Abstract
The biological function of RNA can be modulated by base modifications. Here, we unveiled the occurrence of N4-acetylation of cytidine in plant RNA, including mRNA, by employing LC-MS/MS and acRIP-seq. We identified 325 acetylated transcripts from the leaves of 4-week-old Arabidopsis (Arabidopsis thaliana) plants and determined that 2 partially redundant N-ACETYLTRANSFERASEs FOR CYTIDINE IN RNA (ACYR1 and ACYR2), which are homologous to mammalian NAT10, are required for acetylating RNA in vivo. A double-null mutant was embryo lethal, while eliminating 3 of the 4 ACYR alleles led to defects in leaf development. These phenotypes could be traced back to the reduced acetylation and concomitant destabilization of the transcript of TOUGH, which is required for miRNA processing. These findings indicate that N4-acetylation of cytidine is a modulator of RNA function with a critical role in plant development and likely many other processes.
The base modification N4-acetylation of cytidine in RNA is essential for microRNA biogenesis as well as leaf and embryo development in plants.
IN A NUTSHELL.
Background: Eukaryotic mRNA harbors more than 10 different chemical modifications, most of which are involved in many cellular activities such as RNA processing, RNA degradation, and translation. These chemical modifications are installed by “writer” proteins on pre-mRNA. N4-acetylcytidine (ac4C), the only known type of RNA acetylation modification, has recently been reported in mammalian mRNA. This modification is introduced by a single-copy acetyltransferase NAT10 and promotes translation efficiency.
Question: Does ac4C modification widely exist in plant mRNA and what are the biological functions of this chemical modification?
Findings: Our data demonstrate the wide occurrence of ac4C modification on RNAs from several vascular plants. Employing acRIP-seq, we mapped the distribution of ac4C modifications in Arabidopsis thaliana. We also show that Arabidopsis RNAs are acetylated by N-ACETYLTRANSFERASEs FOR CYTIDINE IN RNA 1 (ACYR1) and ACYR2, which are homologs of human NAT10. A double-null mutation is embryo lethal, while abolishing 3 of the 4 ACYR alleles leads to defects in vegetative development. This phenotypic alteration is caused by the accelerated degradation of TOUGH transcript, a component of miRNA biogenesis, when its ac4C levels are reduced.
Next steps: The identification and characterization of ac4C in plant RNA give rise to the field of RNA modification and open up many research questions. Are any other proteins involved in ac4C formation and/or removal? Does ac4C modification participate in other developmental processes and/or environmental stimuli? What are the molecular mechanisms of ac4C modification regulating the target transcripts?
Introduction
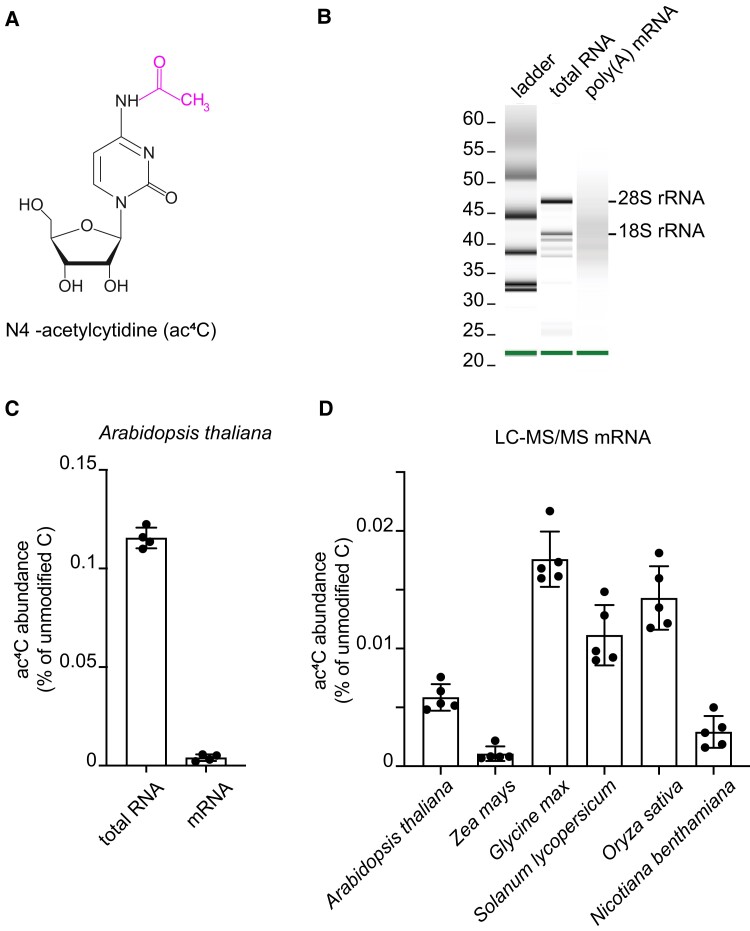
Over 150 chemical nucleoside modifications have been discovered in different RNA species of eukaryotes. Most are present in noncoding RNAs, such as transfer RNA and ribosomal RNA, but mRNA also contains modified bases such as N6-methyladenosine (m6A), N1-methyladenosine, N7-methylguanosine, 5-methylcytidine, N4-acetylcytidine (ac4C), and pseudouridine (Frye et al. 2016; Frye et al. 2018; McCown et al. 2020). These mRNA modifications are installed by “writer” proteins on pre-mRNA in the nucleus, and some modifications can be dynamically removed by “eraser” proteins. The mRNA modifications influence gene expression by regulating pre-mRNA splicing, mRNA structure, transport, stability, and translation efficiency (Shen et al. 2019; Wiener and Schwartz 2021). Often “reader” proteins are involved in decoding the modification signature. Among the different types of mRNA modifications, ac4C is the only acetylation modification (Fig. 1A) (Arango et al. 2018). ac4C was recently identified in mammalian mRNA (Arango et al. 2018), where it is introduced by the ac4C writer N-ACETYLTRANSFERASE 10 (NAT10). Acetylation of mRNA not only occurs in mammals but also has been reported in archaea (Sas-Chen et al. 2020). RNA immunoprecipitation with ac4C antibodies followed by deep sequencing (acRIP-seq) identified thousands of ac4C sites in the mRNA of HeLa cells, with most mapping near the translation start sites within the coding sequences, promoting mRNA stability and translation initiation in this human cell line (Arango et al. 2022). In plants, acetylation of cytidine within RNA, especially mRNA, remains poorly understood.
Figure 1.
Presence of ac4C in plant RNAs. A) Chemical structure of N4-acetylcytidine. B) The integrity of the purified RNA samples and the efficient removal of rRNA from mRNA were analyzed using Bioanalyzer 2100. C) LC-MS/MS analysis of the ac4C/C ratio in total RNA and mRNA of 2-week-old seedlings. Error bars are SD (n = 4 biological replicates; biological replicates are parallel measurements of biological distinct samples). D) LC-MS/MS analysis of the ac4C/C ratio in mRNA of leaves from 3-week-old seedlings of different plant species. Error bars are SD (n = 5 biological replicates; biological replicates are parallel measurements of biological distinct samples).
MicroRNAs (miRNAs) are small endogenous regulatory RNAs that are ∼21 nt in length and affect the translation or stability of target mRNAs (Jones-Rhoades et al. 2006; Voinnet 2009; Dolata et al. 2018). Mature miRNAs are derived from long primary transcripts that are produced by RNA polymerase II and further processed by the dicing complex, which in Arabidopsis (Arabidopsis thaliana) contains DICER-LIKE1 (DCL1), HYPONASTIC LEAVES1 (HYL1), and SERRATE (SE) as the core proteins (Yu et al. 2017). Several other proteins, including TOUGH (TGH), are associated with the dicing complex and co-regulate the production of miRNAs in response to developmental and environmental signals (Ren et al. 2012; Samad et al. 2017; Dolata et al. 2018). The TGH protein consists of 2 conserved functional domains with a role in RNA binding and RNA processing (Calderon-Villalobos et al. 2005). In Arabidopsis, loss of TGH function suppresses the activity of DCL1 and the interaction between pri-miRNA and HYL1. This prevents the accumulation of mature miRNAs (Ren et al. 2012), resulting in severe defects in leaf development (Calderon-Villalobos et al. 2005).
Here, we utilized mass spectrometry and transcriptome-wide approaches to investigate the occurrence, distribution, and function of ac4C in plant mRNAs. acRIP-seq data revealed that ac4C modifications are widespread within the Arabidopsis transcriptome. In protein-coding transcripts, they are enriched in the coding sequences. We identified 2 cytidine-N-acetylases required for acetylating Arabidopsis RNA in vivo and investigated the molecular and phenotypic consequences of altering cytidine-N-acetylation by genetic manipulation. While total loss of ac4C was lethal, reduced N-acetylation of cytidine compromised rosette development. A correlation analysis of acetylation states and transcript abundances provided a plausible connection between reduced transcript acetylation and defects in the miRNA processing machinery. As a result, TGH, which is involved in miRNA biogenesis, is deregulated, providing a functional link to the phenotypic consequences of reduced cytidine acetylation.
Results
Plant mRNAs are acetylated on cytidines
To investigate whether ac4C is present in plant mRNAs, we isolated mRNA from total RNA of 2-week-old Arabidopsis seedlings using 2 rounds of oligo dT purification. The mRNA purity was assessed using a bioanalyzer (Fig. 1B) and by detecting the level of 18S rRNA contamination relative to the ACTIN2 transcript level in the mRNA using RT-qPCR assays (Supplemental Fig. S1). We determined the amount of ac4C in mRNA and total RNA by completely RNA digestion to nucleosides followed by liquid chromatography coupled to quadrupole mass spectrometry (LC-MS/MS). For both total RNA and mRNA, we observed peaks in the respective analyses matching the retention time of the N4-acetylcytidine standard (2.29 min) with similar MS/MS profiles showing mass transitions of m/z 286.3 to 112.0 and more prominently to 153.9 (Supplemental Fig. S2). This demonstrates that ac4C was present in these RNA preparations. A quantitative evaluation revealed that approximately 29-fold more ac4C was present in total RNA (0.115 ± 0.005% ac4C relative to unmodified C) than in the mRNA preparation (0.004 ± 0.0017%, Fig. 1C). We confirmed the purity of the mRNA by showing that 3-methyluridine (m3U), a modification that only occurs in rRNA (McCown et al. 2020), was not detectable in our mRNA preparation (Supplemental Fig. S3). Furthermore, given that the abundance of 18S rRNA was reduced more than 1,000-fold in the mRNA sample versus total RNA (Supplemental Fig. S1) but that there was only 29-fold less ac4C in the mRNA (Fig. 1C), the detected ac4C in the mRNA could not have resulted from contamination with noncoding RNAs. Taken together, these data demonstrate that N4-acetylation of cytidine is a bona fide mRNA modification in Arabidopsis.
