Abstract
Determining the drivers of non-native plant invasions is critical for managing native ecosystems and limiting the spread of invasive species1,2. Tree invasions in particular have been relatively overlooked, even though they have the potential to transform ecosystems and economies3,4. Here, leveraging global tree databases5–7, we explore how the phylogenetic and functional diversity of native tree communities, human pressure and the environment influence the establishment of non-native tree species and the subsequent invasion severity. We find that anthropogenic factors are key to predicting whether a location is invaded, but that invasion severity is underpinned by native diversity, with higher diversity predicting lower invasion severity. Temperature and precipitation emerge as strong predictors of invasion strategy, with non-native species invading successfully when they are similar to the native community in cold or dry extremes. Yet, despite the influence of these ecological forces in determining invasion strategy, we find evidence that these patterns can be obscured by human activity, with lower ecological signal in areas with higher proximity to shipping ports. Our global perspective of non-native tree invasion highlights that human drivers influence non-native tree presence, and that native phylogenetic and functional diversity have a critical role in the establishment and spread of subsequent invasions.
Subject terms: Invasive species, Biogeography, Forest ecology
Analysis combining multiple global tree databases reveals that whether a location is invaded by non-native tree species depends on anthropogenic factors, but the severity of the invasion depends on the native species diversity.
Main
Plant invasions have multifaceted impacts on ecosystems and human wellbeing across the globe1–3,8. It is expected that plant invasions will continue to increase in the coming decades owing to human-assisted introduction and naturalization of these species, with ever-growing impacts on biodiversity within native forest ecosystems1,9,10. These invasions will undoubtedly also have considerable economic impacts in managed landscapes by disrupting timber production, agriculture and human livelihoods11–17. In particular, non-native trees represent an important and increasing concern globally, as they are often actively planted far outside their native ranges for forestry, reforestation, residential, or ornamental purposes4,18. Along with the passive spread of non-native species, the active propagation of trees by humans can often result in an increased potential to become problematic invaders4,19–21. Given the prominent roles of trees in shaping the structure and functioning of ecosystems, such tree invasions have the capacity to alter plant composition, productivity, biodiversity and the services provided to humans1,4,22. Previous research in invasion ecology has expanded our understanding of community-level properties that influence ecosystem susceptibility to invasion23–25, as well as traits that make plant species more likely to become invasive26–30. However, most work has been restricted to local and regional scales31,32, with contrasting ecological mechanisms affecting invasion success in different regions. We thus lack a global unified theory of the human and ecological drivers of tree species invasions33. Developing an integrated global understanding of ecological and anthropogenic forces that drive non-native tree invasions is critical to improve decision making in conservation and management.
Countless ecological mechanisms have been proposed to explain the susceptibility of different ecosystems to invasion by non-native species in different locations. Traditionally, more diverse or ecologically complex systems are thought to exhibit ‘biotic resistance’ to invasion23,34–39. This hypothesis is based on the assumption that greater diversity in the native community fills the available ecological niches and reduces available resources, limiting niche space to novel species. However, most work has focused on testing this hypothesis using species richness as an indicator of niche filling23,35, which may not fully capture the proportion of niches that are filled in the native community. Instead, more informative metrics for niche filling may be phylogenetic or functional diversity. Phylogenetic diversity accounts for evolutionary similarity and represents a reasonable proxy for similarity between taxa, whereas functional diversity directly addresses the underlying mechanism of biotic resistance (that is, the breadth of ecological niches filled), but may be more difficult to measure. Conversely, there is also evidence for the opposite pattern in some ecosystems, whereby a more diverse community is indicative of a more favourable habitat, where a wide range of invasive species might survive. This ‘biotic acceptance’25,40,41 hypothesis leads to the expectation that highly diverse sites are optimal for many plant species and could promote invasion of non-native species. Nonetheless, we still lack a unified understanding of the relative importance of these two competing processes, and their variation across the globe, leading to ongoing calls to resolve this ‘invasion paradox’25.
Invasion success is also likely to depend on the ecological strategy of the invading species relative to the recipient native community. One school of thought is that environmental constraints are the primary drivers of plant species distributions. Therefore, to be successful, invasive species ought to be similar to native species that are adapted for that region, especially in extreme environments42. Under this ‘environmental filtering hypothesis’43,44 (or ‘preadaptation hypothesis’), invasive species will be more successful if their traits mirror those of the native community45. For example, to be successful in a harsh desert environment, non-native plants would need to be ecologically similar to native plants to survive, possessing traits that protect them against high heat and water loss. By contrast, the ‘limiting similarity hypothesis’ (also known as ‘Darwin’s naturalization hypothesis’) postulates that invasive species need to be ecologically distinct from native species to avoid niche overlap46–49. Here, invaders are thought to be more successful if they can fill unique niche spaces that are not already used by the native community, reducing competition and enabling their establishment. These two processes suggest contrasting mechanisms for how species invade: either species invade by being similar or dissimilar to the native community (Darwin’s naturalization conundrum24,50). It is possible that the relative importance of these opposing ecological mechanisms varies under different environmental conditions, with greater importance of environmental filtering in harsh conditions and greater niche differentiation in more moderate environments51,52. Such regional variation in the relative importance of these mechanisms might help to explain the opposing responses observed across studies. However, until now, we lack a broad-scale analysis of these different invasion mechanisms that can help us to see past the idiosyncrasy of local-scale observations to identify unifying trends.
A key challenge hindering a global consensus of the ecological patterns and mechanisms underpinning plant invasion is that these processes are likely strongly influenced by anthropogenic activity, which may dampen the signal of ecological drivers. Humans drive contemporary plant invasions through highly efficient transport—both intentional and accidental—of non-native plants, with proximity to ports and airports being associated with increased invasion11,53,54. A constant influx of non-native species may override a native community’s ability to resist invasion55 (biotic resistance) and obscure the impacts and importance of specific ecological drivers, such as native diversity, particularly at early stages of invasion. That is, with increased propagule pressure of non-natives species exerted by humans, the relative importance of ecological drivers may be reduced. Moreover, sites with high levels of non-native propagule pressure due to human activity are also likely to be heavily disturbed, compounding this anthropogenic influence. Accounting for human global change drivers may be particularly important when considering the role of invasion strategy, with the potential for anthropogenic drivers and human propagule pressure to overwhelm the impact of ecological drivers. This could occur through an increase in the frequency and magnitude of introductions, which would be expected to increase stochastic variation and dampen ecological signals. So far, these hypotheses have been tested only at local and regional scales, with few studies integrating ecological and anthropogenic drivers of invasion at the global scale to disentangle the relative importance of human activity, environmental conditions and biological diversity33.
Here, by combining global datasets of local-scale forest inventories, native status, environmental climate variables and anthropogenic drivers, we test for the relative importance of ecological and anthropogenic influence on non-native tree invasion. Using this large-scale approach, we search for a unifying perspective of the environmental and anthropogenic contexts driving non-native invasion and invasion severity, via both relative richness and abundance of non-natives, as well as invasion strategy. We consider three hypotheses: (H1) greater native diversity reduces non-native invasion23; (H2) high levels of environmental filtering in extreme environmental conditions leads to similarity of non-natives with the surrounding natives, and moderate conditions are associated with greater levels of niche differentiation and dissimilarity24; and (H3) human drivers, specifically proximity to ports and areas of high human population density, will mediate and potentially override these ecological relationships56. We explore these hypotheses through the lens of different biodiversity metrics (phylogenetic diversity, functional diversity and species richness), providing a comprehensive view of the interactions between ecological processes and human influence on invasion. Addressing these hypotheses is important to highlight generalizations in the field for prevention and management of non-native tree invasions, which is key to mitigating the potential severe ecological and socio-economic toll of these invasions.
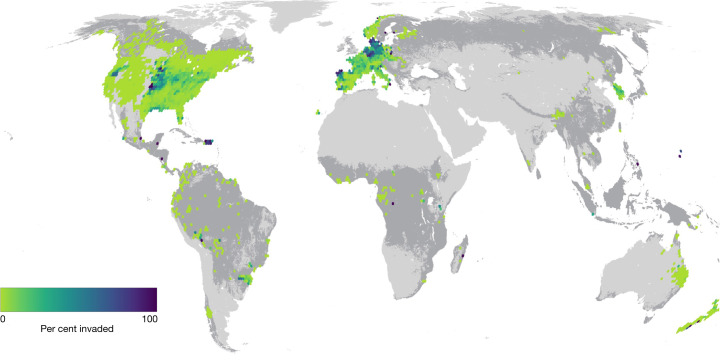
Using the Global Forest Biodiversity Initiative database7, we determined native tree status (native or non-native) according to the Global Naturalized Alien Flora6 and the KEW Plants of the World databases5. This dataset encompassed 471,888 plots, of which 4.9% of plots were invaded, or contained at least one non-native tree species (Fig. 1 and Supplementary Table 1a). Moreover, this dataset contained a larger proportion of invaded plots in tropical (15.2%) than in temperate systems (5.2 %). Overall, 249 individual non-native tree species were identified, with the most frequent being Robinia pseudoacacia, Pinus sylvestris, Maclura pomifera, Picea abies and Ailanthus altissima labelled as non-native in 3,976, 2,603, 2,493, 2,468 and 1,597 plots, respectively (Supplementary Table 2). Regions with the greatest likelihood of being invaded include North America, Europe and East Asia (Extended Data Fig. 1), consistent with previous findings10,57 (but see ref. 58). To test for drivers of non-native tree invasion and invasion strategy, we used a down-sampled version of the dataset consisting of 17,738 forest plots, distributed across 14 biomes proportional to their global land cover.
Fig. 1. Distribution of the study data.
Distribution of the full study dataset, coded for non-native severity (n = 471,888 plots). The map shows average per cent invasion across a 1-degree hexagonal grid, from non-invaded (0%) pixels in green to completely invaded (100%) pixels in purple. Plots are considered invaded if there is any non-native tree present.
