Abstract
Tomato (Solanum lycopersicum L.) is an important grown vegetable in Vietnam. Bacterial wilt caused by Pseudomonas solanacearum has been considered to be an important disease resulting in a harvest loss up to 90% and significant economic loss to farmers. In this study, two bacteriophages DLDT_So2 and BHDT_So9 specific to P. solanacearum were isolated. Morphological analysis indicated that DLDT_So2 and BHDT_So9 had podovirus morphology and were classified into Autographiviridae family. The latent period and burst size of DLDT_So2 was found to be approximately 120 min and 20.0 ± 2.4 virions per infected cell. Meanwhile, the latent period of BHDT_So9 was 140 min with a burst size of 11.5 ± 2.8 virions per infected cell. Of the 23 bacterial strains tested, the phages infected 7/11 strains of P. solanacearum and none of the other bacteria tested were susceptible to the phages. Stability of the phages at different temperatures, pHs, solvents was also investigated. The genomes of DLDT_So2 and BHDT_So9 are 41,341 bp and 41,296 bp and long with a total GC content of 63%, contains 48 and 46 predicted protein-encoding CDSs. No virulence or antibiotic resistance genes were found in the genomes, suggesting they would be useful biocontrol agents against P. solanacearum. Classification of the phage using average nucleotide identity, phylogenetic analysis was also carried out. The two phages represented new species when they had overall average nucleotide identity of < 95%. This is first report of the isolation and characterization of P. solanacearum-specific phages from tomato farms in Vietnam.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12088-023-01090-9.
Keywords: Bacteriophage, Tomato, Bacterial wilt, Characterization, Vietnam
Introduction
Tomato (Solanum lycopersicum L.) is the second most grown vegetable globally with 182.3 million tons in 2017 [1]. In Vietnam, the tomato growing area is approximately 23,000 hectares [2]. Tomato has become a popular food with high nutritional value [2].
As an important crop plant, tomato is a subject of genetic research with respect to characteristics such as fruit quality, yield, or tolerance to biotic and abiotic stresses [1]. Cultivation methods such as fertilizing and intensive farming are continuously improved to increase yield and product quality [2]. In the cultivation and post-harvest storage, tomato is attacked by more than 200 different diseases caused by a wide range of agents belonging to the group of molds, nematodes, bacteria, and viruses. They not only reduce the yield and quality of tomato but also risk human health and economic benefits [1]. Among the various diseases of tomato, bacterial wilt caused by Pseudomonas solanacearum (also known as Ralstonia solanacearum) is considered to be an important disease in tomato growing areas in tropical and subtropical regions [3]. P. solanacearum also infects more than 200 other plant species belonging to over 50 botanical families [4]. It has been ranked in the second place in the top 10 list of more devastating plant pathogenic bacteria [5]. P. solanacearum is well adapted to soil and survives in the rhizosphere. It infects the host through root wounds, enters the xylem tissue, and synthesizes extracellular polysaccharides that interfere with water transfer in the plant causing wilting or death of the host [6]. The bacterial wilt causes a harvest loss up to 100% in banana, 90% in tomato and potato and a significant economic loss to farmers [7]. Currently, the control of tomato diseases is combated by different approaches such as crop rotation, breeding of resistant varieties, and the use of chemical pesticides [8]. Use of chemical pesticides has been the most frequent method to control tomato diseases in Vietnam. However, improper usage of chemical pesticides can greatly affect people’s health, the long-term sustainability of tomato farming, due to the negative biological impact within the farming environment [9]. Due to these adverse impacts, biological control is an alternative approach in agriculture due to its sustainability, efficiency, and more environmentally friendly [10]. However, bacteria and fungi, which are commonly used in biological control, are often ineffective against bacterial pathogens in the rhizosphere [3].
Bacteriophage (phage) therapy is a method that is showing effectiveness in the control of rhizosphere bacterial targets due to its specificity, safety, and economic efficiency [3]. Lytic phages have been shown to be effective and safe when used to control bacterial diseases in crops while they are safe for animals and the environment. Many previous studies have demonstrated that bacteriophages can effectively control bacterial wilt disease in tomato [11].
In this study, two phages specific to P. solanacearum were isolated. Their characteristics, including latent period, burst size, host range and genomic information were determined. This is first report of the isolation and characterization of P. solanacearum-specific virulent phages from tomato farms in Vietnam.