We also investigated if ac4C is present in leaf mRNA from several other plant species. The modification was detected in all species examined, i.e. maize (Zea mays, 0.0011 ± 0.0006% ac4C relative to unmodified C), soybean (Glycine max, 0.0176 ± 0.0024%), tomato (Solanum lycopersicum, 0.0111 ± 0.0026%), rice (Oryza sativa, 0.0143 ± 0.0027%), and Nicotiana benthamiana (0.00292 ± 0.0014%) (Fig. 1D). Notably, the ac4C abundance in different species was not correlated with their genome sizes. These data demonstrate that the ac4C modification of mRNA occurs widely in plants.
Arabidopsis possesses 2 cytidine N-acetyltransferases
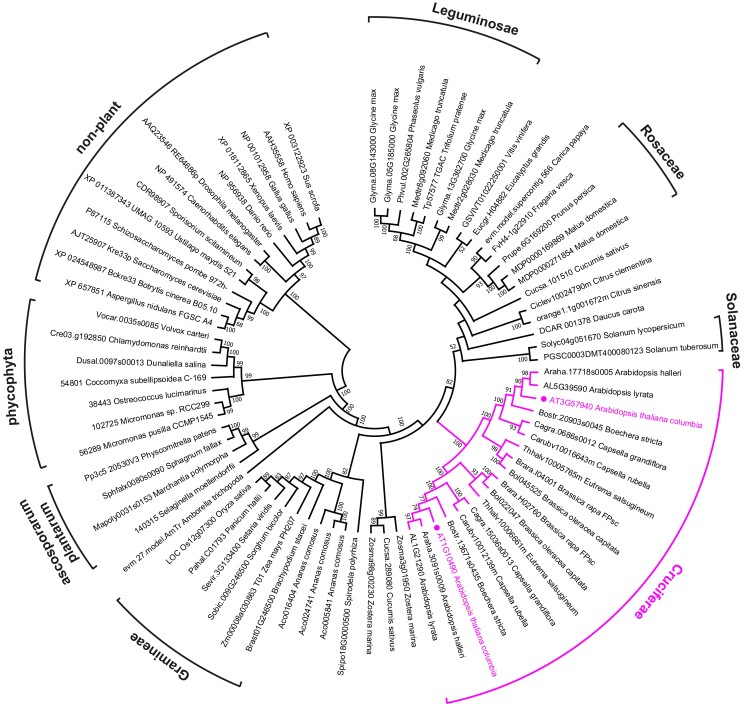
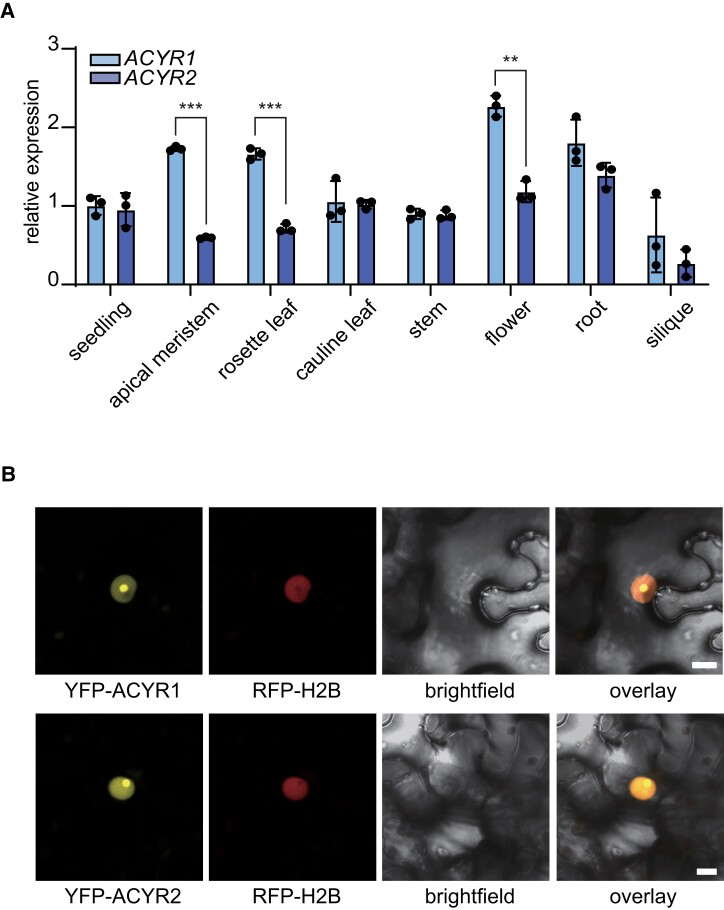
In mammalian cells, the acetyltransferase NAT10 installs ac4C on noncoding RNA (ncRNA) and mRNA (Arango et al. 2018). We used the protein sequence of human NAT10 as a BLAST query against predicted plant proteomes deposited at Phytozome (https://phytozome-next.jgi.doe.gov/), resulting in the identification of NAT10 homologs from 46 plant species (Supplemental Data Set 1). For plants, we suggest naming these enzymes N-ACETYLTRANSFERASEs FOR CYTIDINE IN RNA (ACYR). In addition, we searched the Concise Protein Database at the National Center for Biotechnology Information, which contains data from fully sequenced genomes, using human NAT10 as query and selected sequences from 13 nonplant organisms (Supplemental Data Set 1). Notably, NAT10/ACYR is a single-copy gene in all selected nonplant species and also in many plant species. However, the genomes of Cruciferae encode 2 ACYR proteins that are also phylogenetically well separated (Fig. 2 and Supplemental Data Set 1), indicating that they may have distinct functional roles. In Arabidopsis, we named these 2 proteins ACYR1 and ACYR2, encoded at the loci At1g10490 and At3g57940, respectively. Despite the phylogenetic separation, ACYR1 and ACYR2 are quite similar, with over 84% identical amino acids in their protein sequences (Supplemental Fig. S4). RT-qPCR analysis revealed that both genes are expressed in all tissues investigated, with slightly higher expression of ACYR1 in the apical meristem, rosette leaves, and flowers (Fig. 3A), which is in agreement with publicly available transcriptome data (Arabidopsis eFP browser: https://bar.utoronto.ca/efp//cgi-bin/efpWeb.cgi). Given the sequence similarity, the overlapping expression domains, and the finding that most plants just have 1 ACYR gene, it appears possible that the functions of ACYR1 and ACYR2 are at least partially redundant.
Figure 2.
Sequence clades of eukaryotic NAT10/ACYR enzymes. A neighbor-joining tree was constructed with NAT10 sequences from 46 phylogenetically distant plants and 13 nonplant organisms. A total of 1,000 bootstraps were performed, and branches with bootstrap values below 50% were collapsed.
Figure 3.
ACYR1 and ACYR2 are expressed in all tissues and localize to the nucleus. A) Relative expression quantified by RT-qPCR of ACYR genes using the primers Cp175 and Cp176 or Cp177 and Cp178 in different Arabidopsis tissues. Error bars are SD (n = 3 biological replicates; biological replicates are parallel measurements of biological distinct samples). Asterisks indicate the significant differences (**P < 0.01 and ***P < 0.001, one-way ANOVA). B) Confocal fluorescence microscopy images of N. benthamiana cells from the lower leaf epidermis transiently coproducing RFP-H2B and ACYR1 fused to an N-terminal YFP tag (YFP-ACYR1). Channels (from left to right): YFP, RFP, brightfield, and overlay of YFP, RFP, and brightfield. All infiltrated cells exhibited the same pattern. Bar = 10 μm. Lower panels show the results for N-terminal YFP-tagged ACYR2.
Subcellular localization analysis revealed that N-terminal yellow fluorescent protein (YFP) fusions with ACYR1 or ACYR2 predominantly accumulated in the nucleus and were enriched in a sub-nuclear localization, possibly the nucleolus (Fig. 3B), which is in line with the notion that most RNA modifications are installed before nuclear export.
Arabidopsis ACYRs are required for gametophytic transmission and vegetative growth
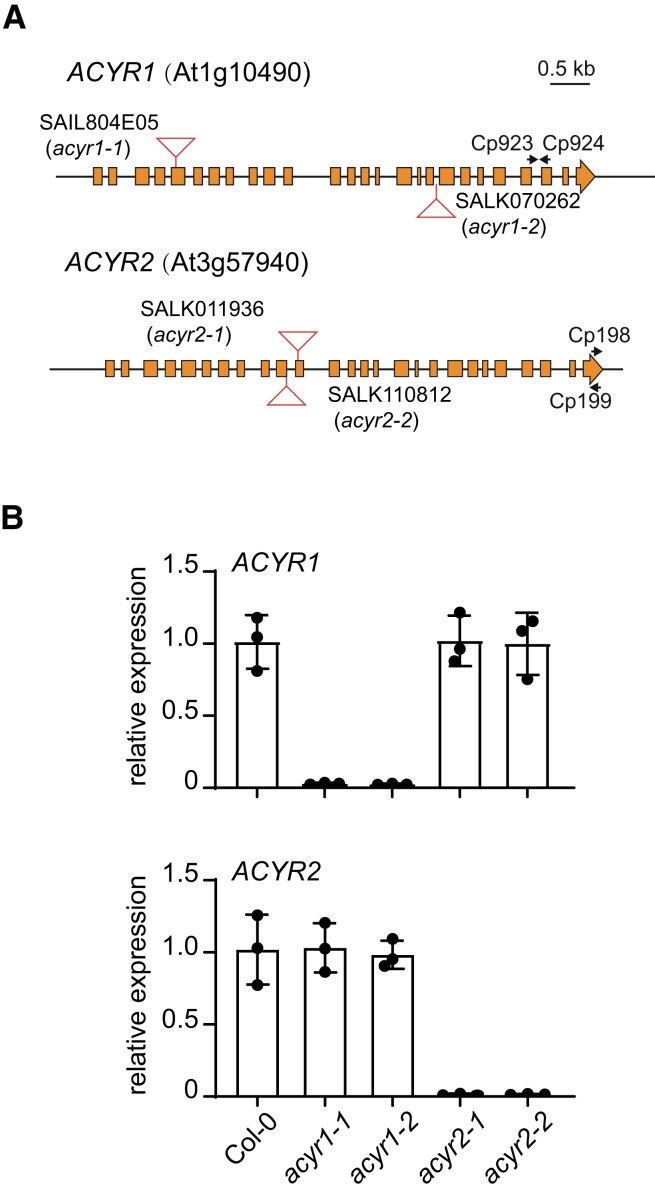
We isolated 2 independent homozygous T-DNA insertion mutants for each gene. RT-qPCR and nonquantitative RT-PCR analyses using primers flanking the insertion site showed that intact mRNA was absent in both homozygous mutants (acyr1 and acyr2) and that the abundance of the defective mRNA was strongly reduced compared to the wild type (Fig. 4 and Supplemental Fig. S5). A mutation in only 1 ACYR gene had no influence on the expression of the other gene (Fig. 4B).
