Abstract
Members of the plant mycobiota are all associated to varying degrees with the development of plant diseases. Although many reports on the plant mycobiota are well documented, the relationships between mycobiota of Rosa roxburghii and plant diseases are poorly understood. Mutual interactions and extent of the roles of microbial communities associated with R. roxburghii and the source of pathogens are still unclear, and more research is needed on the health benefits of this ecologically important population. Using high-throughput sequencing, we analyzed the mycobiota composition and ecological guilds of the rhizosphere, root, and phyllosphere of healthy and diseased R. roxburghii from the Tianfu R. roxburghii Industrial Park in Panzhou city, Guizhou province. Analysis of community composition showed that the relative abundance of pathogens of leaf spot, including Alternaria, Pestalotiopsis and Neofusicoccum in the phyllosphere of diseased plant (LD), were 1.15%, 0.15% and 0.06%, and the relative abundance of Alternaria and Pestalotiopsis were 0.96% and 0.58% in healthy plant (LH). The alpha diversity indices indicated that fungal diversity was higher in healthy plants compared to diseased plants in each compartment. The alpha diversity index of fungi in the phyllosphere (LH) of healthy R. roxburghii, including Shannon, Chao-1, and Faith-pd indices, was 1.02, 81.50 and 10.42 higher than that of the diseased (LD), respectively. The fungi in the rhizosphere of healthy was 1.03, 59.00 and 5.56 higher than the diseased, respectively. The Shannon index of fungi in the root of healthy was 0.29 higher than that of diseased. Principal Coordinate analysis and ANOSIM results showed that there were significant differences in mycobiota composition between healthy and diseased phyllospheres (P < 0.05), as well as rhizosphere fungal community, while there was no significant difference between healthy and diseased roots (P > 0.05). Linear discriminant analysis effect size revealed that, at different taxonomic levels, there were significantly different taxa between the healthy and diseased plants in each compartment. The ecological guilds differed between healthy and diseased plants according to the FUNGuild analysis. For example, of healthy compared to diseased plants, the percentages of “lichenized-undefined saprotroph” were increased by 2.34%, 0.44%, and 1.54% in the phyllosphere, root, and rhizosphere, respectively. In addition, the plant pathogens existed in each compartment of R. roxburghii, but the percentages of “plant pathogen” were increased by 1.16% in the phyllosphere of diseased compared to healthy plants. Together, the ecological guild and co-occurrence network indicated that the potential pathogens of leaf spot were mainly found in the phyllosphere. This study explained one of pathogen origin of leaf spots of R. roxburghii by the microbial community ecology, which will provide the new insights for identification of plant pathogens.
Keywords: Holobiont, Rosa roxburghii, Co-occurrence network, Plant mycobiota, Plant disease
Introduction
Rasa roxburghii is a perennial deciduous shrub of the family Rosaceae. In China, it is mainly distributed in the southwest, with the most extensive cultivation and highest production in Guizhou Province [1]. With the intensive development of the R. roxburghii industry in various planting areas of Guizhou, diseases in this population of plants are becoming more and more serious. Leaf spot is one of the common diseases and can lead to serious premature leaf shedding and reductions in plant growth and fruiting, resulting in large areas of yield reduction or even no harvest [1]. Pathogenic fungi, such as Pestalotiopsis microspora, Neofusicoccum parvum and Alternaria alternata can cause leaf spot disease in R. roxburghii [2]. The current leaf spot control strategies are mainly based on the selection of resistant varieties and the use of chemicals [3–6]. In recent years, biological control has paid more and more attention due to the harm of chemical pesticides to human health and the ecological environment. However, at present, the high-efficiency biocontrol microorganisms screened in the laboratory may be largely affected by the environment, and the application effect in the field environment is not stable [1, 7]. Actually, plants under natural environment facing various pathogens, tend to produce defense to maintain their fitness and minimize pathogenic damage [8]. We should maximize the efficiency of disease control based on the biological regulatory mechanisms of plants. However, biological control of leaf spot of R. roxburghii has not been widely implemented. The identification of the causal pathogens of leaf spot and the details of colonization, infection, and disease development is key to develop effective biological control strategies.
The tissues and rhizosphere soil of plants are colonized by a variety of microorganisms, which altogether form the host–microbiome holobionts as a result of long-term co-evolution [9]. The plant microbiome has been shown to have an important impact on the physiology and health of the host, and the evolution and ecology of the plant host can only be understood in the context of symbiotic functions [10, 11]. For example, plant-associated microorganisms may determine plant fate by affecting plant adaptation and growth, protecting plants from herbivores, or by driving the evolution of disease resistance [12–14]. The impact of the plant microbiome on host health and fitness depends not only on the microorganisms present in the community, but also on how they interact with each other. The plant microbiome is not equal to the sum of all the individual microbial components. They tend to be structured with microbes interacting strongly and frequently with each other and form interconnected microbial networks [15]. In microbial networks, certain microbes often co-occur with other taxa, and they may interact with many others, playing a key role in the microbial community. These interactions may potentially affect the adaptability of various components, and even directly affect soil fertility and plant health. It is critical to fully exploit the potential of beneficial plant-microbial interactions [16, 17].
Normally, microorganisms in different compartments of the plant exchange material and energy with the host plant, reaching a state of equilibrium through interactions, which may lead to disease when the steady state is disrupted, such as ecological dysregulation due to the proliferation of pathogens or the loss of microbial diversity [18, 19]. A recent study showed that a quadruple arabidopsis mutant (min7 fls2 efr cerk1, abbreviated as mfec) in the leaves of Arabidopsis thaliana has defects in certain pattern-triggered immune and cell surface component structuring genes. It assembles a more numerous but less diverse bacterial community in the phyllosphere, which in turn induces disease symptoms (leaf yellowness and necrosis) [20].
The plant microbiome comes from a wide range of sources. Microbes can be transmitted vertically between plant generations or within different development stages via seeds [21, 22]. Microbes can also be transmitted horizontally within plant tissues (e.g. rhizosphere microbes can enter roots, via emerging root sites or injury sites generated by soil herbivores or other pathogens, and migrate to the above-ground parts of the plant via xylem and phloem) or be obtained from external sources such as the atmosphere, water, soil, animals, and other plants [23–27]. Microbes in the soil and leaves/roots are known to overlap, that is, the same microbial taxa appear repeatedly in different compartments of the plant [28, 29]. Plant pathogens may also overlap in different compartments of the plant, leading to the occurrence of plant diseases. It is important to pay attention to the dynamic changes of plant pathogens [30]. Therefore, this paper studied leaf spot disease from the perspective of fungi.
In this study, healthy and leaf spot-diseased R. roxburghii cv. Guinong No. 2 were selected from the Tianfu R. roxburghii Industrial Park in Panzhou City, Guizhou Province. High-throughput MiSeq sequencing based on the ITS1 locus was used to sequence the mycobiota inhabiting in the phyllosphere, root, and rhizosphere of R. roxburghii. We then used FUNGuild and a co-occurrence network analysis to reveal the mycobiota composition and its relationship with leaf spot of R. roxburghii.