Materials and Methods
Bacterial Strains
Eleven P. solanacearum strains were isolated from diseased tomato leaves collected from Duc Trong district, Lam Dong province, Vietnam. Among them, the strain P. solanacearum PS025 that caused the disease rate of 93.3% in tomatoes in the 7-day pathogenicity trial at a dose of 108 CFU mL−1 (data not shown) was used as a host for bacteriophage isolation. The bacteria were recovered from an ultra-freezer in tryptone soya agar medium (TSA, Merck) at 30 °C for 24 h. Other bacterial strains were from the laboratory bacterial collection.
Phage Isolation
Soil samples were obtained from tomato fields in Duc Trong district, Lam Dong province, Vietnam, and transferred to the laboratory under cold conditions for phage isolation. Five grams of soil were placed in a falcon tube. Five mL of distilled water and chloroform 5% (w/v) were added into the tube. The mixture was then mixed by vortexing for 5 min and centrifuged at 2432×g for 10 min at room temperature to obtain the supernatant and discard the soil and the chloroform layers. The supernatant was subject to further centrifugation at 9727×g for 5 min at 4 °C to remove the remaining chloroform. The resulting supernatant was passed through a 0.22 μm pore size filter, and the filtrate was used in phage enrichment and plaque assay. In the first stage of phage enrichment, the filtrate was added to 100 µL of log-phase P. solanacearum PS025 culture in a falcon tube containing 9 mL of TSB medium. After shaking at 150 rpm for 24 h at 30 °C, the culture was centrifuged at 9727×g, 5 min at 4 °C. The resulting supernatant was passed through a 0.22-μm pore size filter, and the filtrate was subjected to a plaque assay. A mixture of 100 μL of filtrate and 200 μL of log-phase P. solanacearum PS025 culture was added to 3 mL of molten 0.5% TSA (maintained at 42 °C) and poured over a 1.5% Luria–Bertani (LB) agar plate. A single transparent plaque was picked from the plate and subjected to purify the phage [12]. The morphologies of bacteriophages were examined as described previously [12]. The host range of phages were evaluated using various bacterial isolates (Table S1), the susceptibility of which was examined using a drop plaque assay [13].
One-Step Growth Curve and Phage Stability Test
The one-step growth curves of two phages were determined with slight adjustments based on previous research [12]. Stability of phages for temperature, pH, and organic solvents were carried out by the method described previously [14].
Phage Genome Sequencing and Analysis
Phage genomic DNA was extracted using the method described previously [13]. Phage genome was used as input for library preparation using NEBnext Ultra II DNA Library Prep Kit for Illumina and sequencing was performed on an Illumina NextSeq550 (150 bp paired end) at the KTEST company (Ho Chi Minh City, Vietnam). The nucleotide sequences were submitted to NCBI under accession number such as OP087422 for phage BHDT_So9 and OP087423 for phage DLDT_So2. The read data in FASTQ format files of two phages was assessed by FASTQC v0.11.9 [15] to determine sequence quality before further analysis. De novo assembly was performed by using Unicycler v0.5.0 [16] with conservative mode. Evaluation of complete contig assemblies was carried out by QUAST v5.0.2 [17]. BLAST search against the NCBI database was used to identify contigs of viral origin. Prediction of open reading frame (ORF) and annotation were performed by RAST [18] with Virus Domain and Genetic code 11. The GenBank files generated by RAST were then manually examined to validate identified ORFs if one of following conditions was satisfied: matching of a BLAST search with a gene of known function from a curated annotation of the closest viral origin, Ralstonia phage RsoP1IDN (NC_047930.1); identification of a Pfam family; having a domain hit by CDD. The presence of potential antimicrobial resistance determinants was investigated using ResFinder v4.1 [19]. Bacteriophage lifestyle was predicted by using BACPHILP [20] to test whether two phage genomes were likely to be virulent (lytic) or temperate (lysogenic). Detection of tRNA was performed using tRNAscan-SE v2.0 [21].