Figure 4.
Characterization of plants varying in ACYR1 or ACYR2 expression. A) Genomic organization of the ACYR1 (At1g10490) and ACYR2 (At3g57940) loci and positions of the T-DNA insertions (triangles) in acyr1-1 (SAIL804E05), acyr1-2 (SALK070262), acyr2-1 (SALK011936), and acyr2-2 (SALK110812). Coding sequences are shown as boxes. B) Relative expression of ACYR1 or ACYR2 quantified by RT-qPCR using the primers Cp923 and Cp924 or Cp198 and Cp199 in wild type and single mutants of ACYR1 and ACYR2. cDNA was reverse transcribed using oligo dT. The TIP41-like gene was set as internal reference gene. The calculation was based on the 2−ΔΔCT method. Error bars are SD (n = 3 biological replicates; biological replicates are parallel measurements of biological distinct samples).
We attempted to generate acyr1 acyr2 double mutant lines by crossing acyr1-2 and acyr2-1. Despite several attempts, double mutants could never be recovered. Nonetheless, plants homozygous for the T-DNA insertion at 1 locus and heterozygous at the other (hemizygous ACYR double mutants) could be recovered (acyr1-2/acyr1-2 acyr2-1/ACYR2 and acyr1-2/ACYR1 acyr2-1/acyr2-1; referred to as acyr1 acyr2/+ and acyr1/+ acyr2 hereafter for simplicity). The transcript levels of ACYR1 or ACYR2 were significantly lower in seedlings of both hemizygous ACYR double mutants compared to the wild type (Supplemental Fig. S6). In line with the absence of a homozygous double mutant in the progeny of the segregating populations, the segregation ratios in the progeny of self-pollinated hemizygous ACYR double mutant plants (either acyr1 acyr2/+ or acyr1/+ acyr2) were abnormal. Instead of the theoretical 1:2:1 genotype ratio for the hemizygous locus, the observed ratio was closer to ∼1:1:0 in the progeny of acyr1 acyr2/+ and ∼20:1:0 in the progeny of acyr1/+ acyr2 plants. Taken together, these data indicate that ACYR1 and ACYR2 are at least partially redundant and are together essential during gametogenesis and/or embryo development. In addition, both hemizygous double mutant plants produced shorter siliques than the wild type (Fig. 5A to C and Supplemental Table S1), hinting at a problem in seed development. We observed approximately 35.8% and 50.8% aborted seeds in the siliques formed by acyr1 acyr2/+ and acyr1/+ acyr2 plants, respectively, compared to less than 5% in the wild type (Col-0) and the single mutants (Supplemental Table S1). These results indicate that gametophytic transmission is affected in these hemizygous ACYR double mutant plants.
Figure 5.
Loss of ACYR function suppresses seed development and vegetative growth. A) Developing seeds in the siliques of wild type, acyr1-2, acyr2-1, acyr1 acyr2/+, and acyr1/+ acyr2 at 10 days after fertilization (DAF). Scale bar = 1 mm. Arrows indicate undeveloped embryos. B) Silique length of different genotypes at 10 DAF. Error bars are SD (n = 20 siliques from 5 independent plants for each genotype). In each boxplot, horizontal line, median; edges of boxes, 25th (bottom) and 75th (top) percentiles; whiskers, minimum and maximum silique length, respectively. The values of the mean and the SD are shown above each data display. Different letters indicate significant differences at P < 0.05 (one-way ANOVA along with two-sided Tukey's pairwise comparisons). C) Undeveloped and intact embryo number per silique of different genotypes. Error bars are SD (n = 20 siliques from 5 independent plants for each genotype). The values of the mean and the SD are shown above each data display. Different letters indicate significant differences at P < 0.05 (one-way ANOVA along with two-sided Tukey's pairwise comparisons). D) Rosettes and rosette leaves of the wild type, the ACYR single mutants, the 2 hemizygous ACYR double mutants, and the complementation line at 39 days after imbibition (dai). Bar = 5 cm. E) Number of rosette leaves of the different genotypes at 39 dai. In each boxplot, horizontal line, median; edges of boxes, 25th (bottom) and 75th (top) percentiles; whiskers, minimum and maximum leaf number, respectively. Error bars are SD (n = 16 independent plants). The values of the mean and the SD are shown above each data display. Different letters indicate significant differences at P < 0.05 (one-way ANOVA along with two-sided Tukey's pairwise comparisons). F) Rosette diameters of plants of different genotypes (n = 10 independent plants) grown 39 dai under long-day conditions. Error bars are SD. Different letters indicate significant differences at P < 0.05 (one-way ANOVA along with two-sided Tukey's pairwise comparisons).
To comprehensively investigate the influence of ACYR-mediated RNA acetylation on plant growth, we generated a complementation line by introducing an ACYR1-Strep transgene encoding a Strep tag fused to the C-terminus of ACYR1 into the acyr1 acyr2/+ background and named it acyr1 acyr2/+ ACYR1-Strep. In this line, the transgene was expressed at a level comparable to ACYR1 in Col-0. Moreover, ACYR1-Strep protein accumulated in transgenic seedlings, as revealed by immunoblot analysis using an anti-Strep antibody (Supplemental Fig. S7).
Next, we performed phenotypic analyses of Col-0, the acyr1 and acyr2 single mutants, the 2 hemizygous ACYR double mutants, and the complementation line grown under long-day conditions. The acyr1 and acyr2 single mutants were phenotypically indistinguishable from the wild type (Fig. 5D and Supplemental Fig. S8). By contrast, both hemizygous ACYR double mutants produced significantly fewer leaves and had smaller rosettes compared to Col-0, the single mutants, and the complementation line (Fig. 5D to F). The effect became apparent starting 20 days after seed imbibition (dai) based on rosette diameter (Fig. 5F). The hemizygous ACYR double mutants produced approximately 12 to 13 rosette leaves before flowering at 39 dai, while Col-0 and the single mutants produced 17 to 18 rosette leaves under the same conditions (Fig. 5D and E). Importantly, this phenotype was complemented by the ACYR1-Strep transgene in the acyr1 acyr2/+ background (Fig. 5), showing that the defects in the ACYR genes caused the phenotypic alteration.
Arabidopsis RNA is acetylated by ACYRs
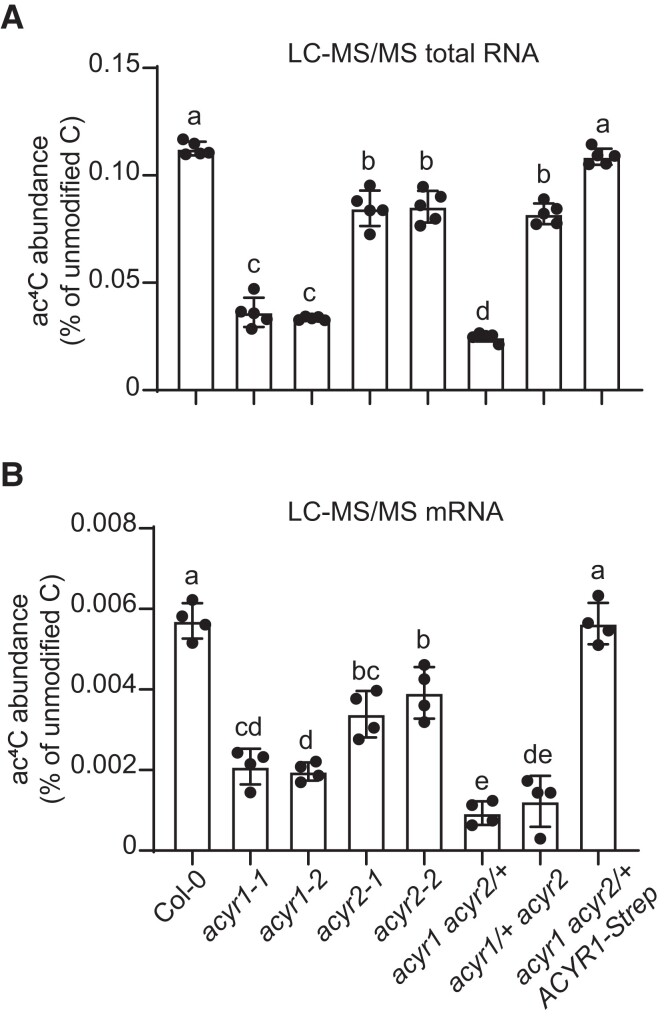
To explore the consequences of the loss of ACYR1 and ACYR2 on cytidine acetylation of RNA, we measured ac4C abundance in total RNA and mRNA from rosette leaves of 4-week-old Col-0, acyr1, acyr2, the 2 hemizygous ACYR double mutants, and the complementation line. Compared to the wild type, total RNA of acyr1 plants contained over 3.5-fold less ac4C, while in acyr2 plants, the ac4C abundance was approximately 1.3-fold lower (Fig. 6A). In the acyr1 acyr2/+ line, the amount of ac4C was even slightly more reduced than in acyr1. In the mRNA, the ac4C/C ratios of all mutants were also smaller than that of the wild type (Fig. 6B). Interestingly, both hemizygous ACYR double mutant lines contained markedly less ac4C in mRNA than the respective single mutants (Fig. 6B). Moreover, the ac4C/C ratios in total RNA and mRNA of the complementation line were similar to those in Col-0 (Fig. 6), indicating that the fusion protein was functional and that the observed molecular phenotype was indeed caused by the loss of ACYR1 function. These data suggest that (1) both ACYR1 and ACYR2 are required for cytidine N4-acetylation of ncRNA and mRNA in Arabidopsis, (2) most ac4C modifications are written by ACYR1, and (3) the redundancy of both writers is likely less pronounced for mRNA acetylation than for ncRNA acetylation, i.e. there are more sites on mRNA where ac4C is preferentially introduced by only 1 of the enzymes. This became apparent when the gene dosage was lowered, as in the hemizygous double mutants, because the lower target affinity of either ACYR1 for preferential ACYR2 targets or vice versa could no longer be masked by high enzyme abundance.