Materials & Methods
Sample Collection
In August 2020, eight 10 × 10 m2 plots were randomly set up in Tianfu Rosa roxburghii base in Panzhou City, Guizhou Province. The healthy and diseased plants (leaves and roots) and soil (rhizosphere soil) in each sample were collected and mixed by five-point method. A total of 24 samples were put into aseptic plastic bag, and the sampling time and number were recorded and brought back to the laboratory for timely processing. All the treated samples were stored in − 80 ℃ cryopreservation for DNA extraction. The sample codes were shown in Table 1. The sampling method was according to our previous culturable experiment, and the current work is a follow-up study to explore the response of the phyllosphere, root and rhizosphere mycobiota of R. roxburghii to the leaf spot and the source of potential pathogens [31]. The process for obtaining the rhizosphere soil is as follows: first removing the topsoil and finding the plant roots, then cuting off the plant roots with a sterilized shovel and gently shaking off the soil on the roots, that is, the rhizosphere soil [32, 33].
Table 1.
Definition of the sample code
| Code | Definition |
|---|---|
| LD | The phyllosphere fungi of infected R. roxburghii |
| LH | The phyllosphere fungi of healthy R. roxburghii |
| RD | The root fungi of infected R. roxburghii |
| RH | The root fungi of healthy R. roxburghi |
| RSD | The rhizosphere fungi of infected R. roxburghii |
| RSH | The rhizosphere fungi of healthy R. roxburghii |
DNA Extraction, PCR Amplification, and Sequencing
Tissue samples (roots and leaves) of R. roxburghii were ground under liquid nitrogen using a mortar and pestle to accelerate the grinding process. Phyllosphere, root and rhizosphere microbial DNA were extracted from 0.2 g of leaf powder, root powder and soil, respectively, using a total DNA extraction kit (FastDNA® SPIN Kit for Soil) according to the manufacturer’s instructions. The ITS rDNA was amplified with primer pairs ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3'), ITS2R (5′-GCTGCGTCTTCATCGATGC-3′), by an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, USA). The conditions and mixtures for PCR amplification referred to our previous method [34]. PCR products were subjected to high-throughput sequencing using an Illumina MiSeq platform by Shanghai Meiji Biomedical Technology Co. (Shanghai, China). The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRR15096517).
Raw Data Processing
The raw ITS sequencing reads were demultiplexed, quality-filtered by fastp version 0.20.0 and merged by FLASH version 1.2.7 with the following criteria: (i) the 300 bp reads were truncated at any site receiving an average quality score of < 20 over a 50 bp sliding window, and the truncated reads shorter than 50 bp were discarded, reads containing ambiguous characters were also discarded; (ii) only overlapping sequences longer than 10 bp were assembled according to their overlapped sequence. The maximum mismatch ratio of overlap region is 0.2. Reads that could not be assembled were discarded; (iii) Samples were distinguished according to the barcode and primers, and the sequence direction was adjusted, exact barcode matching. The operational taxonomic units (OTUs) were then determined based on 97% similarity using UPARSE 7.1. To obtain species annotation information for each OTU, representative OTU sequences were compared to the sequences in the Unite 7.0 fungal database (http://unite.ut.ee/index.php) and subjected to taxonomic assignment using the Ribosomal Database Project (RDP) Bayesian classifier 2.2 (http://rdp.cme.msu.edu/) with a confidence threshold of 0.7 [35–37].
Diversity Analysis
The alpha diversity indices (Shannon’s index, Faith’s phylogenetic diversity) were calculated and compared between samples using one-way analysis of variance (ANOVA) [38]. The statistical analyses were conducted in Excel 2010 (Microsoft Corporation., Redmond, Washington) and SPSS 26.0 (SPSS Inc., Chicago, IL).
The beta diversity (Bray Curtis distance) for all samples were calculated using QIIME2. For constrained PCoA analysis from the vegan package in R was used [39]. The ANOSIM (analysis of similarity) was performed with 999 permutations using the function vegan package in R [40].
Analysis of Differences in Community Composition
To comprehensively evaluate the R. roxburghii mycobiota composition and relative abundances of fungal orders and genera, the abundant, intermediate, and rare taxa in each sample were determined based on the relative abundance thresholds of > 1%, 0.01–1%, and < 0.01%, respectively [41–43]. Bar graphs and Wayne plots were plotted using Excel 2010 and Lianchuan Bioplatform (https://www.omicstudio.cn/index), respectively.
According to the OTU abundance matrix, the taxa with significantly differences were determined by the online analysis software LEfSe (http://huttenhower.sph.harvard.edu/galaxy/) [44]. The linear discriminant analysis (LDA) score was more than 4, and the Kruskall-Wallis test value was less than 0.05.
Analysis of Ecological Guilds
The FUNGuild database (https://github.com/UMNFuN/FUNGuild) was used to determine the ecological guilds in each sample [45]. The relative abundance of each guild is determined based on the number of sequences in each taxon as a percentage of the total number of sequences in each sample and then plotted using Excel.
Co-occurrence Network Analysis
A co-occurrence network of the mycobiota in each sample was constructed using Gephi 0.9.2 [46]. Based on the relative abundance information, six samples of LH, LD, RSH, RSD, RH and RD were analyzed by co-occurrence network. The nodes represent samples and taxa, the edges between the nodes represent the relationships between the samples and taxa, and the thickness of the edges represent the abundance of the taxa in the samples.
Results
Fungal Community Composition and Diversity in Various Compartments of Healthy and Diseased R. roxburghii
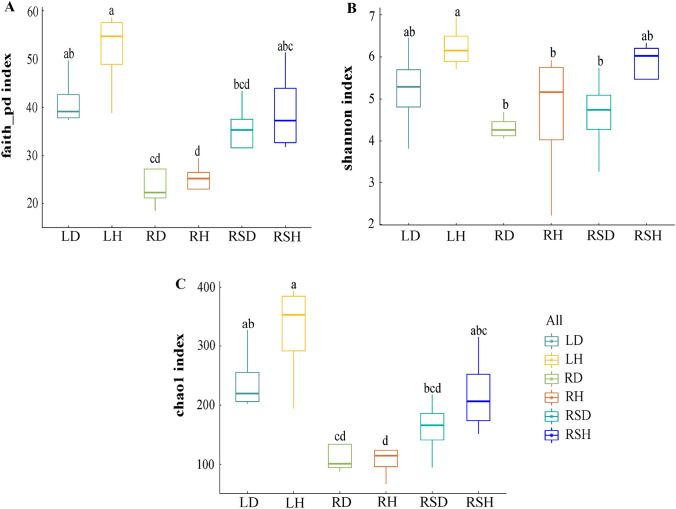
Based on the Illumina Miseq sequencing for each sample, approximately 27,009–46,459 high-quality sequences were detected from each sample, and a total of 1184 OTUs were obtained from all the samples. These OTUs belonged to 11 phyla, 30 classes, 83 orders, 188 families, 283 genera, and 296 species. The alpha diversity indices (Shannon, Chao-1, and Faith-pd) of various compartments of healthy and diseased plants are shown in Fig. 1. The results showed that the Shannon, Chao-1, and Faith-pd indices of fungi in the phyllosphere (LH), root (RH), and rhizosphere (RSH) of healthy R. roxburghii were generally higher than the corresponding indices of diseased plants in the phyllosphere (LD), root (RD) and rhizosphere (RSD), respectively, with higher indices corresponding to more diverse fungal communities. However, none of the differences was significant. Also, the Chao-1 and Faith-pd indices were lower in RH than RD. On the compartments of R. roxburghii, the whole, healthy plants showed higher fungal community diversity than diseased plants.
