Abstract
Summary
Bacterial Healthcare-Associated Infections (HAIs) are a major threat worldwide, which can be counteracted by establishing effective infection control measures, guided by constant surveillance and timely epidemiological investigations. Genomics is crucial in modern epidemiology but lacks standard methods and user-friendly software, accessible to users without a strong bioinformatics proficiency. To overcome these issues we developed P-DOR, a novel tool for rapid bacterial outbreak characterization. P-DOR accepts genome assemblies as input, it automatically selects a background of publicly available genomes using k-mer distances and adds it to the analysis dataset before inferring a Single-Nucleotide Polymorphism (SNP)-based phylogeny. Epidemiological clusters are identified considering the phylogenetic tree topology and SNP distances. By analyzing the SNP-distance distribution, the user can gauge the correct threshold. Patient metadata can be inputted as well, to provide a spatio-temporal representation of the outbreak. The entire pipeline is fast and scalable and can be also run on low-end computers.
Availability and implementation
P-DOR is implemented in Python3 and R and can be installed using conda environments. It is available from GitHub https://github.com/SteMIDIfactory/P-DOR under the GPL-3.0 license.
1 Introduction
Bacterial infections are a constant threat to public health worldwide. When dealing with Healthcare-Associated Infections (HAIs) and outbreaks, timely epidemiological investigation is pivotal to establish effective infection control measures (Harris et al. 2013, Jiang et al. 2015, Raven et al. 2017, Balloux et al. 2018). Despite their wide use, conventional molecular typing techniques, such as Pulsed Field Gel Electrophoresis (PFGE) and Multi-Locus Sequence Typing (MLST), have a lower discriminatory capability in comparison to the modern Whole Genome Sequencing (WGS)-based typing, while maintaining similar costs and timescales.
Over the past few years, WGS-based typing has been increasingly adopted, first for research purposes and then as a routinary screening tool for infectious disease epidemiology in hospitals and public health settings. This approach leverages in silico techniques for isolates typing, antimicrobial profile determination, and outbreak reconstruction (Harris et al. 2013, Jiang et al. 2015, Onori et al. 2015, Raven et al. 2017, Balloux et al. 2018, Ferrari et al. 2019, Sherry et al. 2019). Several computational methods to analyze such datasets have been developed, which include database design (Zhou et al. 2020, Lam et al. 2021), epidemiological models via Bayesian inference (Jombart et al. 2014, De Maio et al. 2016, Campbell et al. 2018), network analysis (Worby et al. 2014, 2017), and phylogeny (Didelot et al. 2021).
Relationships among strains are mainly inferred using Single-Nucleotide Polymorphisms (SNPs) or k-mers. When reconstructing outbreaks, strains isolated from different sources (e.g. patients, fomites) and having SNP-distances below specific thresholds can be considered part of the same transmission cluster. The network of these genetically correlated strains can be used to reconstruct the pathogen transmission route. Although threshold-based methods are largely applied in genomic epidemiology (Dallman et al. 2015, Octavia et al. 2015, Hatherell et al. 2016, David et al. 2019), they lack standardization (Duval et al. 2023). Indeed, threshold values can vary across bacterial species/clones because of their different genomic architectures (e.g. mutation rate, recombination). Also the duration of the epidemic event analyzed can influence the genetic variability in the bacterial population: a SNP-distance threshold set to disentangle a short outbreak can be inappropriate for a long-term genomic surveillance study (Duval et al. 2023). Furthermore, SNP distances can be affected (even by tenths or hundreds) by the SNP calling approach (e.g. mapping reads or aligning assembled contigs) and by the reference genome and software used. Finally, the sole use of genomic data without the inclusion of other information like clinical metadata (e.g. sample date/type, hospitalization ward) limits the comprehension of epidemic events (Jombart et al. 2014, De Maio et al. 2016, Stimson et al. 2019, Didelot et al. 2021, Duval et al. 2023).