Figure 6.
RNA acetylation by ACYR1 and ACYR2 in vivo. A) ac4C/C ratio in total RNA from 4-week-old rosette leaves of wild type, acyr1-1, acyr1-2, acyr2-1, acyr2-2, acyr1 acyr2/+, and acyr1/+ acyr2 and the complementation line analyzed by LC-MS/MS. Error bars are SD (n = 5 biological replicates; biological replicates are parallel measurements of biological distinct samples). Different letters indicate significant differences at P < 0.05 (one-way ANOVA along with two-sided Tukey's pairwise comparisons). B) Same as in (A) but for mRNA. Error bars are SD (n = 4 biological replicates; biological replicates are parallel measurements of biological distinct samples).
Transcriptome-wide mapping of ac4C sites in Arabidopsis using acRIP-seq
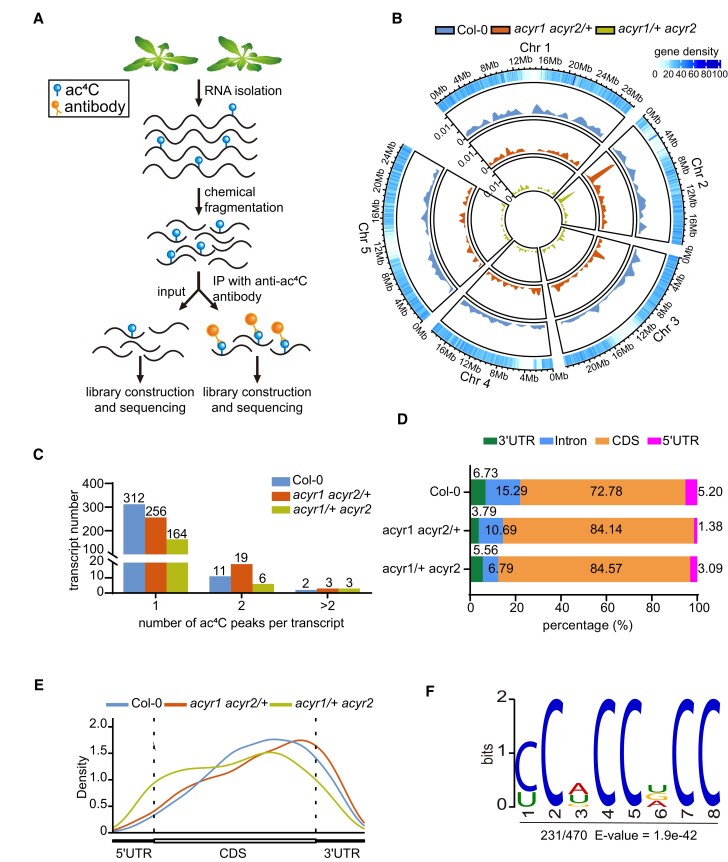
To explore the impact of ACYR activities on the distribution of ac4C in RNA species, we performed acetylated RNA immunoprecipitation and sequencing (acRIP-seq) using RNA extracted from rosette leaves of 4-week-old plants of the wild type and the 2 hemizygous ACYR double mutants, with 2 biological replicates per genotype. RNA was fragmented into oligonucleotides with a length of ∼150 nucleotides (input), followed by immunoprecipitation using an anti-ac4C antibody. The purified RNA was used for library construction and sequencing (Fig. 7A). Adaptors and low-quality reads were removed from the raw data, and the clean reads were mapped to the Arabidopsis TAIR10 genome. We called ac4C peaks with the MACS2 algorithm (Zhang et al. 2008), with an estimated false discovery rate (FDR) of <0.05 and an absolute fold enrichment of >1. Correlation analysis between biological repeats confirmed that the acRIP-seq data were robust (Supplemental Fig. S9). When ac4C peaks were present in both biological replicates, we classified them as “confident peaks” (Supplemental Fig. S10).
Figure 7.
Transcriptome-wide mapping of ac4C in Arabidopsis. A) Scheme of the acRIP-seq procedure. Each genotype was performed with 2 biological replicates. Rosette leaves from 3 individual 4-week-old plants were pooled as 1 biological replicate. B) Circos plot of the ac4C peaks in the Arabidopsis genome. The 4 rings from the outside to the inside show (first) the genomic positions with gene density, (second) the ac4C peak density for Col-0, (third) for acyr1 acyr2/+, and (fourth) for acyr1/+ acyr2. Peak densities are drawn to the same scale. C) Numbers of transcripts bearing 1, 2, or >2 ac4C peaks. D) Bar chart presenting the distribution of ac4C peaks within 7 nonoverlapping segments of the protein-coding transcript in Col-0, acyr1 acyr2/+, and acyr1/+ acyr2 lines. E) Metagene profile presenting the distributions of ac4C peaks identified in Col-0, acyr1 acyr2/+, and acyr1/+ acyr2 lines across the indicted mRNA segments. F) Sequence logo representation of the consensus ac4C motif identified in Col-0 by MEME suite. The number of motif occurrences relative to the total number of ac4C peaks and the corresponding E-value are shown under the logo.
In general, ac4C modifications were more abundant in Col-0 than in the 2 hemizygous ACYR double mutant lines in terms of the quantity of modified fragments mapping to the same genomic location (Fig. 7B), which is in agreement with the results from mass spectrometry (Fig. 6B). Also, in terms of the number of genomic locations carrying a modification, there were more in Col-0 than the double mutants. In total, we identified 470 confident peaks for ac4C associated with 325 transcripts in Col-0, 329 confident peaks associated with 278 transcripts in acyr1 acyr2/+, and 213 confident peaks associated with 173 transcripts in acyr1/+ acyr2 (Fig. 7C and Supplemental Data Set 2). Employing the DAVID Bioinformatics Resources (https://david.ncifcrf.gov/), Gene Ontology (GO) analysis revealed that the 325 ac4C-containing transcripts in Col-0 were enriched in transcripts related to photosynthesis and cold acclimation, indicating that ac4C modification in Arabidopsis may play an essential role in these biological processes (Supplemental Fig. S11). Compared with Col-0, 679 and 607 ac4C peaks differed in acyr1 acyr2/+ and acyr1/+ acyr2, respectively, in terms of presence or absence in the respective genotypes (Supplemental Fig. S12). Often the same peaks were different irrespective of which mutant was compared with the wild type (in 531 cases). Of the ac4C peaks that were present in both the respective hemizygous double mutant and the wild type, 144 and 179 transcripts differed by at least a log2 fold change of ± 2 in acyr1 acyr2/+ versus Col-0 and acyr1/+ acyr2 versus Col-0, respectively (Supplemental Fig. S13). A total of 114 transcripts fulfilling this criterion were identical in both comparisons. These data together indicate that there is a significant functional overlap between ACYR1 and ACYR2 (Supplemental Figs. S12 and S13).
We compared the distribution of ac4C peaks along protein-coding transcripts relative to gene features, using a single splice variant for each gene. The location of peak center points was used to assign each ac4C peak to 1 of 4 nonoverlapping regions: 5′-untranslated region (UTR), coding sequence (CDS), intron, and 3′-UTR. Most ac4C peaks were located in the CDS: 72.8% in Col-0 and even more (∼85%) in the hemizygous ACYR double mutants because acetylation modifications in other regions were suppressed by lowering the ACYR gene dosage in the mutants (Fig. 7D). In Col-0, introns, which were occasionally present due to alternative splicing or sequencing of pre-mRNAs, were approximately ten times less acetylated than exon sequences when the data were normalized to the respective amounts of intron/exon sequence in the sample. A metaplot of ac4C peak distribution across the 5′-UTR, CDS, and 3′-UTR features of mRNAs confirmed that the ac4C modifications were more abundant in the CDS with a slight bias toward the 3′-region of the CDS (Fig. 7E). Notably, the ac4C distribution in acyr1 acyr2/+ was similar to that in Col-0, whereas in acyr1/+ acyr2, the bias toward the 3′-region was not observed, maybe because ACYR2 is especially important for acetylation in that region of the CDS. Taken together, these data show that ACYR1 and ACYR2 preferentially acetylate the CDS in protein-coding transcripts.
To determine whether there are conserved motifs in the identified ac4C peaks, we performed an unbiased search in regions surrounding ac4C peak summits using the MEME (Multiple EM for Motif Elicitation) suite (Ma et al. 2014) (Fig. 7F and Supplemental Table S2). The most markedly enriched motif (approximately 50%) was a C-rich motif, CCWCCDCC (W = A or U or G; D = U or G or A), among confident ac4C peaks in Col-0 (Fig. 7F), resembling the conserved motif identified in mammalian cells (Arango et al. 2018).