Fig. 1.
Comparison of alpha fungal diversity in various components of healthy and diseased Rosa roxburghii (the degree of replication n = 4). A Comparison of Faith-pd index in various components of healthy and diseased R. roxburghii. B Comparison of Shannon index in various components of healthy and diseased R. roxburghii; C Comparison of Chao1 in various components of healthy and diseased R. roxburghii. Note: LD: The phyllosphere fungi of infected R. roxburghii; LH: The phyllosphere fungi of healthy R. roxburghii; RD: The root fungi of infected R. roxburghii; RH: The root fungi of healthy R. roxburghii; RSD: The rhizosphere fungi of infected R. roxburghii; RSH: The rhizosphere fungi of healthy R. roxburghii
In addition, the alpha diversity indices were higher in the phyllosphere (LH, LD) than in the rhizosphere (RSH, RSD), which were higher than in the root (RH, RD), except that the Shannon index was higher in RSH than LD. In other words, the alpha diversity of fungal communities in different varied greatly.
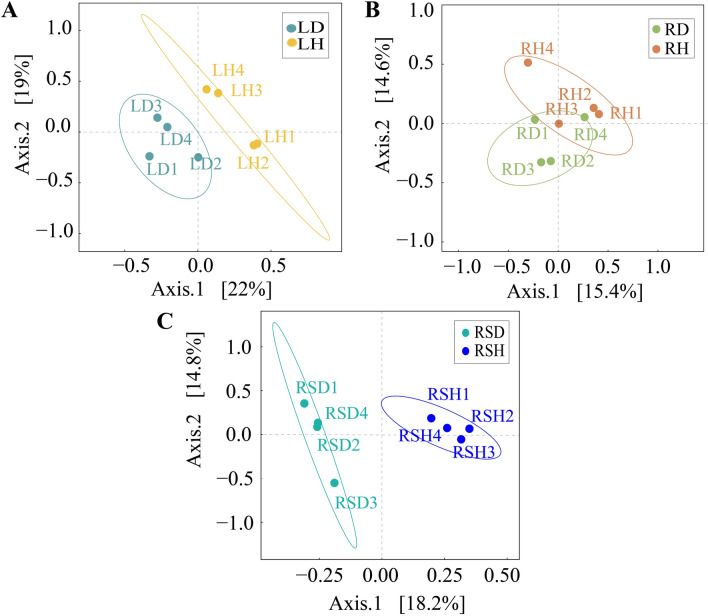
Bray Curtis distance was calculated based on OTU abundance information, and principal coordinate analysis was conducted for each sample to compare the composition of different fungal communities in different compartments of R. roxburghii (Fig. 2). The results showed that in phyllosphere (Fig. 2A), the principal coordinate explained 41% of the community variation, with the abscissa explaining 22% of the relationship value and the ordinate explaining 19%. There was no similarity between the two groups. The results of ANOSIM analysis (Table 2) further indicated that there were statistically significant differences between healthy and diseased phyllospheres (R = 0.65, P = 0.03). In root (Fig. 2B), the total explanatory degree was 30%, in which 15.4% of the relationship value was explained by the abscissa and 14.6% by the ordinate. There was no significant difference between healthy and diseased roots (R = 0.05, P = 0.25). In rhizosphere (Fig. 2C), abscissa explained 18.2% of the relationship value, ordinate explained 14.8% of the relationship value, and the total explanation was 33%. There was no similarity between the two groups, and ANOSIM analysis between healthy and diseased rhizospheres reached a significant level (R = 0.77, P = 0.03).
Fig. 2.
Comparison of beta fungal diversity in various components of healthy and diseased Rosa roxburghii. A PCoA analysis in phyllosphere of healthy and diseased R. roxburghii. B PCoA analysis in root of healthy and diseased R. roxburghii; C PCoA analysis in rhizosphere of healthy and diseased R. roxburghii
Table 2.
ANOSIM analysis among each samples
| Group | R-value | P-value |
|---|---|---|
| LD-LH | 0.65 | 0.03 |
| RD-RH | 0.05 | 0.25 |
| RSD-RSH | 0.77 | 0.03 |
Fungal Community Composition in Various Compartments of Healthy and Diseased R. roxburghii
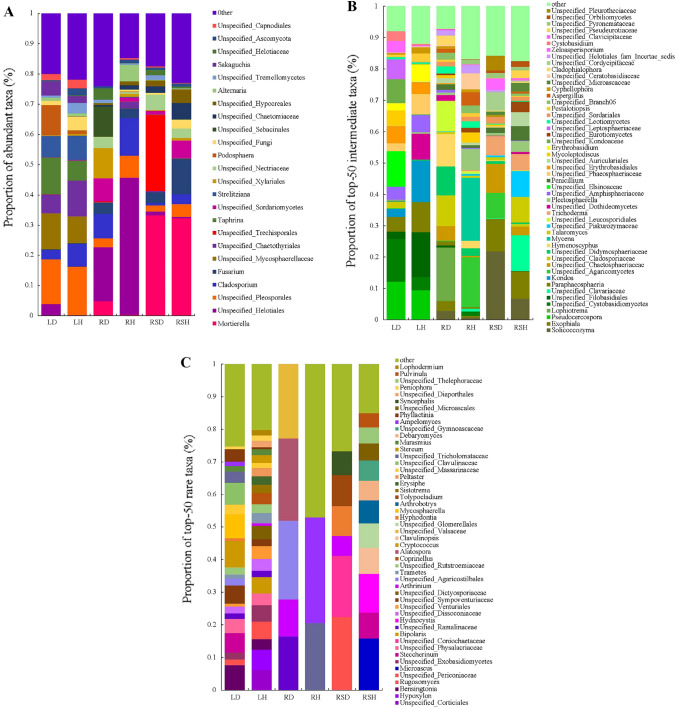
At the genus level, there were differences in abundant taxa in various compartments of healthy and diseased R. roxburghii (Fig. 3A). The dominant genera in LD were, in order, Unspecified_Pleosporales (14.83%), Taphrina (12.25%), and Unspecified_Mycosphaerellaceae (12%); those in LH were Unspecified_Pleosporales (15.91%), Unspecified_Chaetothyriales (11.86%), and Unspecified_Mycosphaerellaceae (8.98%); those in RD were Unspecified_Helotiales (17.85%), Unspecified_Sebacinales (10.01%), and Unspecified_Xylariales (9.94%); those in RH were Unspecified_Helotiales (45.47%), Cladosporium (12.42%), and Unspecified_Pleosporales (7.23%); those in RSD were Mortierella (33.34%), Unspecified_Trechisporales (25.18%), and Unspecified_Nectriaceae (5.07%); and those in RSH were Mortierella (32.36%), Fusarium (11.66%), and Unspecified_Sordariomycetes (5.73%).
Fig. 3.