Multiple whole-genome alignment of two phages from Vietnam and other nine viral origins belong to the Okabevirinae subfamily (ICTV-2020) were generated using progressiveMauve and then converted back to fasta format using the xmfa2fasta.pl script. Single nucleotide polymorphism (SNP) sites were identified using snp-sites v2.5.1 [22] to generate SNP-based alignment. Distances between each pair of phage isolates were calculated using snp-dists (0.8.2). SNP-based phylogenetic tree of eleven Okabevirinae subfamily phage isolates was reconstructed using Fasttree v2.1.11 [23] with the GTR + CAT model. Circular representations of phage genomes were created using BRIG v0.95 [24].
Results and Discussion
Lytic Activity and Morphology of Phages
Two bacteriophages, DLDT_So2 and BHDT_So9, were isolated from different soil samples in Da Loan village and Bac Hoi village (Duc Trong district, Lam Dong province), respectively. After purification, both phages showed round and clear plaques (~ 8 mm in diameter) (Fig. 1A1 and B1). For DLDT_So2, the latent period was found to be approximately 120 min with a burst size of 20.0 ± 2.4 phage per infected cell. Meanwhile, the latent period of BHDT_So9 was 140 min with a burst size of 11.5 ± 2.8 phage per infected cell (Fig. 2A and B).
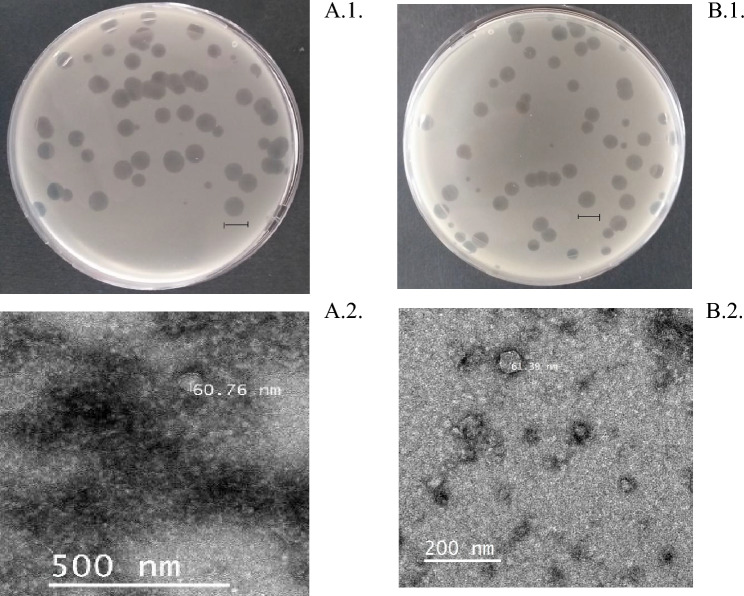
Fig. 1.
Top agar overlay showing plaque morphology of phage DLDT_So2 (A1) and BHDT_So9 (B1); the scale bar indicates 1 cm. Electron micrograph of phage DLDT_So2 (A2) and phage BHDT_So9 (B2); the bar represents 500 nm or 200 nm, respectively
Fig. 2.
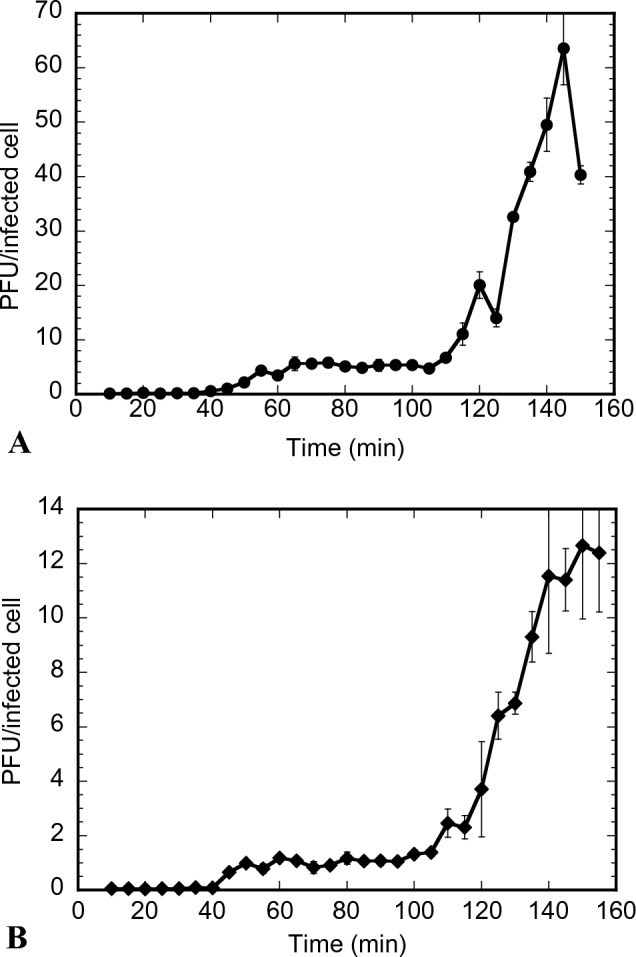
One-step growth curves of phage DLDT_So2 (A) and phage BHDT_So9 (B). Error bars indicate 95% confidence intervals for the averaged values (n = 3)
It revealed that both phage DLDT_So2 and phage BHDT_So9 have an icosahedral head of approximately 60 nm in diameter (Fig. 1A2 and B2). Two phages morphologically are podoviruses and are classified into the Autographiviridae family [25].