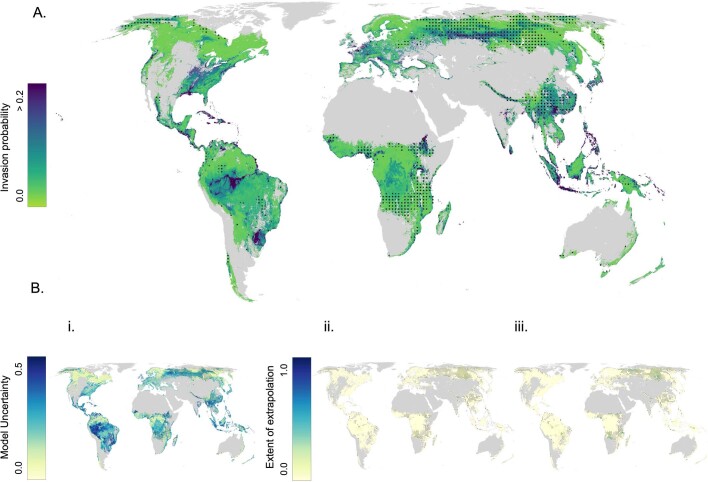
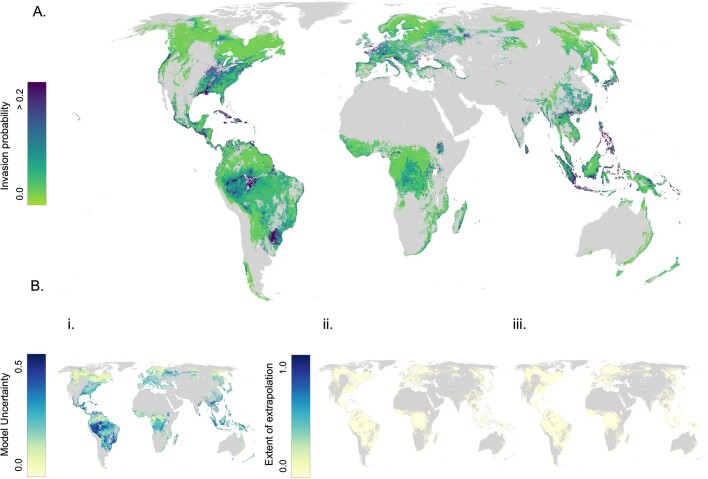
Extended Data Fig. 1. Map of non-native invasion probability.
Map showing probability of non-native tree presence based on the probability output of the random forest classifier (A, total n = 368,030 plots, n per iteration = 10,000 plots) alongside maps showing uncertainty in predictions (B) including local uncertainty of invasion probability via bootstrapped coefficient of variation (i) and extent of extrapolation as percentage of bands outside univariate (ii) and multivariate (ii) training range. Regions outside the Area of Applicability are indicated with dots.
We calculated three metrics of invasion: (1) presence of non-natives in the plot (‘non-native presence’); (2) relative proportion of non-native species richness to total tree richness (‘non-native richness’); and (3) relative proportion of non-native species basal area to total tree basal area (‘non-native abundance’). The first metric (non-native presence) is simply a measure of the presence or absence of invasion, whereas the latter two metrics (relative abundance and richness) provide insight into the subsequent severity of the invasion.
To test how hypothesized human and environmental drivers affected the probability a forest plot was invaded or the invasion severity within invaded plots, we built generalized linear models (GLMs) and random forest models using either phylogenetic or functional diversity metrics (both as richness and redundancy) as predictor variables (Extended Data Fig. 3). For both functional and phylogenetic diversity, we used random forest models to determine variable importance and for visualization purposes, whereas GLMs were used to test for significance and directionality of relationships. Our models also included human drivers (distance to shipping ports (hereafter referred to as ports) and population density) and accounted for several additional soil chemical and climate variables. Next, to test whether non-native tree species invade by being similar or dissimilar to the native community (termed ‘invasion strategy’), we again built models predicting non-native similarity from either native phylogenetic or functional diversity metrics, along with the same environmental and human impact variables. The non-native invasion strategy was defined as the change in redundancy due to addition of non-native trees, with values below zero and values above zero indicating invasion via similarity and dissimilarity, respectively, to the native community.
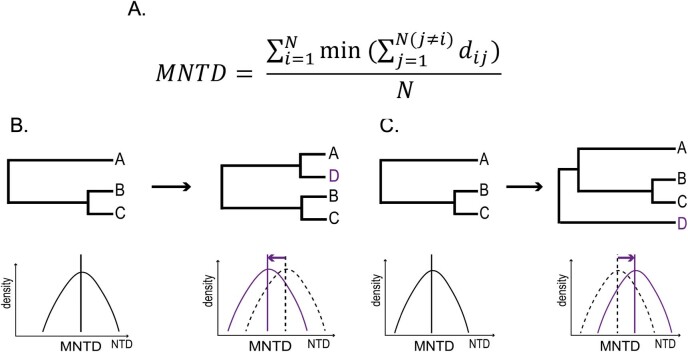
Extended Data Fig. 3. Mean nearest taxon distance (MNTD).
Mean nearest taxon distance is the average distance to nearest neighbor by branch length on the tree, which represents redundancy in the community (A). For each species i, the sum of all shortest distances d to each other taxa j is calculated; these values are then averaged across the total species in the tree (N). If invasion occurs via non-natives being similar to the native community, this would lead to the expectation that MNTD decreases, increasing redundancy (B). Conversely, if non-native invasion occurs via non-natives being dissimilar to the native community, this would lead to the expectation that MNTD increases, reducing redundancy (C). Taxon D represents a non-native addition to the community.
Diversity limits invasion severity
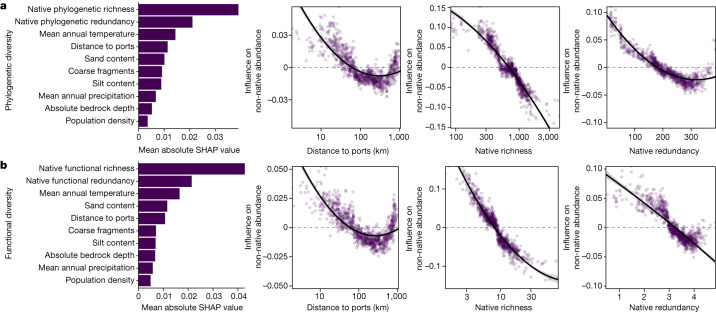
We found that anthropogenic drivers were more important than local native tree diversity in determining non-native invasion (presence) globally (H3), whereas native diversity— both phylogenetic and functional—was most important in determining invasion severity (H1; Fig. 2 and Supplementary Tables 3 and 4; phylogenetic diversity random forest area under the curve (AUC) = 0.634, functional diversity random forest AUC = 0.631). These results indicate the importance of human-induced propagule pressure in initiating invasion of forests and of native biodiversity moderating the severity of the invasion. We found that forest plots closer to ports are more likely to be invaded (Supplementary Tables 3 and 4; linear model P < 0.001). Notably, these results are consistent whether we analyse all data together at the global level or separate data into either the temperate and tropical bioclimatic zones (Supplementary Tables 3 and 4). By contrast, we did not find that human population density was consistently related to non-native presence, with results being variable across diversity metrics and bioclimatic zones considered (Supplementary Tables 3 and 4). However, population density was always positively correlated with invasion probability; population density may be a weaker predictor as it only measures human presence, which is not necessarily related to propagule pressure.
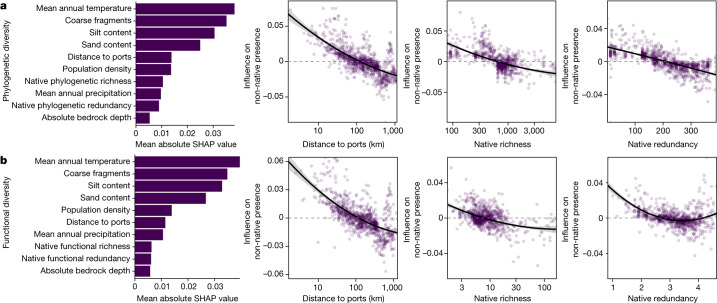
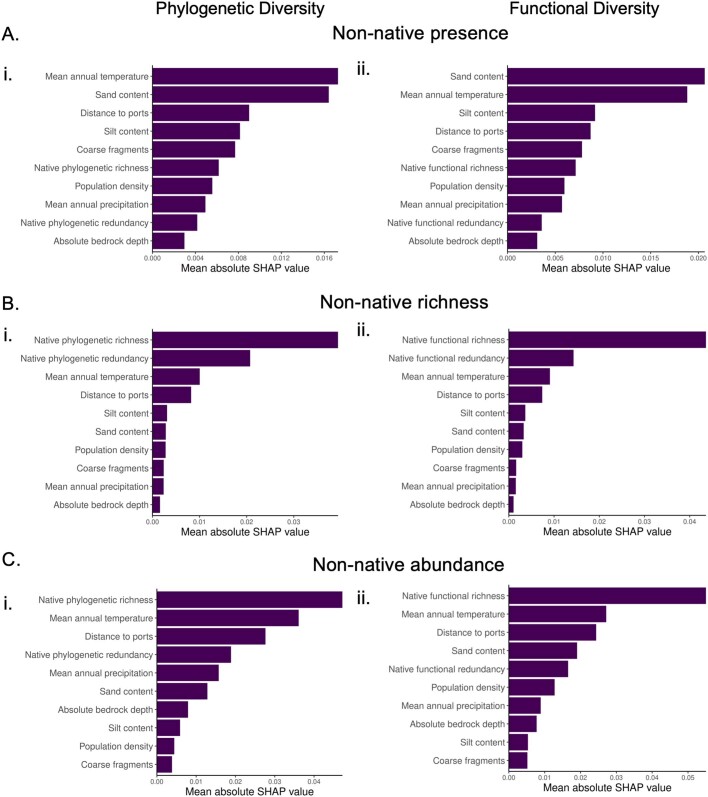
Fig. 2. Anthropogenic drivers are more important than native diversity in determining invasion occurrence.
a,b, Importance (Shapley additive explanations (SHAP) values) of all variables included in random forest models ordered from greatest to least important, alongside influence of distance to ports, native richness and native redundancy on non-native presence (whether a plot is invaded or not) for global models of phylogenetic (a) and functional (b) diversity (phylogenetic diversity, n = 17,640 plots; functional diversity, n = 17,271 plots). All results shown are from random forest models. Note that y-axis ranges differ among panels, with the variable importance plots representing the corresponding magnitude. Error bands represent 95% confidence intervals.
Proximity to ports has long been known to influence invasion11,53,54, with locations closer to a port being likely to experience greater propagule pressure. Moreover, proximity to ports may serve as a proxy for residence time, where plots closer to ports are more likely to have longer exposure to non-native propagule pressure, thus increasing the likelihood of invasion56. Yet, at far enough distances, stochastic processes and historical land-use patterns may begin to weaken the role of ports (Fig. 3, distances greater than 500 km). For example, the third most frequent non-native tree in our dataset, M. pomifera, is widely naturalized throughout the interior of North America, where it has been used for various agricultural purposes dating back to the 1850s59. Such results highlight the idiosyncratic use of trees across the globe, leading to unique invasion trends relative to herbaceous plants. Nevertheless, at more local scales, this strong signal of anthropogenic activity and associated propagule pressure relative to native diversity driving non-native presence is in agreement with previous work that considers invasion across stages56 and recent assessments of regional and global tree invasion57,60, and highlights the prominent role of humans in reshaping biological communities.