Community Composition and relative abundance of fungal community at genus level in each sample. A Community composition and proportion of abundant taxa at the genus level; B Community composition and proportion of the top-50 intermediate taxa at the genus level; C, Community composition and proportion of the top-50 rare taxa at the genus level
There were also great differences in rare genera in various compartments of healthy and diseased R. roxburghii (Fig. 3B). The proportions of rare taxa were 0.18% in LD (36 genera, including Bensingtonia, Neofusicoccum and Steccherinum), 0.46% in LH (51 genera, including Unspecified_Corticiales, Hypoxylon and Bensingtonia), 0.06% in RD (5 genera, including Unspecified_Ramalinaceae, Arthrinium and Unspecified_Agaricostilbales), 0.03% in RH (5 genera, including Unspecified_Tricholomataceae and Ampelomyces), 0.16% in RSD (17 genera, including Arthrinium, Rugosomyces and Unspecified_Physalacriaceae), and 0.14% in RSH (17 genera, including Microascus, Hydnocystis and Steccherinum).
Furthermore, there were also great differences in intermediate genera in various compartments of healthy and diseased R. roxburghii (Fig. 3C). The proportions of intermediate taxa were19.76% in LD (69 genera, including Pestalotiopsis, Unspecified_Elsinoaceae and Pseudocercospora), 21.39% in LH (81 genera, including Pestalotiopsis, Unspecified_Dothideomycetes and Kondoa), 23.98% in RD (79 genera, including Lophiotrema, Hymenoscyphus and Pestalotiopsis), 17.70% in RH (74 genera, including Pestalotiopsis, Mycena and Plectosphaerella), 17.27% in RSD (73 genera, including Solicoccozyma, Exophiala and Pestalotiopsis), and 22.81% in RSH (93 genera, including Unspecified_Clavariaceae, Talaromyces and Pestalotiopsis).
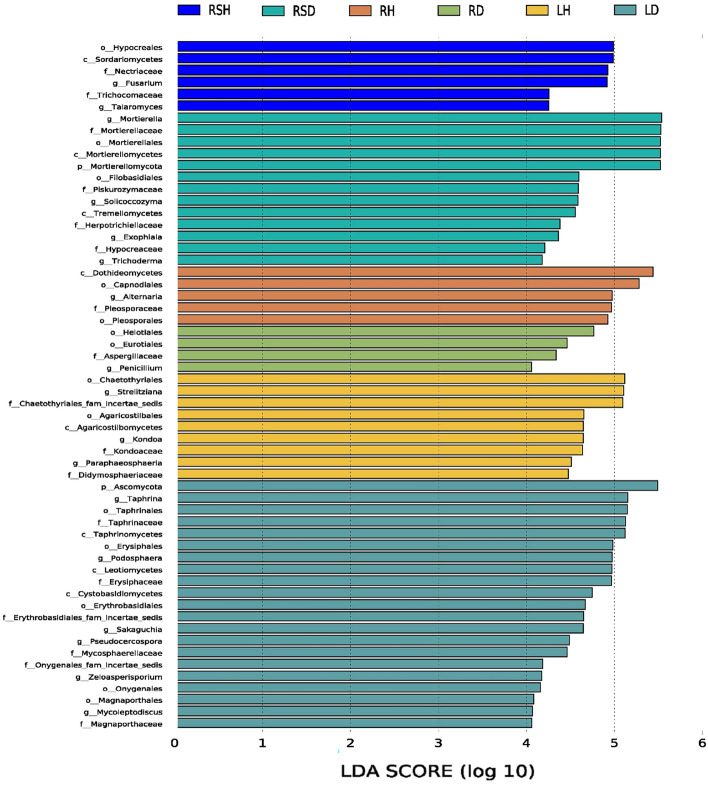
Linear discriminant analysis effect size (Fig. 4) was used to conduct preliminary analysis on the biomarkers with statistical differences in different compartments of healthy and diseased R. roxburghii. The results showed that there were 58 taxa at different taxonomic levels with significant differences among the samples (log LDA > 4, p < 0.05). Among them, 21 taxa were significantly different in LD, including Ascomycota, Taphrina (from class to genus) and Erysiphales, and 9 taxa in LH, including Chaetothyriales, Strelitziana and Agaricostilbales (from class to order). Four taxa played an important role in RD, including Helotiales, Eurotiales and Aspergillaceae; and 5 taxa in RH, including Dothideomycetes, Capnodiales and Alternaria. Thirteen taxa were significantly different in RSD, including Mortierella (from order to genus), Trichoderma and Hypocreaceae, and 6 taxa in RSH, including Hypocreales、Sordariomycetes and Nectriaceae. All of the above taxa contributed significantly to the differences in fungal community composition between healthy and diseased R. roxburghii.
Fig. 4.
LEfSe analysis on the significance of fungal community difference between healthy and diseased R. roxburghii
Fungal Ecological Guilds in Various Compartments of Healthy and Diseased R. roxburghii
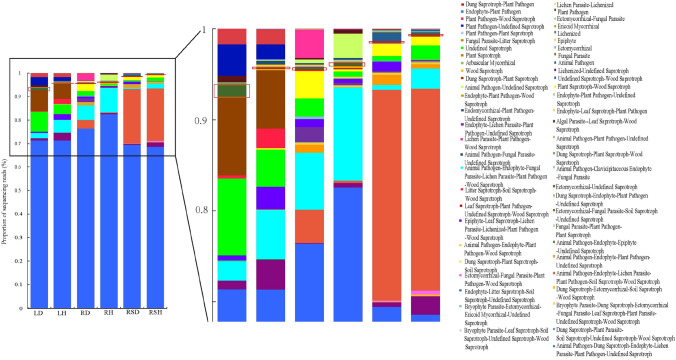
According to the results of FUNGuild annotation, after disregarding the Unspecified guilds, the fungi in various compartments of healthy and diseased R. roxburghii involved 57 ecological guilds such as “plant pathogen”,“plant pathogen-wood saprotroph”, “endophyte-plant pathogen”,“dung saprotroph-plant pathogen”, “dung saprotroph-plant pathogen”, “endophyte-plant pathogen-wood saprotroph”, “endophyte-plant pathogen-undefined saprotroph” and “endomycorrhizal-plant pathogen-undefined saprotroph”.Among them, the plant pathogens (red boxes in Fig. 5) were mainly Pestalotiopsis, Pseudocercospora, Neofusicoccum, Thanatephorus, Taphrina, and Alternaria. The plant pathogens were present in all compartments of healthy and diseased plants, exhibiting the following order: LD (1.32%), LH (0.16%), RD (0.19%), RH (0.35%), RSD (0.12%), and RSH (0.14%).
Fig. 5.
Fungal ecological guilds based on FUNGuild analysis. The boxes indicate plant pathogen
In LD compared to LH, “plant pathogen”, “plant pathogen-undefined saprotroph” and “leaf saprotroph-plant pathogen-undefined saprotroph-wood saprotroph” were increased by 1.16%, 1.79%, and 0.69%, respectively. In RD compared to RH, “animal pathogen-endophyte-lichen parasite-plant pathogen-soil saprotroph-wood saprotroph”, “lichen parasite-plant pathogen-wood saprotroph” and “endophyte-lichen parasite-plant pathogen-undefined saprotroph” were increased by 3.42%, 3.12%, and 1.40%, respectively. In RSD compared to RSH, “animal pathogen-endophyte-lichen parasite-plant pathogen-soil saprotroph-wood saprotroph”, “epiphyte-leaf saprotroph-lichen parasite-lichenized-plant pathogen-wood saprotroph” and “animal pathogen-endophyte-plant pathogen-undefined saprotroph” were increased by 0.96%, 1.05%, and 0.66%, respectively. In summary, the proportion of plant pathogen-related taxa was higher in diseased plants compared to healthy plants in each compartment.