ACYR-mediated ac4C modifications increase miRNA abundance by stabilizing TGH mRNA
To further investigate the molecular link between reduced ac4C modifications and the developmental alterations of acyr1 acyr2/+ and acyr1/+ acyr2 plants, we performed transcriptome deep sequencing (RNA-seq) of rosette leaves from 4-week-old Col-0 plants and the 2 hemizygous ACYR double mutants with 3 biological replicates each (Supplemental Data Set 3). To explore possible correlations between the RNA-seq data and acRIP-seq data, the log2 fold changes of both parameters comparing Col-0 and acyr1 acyr2/+ or acyr1/+ acyr2 were plotted against each other for transcripts with a log2 fold change of ± 0.2 (Fig. 8A, Supplemental Fig. S14, Supplemental Data Set 4). Compared to Col-0, we identified 67 transcripts in the acyr1 acyr2/+ mutant with less ac4C modification, including 26 with increased (Fig. 8A, green) and 41 with decreased (Fig. 8A, blue) abundance. Likewise, we identified 59 transcripts with lower levels of ac4C modification, including 20 that were more abundant (Supplemental Fig. S14, green) and 39 that were less abundant (Supplemental Fig. S14, blue) in the acyr1/+ acyr2 mutant relative to Col-0 (Supplemental Data Set 4).
Figure 8.
ACYR-mediated ac4C modifications enhance TGH transcript levels. A) Correlation between the level of gene expression and ac4C abundance in the acyr1 acyr2/+ mutant compared to Col-0. Padj <0.05, log FC (fold change) >0.2 or <−0.2. The numbers of peaks and the associated transcripts are labeled in each quadrant. B) ac4C modification of the TGH transcript is reduced in both hemizygous ACYR double mutants. The top panel shows the ac4C density along the TGH transcript in ac4C-IP and input samples from Col-0 and the 2 hemizygous double mutants. The gene structure of TGH is displayed below the ac4C density maps, with exons depicted as blue boxes. The ac4C modification site is indicated on the gene structure. The bottom panel shows acRIP-qPCR results. Fragments amplified are indicated and numbered below the TGH gene structure. Different letters indicate significant differences at P < 0.05 (one-way ANOVA along with two-sided Tukey's pairwise comparisons). Error bars are SD (n = 3 biological replicates; biological replicates are parallel measurements of biological distinct samples). C) Relative TGH transcript levels in Col-0, the 2 hemizygous ACYR double mutants, and the complementation line determined by RT-qPCR. Error bars are SD (n = 3 biological replicates; biological replicates are parallel measurements of biological distinct samples). Different letters indicate significant differences at P < 0.05 (one-way ANOVA along with two-sided Tukey's pairwise comparisons).
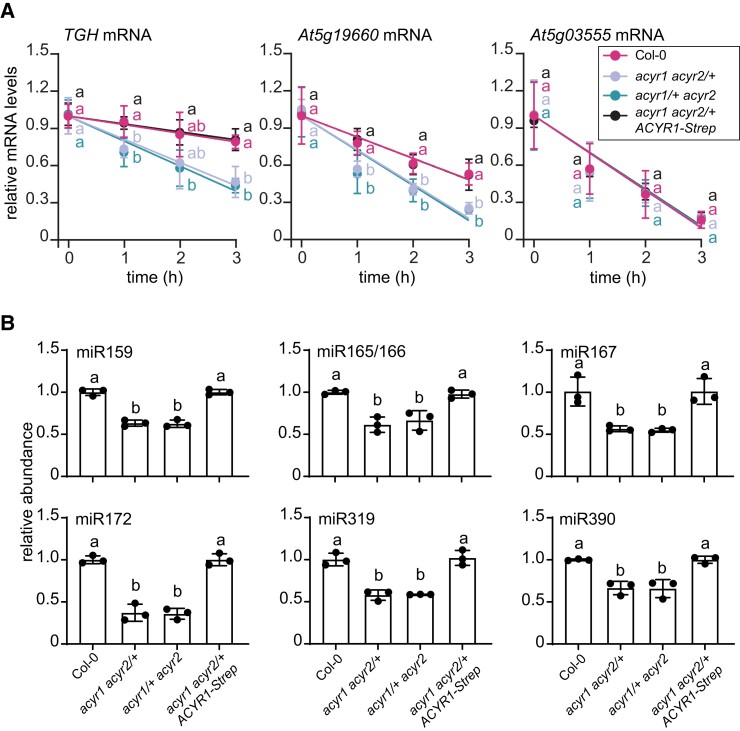
Of the transcripts that were differentially acetylated and changed in abundance in the hemizygous double mutants versus Col-0, several were found in both hemizygous double mutant lines (Supplemental Fig. S15). Cytidine acetylation of the TOUGH transcript (TGH, At5g23080) was strongly reduced (Fig. 8B top panel), and its abundance was significantly reduced in both hemizygous ACYR double mutants relative to Col-0 (Supplemental Data Set 4). These results were also confirmed by acRIP-qPCR and RT-qPCR assays (Fig. 8B bottom panel and 8C). Transcription inhibition assays showed that the TGH transcript was less stable in the 2 hemizygous ACYR double mutants compared to Col-0 and the complementation line, whereas the stability of a nonacetylated control mRNA (PLUTO, At5g03555) was not influenced by the different genotypes (Fig. 9A). Notably, the stability of another ac4C-containing transcript (S1P, At5g19660) was also reduced in the 2 hemizygous ACYR double mutants compared to Col-0 and the complementation line, indicating that mRNA acetylation can apparently promote RNA stability.
Figure 9.
ac4C modifications stabilize the TGH transcript and influence the amounts of several miRNAs. A) Transcript abundance (quantified by RT-qPCR) of TGH, and S1P (positive control) and PLUTO (negative control) in 20-day-old seedlings of Col-0, the hemizygous ACYR double mutants, and the complementation line when de novo transcription was blocked by treatment with actinomycin D for up to 3 h. Error bars are SD (n = 3 biological replicates * 2 technical replicates). Biological replicates are parallel treatments of biological distinct samples, and technical replicates are repeated measurements of the same sample. Different letters indicate significant differences at P < 0.05 (one-way ANOVA along with two-sided Tukey's pairwise comparisons). B) Relative abundances of miR159, miR165/166, miR167, miR172, miR319, and miR390 detected by RT-qPCR in 24-day-old seedlings of the wild type (Col-0), the 2 hemizygous mutants, and the complementation line. Error bars are SD (n = 3 biological replicates; biological replicates are parallel measurements of biological distinct samples). Different letters indicate significant differences at P < 0.05 (one-way ANOVA along with two-sided Tukey's pairwise comparisons).
The null mutation of TGH results in smaller plants with fewer rosette leaves in Arabidopsis (Calderon-Villalobos et al. 2005), which resembles the vegetative growth defects seen in both hemizygous ACYR double mutants. TGH functions in the early steps of miRNA biogenesis. Its mutation leads to the reduced accumulation of several miRNAs, including miR159, miR165/166, miR167, miR172, miR319, and miR390 (Ren et al. 2012). To assess whether the ACYR enzymes modulate vegetative growth by influencing miRNA production through the acetylation of TGH mRNA, we quantified several miRNAs in rosette leaves of 4-week-old Col-0, the 2 hemizygous ACYR double mutants, and the complementation line. Indeed, all miRNAs known to be dependent on TGH activity were less abundant in the 2 hemizygous ACYR double mutants compared to Col-0 and the complementation line (Fig. 9B).
In conclusion, these data show that ac4C modification prolongs the lifetime and thereby increases the abundance of TGH transcripts. This promotes the formation of several miRNAs, which represent an important factor in vegetative development (Fig. 10).
Figure 10.
Proposed model showing how ACYR-dependent ac4C modification modulates vegetative growth through TGH-mediated miRNA processing. In the wild type of Arabidopsis thaliana, TGH transcripts are acetylated by ACYR proteins for promoting RNA stability, which is essential for miRNA homeostasis and vegetative growth. While in the acyr1 acyr2/+ and acyr1/+ acyr2 mutants, ac4C modification in TGH is highly reduced, leading to the decreased miRNA abundance and defects in leaf development.
Discussion
It has been noted that 18S rRNA of Arabidopsis and several other eukaryotes carries ac4C modifications (Sharma et al. 2015). However, while this modification in mRNA has been described in mammals (Arango et al. 2018), little is known about its occurrence in the mRNA of other eukaryotes, including plants. Notably, the abundance and distribution of ac4C modifications in plants are markedly different from those in mammalian cells. Here we found that in Arabidopsis, only 0.115 ± 0.005% and 0.004 ± 0.0017% cytidine nucleosides were N4-acetylated in total RNA and mRNA, respectively, which is far less than the ∼0.5% and 0.2% in total RNA and mRNA of human cells, respectively (Arango et al. 2018). Compared to 4,250 ac4C peaks identified in HeLa cells, we only mapped 470 ac4C peaks associated with 325 transcripts in Arabidopsis. Additionally, we observed that ac4C modifications are distributed along the CDS of the mRNAs, with some slight enrichment in the 3′-regions, whereas in HeLa cells, ac4C was more abundant close to the 5′-region of the CDS, suggesting that the biological function of ac4C may differ between plants and mammals.
RNA methylation influences several RNA metabolic processes including splicing, stability, translocation, and translation (Gilbert et al. 2016; Yang et al. 2017; Frye et al. 2018). In mammalian cells, RNA acetylation increases RNA stability and stimulates translation efficiency (Arango et al. 2018). In plants, we showed that even partial reduction in ac4C levels in certain target transcripts can significantly accelerate their degradation (Fig. 9A), suggesting that ac4C modification could play a role in promoting mRNA stability. However, not always is a lower degree of C acetylation associated with lower transcript abundance—even the contrary can be true (Fig. 8, Supplemental Fig. S14). The molecular mechanisms behind such destabilizing or stabilizing functions remain unclear, but ac4C reader proteins could be involved. If such readers exist, it would be interesting to identify them and elucidate how they work. In addition, it needs to be investigated whether ac4C in plant mRNAs influences translation or other process such as splicing and the nuclear export of RNA.
In plants, m6A is introduced by an evolutionarily conserved methyltransferase complex comprising 2 methyltransferases, mRNA adenosine methylase (MTA) and MTB, and several accessory subunits including FIP37, VIR, and HAKAI (Růžička et al. 2017; Shen 2023) and is dynamically removed by demethylase such as ALKBH10B (Duan et al. 2017) and ALKBH9B (Martínez-Pérez et al. 2017; Tang et al. 2022). Here, we showed that most ac4C modification sites in plant RNAs are installed by the mammalian NAT10 homologs ACYR1 and ACYR2. Interestingly, the mutation of Arabidopsis ACYRs did not completely eliminate ac4C modifications (Fig. 5B and C). This result can be explained either by the only partial loss of ACYR activity in our mutants or by the activity of some currently unknown cytidine acetyltransferases. The NAD-dependent protein deacetylase sirtuin-7 (SIRT7), 1 of the 7 mammalian sirtuins, was recently described as an rRNA deacetylase that directly interacts with NAT10 (Xu et al. 2021), indicating that ac4C modifications (such as m6A) are dynamic and could be regulated by writer and easer proteins. However, whether Arabidopsis SRT1 (encoded by At5g55760) and SRT2 (encoded by At5g09230), 2 mammalian SIRT7 homologs, are the ac4C easers remains unknown.