Host Range of Phages
The host range of DLDT_So2 and BHDT_So9 was determined using 23 bacterial isolates (Table S1). The phages formed clear zones in cultures of 7/11 strains of P. solanacearum. The two phages showed a broader host range at the species level compared to another P. solanacearum phage RsoP1IDN [26] which had the highest similarity in genome structure with two phages in the current study. None of the other bacteria species tested were susceptible to the phages.
Stability of the Phages
Phages DLDT_So2 and BHDT_So9 were found to be relatively thermostable between 4 and 50 °C, even though the concentration of phages after 1 h incubated at different temperatures tended to decrease gradually when the temperature increased (Fig. 3A). DLDT_So2 phage concentration fluctuated with the highest value of 8.1 log10 PFU/mL at 4 °C and the lowest of 7.92 log10 PFU/mL at 50 °C. Concentration reduction was observed in the temperature range from 20 to 30 °C. Similarly, BHDT_So9 concentration is highest at 4℃ (8.00 log10 PFU/mL) and lowest at 50 °C (7.71 log10 PFU/mL). At the temperature ranges of 4–20 °C and 30–37 °C, there are slight decreases in the phage concentration of approximately 0.1 log10 PFU/mL. Compared with other phages of P. solanacearum, such as PE204 isolated from rhizosphere soils by [27], the results showed comparable thermal stability over the temperature range 15–50 °C. The observed temperature stability makes the phages useful for therapeutic used in Vietnam. Phages were isolated from Lam Dong province, which, accounts for one-third of the total tomato growing area of the country (7000—8000 hectares). Lam Dong has climate which is very suitable for tomato plants to grow and develop all year round. The coldest month and the warmest month are January and April with an average maximum temperature of 21 °C (69 °F) and 25 °C (77 °F), respectively [28]. Therefore, in terms of temperature, the phages are expected to be stable when they are applied to tomato farms in Lam Dong province.