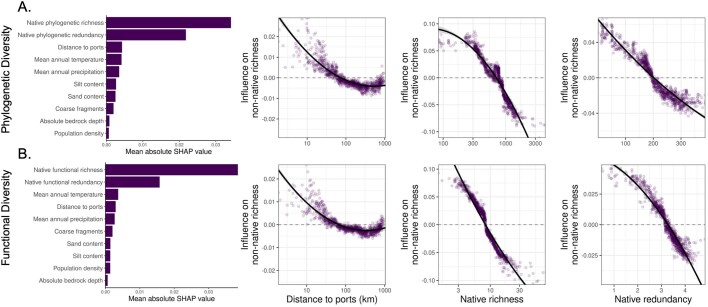
Fig. 3. Native diversity is the most important driver of invasion severity.
a,b, Importance (Shapley additive explanations (SHAP) values) of all variables included in random forest models ordered from greatest to least important, alongside influence of distance to ports, native richness and native redundancy on invasion severity for global models of phylogenetic (a) and functional (b) diversity (phylogenetic diversity, n = 3,498 plots; functional diversity, n = 3,368 plots). Plots are shown for the severity of invasion measured as non-native species abundance (proportion of basal area with non-native plant species); plots for non-native species richness (proportion of non-native plant species) are shown in Extended Data Fig. 4. All results shown are from random forest models. Note that the y-axis ranges differ among panels, with the variable importance plots representing the corresponding magnitude. Error bands represent 95% confidence intervals.
Although proximity to ports determined the probability a forest plot was invaded, native tree communities with higher phylogenetic and functional diversity exhibited lower invasion severity (Fig. 3, Extended Data Fig. 4 and Supplementary Tables 3 and 4; phylogenetic diversity random forest non-native richness R2 = 0.68, phylogenetic diversity random forest non-native abundance R2 = 0.14, functional diversity random forest non-native richness R2 = 0.69 and functional diversity random forest non-native abundance R2 = 0.07; GLM phylogenetic and functional diversity P < 0.001). Additionally, distance to ports was no longer significant in linear models predicting invasion severity (Supplementary Tables 3 and 4) for both phylogenetic (P = 0.16 and 0.28 for non-native richness and abundance, respectively) and functional diversity models (P = 0.63 and 0.86 for non-native richness and abundance, respectively), and showed reduced variable importance in the random forest models (Fig. 3 and Extended Data Fig. 4). When investigating these patterns using conventionally analysed species richness instead of phylogenetic or functional richness, we find similar qualitative results (Supplementary Table 5, random forest non-native richness R2 = 0.71 and random forest non-native abundance R2 = 0.14), suggesting that species diversity may be a useful proxy for projecting invasion severity in the absence of functional and phylogenetic information. Our results are consistent with the hypothesis of biotic resistance (H1), where increased native diversity reduces invasion success, which is probably driven by the native community utilizing more available niche spaces23,34–36,61. These results are also consistent with work investigating tree migration drivers that suggests that migration is slower into more diverse communities owing to greater resource use (fewer available niches) in these systems57.
Extended Data Fig. 4. Native diversity mediates degree of non-native invasion.
Variable importance (SHAP values) of all variables included in random forest models, ordered from greatest to least importance alongside influence of distance to ports, native richness and native redundancy on invasion severity (proportion of non-native plant species) for (A) phylogenetic diversity and (B) functional diversity global models (phylogenetic n = 3,498 plots; functional n = 3,368 plots). All results shown are from random forest models. Note that y-axis ranges differ among panels, with the variable importance plots representing the corresponding magnitude.
Overall, these results show that anthropogenic drivers, particularly distance to shipping centres (ports), are more important in determining which locations will experience non-native invasions compared with traditionally studied native diversity (H3). However, it is the intrinsic ecological drivers, including native tree community phylogenetic and functional diversity (richness and redundancy), that are more important in determining invasion severity (H1). Repeated human introduction of plant species has a more important role in the initial invasion process, but invasion severity is predominantly a result of native intrinsic diversity. Notably, both distance to ports and native diversity show patterns of saturation of effects, suggesting a threshold at which plots that are far enough from ports, or high enough in native diversity, will not benefit from further distance or diversity with regard to reduced invasion or invasion severity. Although our focus here is on the relative importance of human versus biotic drivers of introduction, we find that environmental variables—especially mean annual temperature—correlate strongly with patterns of non-native invasion, which may reflect resource availability26, belowground microorganism composition30 or potential climate compatibility between donor and recipient ranges62. Together, our results suggest that locations near human activity are more likely to experience non-native invasions in part due to increased propagule pressure, whereas those with lower diversity are more likely to experience more severe non-native invasions once non-natives are present. These results may suggest that managing forests to maintain high native tree diversity may be a good strategy to buffer communities against invasion, particularly for locations that are far from human activity.
Evidence for environmental filtering
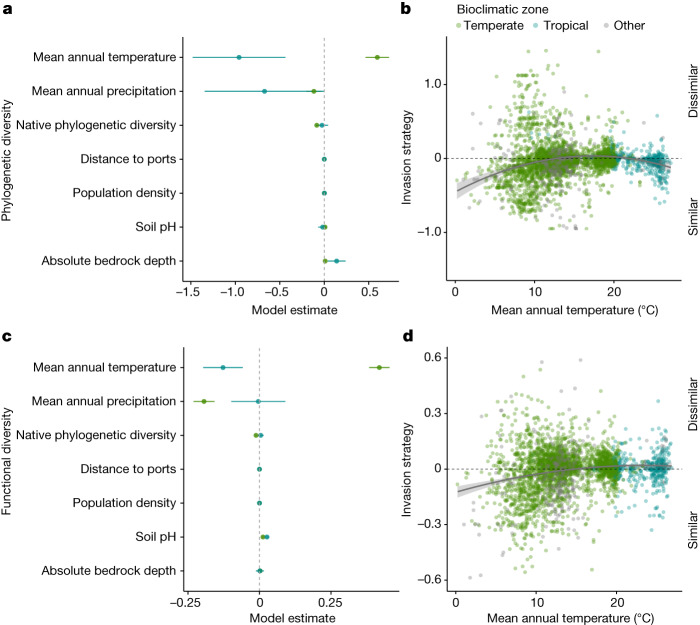
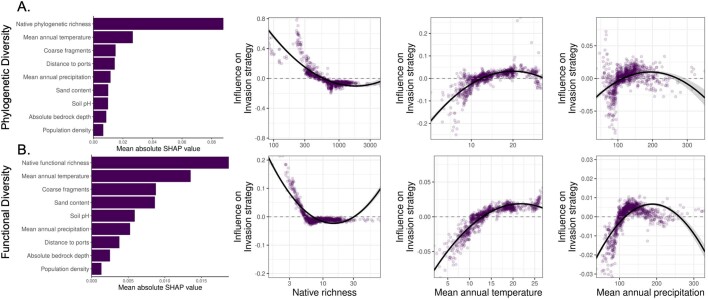
When considering a range of climate, soil and anthropogenic variables, we find evidence for environmental filtering as a driver of invasion strategy, in particular, with respect to mean annual temperature and precipitation. In all global models, temperature was important for predicting tree invasion strategy (Fig. 4, Extended Data Fig. 5 and Supplementary Table 6; phylogenetic diversity random forest R2 = 0.084, functional diversity random forest R2 = 0.099; H2), with our global analysis indicating that non-native trees were more similar to the native community in environments at cold and hot temperature extremes (Fig. 5 and Supplementary Table 6, P < 0.001). That is, in order to invade into a cold or hot environment, non-native plants are more successful if they share similar traits with native plants to survive in these harsher temperature conditions. By contrast, at locations with moderate temperatures, non-natives are neither more nor less similar to native communities, potentially because these less harsh environmental conditions allow a wider range of life strategies to coexist51. For functional diversity, invasion strategy at high temperatures is relatively neutral, with the line approaching a value of zero, suggesting that although phylogenetically similar, these communities show some level of functional divergence, highlighting the importance of including functional diversity in future studies. When separating the data into temperate and tropical systems, we found divergent temperature patterns (Supplementary Table 6; temperate P < 0.001, tropical P = 0.01). In temperate systems, non-native trees were more likely to be similar to the native tree community in colder environments relative to hot environments, in line with previous results in temperate North America63. In tropical systems, we found the opposite pattern, with non-native trees being more likely to be similar to the native tree community in hotter tropical environments. At the lowest temperatures, non-natives invading through similarity were primarily gymnosperms (fir, spruce and pine species) invading into native communities containing species in the same genus; by contrast, at the highest temperatures, non-natives invading through similarity were angiosperms, with a high prevalence of palms and legumes. Further, we detect a similar pattern of environmental filtering for mean annual precipitation when analysing phylogenetic and functional diversity with random forest models, where lower or higher precipitation is associated with non-native invasion through similarity (Extended Data Fig. 5). This suggests that the most likely invaders at low or high temperature or precipitation may be ecologically similar to the host communities, which could inform invasion risk checklists at ports.
Fig. 4. Environmental filtering at temperature extremes.
a,c, Estimates of overlapping variables included in temperate and tropical GLM models (forest plot) for phylogenetic (a) and functional (c) diversity models (phylogenetic diversity, n = 3,498; functional diversity, n = 3,368). Values to the left of the zero line indicate negative model estimates, and those to the right indicate positive estimates. b,d, Relationship between mean annual temperature and invasion strategy for phylogenetic (b) and functional (d) diversity models, showing that at extreme temperatures invasion occurs through similarity (Supplementary Table 7; phylogenetic diversity: P(1) = 9.69 × 10−14, P(2) = 2.13 × 10−11; functional diversity: P(1) < 2 × 10−16, P(2) = 1.07 × 10−4, where P(1) and P(2) represent each temperature and temperature squared P values, respectively). Note for functional diversity, this pattern only holds at low temperatures. Error bars and bands represent standard error.
Extended Data Fig. 5. Variable importance for non-native invasion strategy.
Variable importance (SHAP values) of all variables included in random forest models, ordered from greatest to least importance alongside influence of native richness, mean annual temperature and mean annual precipitation on invasion strategy for (A) phylogenetic diversity and (B) functional diversity global models (phylogenetic n = 3,498 plots; functional n = 3,368 plots). All results shown are from random forest models. Note that y-axis ranges differ among panels, with the variable importance plots representing the corresponding magnitude. Error bands represent 95% confidence intervals.