Most of the software available for WGS-based epidemiological investigation is not user-friendly (De Maio et al. 2016, Campbell et al. 2018, Zhou et al. 2020, Didelot et al. 2021), not free (e.g. SeqSphere+ Ridom GmbH software), and/or does not encompass all the analyses required for a comprehensive study (De Maio et al. 2016, Zhou et al. 2020, Didelot et al. 2021). Most of the methods require the user to have a computational background, as they are composed of multiple command-line tasks that must be serially performed in succession, and often require format changes. This prevents most clinicians from performing genomic investigations in first person and limits their understanding of the results. Consequently, it also hampers them from making epidemiological conclusions in light of both clinical information and of their past experience on the field, which in turn would enable them to provide valuable feedback to developers. On the other hand, online tools are available, which are accessible to a wider usership, but lack the tunability that is required for most epidemiological investigations (e.g. Trifinopoulos et al. 2016) and are restricted to single tasks (e.g. phylogeny).
To answer the need for a comprehensive, tunable, and user-friendly tool, we developed P-DOR, a bioinformatic pipeline for rapid WGS-based bacterial outbreak detection and characterization. P-DOR integrates genomics and clinical metadata and uses a curated global genomic database to contextualize the strains of interest within the appropriate evolutionary frame. P-DOR is available at https://github.com/SteMIDIfactory/P-DOR.
2 P-DOR workflow
The inputs for the core P-DOR analysis are: (i) a folder containing the query genome assemblies; (ii) a reference genome for SNP extraction; (iii) a sketch database file in the Mash format (Ondov et al. 2016); and (iv) a table containing the patient metadata (i.e. hospitalization ward, date of admission and discharge). This last input is not mandatory, but when provided, it will be integrated in the analysis to add further clues on the epidemic event. The query genomes of the study must be in FASTA format and can be complete or draft assemblies.
Sketch files contain the genomic information of the strains from a Source Dataset (SD) chosen by the user. A sketch file is a vastly reduced representation of the genomes, which is produced via the MinHash algorithm to allow fast distance estimation using low memory and storage requirements. Regularly updated sketches for each of the ESKAPE members (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) are available in the P-DOR repository. Personalized SD sketches can alternatively be generated by the user using the “makepdordb.py” script. This script can automatically download the high-quality genomes of a species from the BV-BRC collection (Davis et al. 2020) or build a custom SD sketch starting from any set of genomes. The sketch files are used to compute the k-mer distances between each query genome and the SD genomes. Then, for each query genome the n most similar SD genomes are selected and joined in a Background Dataset (BD). Lastly, the query genomes are joined with the BD to obtain the Analysis Dataset (AD). Optionally, the entire AD can be scanned for the presence of antimicrobial resistance and virulence genes using AMRFinderPlus (Feldgarden et al. 2021).
After that, each genome of the resulting AD is aligned to the reference genome using Nucmer, the alignment obtained is polished using “Delta-Filter” and SNPs are called using “show-snps”, all the commands being part of the Mummer4 package (Marçais et al. 2018). Lastly, the SNPs of all AD genomes are combined into a coreSNPs alignment using a Python script (Ferrari et al. 2019). Then, the coreSNPs alignment is used to infer a Maximum Likelihood (ML) phylogeny (correcting for the SNP ascertainment bias) via the IQ-TREE (Nguyen et al. 2015) software, which includes a prior step for the selection of the substitution model.