Fig. 3.
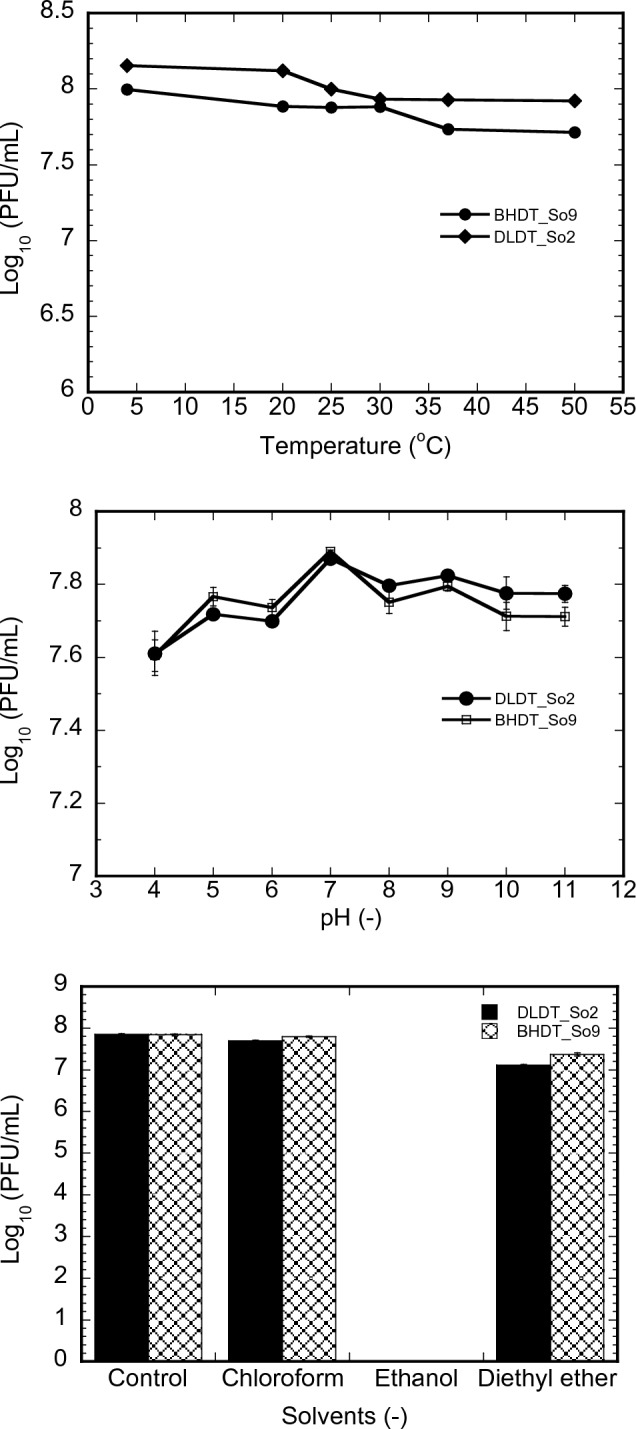
A Effect of thermal treatment on the viability of the phages; B Stability of the phages incubated at various pHs; C Viability of the phages in the presence of various organic solvents; Error bars indicate 95% confidence intervals for the averaged values (n = 3)
The pH stability analysis illustrated that both phages were stable over a wide pH range (4 to 11) when incubated for 24 h at 30 °C (Fig. 3B). Phages DLDT_So2 and BHDT_So9 were stable in pH 7–11 and 5–9, respectively. A drop in stability below pH 5 was observed resulting in no viable phages at pH 3. The stability of two phages at a low pH value remains comparable to or better than the previously published P. solanacearum’s bacteriophages [11, 27]. The persistence of the two phages in different pH also suggests that DLDT_So2 and BHDT_So9 are feasible for successful application under diverse field conditions in Vietnam.
The results of the organic solvent factor reveal that phages DLDT_So2 and BHDT_So9 were stable in chloroform with no concentration differences compared to the control (Fig. 3C). However, the resistance of both phages is slightly declined in diethyl ether solvent. The concentration differences compared to the control are 0.74 log10 PFU/mL for DLDT_So2 and 0.47 log10 PFU/mL for BHDT_So9. Furthermore, both phages were inactivated when incubated in ethanol solvent for 1 h.
Genome Characteristics and Phylogenetic Analysis
Each of two phage assemblies generated one complete contig, with estimated read depth of 2051x (BHDT_So9) and 2232x (DLDT_So2). The two phage genomes are 41,296 bp and 41,341 bp long with a total GC content of 63%, containing 46 and 48 predicted protein CDSs. By using BLAST, closest origins were identified as Higashivirus genus as Ralstonia phage RsoP1IDN—NC_047930.1 (~ 91% coverage, 93.90% identity for both BHDT_So9 and DLDT_So2). They represented new species, based on current standards [25].