Additionally, “lichenized-undefined saprotroph” was higher in LH, RH, and RSH compared to LD, RD, and RSD, respectively, by 2.34%, 0.44%, and 1.54%, respectively. In contrast, saprotrophs such as “dung saprotroph-plant saprotroph” and “undefined saprotroph” accounted for relatively high proportions in LD compared to LH, increasing by 2.16% and 4.40%, respectively.
Co-occurrence Network Analysis
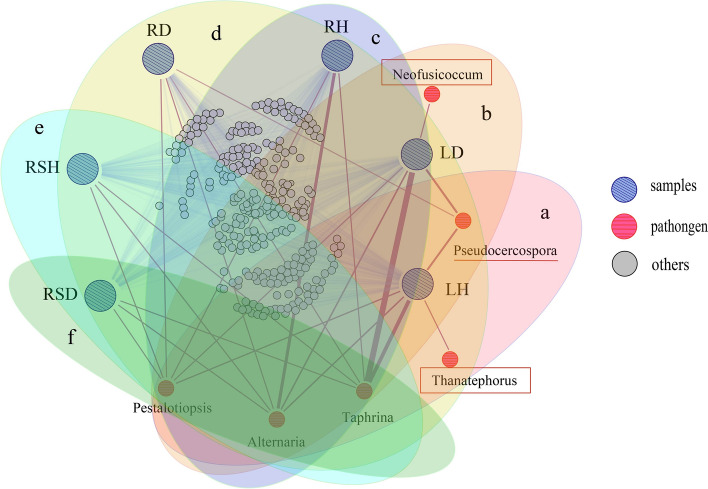
To further determine the potential source of the pathogens of leaf spot of R. roxburghii, this study combined the three leaf spot pathogens (Alternaria, Pestalotiopsis, Neofusicoccum) identified by our previous research with the potential plant pathogens (Taphrina, Thanatephorus, Pseudocercospora) annotated in the ecological guilds, and performed a co-occurrence network analysis of the mycobiota using Gephi software [2]. The co-occurrence network diagram showed that there were 5 potential pathogens in LH (Fig. 6a), including Taphrina (6.55%), Pseudocercospora (2.02%), Alternaria (0.96%), Pestalotiopsis (0.58%) and Thanatephorus (< 0.01%), and 5 potential pathogens in LD (Fig. 6b), including Taphrina (12.25%), Pseudocercospora (2.40%), Alternaria (1.15%), Pestalotiopsis (0.15%) and Neofusicoccum (< 0.01%). Potential pathogens were also found in root and rhizosphere. There were 3 potential pathogens in RH (Fig. 6c), including Taphrina (0.03%), Alternaria (5.55%) and Pestalotiopsis (0.17%), and 4 in RD (Fig. 6d), including Taphrina (0.02%), Pseudocercospora (< 0.01%), Alternaria (0.13%) and Pestalotiopsis (0.16%). RSH included 3 potential pathogens (Fig. 6e), that were Taphrina (0.03%), Alternaria (0.28%) and Pestalotiopsis (0.08%), and RSD included 3 potential pathogens (Fig. 6f), which were Taphrina (0.02%), Alternaria (0.12%) and Pestalotiopsis (< 0.01%). That is to say, there were differences in the distribution of pathogens in different compartments of R. roxburghii, and the types of pathogens and potential pathogens distributed in the phyllosphere were the most, accounting for the highest proportion. From this, it can be inferred that the potential pathogens in our sampling area mainly came from phyllosphere and existed in all compartments of R. roxburghii.
Fig. 6.
Co-occurrence network diagram of mycobiota in the root, rhizosphere, and phyllosphere of R. roxburghii. Nodes with oblique lines represent samples, nodes with transverse lines represent pathogens, and nodes with no lines represent other taxa. The thickness of the edges represent the abundance of the taxa in the samples. a pathogenic fungi present in LH. b pathogenic fungi present in LD. c pathogenic fungi present in RH. d pathogenic fungi present in RD. e pathogenic fungi present in RSH. f pathogenic fungi present inRSD
Discussion
Generally speaking, leaf spot of R. roxburghii is postulated to be caused by a variety of pathogenic fungi [1, 47, 48]. Plant pathogens, including Pestalotiopsis, Neofusicoccum and Alternaria, have been reported to cause leaf spot of R. roxburghii [2]. Pseudocercospora, Thanatephorus and Taphrina, as pathogens of leaf spot with a wide host range, may also be the potential pathogens of leaf spot disease of R. roxburghii [49–51]. In the study, the results of ecological guilds and co-occurrence network showed that the above potential leaf spot pathogens mainly existed in the phyllosphere of R. roxburghii, but they were found in all compartments of the plant. This is similar to our previous results based on culturable experiments, that is, the phyllosphere, including the diseased and healthy phyllosphere, is a major potential reservoir for the pathogens that cause leaf spot of R. roxburghii due to air pollution and insect-mediated transmission [31]. In addition, the co-occurrence network also found that there were multiple pathogenic fungi in the phyllosphere of R. roxburghii, such as Taphrina, Alternaria and Pestalotiopsis, which often caused leaf spot disease [21, 50, 52, 53]. Whether this may suggest that the leaf spot of R. roxburghii may also be the result of mixed infections by multiple pathogens. Of course, this hypothesis still needs to be verified by further experiments.
Plant-associated microbiome assembly is regulated by host factors, and plant health is often closely related to its microbial community composition [54–56]. The microbial community of diseased plants may be altered compared to healthy plants. This alteration is similar to the dysbiosis in the gastrointestinal tract of patients with inflammatory bowel disease, and it leads to the development of associated plant diseases which may account for the significant difference in fungal community composition between healthy and diseased R. roxburghii in our study. In general, plant microbial communities often have a small number of high-abundance dominant species co-existing with a large number of low-abundance rare species [57, 58]. The functions of rare microorganisms have often been overlooked and underestimated in previous studies because changes in the abundance of dominant microorganisms tend to mask the population dynamics of low-abundance taxa [59, 60]. In this study, not only the dominant taxa were considered, but the rare taxa were also given extra attention.
For example, Cladosporium was a abundant genus in all samples, and its abundance in various compartments was higher in healthy compared to diseased plants (P > 0.05). This is consistent with the results of our previous culturable study, that is, Cladosporium only exist in the root epiphytic microbiota of healthy R. roxburghii [2]. These fungi are able to synthesize a lot of different bioactive compounds, and some of their metabolites show antagonistic activity against certain pathogens. For example, four compounds produced by Cladosporium cladosporioides (isocladosporin, cladosporin, 5-hydroxyasperentin, and cladosporin-8-methyl ether) have large inhibitory effects on seven pathogens of strawberry anthracnose, with isocladosporin significantly inhibiting the growth of the pathogens Colletotrichum acutatum, Colletotrichum fragariae, and Colletotrichum gloeosporioides [61]. The higher abundance of Cladosporium in healthy plants compared to diseased plants may be related to the antagonistic activity of its secondary metabolites against leaf spot pathogens. Hypoxylon was a rare genus, which exists only in the phyllosphere of healthy plants, but not in diseased plants. This is consistent with the culturable results that the proportion of Hypoxylon in the phyllosphere epiphytic microbiota of healthy R. roxburghii is higher than that of diseased R. roxburghii. Hypoxylon can produce bioactive products and have great potential in resistance to human diseases and plant diseases [62]. For example, the ethyl acetate extract of Hypoxylon had high inhibitory activity against Sphaeropsis sapinea [63].