NAT10/ACYR is a single-copy gene in many organisms, including most plants, but interestingly, there are 2 copies in the Brassicaceae and thus also in Arabidopsis. Sequence analysis suggested that both isogenes may have distinct functions in Arabidopsis, but functional analysis indicated that ACYR1 and ACYR2 are both required for full acetylation of coding and noncoding RNAs and have an overlapping substrate spectrum, although with different preferential substrates. Despite the apparent redundancy, it appears that the more abundant rRNAs are to a greater extent acetylated by ACYR1, explaining the stronger negative impact on the total acetylation level when ACYR1 was mutated. 18S rRNA is highly acetylated, which could explain why YFP-labeled ACYR enzymes accumulated in a nuclear subcompartment that might be the nucleolus (Fig. 3B).
Loss of function in either ACYR1 or ACYR2 reduced ac4C modification levels without affecting plant growth, while the complete loss of both ACYR1 and ACYR2 function in Arabidopsis led to embryonic lethality, pointing to the functional redundancy between ACYR1 and ACYR2. Similarly, homozygous nat10 mutants are lethal before embryonic day E14.5 in mice (Mus musculus) (Balmus et al. 2018), indicating that ACYR-mediated ac4C modification plays an essential role in eukaryotic development.
The segregation ratios measured from the progeny of the hemizygous mutants suggest that both the female and the male gametophytes suffer when there is not enough cytidine-N-acetylation. If the acyr1 acyr2 ovules died but the corresponding double mutant pollen were viable, one would expect a 1:1:0 ratio of wild type/heterozygous/mutant genotypes for the respective hemizygous allele, and in the silique, ∼0% of positions would be empty. When selfing the acyr1 acyr2/+ hemizygous line, we observed a 1:1:0 ratio but only ∼35% empty positions in the silique. This indicates that the acyr1 acyr2 ovule suffers and many die but that some fertilized acyr1 acyr2 ovules survive, and those that receive an ACYR2 (+) allele from the pollen can sometimes generate a viable seed. When the acyr1/+ acyr2 hemizygous line was selfed, a 20:1:0 ratio with 50% empty siliques was observed, indicating that ACYR2 has a more important role for the gametes than ACYR1. It appears that pollen viability is also strongly compromised in plants in which the ACYR1 gene dosage is lowered in an acyr2 null background, explaining the skewed allele frequency toward the wild type. The complete depletion of TGH in plants does not affect seed production (Calderon-Villalobos et al. 2005), indicating that the defective acetylation of transcripts other than TGH must be responsible for the observed partial infertility. The reduction of ac4C levels in nonpolyadenylated RNA in the 2 hemizygous ACYR double mutants may also contribute to the pleiotropic phenotypic alterations observed in these mutants.
This transcriptome-wide description of RNA ac4C modification in plants is only the tip of the iceberg. Investigating ac4C function in detail for individual transcripts will be possible by mutating the acetylation sites without affecting the coding potential in the case of mRNAs. It is likely that dynamic ac4C writing and perhaps erasing occur to transmit regulative cues. It will be interesting to investigate whether additional regulatory components promote or prevent access of the ACYR proteins to their in vivo targets.
Materials and methods
Plant material and cultivation
The Arabidopsis thaliana ecotype Col-0 was chosen as the wild type. T-DNA insertion mutants of Arabidopsis from the SAIL collection (SAIL804E05, acyr1-1) (Sessions et al. 2002) and the SALK collection (SALK070262, acyr1-2; SALK011936, acyr2-1; and SALK110812, acyr2-2) (Alonso et al. 2003) were obtained from the Nottingham Arabidopsis Stock Centre. Hemizygous ACYR double mutants (acyr1 acyr2/+ and acyr1/+ acyr2) were obtained by crossing acyr1-2 (as the pollen donor) and acyr2-1. The T-DNA insertions were confirmed using the primer pairs 1197/Cp2, N61/Cp3, N61/Cp7, and N61/Cp8 for acyr1-1, acyr1-2, acyr2-1, and acyr2-2, respectively, and the primer pairs Cp1/Cp2, Cp3/Cp4, Cp7/Cp10, and Cp7/Cp10 for the wild-type alleles of acyr1-1, acyr1-2, acyr2-1, and acyr2-2, respectively (Supplemental Table S3).
Plants were cultivated on agar plates containing half-strength Murashige and Skoog (1/2 MS) medium or full nutrient soil under long-day conditions (16 h light of 75 μmol m−2 s−1 intensity provided by white LED lamps at 22 °C and 8 h dark at 20 °C, 75% relative humidity). For quantification of ACYR1 or ACYR2 expression, wild-type seedlings and apical meristems were sampled from 2-week-old plants, while rosette leaves, cauline leaves, stems, flowers, roots, and siliques were randomly collected from 6-week-old plants.
RNA extraction, reverse transcription, and quantitative RT-PCR analysis
Total RNA was isolated from different tissues or plants using TRIzol reagent (Monad) and treated with DNase (4 × gDNA wiper mix; Vazyme Biotech Co. Ltd.), and cDNA was prepared using HiScript II Q Select RT Super Mix (Vazyme Biotech Co. Ltd.). Random primers were used for reverse transcription in the experiment evaluating rRNA contamination in mRNA, and oligo dT was used for the other experiments. Primer pairs Cp28/Cp29 and Cp32/Cp33 were employed for ACYR1 and ACYR2 cloning, introducing the Ndel/XmaI and XmaI/BamHI sites, respectively. The resulting PCR products were inserted into the vector pXNS1pat-YFP (V99) and pXNScpmv-HA-Strep (V89) (Chen and Witte 2020) for subcellular localization analyses and the generation of the complementation lines, respectively.
To analyze mRNA abundance, quantitative reverse transcription PCR (qRT-PCR) was performed with QuantStudio 1 (Thermo Fisher Scientific) using Hieff qPCR SYBR Green Master Mix (Yeasen Biotechnology) according to the manufacturer's instructions. Transcript abundance of ACYR1, ACYR2, TGH, S1P, PLUTO, and 18S rRNA was analyzed by employing the primer pairs Cp923/Cp924 or Cp175/Cp176, Cp198/Cp199 or Cp177/Cp178, Cp211/Cp212, Cp619/Cp620, Cp715/Cp716, and Cp448/Cp449, respectively. ACTIN2 was amplified with the primer pairs Cp204/Cp205 as an internal reference gene for quantifying rRNA contamination in mRNA, and the TIP41-like gene (Czechowski et al. 2005) was amplified with the primer pair Cp307/Cp308 as an internal reference gene for the transcript stability and other qRT-PCR analyses. The calculation was based on the 2−ΔΔCT method (Livak and Schmittgen 2001). To measure gene-specific mRNA levels in the mutants and the complementation line, cDNAs were prepared from seedlings of each genotype, and primer pairs Cp28/Cp29 and Cp32/Cp33 were used for the quantification of ACYR1 and ACYR2 transcripts.
To detect mature miRNAs, first-strand cDNA of individual miRNA was synthesized from 1 μg total RNA using an miRNA 1st Strand cDNA Synthesis Kit (by stem-loop; Vazyme Biotech Co. Ltd.) according to the manufacturer's instructions. Stem-loop RT primers were designed as described previously (Chen et al. 2005). Approximately 50 nM of TAFII15-StLp-cDNA_rv (Cp421) (Werner et al. 2021) was added as an internal control for qRT-PCR analysis, and 50 mM of the respective miRNA-specific stem-loop primer Cp479, Cp483, Cp487, Cp386, Cp489, or Cp491 was used for cDNA synthesis of miR159, miR165/166, miR167, miR172, miR319, and miR390, respectively. A 5-fold dilution of cDNA was used for qRT-PCR. The primer pair Cp387/Cp478, Cp387/Cp482, Cp387/Cp486, Cp387/Cp390, Cp387/Cp488, or Cp387/Cp490 was employed in qRT-PCR analyses of mature miRNAs of miR159, miR165/166, miR167, miR172, miR319, or miR390, respectively. TBP-ASSOCIATED FACTOR Ⅱ 15 (TAFII15) (Werner et al. 2021) amplified by primers Cp395/Cp396 was used as reference genes. The sequences of primers used in RT-qPCR analyses are listed in Supplemental Table S3.
To detect mRNA modifications, mRNA was isolated from total RNA for digestion using the PolyATtract mRNA Isolation Systems III (Promega). For mRNA preparation, 2 rounds of purification were performed. The RNA was quantified using a NanoDrop spectrophotometer (Thermo Scientific), and the purity and integrity of the RNA were assessed by applying 100 ng of RNA per sample on a Bioanalyzer 2100 system (Agilent).
Quantitative analyses of the ac4C/C ratio in RNA by LC-MS/MS
RNA samples were fully digested into single nucleosides as described previously (Chen et al. 2018; Chen and Witte 2020) with minor modifications. Approximately 800 ng of total RNA or mRNA isolated from 2-week-old seedlings of Arabidopsis thaliana (Col-0), Zea mays (Cultivar B73), Solanum lycopersicum (Micro-Tom), Oryza sativa (Nipponbare), Nicotiana benthamiana, and Glycine max (Williams 82) were individually digested by benzonase (0.4 unit; Sigma-Aldrich), phosphodiesterase I (0.004 units; Sigma-Aldrich), and shrimp alkaline phosphatase (0.04 unit; NEB) in 50 μL of buffer containing 10 mM Tris–HCl, pH 7.9, 1 mM MgCl2, and 0.1 mg mL−1 BSA. After incubation at 37 °C for 10 h, the samples were filtered in ultrafiltration tubes (3 kDa cutoff; Pall), and 2 μL aliquots were analyzed by an Agilent 1290 HPLC system coupled to a Sciex 6500 Qtrap mass spectrometer. The nucleosides were separated on a WATERS T3 column (2.1 × 100 mm; particle size 1.8 µm) at a flow rate of 0.3 mL min−1. Solvent A was 0.1% formic acid, and Solvent B was acetonitrile. The gradient was 0 min, 5% B; 0.5 min, 5% B; 3 min, 90% B; 4 min, 90% B; 4.1 min, 5% B and 7 min, 5% B. The mass transitions m/z 286.3 to 112 for N4-acetylcytidine and m/z 244.3 to 111.9 for cytidine were monitored. Standard solutions of cytidine (1, 5, 25 50, 100, 200, 400, 2,000, and 10,000 ng mL−1) and N4-acetylcytidine (0.005, 0.01, 0.05, 0.1, 0.5, 1, 5, 25, and 50 ng mL−1) were used for quantification. The ratio of N4-acetylcytidine to cytidine was calculated based on the calibrated concentrations.