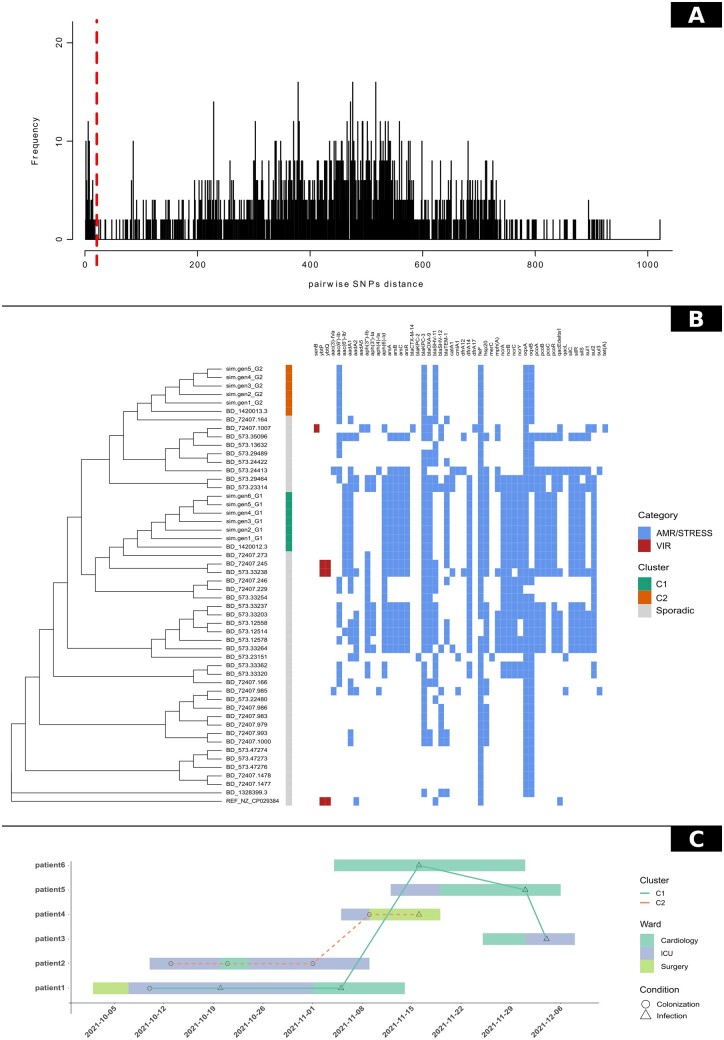
To infer relationships among the strains, epidemiological clusters are inferred on the basis of coreSNPs distances using a threshold value. The user can manually set the SNP threshold parameter according to previous studies in the literature, or by visualizing the SNP pairwise distances distribution plot provided among the outputs of P-DOR (Fig. 1A). The user should analyze the results, possibly tweak the threshold parameter and run P-DOR again.
Figure 1.
The P-DOR outputs of the test analysis. (A) Phylogenetic tree of the Analysis Dataset (AD). The first column shows epidemiological clusters of strains with SNP distances below the threshold set by the user. In addition, a heatmap representing the detection of resistance and virulence determinants is shown next to the tree for a better representation of the epidemic event. (B) Distribution of SNP distances calculated between all permutations of genome pairs in the AD. (C) Timeline depicting the movements of the patients during the hospitalization. Points indicate outbreak genomes and are shaped according to the isolation source of the corresponding strain. Samples are linked if their genetic distance in terms of SNPs is below the threshold.
3 Output
The main outputs of P-DOR are: (i) a SNP-based phylogenetic tree; (ii) a heatmap reporting the phylogenetic tree and presence/absence of resistance and virulence factors; (iii) a heatmap showing the coreSNP distance matrix; (iv) the histogram of the distribution of the SNP distances; and (v) a graph visualization of the epidemiological clusters, in which each pair of strains (nodes) is connected if the coreSNP distance between them is below the threshold. The epidemiological clusters are also highlighted on the phylogenetic tree. In addition, if patients metadata are provided, P-DOR creates a spatio-temporal representation of the outbreak and outputs a patients timeline plot where strains are placed based on the date of isolation and are connected on the basis of epidemic clusters.
4 Performance test
To test P-DOR, we simulated the genomic sequences of 11 K.pneumoniae isolates, involved in a complex epidemic event, with 2 distinct bacterial strains (both belonging to Sequence Type 258) circulating in a hospital in the same period. In detail, we obtained six simulated sequences starting from genome NJST258_1 and 5 sequences from genome NJST258_2 using the software simuG (Yue and Liti 2019). The genomes were simulated in a hierarchical way: i.e. each generated genome differs one to five SNPs from the parent. The simulated dataset was used as query to P-DOR. The SD was obtained from BV-BRC on 16 April 2023 using the script makepdordb.py. The BD was built selecting the 20 closest SD genomes for each query genome, as described above. After analyzing the distribution of SNP distances, the threshold was set at 21 SNPs (Fig. 1A). Finally, the complete genome of strain HS11286 (NZ_CP029384.2) was used as a reference for SNP calling. The phylogeny obtained correctly determined the presence of two outbreaking strains (Fig. 1B). P-DOR divided the simulated genomes into two monophyletic clusters labeled C1 (green) and C4 (orange). Both clusters also include a background genome (BV-BRC codes 1420013.3 and 1420012.3), which correspond to isolates NJST258_2 and NJST258_1, i.e. the genomes used as starting points for the generation of the simulated sequences. These results demonstrate the capability of the P-DOR pipeline to select a background suitable for epidemiological investigations and to identify outbreak clusters. They also show that P-DOR can identify the putative source of each epidemic cluster, when it is available in the SD. Supplementary Fig. S1 shows the SNP distances among all genomes in analysis and further confirms the results observed in the phylogeny (Fig. 1B). Furthermore, the timeline (Fig. 1C) can be used to hypothesize the chain of transmission based on the dates of isolation and the epidemiologic classification of the isolates.