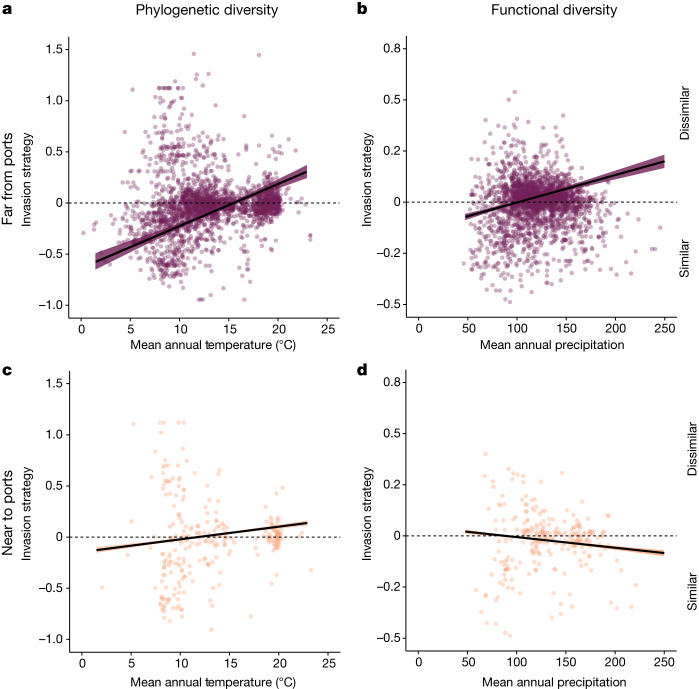
Fig. 5. Proximity to ports weakens environmental filtering in the temperate bioclimate zone.
a,b, In temperate plots far from ports, temperature is positively correlated with an invasion strategy of increasing dissimilarity for phylogenetic (a) and functional (b) diversity (phylogenetic diversity: n = 2,710 plots, P = 6.37 × 10−6; functional diversity: n = 2,603, P < 2 × 10−16). c,d, This relationship between temperature and invasion strategy weakens for phylogenetic (c) and functional (d) diversity with proximity to ports (Supplementary Table 7; phylogenetic diversity: P = 0.0001; functional diversity: P = 2.71 × 10−13). Lines and points represent the lowest (c,d) and highest (a,b) 10% of data. Error bands represent standard error.
Within the temperate bioclimatic zone, we found evidence that anthropogenic activity weakened the environmental filtering pattern for phylogenetic and functional diversity seen for temperature and precipitation, respectively (H3). In particular, proximity to ports modified the signal of environmental filtering due to temperature, weakening the influence of temperature on invasion strategy with respect to phylogenetic similarity (Fig. 5 and Supplementary Table 6; P < 0.001). Colder ecosystems show evidence of environmental filtering of invasion; however, increased proximity to ports reduces the prevalence of this strategy. We suggest that this may be due to increased introductions around shipping ports, which would increase stochastic variation and dampen ecological strategies. However, we did not detect a similar interaction governing the tropical bioclimatic zone, potentially owing to relatively lower human pressure, and particularly lower ship traffic64, compared to temperate systems. Alternatively, this pattern may also reflect the fact that some temperate plots occur at greater distances to ports than tropical sites (95th percentile of 784 km versus 311 km for temperate and tropical, respectively), increasing statistical power for detecting this trend in temperate regions. Furthermore, proximity to ports also marginally weakened the signal of environmental filtering due to precipitation for functional invasion strategy (Supplementary Table 6; P = 0.07). These results illustrate that human influence can override the ecological factors driving invasion, suggesting that at high enough propagule pressure, the phylogenetic and functional similarity of a non-native becomes less important in predicting its ability to invade a native community. Nevertheless, as our analyses are not causal, these results could also reflect correlations between port locations and invasion strategy. However, when we investigated the same effect with human population density, we did not see this weakening effect, demonstrating that distance to ports seems to be a particularly relevant mediator of these patterns. These results suggest that human activity may overwhelm ecological drivers of non-native invasion strategies and reduce the influence of ecological processes, making inclusion of human impacts critical for studying global invasion strategies.
Collectively, our work integrates biotic and anthropogenic factors across phylogenetic and functional diversity for both invasion presence and invasion severity of non-native tree species worldwide. Although non-native trees have been relatively overlooked relative to herbaceous plants, their large size, long lifespans and important history in forestry, food, reforestation and city landscaping exposes trees to unique ecological and anthropogenic factors that shape their worldwide distributions. Moreover, given that many tree invasions are in their infancy, with substantial ‘invasion debts’ of recent tree plantings3, understanding the ecological drivers promoting spread has the potential to provide real-time feedback for the preventative management of invasive trees. However, there are important considerations when interpreting these findings, many of which could be addressed with increased data resolution and increased sampling within under-sampled geographic regions. First, our analysis is largely observational, whereas community composition would ideally be compared before and after invasion to better understand the causality of the trends observed here. We can gain some insight into this question by conducting a sensitivity analysis on the subset of invaded plots that were measured at multiple time points and that had no initial invasion. Doing so reveals that the reduction in native diversity due to invasion can potentially account for as much as 10.4% (mean of 6.7%) of the observed biotic resistance (Supplementary Table 9), but that the remainder of this effect is attributable to difference in native diversity (that is, biotic resistance) across plots. Additional long-term data on plots that are uninvaded and become invaded will be useful in further addressing the influence of invasion on native diversity. Second, many tree species in our analysis were only identified to genus level or were not present in the master plant phylogeny, which may lead to an underestimation of native diversity or invasive species richness in some plots, particularly in species-rich forests. Indeed, a key challenge in global analyses such as ours is the underrepresentation of certain ecosystems, for example, tropical ecosystems58. This is addressed to some extent by our down-sampling approach, as well as our spatial cross-validation approach (Methods), but ongoing efforts to fund and develop open-access and fair65 tropical forest inventory data are critical for gaining better insight into these ecologically and socially important ecosystems.
Many tree species are intentionally introduced for forestry or wood products and may be managed4, generating variation in the drivers underpinning invasion that are unique to trees. To minimize the influence of heavily managed forests, we included only plots with a minimum of three species and thus our dataset does not include monoculture forestry plantations. In addition, when restricting our analysis to the subset of global plots that occur in protected areas with minimal human footprint, our core results and inferences remain unchanged (Supplementary Table 7). Having additional high-quality data on the human role in invasion, including the type and time of management, and overall level in disturbance regime66, would refine our results and better separate ecological versus human drivers. Future work should also focus on drivers of tree invasion and invasion strategies across scales25,63,67, as patterns may differ at scales larger than the local plot level that we include here, which may be important for regional versus local management of non-native trees. Finally, emerging work shows that the consideration of native range size and change in environment and/or disturbance from donor to recipient community may be more helpful in understanding introduction and invasion success than simply quantifying these variables in the novel, recipient range62,66. Therefore, including the change in environmental and human impact variables would also be a fruitful avenue for future research.
Together, these results provide important unifying insights into the global drivers of non-native tree invasions and the ecological strategies that might be most successful in different regions. The trends and ecological mechanisms identified here can provide tangible guidelines to support forest management of non-native tree invasions around the globe. However, because non-native trees are introduced purposefully for forestry or to support local livelihoods, which can lead to differences in forest management objectives and strategies4, it is critical that local stakeholders are included when making decisions about how to best manage these introductions68,69. Ultimately, this emerging understanding of global tree invasions provides fundamental insights that are needed to understand how forest composition is being reshaped under global change, and for forest management practices to limit the spread and impacts of non-native tree invasions worldwide.
Methods
Tree inventory and non-native status
For tree inventory data, we used the Global Forest Biodiversity Initiative (GFBI) database7, which contains tree-level abundance data for more than 1.2 million forest plots on all continents across the globe, containing more than 31 million unique georeferenced records of tree size and density dating from 1958. Each observation in the dataset consists of a unique tree ID, plot ID, plot coordinates, tree diameter at breast height (DBH), tree-per-hectare expansion factors, year of measurement, and binomial species names. In this study, we applied several filters to these data before analyses. First, where plots had multiple years of data, we kept only the most recent year of census data. We then subset the data to include only plots with at least three species as required for our phylogenetic metrics, excluding monoculture forest plantations from the study.
To assign native status to each tree species (native or non-native, representing naturalized and invasive), we established a consensus status between the Global Naturalized Alien Flora (GloNAF)6 and the KEW Plants of the World70 databases. All databases were standardized to The Plant List taxonomy71. The GloNAF database contains detailed, georeferenced information on the naturalized status of more than 10,000 plant species in each of 1,029 regions across the globe representing countries or federal states; the KEW database outlines native ranges of vascular plant species for over 1.2 million plant species70. The GFBI and GloNAF datasets were joined by matching each unique species by location in GFBI to a GloNAF region polygon and species status. Then, for each GFBI plot, we extracted the GloNAF region identifier using Google Earth Engine72. This process was then repeated for the KEW database. We then filtered out plots that included any species with disagreement between GloNAF and KEW databases (that is, conflicting native status), and only included trees with a minimum diameter of 5 cm and a minimum height of 1.3 m to allow for DBH measurements. All trees identified as ‘non-native’ were verified to be listed in the BGCI Tree List, which defines a tree as, “A woody plant with usually a single stem growing to a height of at least two metres, or if multi-stemmed, then at least one vertical stem five centimetres in diameter at breast height”73. Note that this is an inclusive definition which includes monocots and tree ferns, as well as species that can occur both as tall single-stem and shrub-like multi-stem phenotypes.
To account for unequal representation of plots across biomes (Fig. 1), we used a reduced version of this database, down-sampled to a number of plots proportional to the land area covered by each of 14 biomes (Supplementary Table 1), while conserving as many tropical plots as possible. This ensured that we were not overrepresenting historically oversampled biomes, particularly in temperate regions. In addition, we preferentially retained invaded plots during this down-sampling to ensure adequate representation of invaded plots in the final dataset, with a maximum of half of the plots within a biome being invaded. This oversampling of invaded plots allowed for adequate representation of invaded and non-invaded plots in our analyses of non-native presence, and allowed sufficient data for our analyses of invasion severity, as these analyses only used data from plots that had non-native species invasions. Results were not qualitatively different if we did not preferentially retain invaded plots in our down-sampling (Extended Data Fig. 6 and Supplementary Table 8). Note also that the global mapping used the full dataset, with no subsampling. Prior to analyses, we also collapsed locations with multiple replicate plots and removed plots where phylogenetic of functional diversity could not be calculated for both native and full communities due to less than three species being present (see below).