Subcellular localization and immunoblot analysis
YFP-ACYR1 or YFP-ACYR2 was transiently coexpressed with the nucleus marker protein RFP-H2B (CFP fused to histone 2B) (Martin et al. 2009) in N. benthamiana leaves for 5 d. The samples were analyzed using a ZEISS LSM 980 with Airyscan2 microscope equipped with an HC PLAPO CS2 40 × 1.0 water immersion objective (ZEISS Microsystems). The acquired images were processed using ZEN 2.3 (blue edition) software (Carl Zeiss Microscopy GmbH).
Monoclonal anti-Strep antibody from mouse (1:1000; Abcam, ab184224) was used together with antimouse IgG conjugated to horseradish peroxidase (HRP; 1:1000; Solarbio SE131) to detect Strep on immunoblots employing a WesternBright™ ECL kit (Advansta, Lot: 220616-60).
Analysis of transcript stability
For the transcription inhibition assays, 20-day-old seedlings of the wild type (Col-0), acyr1 acyr2/+, or acyr1/+ acyr2 mutants were transferred to a 1/2 MS medium containing 100 μM actinomycin D (AbMole BioScience). Six seedlings of each genotype were collected at 0, 1, 2, and 3 h after treatment. Total RNA was then extracted for reverse transcription and quantification of gene expression levels. 18S rRNA was used as an internal reference gene.
RNA-seq
Triplicate biological replicates of total RNA were isolated from 4-week seedlings of wild type (Col-0), acyr1 acyr2/+, and acyr1/+ acyr2 mutants as described above. Rosette leaves from 3 individual plants were pooled as 1 biological replicate. The purity and integrity of the RNA were assessed by a NanoDrop spectrophotometer (Thermo Scientific) and a Bioanalyzer 2100 system (Agilent). mRNA was isolated from total RNA as described above. A total amount of 1 μg RNA per sample was applied as input material for RNA sample preparation. The RNA library was generated by Novogene Co. Ltd. using a NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB) following the manufacturer's recommendations, and index codes were added to attribute sequences to each sample. Transcriptome sequencing (RNA-seq) was performed on an Illumina NovaSeq platform. Clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N, and low-quality reads from raw data (raw reads). The resulting reads were aligned to the Arabidopsis reference genome (TAIR10) using Hisat2 v2.0.5 (Mortazavi et al. 2008). The mapped reads of each sample were assembled by StringTie (v1.3.3b) (Pertea et al. 2015) using a reference-based approach. FeatureCounts v1.5.0-p3 was used to calculate FPKM of each gene (Liao et al. 2014). Differential expression analysis was performed using the DESeq2 R package (1.20.0) (Love et al. 2014). Genes with an adjusted P-value <0.05 were assigned as differentially expressed.
acRIP-seq and validation
The acRIP-seq analyses were performed by DIATRE Biotechnology, Shanghai, China, according to a previous description (Arango et al. 2018) with minor modifications. Briefly, total RNA was extracted from the tissues and treated with DNase as described above. The amount and quality of RNA were tested using gel electrophoresis, an Equalbit RNA BR Assay Kit (Vazyme Biotech Co. Ltd.), and an Agilent Bioanalyzer 2100 system. Total RNA samples were fragmented into ∼150 nt using NEBNext Magnesium RNA Fragmentation buffer. Fragmented RNAs were incubated with 5 μg anti-ac4C antibody (Abcam, ab252215) for 2 h at 4 °C in 100 μl IP buffer containing PBS, 0.1% BSA, Triton X-100, and 40 U RNase inhibitor (NEB). The mixture was immunoprecipitated by incubation with Dynabeads Protein A for Immunoprecipitation (Thermo Fisher Scientific) at 4 °C for 2 h. After washing 5 times with IP buffer, the eluted RNA was precipitated by ethanol and used for library construction using a NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB). Sequencing was performed on an Illumina NovaSeq platform. Each genotype was performed with 2 biological replicates. Rosette leaves from 3 individual 4-week-old plants were pooled as 1 biological replicate for total RNA extraction.
acRIP-qRT-PCR was performed to validate the ac4C sequencing results. Immunoprecipitated RNA described as above was reverse transcribed with random hexamers using HiScript II Q Select RT Super Mix (Vazyme Biotech Co. Ltd.). Relative enrichment of the fragments within the target gene was determined by qRT-PCR and calculated by normalizing the amount of the fragment against that of the TIP41-like gene as an internal control and then by normalizing the value for the immunoprecipitated sample against that for the input. The primer pairs Cp702/Cp703, Cp704/Cp705, Cp706/Cp707, Cp708/Cp709, and Cp691/Cp692 were used to amplify fragments No. 1 to 5 of the TGH transcript, respectively.
Peak calling and analyses
Adaptors and low-quality reads were trimmed from raw sequencing reads with trim galore v0.6.6 using default parameters, and the resulting clean reads were mapped to the reference genome (TAIR10.51) using STAR (Dobin et al. 2013). Samtools (Li et al. 2009) were used to sort the SAM files. Peak calling was performed using MACS2 software with default settings (q value <0.05 and fold change >1). Based on the peak center position, peaks were assigned to 5′UTR, CDS, introns, and 3′UTR regions. The distribution of ac4C peaks along mRNAs was analyzed using guitar scripts (Cui et al. 2016). Consensus motifs containing ac4C peaks were identified by MEME-ChIP (Ma et al. 2014).
Phylogenetic analysis
Protein sequences for NAT10 were obtained from 46 plant species and 13 nonplant organisms (accession numbers in Supplemental Data Set 1). A phylogenetic tree was constructed with Mega X (Kumar et al. 2018) using the neighbor-joining method (Saitou and Nei 1987). A total of 1,000 bootstraps were performed. The alignment and tree files are provided in Supplemental File S1 and Supplemental File S2, respectively.
Statistical analysis
Statistical analysis was performed as previously described (Heinemann et al. 2021). R software (version 4.2.1) was used with the CRAN packages multcomp and sandwich to perform two-sided Tukey's pairwise comparisons. Heteroscedasticity of the data set was examined using the sandwich variance estimator (Herberich et al. 2010; Pallmann and Hothorn 2016). Different letters indicate significant differences at P < 0.05. The adjusted P-values of the individual data sets are listed in Supplemental Data Set 5.
Accession numbers
The materials generated in this work are available on reasonable request from the authors. Sequence data related to this article can be found in the Arabidopsis information portal (https://www.arabidopsis.org/) under accession numbers ACYR1 (At1g10490), ACYR2 (AT3G57940), TGH (AT5G23080), S1P (At5g19660), PLUTO (At5g03555), SRT1 (At5g55760), SRT2 (At5g09230), ACTIN2 (At3g18780), TAFII15 (At4g31720), and TIP41-like (At4g34270). acRIP-seq and RNA-seq data in this study have been deposited in the National Center for Biotechnology Information Sequence Read Archive under BioProject PRJNA929652 and PRJNA930058, respectively.
Supplementary Material
Acknowledgments
The authors thank Jialei Wang for her help with mass spectrometry analyses, Christel Schmiechen for her assistance with seedling phenotype analyses, and Markus Niehaus for his help with statistical analyses. This work was financially supported by the Fundamental Research Funds for the Central Universities [grant number YDZX2023013]; the National Natural Science Foundation of China [grant number 31900907]; the Natural Science Foundation of Jiangsu Province, China [grant number BK20190528]; the International Centre for Genetic Engineering and Biotechnology [grant number CRP/CHN20-04_EC]; the Deutsche Forschungsgemeinschaft (DFG) [grant numbers CH2292/1-1 to M.C. and C.-P.W, WI3411/8-1, and INST 187/741-1 FUGG to C.-P.W.]; the Special Fund on Technology Innovation of Carbon Dioxide Peaking and Carbon Neutrality of Jiangsu Province [grant number BE2022306 to J.Y.]; and the start-up fund for advanced talents from Nanjing Agricultural University to M.C.
Contributor Information
Wenlei Wang, College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, P.R. China.
Huijie Liu, College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, P.R. China.
Feifei Wang, College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, P.R. China.
Xiaoye Liu, Department of Criminal Science and Technology, Nanjing Forest Police College, Nanjing 210023, P.R. China.
Yu Sun, College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, P.R. China.
Jie Zhao, College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, P.R. China.
Changhua Zhu, College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, P.R. China.
Lijun Gan, College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, P.R. China.
Jinping Yu, Jiangsu Key Laboratory for the Research and Utilization of Plant Resources, Institute of Botany, Jiangsu Province and Chinese Academy of Sciences (Nanjing Botanical Garden Mem. Sun Yat-Sen), Nanjing 210014, P.R. China.
Claus-Peter Witte, Department of Molecular Nutrition and Biochemistry of Plants, Institute of Plant Nutrition, Leibniz University Hannover, Hannover 30419, Germany.
Mingjia Chen, College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, P.R. China.
Author contributions
M.C. conceived the research. W.W., H.L., F.W., X.L., Y.S., J.Z., C.Z., L.G., and J.Y. performed the experiments. C.-P.W. and M.C. analyzed the data and wrote the manuscript. All authors read and agreed with the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. The purity of the mRNA preparation.
Supplemental Figure S2. Identification of ac4C modification in total RNA and mRNA of Arabidopsis by mass spectrometry.
Supplemental Figure S3. Quantification of ac4C and m3U in total RNA and mRNA.