The analysis was performed on a total of 50 genomes using a maximum of 313 Mb of RAM. The entire process required 1 h and 37 min, using four threads at 2.3 Ghz, 29 min when using 20 threads; these numbers drop respectively to 2.9 and 2.1 min when excluding the time-consuming AMRFinderPlus step (default setting).
We separately tested the performance and accuracy of our SNP-calling approach, which is based on the Mummer4 package (Marçais et al. 2018). We called point mutations using our method, Parsnp (Treangen et al. 2014), and Snippy (github.com/tseemann/snippy); we then inferred the phylogeny of the simulated dataset using all three alignments (IQ-TREE; Nguyen et al. 2015). Using two threads, the P-DOR built-in method yielded 8318 SNPs in 1.7 min, Parsnp yielded 10202 SNPs in 3.1 min, while Snippy took 31.2 min to output 9266 SNPs. We calculated the pairwise cophenetic correlation value among the three phylogenetic trees using the R library dendextend (Galili 2015). Phylogenies were highly concordant (Parsnp versus P-DOR: 0.996; Snippy versus P-DOR: 0.996; Snippy versus Parsnp: 0.999) and the monophyly of outbreak genomes was maintained among the three approaches.
To test the pipeline performance on a large-scale dataset, the P-DOR analysis on the simulated dataset was repeated using 1000 nearest genomes for each genome in the input folder. The AD was composed of 1388 total genomes and the analysis took 7 h and 50 min to complete, using a peak of 4.49 Gb of RAM on 20 threads (avoiding the AMRFinderPlus step; background genome download time excluded). These results show that P-DOR is a fast tool, which can be used in clinical contexts even when high informatic skills or resources are not available. Future efforts will be focused on developing a web-based interface, to further improve the ease of use and removing the need for computational resources.
Supplementary Material
Contributor Information
Gherard Batisti Biffignandi, Department of Biology and Biotechnology, University of Pavia, Pavia, 27100, Italy.
Greta Bellinzona, Department of Biology and Biotechnology, University of Pavia, Pavia, 27100, Italy.
Greta Petazzoni, Department of Medical, Surgical, Diagnostic and Pediatric Sciences, University of Pavia, Pavia, 27100, Italy; Microbiology and Virology Unit, Fondazione IRCCS Policlinico San Matteo, Pavia, 27100, Italy.
Davide Sassera, Department of Biology and Biotechnology, University of Pavia, Pavia, 27100, Italy; Fondazione IRCCS Policlinico San Matteo, Pavia, 27100, Italy.
Gian Vincenzo Zuccotti, Department of Biomedical and Clinical Sciences, Pediatric Clinical Research Center Romeo ed Enrica Invernizzi, University of Milan, Milan, 20157, Italy; Pediatric Department, Buzzi Children’s Hospital, Milan, 20154, Italy.
Claudio Bandi, Department of Biosciences, Pediatric Clinical Research Center Romeo ed Enrica Invernizzi, University of Milan, Milan, 20133, Italy.
Fausto Baldanti, Department of Medical, Surgical, Diagnostic and Pediatric Sciences, University of Pavia, Pavia, 27100, Italy; Microbiology and Virology Unit, Fondazione IRCCS Policlinico San Matteo, Pavia, 27100, Italy.
Francesco Comandatore, Department of Biomedical and Clinical Sciences, Pediatric Clinical Research Center Romeo ed Enrica Invernizzi, University of Milan, Milan, 20157, Italy.
Stefano Gaiarsa, Microbiology and Virology Unit, Fondazione IRCCS Policlinico San Matteo, Pavia, 27100, Italy.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
None declared.
Funding
This work was supported by Ricerca Corrente [924-rcr2018i-34] (Italian Ministry of Health).
Data availability
All data are incorporated into the article and its online supplementary material.