Extended Data Fig. 6. Variable importance for analyses using data down-sampled without preferentially retaining invaded plots.
Variable importance (SHAP values) for all variables included in random forest models, ordered from greatest to least importance for (A) non-native presence, (B) richness, and (C) abundance, each for (i) phylogenetic diversity and (ii) functional diversity global models (presence: phylogenetic n = 18,898; functional n = 18,611, richness: phylogenetic n = 840 plots; functional n = 823 plots, abundance: phylogenetic n = 840 plots; functional n = 823 plots). All results shown are from random forest models with down-sampled data, but without preferentially retaining invaded plots. Error bands represent 95% confidence intervals.
Non-native invasion metrics
We split our invasion metrics into the two broad categories of ‘non-native invasion’ (presence) and ‘invasion severity’. Specifically, using our data, we were able to determine for each plot (1) whether any non-native tree species were present (non-native presence); (2) the proportion of tree species that were non-native relative to total tree species (invasion severity, assessed via non-native richness)23; and (3) the proportion basal area of non-native tree species relative to total tree species basal area (invasion severity, assessed via non-native abundance). These metrics are congruent with recently proposed frameworks for measuring and reporting invasive plant species74,75. The metric of relative introduced species richness may be hypothesized to lead to a bias in detection of biotic resistance, with greater biotic resistance falsely detected in diverse communities, as these communities will have a lower proportion of non-native trees due to the higher denominator (total site diversity). However, use of the binomial approach in our GLM modelling of this proportion, as opposed to direct proportion, overcomes this limitation, as it uses raw counts of proportion, effectively weighting observations by the total species number in the community23.
Climatic and anthropogenic variables
For climatic and anthropogenic variables, we relied on the Global Environmental Composite76,77. This global database contains spatially explicit geographic information system (GIS) layers of more than 260 unique environmental variables, encompassing climate, soil, land cover and land use, plant biomass, topography, human footprint, and disturbance78,79. Climate variables were extracted from the CHELSA (climatologies at high resolution for the earth’s land surface areas) dataset78, whereas soil variables were from the SoilGrids80 dataset. In addition, we created distance measures by calculating the spherical distance to shipping ports81 and airports82. All layers were standardized to a 30 arcsec resolution (~1 km2 at the equator), a resolution at which these variables have been shown to have an influence on plant biogeography and assembly patterns83,84. We chose model variables to represent both climate and soil properties that exhibited low collinearity for each of three datasets: global (all 14 biomes from Supplementary Table 1), temperate (temperate broadleaf, coniferous, grassland biomes) and tropical (tropical moist broadleaf, deciduous broadleaf, coniferous, and grassland biomes). We chose to use distinct variables rather than transforming them into principal component analysis axes for increased interpretability of these variables and their effects. Because variables exhibiting collinearity varied between the three datasets, the resulting models include different variable combinations. For all models, we used mean annual temperature (MAT), mean annual precipitation (MAP), distance to shipping ports81 (hereafter ‘ports’) and human population density85. For the global models, we used the following additional environmental variables: absolute depth to bedrock, coarse fragments, sand content and soil pH. For temperate models, we used absolute depth to bedrock, clay content, and soil pH as additional variables; for tropical models we used absolute depth to bedrock, soil organic content, and soil pH as additional variables. All soil variables used were determined at a depth of 0 cm, or the top layer of soil.
Diversity metrics
We analysed data using either phylogenetic or functional diversity; these two approaches were chosen to be as analogous as possible. Phylogenetic alpha diversity explains the genetic relatedness of species within a community and is often assumed to represent a proxy for functional similarity across species within a community assemblage. Yet, congruency between these two metrics remains under debate86,87 and their role in invasion patterns remains untested; therefore, we focused on two major axes of diversity, explaining richness and divergence in the community across both phylogenetic and functional space88, capturing both evolutionary and ecological processes. For each native and entire tree community (native and non-native species), we calculated Faith’s phylogenetic diversity (phylogenetic richness) and mean nearest taxon distance (MNTD, phylogenetic redundancy; Extended Data Fig. 2). Entire tree community metrics were calculated on all species, whether they were matched to GloNAF and KEW or not; this included tree species which were identified to genus level. Faith’s phylogenetic diversity was calculated as the sum of the branch lengths on the phylogenetic tree of the species in the community; MNTD was calculated as the average distance to the nearest neighbour across the community. These metrics were calculated based on tree placement of taxa in a recently published reference backbone tree for plants89. Out of 13,345 starting taxa, a total of 12,325 were placed on the reference tree, with 4,960 placed at the species level and 7,365 placed at the genus level. We chose MNTD over other available metrics describing community divergence because we were interested in redundancy of the community, and this metric captures this best24,90. To enable a more intuitive understanding of this metric, we transformed each community-level value of MNTD to the maximum MNTD across all communities minus calculated MNTD. This transformed the maximum value to zero and all smaller values transformed to increasingly larger numbers, with higher MNTD values indicating a greater native redundancy, similar to the expected increased redundancy with greater phylogenetic richness (Faith’s phylogenetic diversity). To determine the non-native invasion strategy, or impact of non-natives on native MNTD, we calculated the difference between the native and non-native community relative to the native community alone. We used the following formula for non-native invasion strategy: (entire community MNTD – native community MNTD)/native community MNTD. When non-native invasion strategy was greater than zero, this indicated that the addition of the non-native species resulted in a more dissimilar community, whereas a non-native invasion strategy less than zero corresponded to the opposite.
Extended Data Fig. 2. Map of non-native invasion probability inside the area of applicability.
Map showing probability of non-native tree presence based on the probability output of the random forest classifier (A, total n = 368,030 plots, n per iteration = 10,000 plots) alongside maps showing uncertainty in predictions (B) including local uncertainty of invasion probability via bootstrapped coefficient of variation (i) and extent of extrapolation as percentage of bands outside univariate (ii) and multivariate (ii) training range. Regions outside the Area of Applicability are masked.
For functional diversity, we calculated the analogous metrics using trait distance matrices instead of phylogenetic tree-based distances. We selected eight traits extracted from Maynard et al.83 that represented the major clusters of functional trait diversity, thereby capturing the full spectrum of tree form and function while minimizing correlation between traits. Maynard et al.83 used data from the TRY plant trait database to parametrize machine learning models to estimate the expression of 18 traits as a function of the local environment and/or phylogeny. The observed trait data underlying these models encompassed 491,001 unique observations across 13,189 species from 2,313 genera, with consistent representation across taxonomic orders. The resulting models were then used to generate trait estimates for 52,255 tree species, capturing approximately 80% of documented tree species91. Using this trait database, we were able to assign trait value to 81% of the tree species in GFBI reported to the species level. The eight traits we included in our metrics were chosen to include traits typically associated with plant invasion28,92 including those associated with dispersal, establishment, resource acquisition and competitive ability that represent the major trait clusters encompassing the full dimensionality of trait space from Maynard et al.83 The eight traits included in our study were the following: wood density, root depth, leaf nitrogen, leaf phosphorus, leaf area, tree height, seed dry mass, and bark thickness. All traits were log-transformed and normalized to allow for statistically valid comparisons83. To obtain functional diversity metrics analogous to those used for phylogenetic diversity, we used the dendrogram approach of Petchey and Gaston93. Specifically, for every plot we calculated the species-by-species trait distance matrix encompassing all eight traits, and then used hierarchical clustering to create a functional dendrogram. This dendrogram was subsequently used to calculate ‘functional richness’ (analogous to Faith’s phylogenetic diversity) and ‘functional redundancy’ (MNTD); we use this terminology for functional diversity to maintain naming of variables between phylogenetic and functional diversity analyses. Metrics were calculated in R using packages ape94, tidyverse95, abdiv96, doParallel97, foreach98 and pez99.
Because both functional and phylogenetic diversity metrics have unique limitations, we considered them both here so as to obtain a more robust view of underlying patterns and processes. The benefit of phylogenetic diversity is that it does not rely on imputed data, and thus it provides more consistent results with lower uncertainty. However, phylogenetic diversity is only a loose proxy for functioning, depending on the degree to which the functional traits of interest are phylogenetically conserved. Thus, as a complement of this, we also use imputed trait values to estimate functional diversity, which should better capture underlying functional differences across species, but which is subject to higher uncertainty relative to phylogeny (or measured trait values), and may omit rare and potentially functionally unique species. Thus, by simultaneously considering both functional and phylogenetic diversity and showing that these metrics yield consistent global trends, our approach provides consistent evidence that these patterns are robust to the limitations of either approach taken individually.
Statistical analyses
We combined random forest100 and GLM approaches to answer our focal questions. Specifically, we used random forest models to visualize patterns and determine variable importance, while GLMs were used to assess statistical significance and directionality of patterns. We first tested for environmental and anthropogenic drivers of non-native invasion, including non-native presence and invasion severity (non-native richness, non-native abundance). Our independent variables included either phylogenetic or functional metrics, climate and soil variables, and human impact variables. Next, we tested the impact of these variables on non-native invasion strategy (difference in MNTD due to non-natives). We focused on addressing specific hypotheses related to drivers of non-native invasion and invasion strategy. We acknowledge the importance of other variables, and therefore included them in our models, but do not interpret each variable.
Random forest models and GLMs used the same model designs. Models predicting non-native presence as well as invasion severity, for both non-native richness and abundance, included independent predictor variables of native diversity and native redundancy, as well as climate and human driver variables detailed in ‘Climatic and anthropogenic variables’. For comparison, we repeated these models with native tree species richness in place of both diversity variables (richness and redundancy), as species richness is commonly used in the invasion literature when testing for biotic resistance23,34,35. Finally, we used an adapted version of the random forest models, removing diversity variables, to assess probability of locations with non-native trees globally and generate an associated map (Extended Data Fig. 1).
To account for spatial autocorrelation in the modelling step, we used residual autocovariates (RACs)101,102. First, we used simple linear regression to determine the range of spatial autocorrelation for the models with continuous outcomes (invasion severity and invasion strategy). We then assessed residual spatial autocorrelation using correlelogram plots using the ncf103 package in R, which showed that residual correlation was consistently negligible beyond 250 km, which was also applied to the models with binary outcomes (non-native presence). Using this buffer distance, we generated RAC values using the autocov_dist() function in the spdep package70,104, which determines an inverse distance weighted residual value for each data point in the 250 km neighbourhood. RAC incorporates the spatial signature of the model residuals, relative to the model without any spatial autocorrelation correction, into a variable that is included in each model101,102. The result is an inverse distance weighted residual value for each data point in the 250 km neighbourhood, which we used as continuous predictors in both the linear and random forest models.