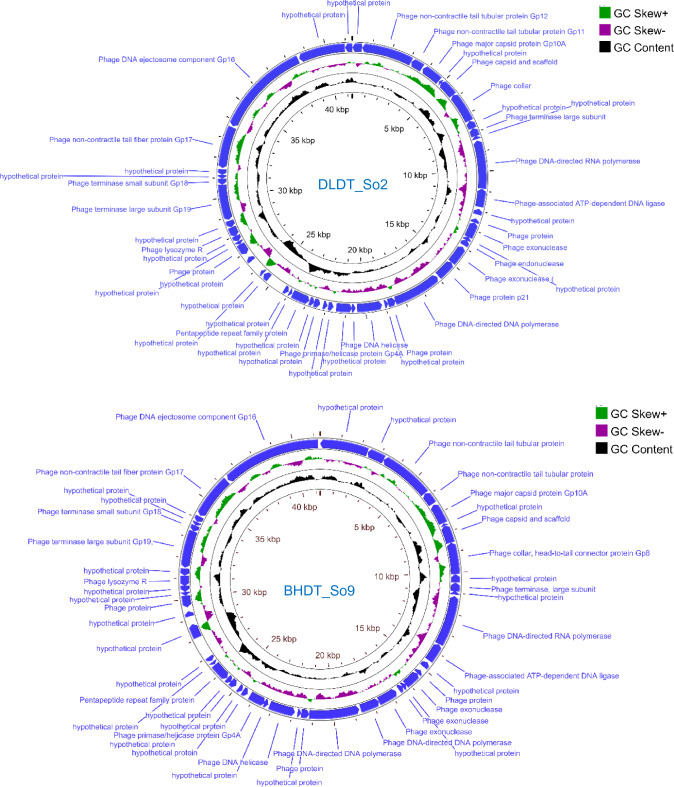
The whole genomes of two phages encoding variety of different functional proteins were annotated (Fig. 4). Manual investigation showed BHDT_So9 and DLDT_So2 had the same 24 genes with known of functional proteins, with synteny between the genomes. Genes related to DNA replication and modification enzymes were detected such as polymerase, exonuclease, endonuclease, helicase, primase, peptidoglycan lytic exotransglycosylase. Furthermore, one lysozyme-encoding gene was also found in these two genomes. Genes encoding structural and packaging proteins was detected including capsid protein, head-to-tail connector, tail fiber, tail tubular. The remaining predicted proteins were categorized as hypothetical. The phage genomes were submitted to NCBI under accession numbers OP087422 and OP087423 for phage BHDT_So9 and phage DLDT_So2 respectively.
Fig. 4.
Genome map of DLDT_So2 (A) and BHDT_So9 (B). Relative orientations of ORFs are indicated blue colors (sense and antisense)
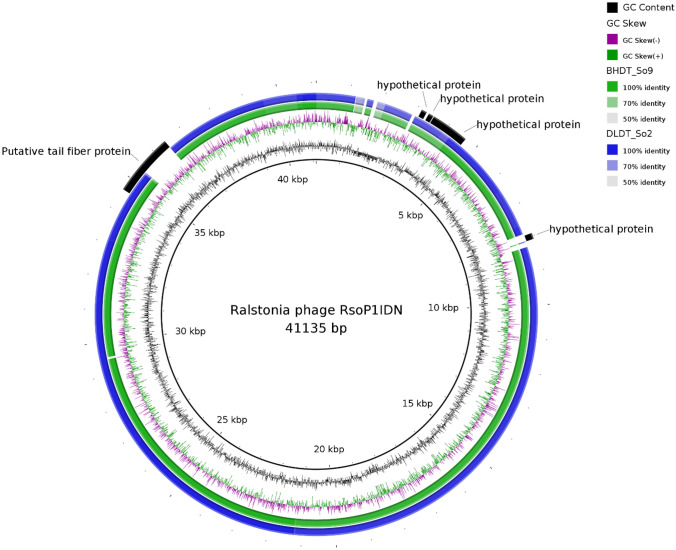
Genome-wide comparison of phages belonging to the genus Higashivirus showed that the most notable difference between two phage genomes from Vietnam and other members of the genus was the putative tail fiber protein (Fig. 5). This protein has the length of 711 aa in two phage genomes from Vietnam, but only 603 aa in RsoP1IDN (locus HOS84_gp34), and it only the first 231 aa are conserved, the rest are not similar. Ralstonia phage RsoP1IDN [NC_047930.1] also had higher similarity in genome synteny to the two phages from Vietnam when comparing with Ralstonia phage RSB1, which agreed with the BLAST result.
Fig. 5.