References
- Balloux F, Brønstad Brynildsrud O, van Dorp L. et al. From theory to practice: translating whole-genome sequencing (WGS) into the clinic. Trends Microbiol 2018;26:1035–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell F, Didelot X, Fitzjohn R. et al. outbreaker2: a modular platform for outbreak reconstruction. BMC Bioinformatics 2018;19:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallman TJ, Ashton PM, Byrne L. et al. Applying phylogenomics to understand the emergence of Shiga-toxin-producing O157:H7 strains causing severe human disease in the UK. Microb Genom 2015;1:e000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David S, Reuter S, Harris SR. et al. ; ESGEM Study Group. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 2019;4:1919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JJ, Wattam AW, Aziz RK. et al. The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res 2020;48:D606–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio N, Wu C-H, Wilson DJ. et al. SCOTTI: efficient reconstruction of transmission within outbreaks with the structured coalescent. PLoS Comput Biol 2016;12:e1005130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Kendall M, Xu Y. et al. Genomic epidemiology analysis of infectious disease outbreaks using TransPhylo. Curr Protoc 2021;1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval A, Opatowski L, Brisse S. et al. Defining genomic epidemiology thresholds for common-source bacterial outbreaks: a modelling study. Lancet Microbe 2023;4:e349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldgarden M, Brover V, Gonzalez-Escalona N. et al. AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep 2021;11:12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari C, Corbella M, Gaiarsa S. et al. Multiple KPC clones contribute to an extended hospital outbreak. Front Microbiol 2019;10:2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili T. dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015;31:3718–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SR, Cartwright EJP, Török ME. et al. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 2013;13:130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatherell H-A, Colijn C, Stagg HR. et al. Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Med 2016;14:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wei Z, Wang Y. et al. Tracking a hospital outbreak of KPC-producing ST11 Klebsiella pneumoniae with whole genome sequencing. Clin Microbiol Infect 2015;21:1001–7. [DOI] [PubMed] [Google Scholar]
- Jombart T, Cori A, Didelot X. et al. Bayesian reconstruction of disease outbreaks by combining epidemiologic and genomic data. PLoS Comput Biol 2014;10:e1003457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MMC, Wick RR, Watts SC. et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 2021;12:4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Delcher AL, Phillippy AM. et al. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol 2018;14:e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A. et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015;32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Octavia S, Wang Q, Tanaka MM. et al. Delineating community outbreaks of Salmonella enterica serovar typhimurium by use of whole-genome sequencing: insights into genomic variability within an outbreak. J Clin Microbiol 2015;53:1063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov BD, Treangen TJ, Melsted P. et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 2016;17:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onori R, Gaiarsa S, Comandatore F. et al. Tracking nosocomial Klebsiella pneumoniae infections and outbreaks by whole-genome analysis: small-scale Italian scenario within a single hospital. J Clin Microbiol 2015;53:2861–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven KE, Gouliouris T, Brodrick H. et al. Complex routes of nosocomial vancomycin-resistant Enterococcus faecium transmission revealed by genome sequencing. Clin Infect Dis 2017;64:886–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry NL, Lane CR, Kwong JC. et al. Genomics for molecular epidemiology and detecting transmission of Carbapenemase-Producing in Victoria, Australia, 2012 to 2016. J Clin Microbiol 2019;57:e00573–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimson J, Gardy J, Mathema B. et al. Beyond the SNP threshold: identifying outbreak clusters using inferred transmissions. Mol Biol Evol 2019;36:587–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Ondov BD, Koren S. et al. The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 2014;15:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen L-T, von Haeseler A. et al. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 2016;44:W232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CJ, Lipsitch M, Hanage WP. et al. Shared genomic variants: identification of transmission routes using pathogen deep-sequence data. Am J Epidemiol 2017;186:1209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CJ, Lipsitch M, Hanage WP. et al. Within-host bacterial diversity hinders accurate reconstruction of transmission networks from genomic distance data. PLoS Comput Biol 2014;10:e1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J-X, Liti G.. simuG: a general-purpose genome simulator. Bioinformatics 2019;35:4442–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Alikhan N-F, Mohamed K. et al. ; Agama Study Group. The EnteroBase user’s guide, with case studies on transmissions, phylogeny, and core genomic diversity. Genome Res 2020;30:138–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are incorporated into the article and its online supplementary material.