Random forest models were used primarily to assess variable importance and influence. Specifically, we used Shapley additive explanations (SHAP) values to infer variable importance in the model outcome105,106. SHAP values are a machine learning analogue of partial regression, quantifying the relative importance of each variable on the outcome, accounting for all other variables in the model. To estimate the SHAP values, random forest models were fit in R using the ranger package107, using default hyperparameters (500 trees, observations sampled with replacement, number of variables per split equal to the square root of the number of predictors, a minimum of 5 observations per node). We then used the fastshap package108 to estimate approximate SHAP values for each predictor, using n = 100 simulations. The overall variable importance was taken as the sum of the absolute value of the SHAP values, and the marginal effect of each variable was visualized by plotting the covariate versus the corresponding SHAP value for each observation.
To account for spatial autocorrelation in the accuracy assessment of random forest models, we implemented spatially-buffered leave-one-out cross-validation (LOO-CV) to obtain conservative lower-bound accuracy measures109. To do this, we first randomly selected a focal observation as the test data, and then we omitted all observations within a 250 km buffer distance around this observation. The remaining data were used to train the model, and the resulting fit was used to predict outcome for the withheld focal observation. This was repeated 500 times for each model, each time selecting a new focal point and predicting its outcome using the 250 km spatially-buffered training set. The resulting accuracy measures were calculated on the set of 500 out-of-fit predictions. For continuous variables, we estimated accuracy using the cross-validated coefficient of determination relative to the one-to-one line (termed VEcv110), denoted simply R2 here, and for binary outcomes we used area under the ROC curve (AUC), which quantifies the ability of the classifier to distinguish between classes, and serves as an assessment of model performance.
To create a global map of invasion probability and its local uncertainty, we used a repeated prediction approach in Google Earth Engine60 (Extended Data Fig. 1a; AUC of spatial cross-validation = 0.84 ± 0.04, mean F1 score of non-native presence = 0.36). This repeated prediction approach used the full dataset without any down-sampling. To our knowledge, no global maps on phylogenetic or functional diversity metrics exist, so we were unable to include these diversity metrics in the random forest model for mapping; therefore, these models include the same covariates as the other models except diversity metrics. We thought it reasonable to exclude diversity metrics in this analysis as distance to ports is the most important driver of invasion probability, while native diversity is less important. After aggregating samples within the 30-arcsec pixels, 368,030 data points remained for our repeated prediction approach. We first trained 50 random forest models on stratified bootstrapped samples with a total of 10,000 data points each, using biome as stratification category; this allowed us to repeatedly predict the probability of non-native presence for each terrestrial pixel on Earth. The resulting 50 predictions were used to create per-pixel mean and coefficient of variation maps of the probability of non-native presence, with probabilities calibrated using Platt scaling111,112. These two maps allow us to investigate the patterns of invasion and the regions of uncertainty in the predictions. Next, the extrapolation extent was estimated as a per-pixel percentage of predictor variables, and interactions of predictor variables, outside of the training range, in univariate and multivariate space, respectively (Extended Data Fig. 1b)60. In addition, to account for gaps in predictor space, we estimated the Area of Applicability113, used to mark regions of extrapolation in this map. All maps are restricted to regions with a minimum of 10% forest cover114.
GLM models were used to estimate statistical parameters and conduct statistical tests. All GLM models included the same variables as those in the random forest models. In the models predicting non-native presence, we used a binomial distribution and logit link. For non-native abundance, we used a beta regression approach to predict the proportion of non-native basal area, as a method of modelling proportions between 0 and 1. We could not use a binomial GLM analogous to that used for non-native abundance because basal area measurements were not whole numbers and we wanted to retain all information in the data. Finally, to account for spatial autocorrelation and non-independently distributed residuals, we employed the inclusion of RACs as described above. These models were repeated separately for temperate and tropical bioclimatic zones, but results were qualitatively similar to the global model, so we report only global results here. All GLM results can be found in Supplementary Tables 3–5. GLMs were run in R (v. 4.2.2)115 using lme4116, lmerTest117, and betareg118, while visualizations for these models used ggplot2119; tidyverse95 was used throughout as well.
Because invasion of non-native species may alter the native diversity of the site into which they invade, we conducted a sensitivity test using plots where we had data across two time points to incorporate this effect. We first took all plots for which we had two time points, where the first time point represented a fully native community (that is, no presence of non-natives; n = 8,221 plots). We then modelled the per cent change of species richness in each plot from this uninvaded first time point to a later time point. Our predictor variables included final invasion status (non-natives present or not) to determine the impact of invasion on per cent change of species richness, along with all climate, soil, and anthropogenic impact variables we included in other global models. We extracted the coefficient of final invasion status (along with upper and lower confidence ranges), which quantifies the per cent change in richness due to invasion, and we used this to update the native species richness of the full global dataset. We then used these coefficients to estimate the pre-invasion native diversity for each plot in the global dataset by adding the corresponding species change resulting from invasion. Finally, we reran our global analysis with this updated pre-invasion native diversity. The relative contribution of native species loss to biotic resistance was calculated by comparing the relative change in the richness coefficient for each of the updated models relative to the original model (Supplementary Table 9).
Non-native invasion strategy was predicted using the difference in redundancy (MNTD) in the tree community due to invasion. We included the same variables as in the previous set of models, except native redundancy, as this is integrated in our response variable and therefore would exhibit high collinearity. In GLM models, we tested for the interaction between MAP and MAT to detect potential non-additive environmental filtering effects of these two dominant climate variables. In addition, we tested for the interaction between each MAP and MAT with distance to ports, to examine whether this important anthropogenic driver modified main ecological relationships. Final reported models are those resulting from a process of first creating a full model with all interactions, and subsequently removing nonsignificant interactions. All GLM results for invasion strategy can be found in Supplementary Table 7.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-023-06440-7.
Supplementary information
This file contains Supplementary Tables 1–9.
Acknowledgements
The authors acknowledge the Bernina Foundation and DOB Ecology for financial support. C.S.D. thanks the Swiss National Science Foundation (Postdoctoral Fellowship #TMPFP3_209925). C.S.D. also acknowledges funding from the Marc R. Benioff Revocable Trust, which, in collaboration with the World Economic Forum, also made this work possible. D.S.M. thanks the Swiss National Science Foundation (Ambizione Grant #PZ00P3_193612). The authors thank L. Mo for assistance in compiling the author list and G. Smith for early discussions about invasion severity. J.C.S. considers this work a contribution to Center for Ecological Dynamics in a Novel Biosphere (ECONOVO), funded by Danish National Research Foundation (grant DNRF173) and his VILLUM Investigator project ‘Biodiversity Dynamics in a Changing World’, funded by VILLUM FONDEN (grant 16549). P. Schall thanks the Deutsche Forschungsgemeinschaft (DFG) Priority Program 1374 Biodiversity Exploratories. G.A. thanks the French National Forest Inventory and the Italian Forest Inventory; G.A. was supported by the Italian National Recovery Plan through the National Biodiversity Future Center. Financial support from Monafor network in Mexico was funded by the National Forestry Commission (CONAFOR), Council of Science and Technology of the State of Durango (COCYTED), the Natural Environment Research Council, UK (NERC; NE/T011084/1), and local support of Ejidos and Comunidades.
Extended data figures and tables
Author contributions
C.S.D., T.W.C., D.S.M. and C.M.Z. contributed the conceptualization of the project. C.S.D. and D.S.M. contributed methodology, investigation and project administration. All authors contributed to data collection and/or curation. C.S.D., D.S.M., N.M.R. and T.L. contributed visualization. T.W.C., D.S.M. and C.S.D. obtained funding and provided supervision. Writing was led by C.S.D. and D.S.M., with review and editing contributed by all other co-authors.
Peer review
Peer review information
Nature thanks Blas Benito, Kevin Potter, Marcel Rejmánek and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Funding
Open access funding provided by Swiss Federal Institute of Technology Zurich.
Data availability
Data used in this study can be found in cited references for the Global Naturalized Alien Flora (GloNAF) database6 (non-native status), the KEW Plants of the World database5 (native ranges) and the Global Environmental Composite63,77 (environmental data layers). Plant trait data were extracted from Maynard et al.78. Data from the Global Forest Biodiversity Initiative (GFBI) database57 are not available due to data privacy and sharing restrictions, but can be obtained upon request via Science-I (https://science-i.org/) or GFBI (gfbinitiative.org) and an approval from data contributors.
Code availability
All code used to complete analyses for the manuscript is available at the following link: https://github.com/thomaslauber/Global-Tree-Invasion. Data analyses were conducted and were visualizations generated in R (v. 4.2.2), Python (v. 3.9.7), Google Earth Engine (earthengine-api 0.1.306), QGIS-LTR (v. 3.16.7) and the ETH Zurich Euler cluster.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
9/26/2023
A Correction to this paper has been published: 10.1038/s41586-023-06654-9
Extended data
is available for this paper at 10.1038/s41586-023-06440-7.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-023-06440-7.
References
- 1.Pyšek P, et al. Scientists’ warning on invasive alien species. Biol. Rev. 2020;95:1511–1534. doi: 10.1111/brv.12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pejchar L, Mooney HA. Invasive species, ecosystem services and human well-being. Trends Ecol. Evol. 2009;24:497–504. doi: 10.1016/j.tree.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 3.Richardson DM, Hui C, Nuñez MA, Pauchard A. Tree invasions: patterns, processes, challenges and opportunities. Biol. Invasions. 2014;16:473–481. [Google Scholar]
- 4.Richardson DM, Rejmánek M. Trees and shrubs as invasive alien species–a global review. Divers. Distrib. 2011;17:788–809. [Google Scholar]
- 5.Plants of the World Online. Royal Botanic Gardens Kewhttp://www.plantsoftheworldonline.org/ (2022).
- 6.van Kleunen M, et al. The Global Naturalized Alien Flora (GloNAF) database. Ecology. 2019;100:e02542. doi: 10.1002/ecy.2542. [DOI] [PubMed] [Google Scholar]
- 7.Liang J, et al. Positive biodiversity-productivity relationship predominant in global forests. Science. 2016;354:aaf8957. doi: 10.1126/science.aaf8957. [DOI] [PubMed] [Google Scholar]
- 8.Vitousek PM, D'Antonio CM, Loope LL, Rejmanek M, Westbrooks R. Introduced species: a significant component of human-caused global change. N. Z. J. Ecol. 1997;21:1–16. [Google Scholar]
- 9.Butchart SH, et al. Global biodiversity: indicators of recent declines. Science. 2010;328:1164–1168. doi: 10.1126/science.1187512. [DOI] [PubMed] [Google Scholar]
- 10.Early R, et al. Global threats from invasive alien species in the twenty-first century and national response capacities. Nat. Commun. 2016;7:12485. doi: 10.1038/ncomms12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lockwood, J. L., Hoopes, M. F. & Marchetti, M. P. Invasion Ecology 2nd edn (John Wiley & Sons, 2013).