Comparative genomic analysis of members belonging to Higashivirus genus. The genomic diagram was drawn with BRIG with specific colors assigned for specific phage genomes. The inner circle is Ralstonia phage RsoP1IDN as the reference. Colored rings indicate regions with high pairwise genomic sequence similarity and present on both reference and compared genomes. White gaps show certain sections not present on compared genomes. Outer most ring fragments colored in black indicate ORFs encoding proteins that are different among members in Higashivirus genus
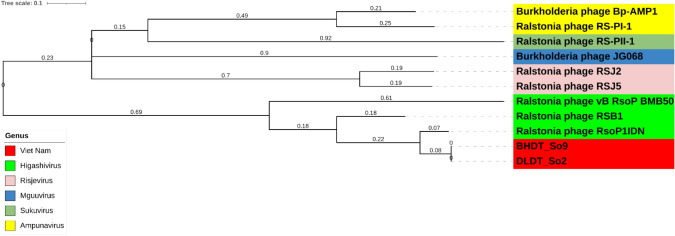
In total 52,451 SNPs were identified from the whole-genome alignment of 11 members of the Okabevirinae subfamily. Based on the SNP distance analysis, the nucleotide similarity level was high between BHDT_So9 and DLDT_So2 with only 30 SNPs, comparing to other Okabevirinae viruses within the dataset (up to 42,950 SNPs). SNP-based phylogenetic tree of whole genomes of Okabevirinae subfamily indicated that the two phages from Vietnam could have emerged from Higashivirus genus clade (Fig. 6). However, BHDT_So9 and DLDT_So2 genomes still had significant genetic divergence from the closest sister phage Ralstonia phage RsoP1IDN by more than 5400 SNPs (Supplement Table S2).
Fig. 6.
Comparative genomic analysis of 11 members of Okabevirinae subfamily using whole-genome alignment. The number on each branch indicates the branch length based on the tree scale. The SNP-based phylogenetic tree was reconstructed using Fasttree. Legend indicates each genus belonging to Okabevirinae subfamily
Temperate phages are not generally recognised as potential phage therapy candidates. Therefore, a significant checkpoint is to identify whether the phage lifestyle is likely to be lytic or lysogenic [29]. Furthermore, identifying the presence of toxins, virulence factors, or antimicrobial resistance genes is also a vital step. Prior to the application of phage to control P. solanacearum infections in tomato, it is essential to have a comprehensive genomic characterization of the phages to be used. No known antibiotic resistant genes or virulence factors were detected in phages BHDT_So9 and DLDT_So2 in the current study. In addition, no known genes were found to be associated with lysogeny, e.g. no integrase was detected. Prediction of phage lifestyle indicated that the two phages BHDT_So9 and DLDT_So2 were likely to be virulent ones. The lytic nature of the phage suggested it might serve as a potential agent against P. solanacearum, an infectious agent in tomatoes in Vietnam.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This research is funded by Vietnam National University Ho Chi Minh City, Vietnam (VNU-HCM) under grant number B2021-20-09.
Declarations
Conflict of interest
The authors declare that they have no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Singh VK, Singh AK, Kumar A. Disease management of tomato through PGPB: current trends and future perspective. 3 Biotech. 2017;7(4):255. doi: 10.1007/s13205-017-0896-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chau MH, Chinh NX. Effect of plant density and fertilizer application rates on growth, fruit yield and quality of tomato (Solanum lycopersicum L.) in greenhouse condition. Asian Plant Res J. 2021;8(3):22–31. doi: 10.9734/aprj/2021/v8i330177. [DOI] [Google Scholar]
- 3.Wang X, Wei Z, Yang K, Wang J, Jousset A, Xu Y, Shen Q, Friman VP. Phage combination therapies for bacterial wilt disease in tomato. Nat Biotechnol. 2019;37(12):1513–1520. doi: 10.1038/s41587-019-0328-3. [DOI] [PubMed] [Google Scholar]
- 4.Hayward AC. Ralstonia solanacearum. In: Lederberg J, editor. Encyclopedia of microbiology. San Diego: Academic Press; 2000. pp. 32–42. [Google Scholar]