Another interesting point is that Exophiala was mainly distributed in the rhizosphere (RSD, RSH), followed by the root (RD, RH) and phyllosphere of healthy plants (LH), but not in the phyllosphere of diseased plants (LD). Exophiala is a common type of dark septate endophyte (DSE), which is a kind of endophytic soil fungi that colonize plant roots without causing obvious negative effects [64]. Exophiala have strong biological adaptability, promote plant growth by improving plant nutrition, and exhibit strong stress tolerance [65, 66]. As Exophiala has a similar niche to the potential leaf spot pathogens, it inhibits the overgrowth of the pathogens through competition for resources such as nutrients and space. This hinders disease development, which may explain why this genus is present in LH but not LD.
The generalized definition of ecological guild refers to a group of related or unrelated species that utilize the same type of ecological resources in a similar way [67]. The focus of the classification on trophic preferences, rather than on species abundance and taxonomic homogeneity, provides a different perspective on community composition [68, 69]. In this study, based on the FUNGuild analysis, it was found that there were differences in the guilds in the various compartments of the mycobiota of healthy and diseased R. roxburghii. The proportions of pathogens were higher in diseased compared to healthy plants in all compartments. This may mean that the health status of R. roxburghii is directly related to the number of pathogens. “Lichenized-undefined saprotroph” accounted for a high proportion of fungi in various compartments of healthy R. roxburghii. These symbiotrophs are the result of long-term plant–environment co-evolution, and the symbiotic relationships may promote plant growth and resistance. In addition, the proportions of plant pathogens and saprotrophs in the phyllosphere were higher in diseased compared to healthy plants. There was evidence of a functional shift between healthy and diseased plants, from colonization by symbiotrophs (e.g., “lichenized-undefined saprotroph”) to colonization by pathotrophs (e.g., “plant pathogen-undefined saprotroph”) and saprotrophs (e.g., “undefined saprotroph”), which may be an important reason for disease in R. roxburghii.
Plant health and pathogen resistance are often closely related to microbial diversity. More complex networks in the plant environment can inhibit pathogen colonization by competing for resources and occupying niches, thereby reducing the risk of pathogen outbreaks [70]. In general, microbial communities with high diversity and abundance levels may enhance the functional redundancy of the inhabited environment, which may help to maintain the health of the host plant [70, 71]. In this study, we found that the alpha diversity indices of the six samples were basically consistent (except that the Chao-1 and Faith-pd indices were lower in RH than RD), that is, the diversity of fungi in various compartments was generally higher in healthy compared to diseased plants. This implies that the degree of plant-associated fungal diversity is positively correlated with the health of R. roxburghii.
Different compartments of the plant act as different microhabitats with diverse morphological characteristics, metabolic activities, and corresponding functions. These distinct microhabitats selectively recruit different microorganisms to colonize, thus leading to differences in microbial diversity in different compartments of the plant [26, 72]. Typically, microbial diversity is more abundant in rhizosphere soils than in non-rhizosphere compartments [73]. However, our results showed that the compartment with the highest fungal diversity was the phyllosphere, followed by the rhizosphere and then the root, which may be due to the fact that the phyllosphere, as a microhabitat with great variability, is susceptible to strong disturbances of fungal communities by environmental factors, so the phyllosphere was more diverse than other compartments [23].
Conclusions
This study revealed that there were differences in microbial composition and ecological functional groups between healthy and diseased R. roxburghii, which provided a theoretical basis for the construction of antagonistic synthetic communities. As for what is a healthy plant microbiome, researchers still do not have a definite conclusion, which is a problem that needs further research in the future. In addition, this study also clarified that R. roxburghii leaf spot is caused by mixed pathogens, which mainly come from phyllosphere. As a reservoir of pathogens of leaf spot disease, phyllosphere can spread to other compartments of the plant through plant tissue or external force, resulting in the recurrence of pathogens in different parts of the plant, so cutting off the spread pathway of pathogens may provide a new idea for disease control.
Acknowledgements
The authors thank the members of the Institute of Fungus Resources, Guizhou University for their guidance on data processing in this paper, and the Chinese Institute of Rosa roxburghii of Guizhou Hongcai Group for providing the samplingfor this study. We also appreciated the Charlesworth Author Services (https://www.cwauthors.com/) and Dr. Sunil (TERI-Deakin Nano Biotechnology Centre, India) for their English editing. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author Contributions
YH: conceptualization and Funding acquisition; YL, WG, ZZ, CD, and QS: data acquisition; YL, and CD: formal analysis; YL: writing the first draft; YH: writing, review, and editing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Program of High-level innovative scientific talents of Guizhou Province ([2020]6005), the National Natural Science Foundation of China (32060011, 32260003), Key-Area Research and Development Program of GuangDong Province (2018B020205003).
Data Availability
The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRR15096517). The datasets generated during and/or analyzed during the current study are available from the corresponding authors upon request.
