endod J. 2020:54(6):916–925. 10.1111/iej.13469 [DOI] [PubMed] [Google Scholar]
- 75. Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015:21(3):172–183. 10.1016/j.molmed.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hajishengallis G, Chavakis T, Lambris JD. Current understanding of periodontal disease pathogenesis and targets for host-modulation therapy. Periodontol 2000. 2020:84(1):14–34. 10.1111/prd.12331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gasiorek A, Dobosz E, Potempa B, Ciaston I, Wilamowski M, Oruba Z, Lamont RJ, Jura J, Potempa J, Koziel J. Subversion of lipopolysaccharide signaling in gingival keratinocytes via MCPIP-1 degradation as a novel pathogenic strategy of inflammophilic pathobionts. mBio. 2021:12(3):e0050221. 10.1128/mBio.00502-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Abusleme L, Hoare A, Hong BY, Diaz PI. Microbial signatures of health, gingivitis, and periodontitis. Periodontol 2000. 2021:86(1):57–78. 10.1111/prd.12362 [DOI] [PubMed] [Google Scholar]
- 79. Diaz PI, Hoare A, Hong BY. Subgingival microbiome shifts and community dynamics in periodontal diseases. J Calif Dent Assoc. 2016:44(7):421–435. 10.1080/19424396.2016.12221035 [DOI] [PubMed] [Google Scholar]
- 80. Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018:16(12):745–759. 10.1038/s41579-018-0089-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hajishengallis G, Chavakis T. Local and systemic mechanisms linking periodontal disease and inflammatory comorbidities. Nat Rev Immunol. 2021:21(7):426–440. 10.1038/s41577-020-00488-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.