- 5.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012;13(6):614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran TM, Jacobs JM, Huerta A, Milling A, Weibel J, Allen C. Sensitive, Secure Detection of Race 3 Biovar 2 and Native U.S. Strains of Ralstonia solanacearu. Plant Dis. 2016;100(3):630–639. doi: 10.1094/PDIS-12-14-1327-RE. [DOI] [PubMed] [Google Scholar]
- 7.Elphinstone JG. The current bacterial wilt situation: a global overview. In: Allen C, Prior P, Hayward A, editors. Bacterial wilt disease and the Ralstonia solanacearum complex. Minnesota: APS press; 2005. pp. 9–28. [Google Scholar]
- 8.Singh S, Gautam RK, Singh DR, Sharma TVRS, Sakthivel K, Roy SD. Genetic approaches for mitigating losses caused by bacterial wilt of tomato in tropical islands. Eur J Plant Pathol. 2015;143(2):205–221. doi: 10.1007/s10658-015-0690-z. [DOI] [Google Scholar]
- 9.Nguyen TM, Le NTT, HaVukaiNeN J, HaNNaway DB. Pesticide use in vegetable production: A survey of Vietnamese farmers’ knowledge. Plant Prot Sci. 2018;54(4):203–214. doi: 10.17221/69/2017-PPS. [DOI] [Google Scholar]
- 10.Kaari M, Joseph J, Manikkam R, et al. Anti-biofilm activity and biocontrol potential of Streptomyces cultures against Ralstonia solanacearum on tomato plants. Indian J Microbiol. 2022;62:32–39. doi: 10.1007/s12088-021-00963-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magar TR, Lee SY, Kim HJ, Lee SW. Biocontrol of bacterial wilt in tomato with a cocktail of lytic bacteriophages. Appl Microbiol Biotechnol. 2022;106(9–10):3837–3848. doi: 10.1007/s00253-022-11962-7. [DOI] [PubMed] [Google Scholar]
- 12.Hoang HA, Yen MH, Ngoan VT, Nga LP, Oanh D. Virulent bacteriophage of Edwardsiella ictaluri isolated from kidney and liver of striped catfish Pangasianodon hypophthalmus in Vietnam. Dis Aquat Organ. 2018;132(1):49–56. doi: 10.3354/dao03302. [DOI] [PubMed] [Google Scholar]
- 13.Tu VQ, Nguyen TT, Tran XTT, et al. 2020) Complete genome sequence of a novel lytic phage infecting Aeromonas hydrophila, an infectious agent in striped catfish (Pangasianodon hypophthalmus. Arch Virol. 2020;165:2973–2977. doi: 10.1007/s00705-020-04793-2. [DOI] [PubMed] [Google Scholar]
- 14.Xuan T, Hoang H, Tam L. Stability and activity of TG25P phage in control of Aeromonas hydrophila in striped catfish pond water. VNUHCM J Sci Technol Dev. 2018;21(2):64–70. doi: 10.32508/stdj.v21i2.429. [DOI] [Google Scholar]
- 15.Andrews S (2010) FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 16.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13(6):e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hockenberry AJ, Wilke CO. BACPHLIP: predicting bacteriophage lifestyle from conserved protein domains. PeerJ. 2021;9:e11396. doi: 10.7717/peerj.11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Page AJ, Keane JA, Delaney AJ, Taylor B, Seemann T, Harris SR, Soares J. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genomics. 2016 doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner D, Kropinski AM, Adriaenssens EM. A roadmap for genome-based phage taxonomy. Viruses. 2021;13:506. doi: 10.3390/v13030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Addy HS, Farid MM, Ahmad AA, Huang Q. Host range and molecular characterization of a lytic Pradovirus-like Ralstonia phage RsoP1IDN isolated from Indonesia. Adv Virol. 2018;163(12):3409–3414. doi: 10.1007/s00705-018-4033-1. [DOI] [PubMed] [Google Scholar]
- 27.Bae JY, Wu J, Lee HJ, Jo EJ, Murugaiyan S, Chung E, Lee SW. Biocontrol potential of a lytic bacteriophage PE204 against bacterial wilt of tomato. J Microbiol Biotechnol. 2012;22(12):1613–1620. doi: 10.4014/jmb.1208.08072. [DOI] [PubMed] [Google Scholar]
- 28.Weather and Climate (2022) https://weather-and-climate.com/average-monthly-Rainfall-Temperature-Sunshine-region-lam-dong-vn, Vietnam (accessed on July 30, 2022).
- 29.Philipson CW, Voegtly LJ, Lueder MR, Long KA, Rice GK, Frey KG, Biswas B, Cer RZ, Hamilton T, Bishop-Lilly KA. Characterizing phage genomes for therapeutic applications. Viruses. 2018;10(4):188. doi: 10.3390/v10040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.