- 12.DiTomaso JM. Invasive weeds in rangelands: species, impacts, and management. Weed Sci. 2000;48:255–265. [Google Scholar]
- 13.Simberloff D. We can eliminate invasions or live with them. Successful management projects. Biol. Invasions. 2008;11:149–157. [Google Scholar]
- 14.Mack RN, et al. Biotic invasions: causes, epidemiology, global consequences, and control. Ecol. Appl. 2000;10:689–710. [Google Scholar]
- 15.Boyd IL, Freer-Smith PH, Gilligan CA, Godfray HC. The consequence of tree pests and diseases for ecosystem services. Science. 2013;342:1235773. doi: 10.1126/science.1235773. [DOI] [PubMed] [Google Scholar]
- 16.Vitousek PM, D’Antonio CM, Loope LL, Westbrooks R. Biological invasions as global environmental change. Am. Sci. 1996;84:468–478. [Google Scholar]
- 17.Diagne C, et al. High and rising economic costs of biological invasions worldwide. Nature. 2021;592:571–576. doi: 10.1038/s41586-021-03405-6. [DOI] [PubMed] [Google Scholar]
- 18.Nunez MA, et al. Exotic mammals disperse exotic fungi that promote invasion by exotic trees. PLoS ONE. 2013;8:e66832. doi: 10.1371/journal.pone.0066832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson DM, Pyšek P. Naturalization of introduced plants: ecological drivers of biogeographical patterns. New Phytol. 2012;196:383–396. doi: 10.1111/j.1469-8137.2012.04292.x. [DOI] [PubMed] [Google Scholar]
- 20.Richardson DM, Pyšek P. Plant invasions: merging the concepts of species invasiveness and community invasibility. Prog. Phys. Geog. 2006;30:409–431. [Google Scholar]
- 21.Seabloom EW, et al. Human impacts, plant invasion, and imperiled plant species in California. Ecol. Appl. 2006;16:1338–1350. doi: 10.1890/1051-0761(2006)016[1338:hipiai]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 22.Dayton, P. K. in Proc. Colloquium on Conservation Problems in Antarctica (ed. Parker, B. C.) 81–96 (Allen Press, 1972).
- 23.Beaury EM, Finn JT, Corbin JD, Barr V, Bradley BA. Biotic resistance to invasion is ubiquitous across ecosystems of the United States. Ecol. Lett. 2020;23:476–482. doi: 10.1111/ele.13446. [DOI] [PubMed] [Google Scholar]
- 24.Cadotte MW, Campbell SE, Li S-P, Sodhi DS, Mandrak NE. Preadaptation and naturalization of nonnative species: Darwin’s two fundamental insights into species invasion. Annu. Rev. Plant Biol. 2018;69:661–684. doi: 10.1146/annurev-arplant-042817-040339. [DOI] [PubMed] [Google Scholar]
- 25.Fridley J, et al. The invasion paradox: reconciling pattern and process in species invasions. Ecology. 2007;88:3–17. doi: 10.1890/0012-9658(2007)88[3:tiprpa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 26.Funk JL. The physiology of invasive plants in low-resource environments. Conserv. Physiol. 2013;1:cot026. doi: 10.1093/conphys/cot026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuñez MA, Dickie IA. Invasive belowground mutualists of woody plants. Biol. Invasions. 2014;16:645–661. [Google Scholar]
- 28.van Kleunen M, Weber E, Fischer M. A meta-analysis of trait differences between invasive and non-invasive plant species. Ecol. Lett. 2010;13:235–245. doi: 10.1111/j.1461-0248.2009.01418.x. [DOI] [PubMed] [Google Scholar]
- 29.Pyšek, P. & Richardson, D. M. in Biological Invasions (ed. Nentwig, W.) 97–125 (Springer, 2007).
- 30.Dickie IA, et al. The emerging science of linked plant–fungal invasions. New Phytol. 2017;215:1314–1332. doi: 10.1111/nph.14657. [DOI] [PubMed] [Google Scholar]
- 31.Diez JM, Sullivan JJ, Hulme PE, Edwards G, Duncan RP. Darwin’s naturalization conundrum: dissecting taxonomic patterns of species invasions. Ecol. Lett. 2008;11:674–681. doi: 10.1111/j.1461-0248.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 32.Ma C, et al. Different effects of invader–native phylogenetic relatedness on invasion success and impact: a meta-analysis of Darwin’s naturalization hypothesis. Proc. R. Soc. B. 2016;283:20160663. doi: 10.1098/rspb.2016.0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Catford JA, Jansson R, Nilsson C. Reducing redundancy in invasion ecology by integrating hypotheses into a single theoretical framework. Divers. Distrib. 2009;15:22–40. [Google Scholar]
- 34.Levine JM, D'Antonio CM. Elton revisited: a review of evidence linking diversity and invasibility. Oikos. 1999;87:15–26. [Google Scholar]
- 35.Tilman D. Community invasibility, recruitment limitation, and grassland biodiversity. Ecology. 1997;78:81–92. [Google Scholar]
- 36.Fine PV. The invasibility of tropical forests by exotic plants. J. Trop. Ecol. 2002;18:687–705. [Google Scholar]
- 37.Elton, C. S. The Ecology of Invasion by Animals and Plants 1st edn (Methuen, 1958).
- 38.Kennedy TA, et al. Biodiversity as a barrier to ecological invasion. Nature. 2002;417:636–638. doi: 10.1038/nature00776. [DOI] [PubMed] [Google Scholar]
- 39.Mungi NA, Qureshi Q, Jhala YV. Role of species richness and human impacts in resisting invasive species in tropical forests. J. Ecol. 2021;109:3308–3321. [Google Scholar]
- 40.Stohlgren TJ, Jarnevich C, Chong GW, Evangelista PH. Scale and plant invasions: a theory of biotic acceptance. Preslia. 2006;78:405–426. [Google Scholar]
- 41.Zeiter M, Stampfli A. Positive diversity–invasibility relationship in species-rich semi-natural grassland at the neighbourhood scale. Ann. Bot. 2012;110:1385–1393. doi: 10.1093/aob/mcs193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zanne AE, et al. Functional biogeography of angiosperms: life at the extremes. New Phytol. 2018;218:1697–1709. doi: 10.1111/nph.15114. [DOI] [PubMed] [Google Scholar]
- 43.Kraft NJ, et al. Community assembly, coexistence and the environmental filtering metaphor. Funct. Ecol. 2015;29:592–599. [Google Scholar]
- 44.Keddy PA. Assembly and response rules: two goals for predictive community ecology. J. Veg. Sci. 1992;3:157–164. [Google Scholar]
- 45.Gallien L, Carboni M, Münkemüller T. Identifying the signal of environmental filtering and competition in invasion patterns–a contest of approaches from community ecology. Methods Ecol. Evol. 2014;5:1002–1011. [Google Scholar]
- 46.Divíšek J, et al. Similarity of introduced plant species to native ones facilitates naturalization, but differences enhance invasion success. Nat. Commun. 2018;9:4631. doi: 10.1038/s41467-018-06995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacArthur R, Levins R. The limiting similarity, convergence, and divergence of coexisting species. Am. Nat. 1967;101:377–385. [Google Scholar]
- 48.Park DS, Potter D. A test of Darwin’s naturalization hypothesis in the thistle tribe shows that close relatives make bad neighbors. Proc. Natl Acad. Sci. USA. 2013;110:17915–17920. doi: 10.1073/pnas.1309948110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaefer H, Hardy OJ, Silva L, Barraclough TG, Savolainen V. Testing Darwin’s naturalization hypothesis in the Azores. Ecol. Lett. 2011;14:389–396. doi: 10.1111/j.1461-0248.2011.01600.x. [DOI] [PubMed] [Google Scholar]
- 50.Darwin, C. On the Origin of Species by Means of Natural Selection, or Preservation of Favoured Races in the Struggle for Life (John Murray, 1859). [PMC free article] [PubMed]
- 51.Hubbell, S. P. The Unified Neutral Theory of Biodiversity and Biogeography (Princeton Univ. Press, 2001). [DOI] [PubMed]
- 52.Pinto‐Ledezma JN, et al. Testing Darwin’s naturalization conundrum based on taxonomic, phylogenetic, and functional dimensions of vascular plants. Ecol. Monogr. 2020;90:e01420. [Google Scholar]
- 53.Theoharides KA, Dukes JS. Plant invasion across space and time: factors affecting nonindigenous species success during four stages of invasion. New Phytol. 2007;176:256–273. doi: 10.1111/j.1469-8137.2007.02207.x. [DOI] [PubMed] [Google Scholar]
- 54.Hulme PE. Trade, transport and trouble: managing invasive species pathways in an era of globalization. J. Appl. Ecol. 2009;46:10–18. [Google Scholar]
- 55.Simberloff D, Von Holle B. Postitive interactions of nonindigenous species: invasion meltdown? Biol. Invasions. 1999;1:21–23. [Google Scholar]
- 56.Pyšek P, Křivánek M, Jarošík V. Planting intensity, residence time, and species traits determine invasion success of alien woody species. Ecology. 2009;90:2734–2744. doi: 10.1890/08-0857.1. [DOI] [PubMed] [Google Scholar]
- 57.Rejmánek M. Invasive trees and shrubs: where do they come from and what we should expect in the future? Biol. Invasions. 2014;16:483–498. [Google Scholar]
- 58.Chong KY, et al. Are terrestrial biological invasions different in the tropics? Annu. Rev. Ecol. Evol. Syst. 2021;52:291–314. [Google Scholar]
- 59.Smith JL, Perino JV. Osage orange (Maclura pomifera): history and economic uses. Econ. Bot. 1981;35:24–41. [Google Scholar]
- 60.Pyšek P, et al. The global invasion success of Central European plants is related to distribution characteristics in their native range and species traits. Divers. Distrib. 2009;15:891–903. [Google Scholar]
- 61.Svenning JC, et al. The influence of interspecific interactions on species range expansion rates. Ecography. 2014;37:1198–1209. doi: 10.1111/j.1600-0587.2013.00574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Amarasekare P, Simon MW. Latitudinal directionality in ectotherm invasion success. Proc. R. Soc. B. 2020;287:20191411. doi: 10.1098/rspb.2019.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park DS, Feng X, Maitner BS, Ernst KC, Enquist BJ. Darwin’s naturalization conundrum can be explained by spatial scale. Proc. Natl Acad. Sci. USA. 2020;117:10904–10910. doi: 10.1073/pnas.1918100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tournadre J. Anthropogenic pressure on the open ocean: The growth of ship traffic revealed by altimeter data analysis. Geophys. Res. Lett. 2014;41:7924–7932. [Google Scholar]
- 65.de Lima RAF, et al. Making forest data fair and open. Nat. Ecol. Evol. 2022;6:656–658. doi: 10.1038/s41559-022-01738-7. [DOI] [PubMed] [Google Scholar]
- 66.Moles AT, et al. Invasions: the trail behind, the path ahead, and a test of a disturbing idea. J. Ecol. 2012;100:116–127. [Google Scholar]
- 67.Peng S, Kinlock NL, Gurevitch J, Peng S. Correlation of native and exotic species richness: a global meta‐analysis finds no invasion paradox across scales. Ecology. 2019;100:e02552. doi: 10.1002/ecy.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dickie IA, et al. Conflicting values: ecosystem services and invasive tree management. Biol. Invasions. 2014;16:705–719. [Google Scholar]
- 69.Finnoff D, Shogren JF, Leung B, Lodge D. Take a risk: preferring prevention over control of biological invaders. Ecol. Econ. 2007;62:216–222. [Google Scholar]
- 70.Bivand, R. & Piras, G. Comparing implementations of estimation methods for spatial econometrics. J. Stat. Softw. 63, 1–36 (2015).