Declarations
Conflict of interest
The authors declare Conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huang WY, Li XQ, Han L, et al. Advances in the researches on the occurrence and control of Rosa roxburghii disease and insect pests. China Plant Prot. 2020;40:24–30. [Google Scholar]
- 2.Liu YX (2022) Analysis of microbial structural characteristics and evaluation of antagonistic effect on leaf spot of Rosa roxburghii. Dissertation, Guizhou University
- 3.Seif AA, Hillocks RJ. Chemical control of Phaeoramularia fruit and leaf spot of citrus in Kenya. Crop Prot. 1997;16:141–145. doi: 10.1016/S0261-2194(96)00086-5. [DOI] [Google Scholar]
- 4.Bot PJ, Arain AR, Jiskani MM, et al. Incidence and chemical control of okra leaf spot disease. Pak J Bot. 2011;44:1769–1774. [Google Scholar]
- 5.Gremillion S, Culbreath A, Gorbet D, et al. Response of progeny bred from Bolivian and north American cultivars in integrated management systems for leaf spot of peanut (Arachis hypogaea) Crop Prot. 2011;30:698–704. doi: 10.1016/j.cropro.2011.02.012. [DOI] [Google Scholar]
- 6.Singh AK. Efficacy of fungicides for the control of leaf spot disease of ginger under the field conditions of Chhattisgarh (India) Acad J. 2015;10:1301–1305. doi: 10.5897/AJAR2013.7708. [DOI] [Google Scholar]
- 7.Wu XQ, Zhou FY, Zhang XJ. Enlightenment from microbiome research towards biocontrol of plant disease. Acta Microbiol Sinica. 2017;57:867–875. doi: 10.13343/j.cnki.wsxb.20170073. [DOI] [Google Scholar]
- 8.Zaynab M, Fatima M, Sharif Y, et al. Role of primary metabolites in plant defense against pathogens. Microb Pathog. 2019;137:103728. doi: 10.1016/j.micpath.2019.103728. [DOI] [PubMed] [Google Scholar]
- 9.Kundu P, Blacher E, Elinav E, et al. Our gut microbiome: the evolving inner self. Cell. 2017 doi: 10.1016/j.cell.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 10.Agler MT, Ruhe J, Kroll S, et al. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016;14:e1002352. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali H, Muhammad A, Sanda NB, et al. Pyrosequencing uncovers a shift in bacterial communities across life stages of Octodonta nipae (Coleoptera: Chrysomelidae) Front microbiol. 2019;10:466. doi: 10.3389/fmicb.2019.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haney CH, Samuel BS, Bush J, et al. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nature Plants. 2015;1:15051. doi: 10.1038/nplants.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen AK, Moran NA. The impact of microbial symbionts on host plant utilization by herbivorousinsects. Mol Ecol. 2014;23(6):1473–1496. doi: 10.1111/mec.12421. [DOI] [PubMed] [Google Scholar]
- 14.Karasov TL, Kniskern JM, Gao L, et al. The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature. 2014;512:436–440. doi: 10.1038/nature13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Heijden MGA, Hartmann M. Networking in the Plant Microbiome. PLOS Biol. 2016;14:e1002378. doi: 10.1371/journal.pbio.1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandenkoornhuyse P, Quaiser A, Duhamel M, et al. The importance of the microbiome of the plant holobiont. New Phytol. 2015;206:1196–1206. doi: 10.1111/nph.13312. [DOI] [PubMed] [Google Scholar]
- 17.Berg G. Beyond borders: investigating microbiome interactivity and diversity for advanced biocontrol technologies. Microb Biotechnol. 2015;8:5–7. doi: 10.1111/1751-7915.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan KA, Al-Ghamdi AA, Ghramh HA, et al. Structural diversity and functional variability of gut microbial communities associated with honey bees. Microb Pathog. 2020;138:103793. doi: 10.1016/j.micpath.2019.103793. [DOI] [PubMed] [Google Scholar]
- 19.Agriosa GN. Plant pathogens and disease: general introduction. Netherlands: Elsevier; 2009. [Google Scholar]
- 20.Liu H, Brettell LE, Singh B. Linking the phyllosphere microbiome to plant health. Trends Plant Sci. 2020;25:841–844. doi: 10.1016/j.tplants.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Naveed M, Mitter B, Reichenauer TG, et al. Increased drought stress resilience of maize through endophytic colonization by Burkholderia phytofirmans PsJN and Enterobacter sp. FD17. Environ Experimen Bot. 2014;97:30–39. doi: 10.1016/j.envexpbot.2013.09.014. [DOI] [Google Scholar]
- 22.Liu J, Nagabhyru P, Schardl CL. Epichlo festucae endophytic growth in florets, seeds, and seedlings of perennial ryegrass (Lolium perenne) Mycologia. 2017;109:1–10. doi: 10.1080/00275514.2017.1400305. [DOI] [PubMed] [Google Scholar]
- 23.Singh BK, Liu H, Trivedi P. Eco-holobiont: a new concept to identify drivers of host-associated microorganisms. Environ Microbiol. 2020;22:564–567. doi: 10.1111/1462-2920.14900. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez RJ, White JF, Arnold AE, et al. Fungal endophytes: diversity and functional roles. New Phytol. 2010;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 25.Osono T. Diversity and ecology of endophytic and epiphytic fungi of tree leaves in Japan: a review. New Delhi: Springer; 2014. [Google Scholar]
- 26.Lemanceau P, Barret M, Mazurier S, et al. Plant communication with associated microbiota in the spermosphere, rhizosphere and phyllosphere. Adv Bot Res. 2017;82:101–133. doi: 10.1016/bs.abr.2016.10.007. [DOI] [Google Scholar]
- 27.Tkacz A, Bestion E, Bo Z, et al. Influence of plant fraction, soil, and plant species on microbiota: a multikingdom comparison. MBio. 2020;11:e02785–e12719. doi: 10.1128/mBio.02785-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarraonaindia I, Owens SM, Weisenhorn P, et al. The soil microbiome influences grapevine-associated microbiota. MBio. 2015;6:e02727–e12714. doi: 10.1128/mBio.02527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finkel OM, Salas-Gonzalez I, Castrillo G, et al. The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLOS Biol. 2019;17:e3000534. doi: 10.1371/journal.pbio.3000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ali H, Muhammad A, Islam SU, et al. A novel bacterial symbiont association in the hispid beetle, Octodonta nipae (Coleoptera: Chrysomelidae), their dynamics and phylogeny. Microb Pathog. 2018;2018:378–386. doi: 10.1016/j.micpath.2018.03.046. [DOI] [PubMed] [Google Scholar]
- 31.Liu YX, Shao QY, Ge W, et al. Comparison of culturable phyllosphere fungal flora between healthy and infected Rosa roxburghii. Mycosystema. 2021;40:2620–2640. doi: 10.13346/j.mycosystema.210043. [DOI] [Google Scholar]
- 32.Riley D, Barber SA. Bicarbonate accumulation and pH changes at the soybean (Glycine max (L.) Merr.) root–soil interface. Soil Sci Soc of Am J. 1969;33:905–908. doi: 10.2136/sssaj1969.03615995003300060031x. [DOI] [Google Scholar]
- 33.Riley D, Barber SA. Salt accumulation at the soybean root–soil interface. Soil Sci Soc of Am J. 1970;34:154–155. doi: 10.2136/sssaj1970.03615995003400010042x. [DOI] [Google Scholar]
- 34.Dong CB, Shao QY, Zhang QQ, et al. Preferences for core microbiome composition and function by different definition methods: Evidence for the core microbiome of Eucommia ulmoides bark. Sci total Environ. 2021;790:148091. doi: 10.1016/j.scitotenv.2021.148091. [DOI] [PubMed] [Google Scholar]