Supplemental Figure S4. Multiple alignment of 2 ACYRs from Arabidopsis and NAT10 from human.
Supplemental Figure S5. Characterization of mutants in ACYR1 or ACYR2.
Supplemental Figure S6. Expression analysis of ACYR1 and ACYR2 in seedlings of Col-0 and both hemizygous mutant lines.
Supplemental Figure S7. Characterization of the complementation line.
Supplemental Figure S8. Vegetative growth of the acyr lines.
Supplemental Figure S9. Pearson correlation analyses of acRIP-seq data from the wild type and the 2 hemizygous mutants.
Supplemental Figure S10. Uniquely and repeatedly identified ac4C-modified transcripts in the 2 replicates performed for each genotype in the acRIP-seq analysis.
Supplemental Figure S11. GO analysis of the ac4C containing transcripts in Col-0.
Supplemental Figure S12. Different and identical ac4C peaks identified in acyr1 acyr2/+ versus Col-0 compared to acry1/+ acyr2 versus Col-0.
Supplemental Figure S13. Venn diagram for different ac4C-modified transcripts identified between acyr1 acyr2/+ versus Col-0 and acyr1/+ acyr2 versus Col-0.
Supplemental Figure S14. Correlation between differential gene expression and ac4C modification level in the acyr1/+ acyr2 mutant compared to Col-0.
Supplemental Figure S15. Venn diagrams for the transcripts with differential abundance and ac4C modification level between acyr1 acyr2/+ versus Col-0 and acyr1/+ acyr2 versus Col-0.
Supplemental Table S1. Quantification of silique length and embryo number in wild type and mutants varying in ACYR expression
Supplemental Table S2. The most abundant consensus motif in Col-0, acyr1 acyr2/+, and acyr1/+ acyr2 lines using MEME suite.
Supplemental Table S3. Primers used in this study.
Supplemental Data Set 1. NAT10 homologs from 46 plant species and 13 nonplant organisms
Supplemental Data Set 2. Confident ac4C peaks in Col-0, acyr1 acyr2/+, and acyr1/+ acyr2.
Supplemental Data Set 3. RNA-seq raw data overview.
Supplemental Data Set 4. Differential ac4C peaks in acyr1 acyr2/+ versus Col-0, and acyr1/+ acyr2 versus Col-0.
Supplemental Data Set 5. Adjusted P-values for all multiple comparisons.
Supplemental File S1. Sequence alignments of NAT10 homologs from 46 plant species and 13 nonplant organisms.
Supplemental File S2. Machine-readable tree file.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003:301(5633):653–657. 10.1126/science.1086391 [DOI] [PubMed] [Google Scholar]
- Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler MD, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018:175(7):1872–1886.e24. 10.1016/j.cell.2018.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango D, Sturgill D, Yang R, Kanai T, Bauer P, Roy J, Wang Z, Hosogane M, Schiffers S, Oberdoerffer S. Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine. Mol. Cell. 2022:82(15):2797–2814.e11. 10.1016/j.molcel.2022.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmus G, Larrieu D, Barros AC, Collins C, Abrudan M, Demir M, Geisler NJ, Lelliott CJ, White JK, Karp NA, et al. Targeting of NAT10 enhances healthspan in a mouse model of human accelerated aging syndrome. Nat Commun. 2018:9(1):1700. 10.1038/s41467-018-03770-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Villalobos LIA, Kuhnle C, Dohmann EMN, Li H, Bevan M, Schwechheimer C. The evolutionarily conserved TOUGH protein is required for proper development of Arabidopsis thaliana. Plant Cell. 2005:17(9):2473–2485. 10.1105/tpc.105.031302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005:33(20):e179. 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Urs MJ, Sánchez-González I, Olayioye MA, Herde M, Witte C-P. M6a RNA degradation products are catabolized by an evolutionarily conserved N6-methyl-AMP deaminase in plant and mammalian cells. Plant Cell. 2018:30(7):1511–1522. 10.1105/tpc.18.00236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Witte C-P. A kinase and a glycosylase catabolize pseudouridine in the peroxisome to prevent toxic pseudouridine monophosphate accumulation. Plant cell. 2020:32(3):722–739. 10.1105/tpc.19.00639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Wei Z, Zhang L, Liu H, Sun L, Zhang S-W, Huang Y, Meng J. Guitar: an R/Bioconductor package for gene annotation guided transcriptomic analysis of RNA-related genomic features. Biomed Res Int. 2016:2016:8367534. 10.1155/2016/8367534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005:139(1):5–17. 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics. 2013:29(1):15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolata J, Taube M, Bajczyk M, Jarmolowski A, Szweykowska-Kulinska Z, Bielewicz D. Regulation of plant microprocessor function in shaping microRNA landscape. Front. Plant Sci. 2018:9:753. 10.3389/fpls.2018.00753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H-C, Wei L-H, Zhang C, Wang Y, Chen L, Lu Z, Chen PR, He C, Jia G. ALKBH10B Is an RNA N6-methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell. 2017:29(12):2995–3011. 10.1105/tpc.16.00912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Harada BT, Behm M, He C. RNA Modifications modulate gene expression during development. Science. 2018:361(6409):1346–1349. 10.1126/science.aau1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T. RNA Modifications: what have we learned and where are we headed? Nat Rev Genet. 2016:17(6):365–372. 10.1038/nrg.2016.47 [DOI] [PubMed] [Google Scholar]
- Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: form, distribution, and function. Science. 2016:352(6292):1408–1412. 10.1126/science.aad8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann KJ, Yang S-Y, Straube H, Medina-Escobar N, Varbanova-Herde M, Herde M, Rhee S, Witte C-P. Initiation of cytosolic plant purine nucleotide catabolism involves a monospecific xanthosine monophosphate phosphatase. Nature Commun. 2021:12(1):6846. 10.1038/s41467-021-27152-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberich E, Sikorski J, Hothorn T. A robust procedure for comparing multiple means under heteroscedasticity in unbalanced designs. PLoS One. 2010:5(3):e9788. 10.1371/journal.pone.0009788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006:57(1):19–53. 10.1146/annurev.arplant.57.032905.105218 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018:35(6):1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009:25(16):2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014:30(7):923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001:25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Noble WS, Bailey TL. Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat Protoc. 2014:9(6):1428–1450. 10.1038/nprot.2014.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 2009:59(1):150–162. 10.1111/j.1365-313X.2009.03850.x [DOI] [PubMed] [Google Scholar]
- Martínez-Pérez M, Aparicio F, López-Gresa MP, Bellés JM, Sánchez-Navarro JA, Pallás V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc Natl Acad Sci U S A. 2017:114(40):10755–10760. 10.1073/pnas.1703139114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown PJ, Ruszkowska A, Kunkler CN, Breger K, Hulewicz JP, Wang MC, Springer NA, Brown JA. Naturally occurring modified ribonucleosides. Wiley Interdiscip Rev RNA. 2020:11(5):e1595. 10.1002/wrna.1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008:5(7):621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Pallmann P, Hothorn LA. Analysis of means: a generalized approach using R. J Appl Stat. 2016:43(8):1541–1560. 10.1080/02664763.2015.1117584 [DOI] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. Stringtie enables improved reconstruction of a transcriptome from RNA-Seq reads. Nat biotechnol. 2015:33(3):290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Xie M, Dou Y, Zhang S, Zhang C, Yu B. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci U S A. 2012:109(31):12817–12821. 10.1073/pnas.1204915109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, Eeckhout D, El-Showk S, Li H, Zhong S, et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017:215(1):157–172. 10.1111/nph.14586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987:4(4):406–425. 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- Samad AFA, Sajad M, Nazaruddin N, Fauzi IA, Murad AMA, Zainal Z, Ismail I. MicroRNA and transcription factor: key players in plant regulatory network. Front Plant Sci. 2017:8:565. 10.3389/fpls.2017.00565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sas-Chen A, Thomas JM, Matzov D, Taoka M, Nance KD, Nir R, Bryson KM, Shachar R, Liman GLS, Burkhart BW, et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature. 2020:583(7817):638–643. 10.1038/s41586-020-2418-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002:14(12):2985–2994. 10.1105/tpc.004630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Langhendries J-L, Watzinger P, Kötter P, Entian K-D, Lafontaine DLJ. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015:43(4):2242–2258. 10.1093/nar/gkv075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. Functional interdependence of m6A methyltransferase complex subunits in Arabidopsis. Plant Cell. 2023:35(6):1901–1916. 10.1093/plcell/koad070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Liang Z, Wong CE, Yu H. Messenger RNA modifications in plants. Trends Plant Sci. 2019:24(4):328–341. 10.1016/j.tplants.2019.01.005 [DOI] [PubMed] [Google Scholar]
- Tang J, Yang J, Lu Q, Tang Q, Chen S, Jia G. The RNA N6 -methyladenosine demethylase ALKBH9B modulates ABA responses in Arabidopsis. J Integr Plant Biol. 2022:64(12):2361–2373. 10.1111/jipb.13394 [DOI] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009:136(4):669–687. 10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- Werner S, Bartrina I, Schmülling T. Cytokinin regulates vegetative phase change in Arabidopsis thaliana through the miR172/TOE1-TOE2 module. Nat Commun. 2021:12(1):5816. 10.1038/s41467-021-26088-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener D, Schwartz S. The epitranscriptome beyond m6A. Nat Rev Genet. 2021:22(2):119–131. 10.1038/s41576-020-00295-8 [DOI] [PubMed] [Google Scholar]
- Xu C, Zhang J, Zhang J, Liu B. SIRT7 Is a deacetylase of N4-acetylcytidine on ribosomal RNA. Genome Instab Dis. 2021:2(4):253–260. 10.1007/s42764-021-00046-x [DOI] [Google Scholar]
- Yang X, Yang Y, Sun B-F, Chen Y-S, Xu J-W, Lai W-Y, Li A, Wang X, Bhattarai DP, Xiao W, et al. 5-methylcytosine Promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017:27(5):606–625. 10.1038/cr.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Jia T, Chen X. The ‘how' and ‘where’ of plant microRNAs. New Phytol. 2017:216(4):1002–1017. 10.1111/nph.14834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-seq (MACS). Genome Biol. 2008:9(9):R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.