- 71.Kalwij JM. Review of ‘The Plant List, a working list of all plant species’. J. Veg. Sci. 2012;23:998–1002. [Google Scholar]
- 72.Gorelick N, et al. Google Earth Engine: planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017;202:18–27. [Google Scholar]
- 73.Beech E, Rivers M, Oldfield S, Smith PP. GlobalTreeSearch: the first complete global database of tree species and country distributions. J. Sustain. For. 2017;36:454–489. [Google Scholar]
- 74.Catford JA, Vesk PA, Richardson DM, Pyšek P. Quantifying levels of biological invasion: towards the objective classification of invaded and invasible ecosystems. Glob. Change Biol. 2012;18:44–62. [Google Scholar]
- 75.Guo Q, et al. A unified approach for quantifying invasibility and degree of invasion. Ecology. 2015;96:2613–2621. doi: 10.1890/14-2172.1. [DOI] [PubMed] [Google Scholar]
- 76.Van Den Hoogen J, et al. Soil nematode abundance and functional group composition at a global scale. Nature. 2019;572:194–198. doi: 10.1038/s41586-019-1418-6. [DOI] [PubMed] [Google Scholar]
- 77.Bastin J-F, et al. The global tree restoration potential. Science. 2019;365:76–79. doi: 10.1126/science.aax0848. [DOI] [PubMed] [Google Scholar]
- 78.Karger DN, et al. Climatologies at high resolution for the earth’s land surface areas. Sci. Data. 2017;4:170122. doi: 10.1038/sdata.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stewart S, et al. Climate extreme variables generated using monthly time‐series data improve predicted distributions of plant species. Ecography. 2021;44:626–639. [Google Scholar]
- 80.Hengl T, et al. SoilGrids250m: Global gridded soil information based on machine learning. PLoS ONE. 2017;12:e0169748. doi: 10.1371/journal.pone.0169748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Global Ports. All Layers and Tables (GLOBAL/GlobalPorts)https://gis.wfp.org/arcgis/rest/services/GLOBAL/GlobalPorts/MapServer/layers (2021).
- 82.Global Airports. The World Bank Data Cataloghttps://datacatalog.worldbank.org/search/dataset/0038117 (2021).
- 83.Maynard DS, et al. Global relationships in tree functional traits. Nat. Commun. 2022;13:3185. doi: 10.1038/s41467-022-30888-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Joswig JS, et al. Climatic and soil factors explain the two-dimensional spectrum of global plant trait variation. Nat. Ecol. Evol. 2022;6:36–50. doi: 10.1038/s41559-021-01616-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Florczyk, A. J. et al. GHSL Data Package 2019: Public Release GHS P2019 (European Commission, Joint Research Centre, 2019).
- 86.Owen NR, Gumbs R, Gray CL, Faith DP. Global conservation of phylogenetic diversity captures more than just functional diversity. Nat. Commun. 2019;10:859. doi: 10.1038/s41467-019-08600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mazel F, et al. Prioritizing phylogenetic diversity captures functional diversity unreliably. Nat. Commun. 2018;9:2888. doi: 10.1038/s41467-018-05126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tucker CM, et al. A guide to phylogenetic metrics for conservation, community ecology and macroecology. Biol. Rev. 2017;92:698–715. doi: 10.1111/brv.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jin Y, Qian HV. PhyloMaker: an R package that can generate very large phylogenies for vascular plants. Ecography. 2019;42:1353–1359. [Google Scholar]
- 90.Strauss SY, Webb CO, Salamin N. Exotic taxa less related to native species are more invasive. Proc. Natl Acad. Sci. USA. 2006;103:5841–5845. doi: 10.1073/pnas.0508073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cazzolla Gatti R, et al. The number of tree species on Earth. Proc. Natl Acad. Sci. USA. 2022;119:e2115329119. doi: 10.1073/pnas.2115329119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pyšek, P. & Richardson, D. M. in Biological Invasions (ed. Nentwig, W.) 97–125 (Springer, 2008).
- 93.Petchey OL, Gaston KJ. Functional diversity (FD), species richness and community composition. Ecol. Lett. 2002;5:402–411. [Google Scholar]
- 94.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 95.Wickham H, et al. Welcome to the Tidyverse. J. Open Source Softw. 2019;4:1686. [Google Scholar]
- 96.Bittinger, K. Abdiv: Alpha and beta diversity measures. R package version 0.2.0 https://cran.r-project.org/web/packages/abdiv/index.html (2020).
- 97.Calaway, R., Analytics, R., Weston, S., Tenenbaum, D. & Calaway, M. doParallel: Foreach parallel adaptor for the ‘parallel’ package. R package version 1.0.17 https://cran.r-project.org/web/packages/doParallel/index.html (2015).
- 98.Microsoft & Weston, S. foreach: Provides foreach looping construct. R package version 1.5.1. https://CRAN.R-project.org/package=foreach (2020).
- 99.Pearse WD, et al. Pez: phylogenetics for the environmental sciences. Bioinformatics. 2015;31:2888–2890. doi: 10.1093/bioinformatics/btv277. [DOI] [PubMed] [Google Scholar]
- 100.Breiman L. Bagging predictors. Mach. Learn. 1996;24:123–140. [Google Scholar]
- 101.Crase B, Liedloff AC, Wintle BA. A new method for dealing with residual spatial autocorrelation in species distribution models. Ecography. 2012;35:879–888. [Google Scholar]
- 102.Portier J, Gauthier S, Robitaille A, Bergeron Y. Accounting for spatial autocorrelation improves the estimation of climate, physical environment and vegetation’s effects on boreal forest’s burn rates. Landsc. Ecol. 2018;33:19–34. doi: 10.1007/s10980-017-0578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bjornstad, O. N. ncf: Spatial covariance functions. R package version 1.3-2 https://cran.r-project.org/web/packages/ncf/index.html (2022).
- 104.Bivand, R. S., Pebesma, E. J., Gómez-Rubio, V. & Pebesma, E. J. Applied spatial data analysis with R (Springer, 2008).
- 105.Lundberg SM, et al. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020;2:56–67. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lundberg, S. M. & Lee, S.-I. in Advances in Neural Information Processing Systems 30 (eds Guyon, I. et al.) (NIPS, 2017).
- 107.Wright, M. N., Wager, S. & Probst, P. Ranger: A fast implementation of random forests. R package version 0.12 (2020).
- 108.Greenwell, B. fastshap: Fast approximate Shapley values. R package version 0.0.7 https://github.com/bgreenwell/fastshap (2020).
- 109.Roberts DR, et al. Cross‐validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography. 2017;40:913–929. [Google Scholar]
- 110.Li J. Assessing the accuracy of predictive models for numerical data: not r nor r2, why not? Then what? PLoS ONE. 2017;12:e0183250. doi: 10.1371/journal.pone.0183250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Platt, J. in Advances in Large Margin Classifiers, Vol. 10 (eds Smola, A. J. et al.) 61–74 (MIT Press, 1999).
- 112.Niculescu-Mizil, A. & Caruana, R. in Proc. 22nd International Conference on Machine Learning 625–632 (Association for Computing Machinery, 2005).
- 113.Meyer H, Pebesma E. Predicting into unknown space? Estimating the area of applicability of spatial prediction models. Methods Ecol. Evol. 2021;12:1620–1633. [Google Scholar]
- 114.FAO. Global Forest Resources Assessment 2020: Main Report (Food and Agriculture Organization of the United Nations, 2020).
- 115.R Core Team. R: A Language and Environment for Statistical Computing. http://www.R-project.org/ (R Foundation for Statistical Computing, 2019).
- 116.Bates, D., Maechler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw.10.18637/jss.v067.i01 (2015).
- 117.Kunzetsova, A., Brockhoff, P. & Christensen, R. lmerTest package: tests in linear mixed effect models. J. Stat. Softw.10.18637/jss.v082.i13 (2017).
- 118.Zeileis, A. et al. betareg: Beta regression. R package version 3.1-4 https://cran.r-project.org/web/packages/betareg/index.html (2021).
- 119.Wickham, H., Chang, W. & Wickham, M. H. ggplot2: Create elegant data visualisations using the grammar of graphics. R package version 2 https://search.r-project.org/CRAN/refmans/ggplot2/html/ggplot2-package.html (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Tables 1–9.
Data Availability Statement
Data used in this study can be found in cited references for the Global Naturalized Alien Flora (GloNAF) database6 (non-native status), the KEW Plants of the World database5 (native ranges) and the Global Environmental Composite63,77 (environmental data layers). Plant trait data were extracted from Maynard et al.78. Data from the Global Forest Biodiversity Initiative (GFBI) database57 are not available due to data privacy and sharing restrictions, but can be obtained upon request via Science-I (https://science-i.org/) or GFBI (gfbinitiative.org) and an approval from data contributors.
All code used to complete analyses for the manuscript is available at the following link: https://github.com/thomaslauber/Global-Tree-Invasion. Data analyses were conducted and were visualizations generated in R (v. 4.2.2), Python (v. 3.9.7), Google Earth Engine (earthengine-api 0.1.306), QGIS-LTR (v. 3.16.7) and the ETH Zurich Euler cluster.