- 35.Chen S, Zhou Y, Chen Y, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1101/274100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mago T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Garrity GM, Tiedje JM, et al. Nave bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuczynski J, Stombaugh J, Walters WA, et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinform. 2011;36:10.7.1–10.7.20. doi: 10.1002/0471250953.bi1007s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vázquez-Baeza Y, Pirrung M, Gonzalez A, et al. Emperor: a tool for visualizing high-throughput microbial community data. GigaSci. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desgarennes D, Garrido E, Torres-Gomez MJ, et al. Diazotrophic potential among bacterial communities associated with wild and cultivated Agave species. FEMS Microbiol Ecol. 2014;90:844–857. doi: 10.1111/1574-6941.12438. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Ye F, Wu S, et al. Biogeographic pattern of bacterioplanktonic community and potential function in the Yangtze River: roles of abundant and rare taxa. Sci Total Environ. 2020;747:141335. doi: 10.1016/j.scitotenv.2020.141335. [DOI] [PubMed] [Google Scholar]
- 42.Jiao S, Lu Y. Abundant fungi adapt to broader environmental gradients than rare fungi in agricultural fields. Global Change Biol. 2020;26:4506–4529. doi: 10.1111/gcb.15130. [DOI] [PubMed] [Google Scholar]
- 43.Galand PE, Casamayor EO, Kirchman DL, et al. Ecology of the rare microbial biosphere of the Arctic Ocean. P Natl Acad Sci USA. 2009;106:22427–22432. doi: 10.1073/pnas.0908284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segata N, Abubucker S, Goll J, et al. Microbial community function and biomarker discovery in the human microbiome. Genome Biol. 2011;12:47. doi: 10.1186/1465-6906-12-S1-P47. [DOI] [Google Scholar]
- 45.Nguyen NH, Song Z, Bates ST, et al. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016;20:241–248. doi: 10.1016/j.funeco.2015.06.006. [DOI] [Google Scholar]
- 46.Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks. In: Proceedings of the international AAAI conference on web and social media. 10.13140/2.1.1341.1520
- 47.Zhong M, Shao ZW, Shi L, et al. Preliminary report on control techniques of powdery mildew and brown spot of Rosa sterilis. Guizhou Sci Technol. 2018;46:48–50. doi: 10.16709/j.cnki.gzlykj.2018.04.010. [DOI] [Google Scholar]
- 48.Zhang XW, Zhang P, Li YQ, et al. Isolation and identification of the pathogen of black spot of Rosa roxburghii. South China Fruits. 2019;48:106–109. doi: 10.13938/j.issn.1007-1431.20180375. [DOI] [Google Scholar]
- 49.Priyatmojo A, Escopalao VE, Tangonan NG, et al. Characterization of a new subgroup of Rhizoctonia solani anastomosis group 1 (AG-1-ID), causal agent of a necrotic leaf spot on coffee. Phytopathology. 2001;91:1054–1061. doi: 10.1094/phyto.2001.91.11.1054. [DOI] [PubMed] [Google Scholar]
- 50.Upadhyay R, Pavgi MS. Some factors affecting incidence of leaf spot of Turmeric by Taphrina maculans Butler. JPN J Phytopathol. 2009;33:176–180. doi: 10.3186/jjphytopath.33.176. [DOI] [Google Scholar]
- 51.Sisterna M, Ronco L. Occurrence of grapevine leaf spot caused by Pseudocercospora vitis in Argentina. Plant Path. 2005;54:247–247. doi: 10.1111/j.1365-3059.2005.01135.x. [DOI] [Google Scholar]
- 52.Garibaldi A, Tabone G, Luongo I, et al. First report of Alternaria alternata causing leaf spot on Hosta fortunei in Italy. J Plant Pathol. 2021;103:719. doi: 10.1007/s42161-021-00796-310.1007/s42161-021-00796-3. [DOI] [Google Scholar]
- 53.Nozawa S, Seto Y, Watanabe K. First report of leaf blight caused by Pestalotiopsis chamaeropis and Neopestalotiopsis sp. in Japanese andromeda. J Gen Plant Pathol. 2019;85:449–452. doi: 10.1007/s10327-019-00868-4. [DOI] [Google Scholar]
- 54.Yang SD, Ren KY, Tan HW. Differences in plant nutrient content, root growth and endophytic bacterial community between infected and non-infected sugarcanes by ratoon stunting disease. J Plant Nutr Fert. 2020;26:1591–1599. doi: 10.11674/zwyf.20144. [DOI] [Google Scholar]
- 55.Rastogi G, Sbodio A, Tech JJ, et al. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012;6:1812–1822. doi: 10.1038/ismej.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou ZX, Jiang H, Yang C, et al. Microbial community on healthy and diseased leaves of an invasive plant Eupatorium adenophorum in Southwest China. J Microbiol. 2010;48:139–145. doi: 10.1007/s12275-010-9185-y. [DOI] [PubMed] [Google Scholar]
- 57.Nemergut DR, Costello EK, Hamady M, et al. Global patterns in the biogeography of bacterial taxa. Environl Microbiol. 2011;13:135–144. doi: 10.1111/j.1462-2920.2010.02315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jousset A, Bienhold C, Chatzinotas A, et al. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 2017;11:853–862. doi: 10.1038/ismej.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hol WH, de Boer W, Aad T, et al. Reduction of rare soil microbes modifies plant-herbivore interactions. Ecol Lett. 2010;13:292–301. doi: 10.1111/j.1461-0248.2009.01424.x. [DOI] [PubMed] [Google Scholar]
- 60.Lynch M, Neufeld JD. Ecology and exploration of the rare biosphere. Nat Rev Microbio. 2015;13:217. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]
- 61.Wang X, Radwan MM, Taráwneh AH, et al. Antifungal activity against plant pathogens of metabolites from the endophytic fungus Cladosporium cladosporioides. J Agr Food Chem. 2013;61:4551–4555. doi: 10.1021/jf400212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferreira A, Rodrigues M, Fortuna A, et al. Huperzine A from Huperzia serrata: a review of its sources, chemistry, pharmacology and toxicology[J] Phytochem Rev. 2016;15:51–85. doi: 10.1007/s11101-014-9384-y. [DOI] [Google Scholar]
- 63.Song RQ, Gao HY. Antifungal activities and stability of extracts from culture liquid of Hypoxylon perforatum to Sphaeropsis sapinea. Acta Microbiol Sinica. 2009;49:910–917. doi: 10.13343/j.cnki.wsxb.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 64.Mandyam K, Jumpponen A. Seeking the elusive function of the root-colonising dark septate endophytic fungi. Stud Mycol. 2005;53:173–189. doi: 10.3114/sim.53.1.173. [DOI] [Google Scholar]
- 65.Lisina TO, Garan'kina NG, Kruglov YV. The effect of soil inoculation with microbial pesticide destructors on plant growth and development. Prikl Biokhim Mikrobiol. 2001;37:322–326. doi: 10.1023/A:1010253823414. [DOI] [PubMed] [Google Scholar]
- 66.Li T, Liu MJ, Zhang T, et al. Improved tolerance of maize (Zea mays L.) to heavy metals by colonization of a dark septate endophyte (DSE) Exophiala pisciphila. Sci Total Environ. 2011;409:1069–1074. doi: 10.1016/j.scitotenv.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 67.Root R. The niche exploitation pattern of the blue-grey gnatcatcher. Ecol Monogr. 1976;37:317–350. doi: 10.2307/1942327. [DOI] [Google Scholar]
- 68.Hawkins CP, MacMahon JA. Guilds: the multiple meanings of a concept. Annu Rev Entomol. 1988;34:23–451. doi: 10.1146/annurev.en.34.010189.002231. [DOI] [Google Scholar]
- 69.Wilson JB. Guilds, functional types and ecological groups. Oikos. 1999;86:507–522. doi: 10.2307/3546655. [DOI] [Google Scholar]
- 70.Barberán A, Bates ST, Casamayor EO, et al. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qin C, Tao J, Liu T, et al. Responses of phyllosphere microbiota and plant health to application of two different biocontrol agents. AMB Express. 2019;9:42. doi: 10.1186/s13568-019-0765-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Herrmann M, Geesink P, Richter R, et al. Canopy position has a stronger effect than tree species identity on phyllosphere bacterial diversity in a floodplain hardwood forest. Micro Ecol. 2020;81:157–168. doi: 10.1007/s00248-020-01565-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schloter M, Nannipieri P, Sorensen SJ, et al. Microbial indicators for soil quality. Biol Fert Soils. 2017;54:1–10. doi: 10.1007/s00374-017-1248-3. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRR15096517). The datasets generated during and/or analyzed during the current study are available from the corresponding authors upon request.