Abstract
The emergence and spread of antibiotic-resistant pathogenic bacteria have necessitated finding new control alternatives. Under these circumstances, lytic bacteriophages offer a viable and promising option. This review focuses on Vibrio-infecting bacteriophages and the characteristics that make them suitable for application in the food and aquaculture industries. Bacteria, particularly Vibrio spp., can produce biofilms under stress conditions. Therefore, this review summarizes several anti-biofilm mechanisms that phages have, such as stimulating the host bacteria to produce biofilm-degrading enzymes, utilizing tail depolymerases, and penetrating matured biofilms through water channels. Additionally, the advantages of bacteriophages over antibiotics, such as a lower probability of developing resistance and the ability to infect dormant cells, are discussed. Finally, this review presents future research prospects related to further utilization of phages in diverse fields.
Keywords: Bacteriophage, Vibrio spp., Antibiotic resistance, Biofilm, Aquaculture
Introduction
Vibrio spp. are Gram-negative, rod-shaped bacteria found in brackish water environments, with their abundance increasing as temperatures rise (Oliver et al., 2018). Vibrio cholerae serogroups O1 and O139 cause acute diarrheal disease cholera (CDC, 2022). Among non-cholera vibrios, Vibrio vulnificus and Vibrio parahaemolyticus are highly pathogenic to humans, with the former having a higher mortality rate (Kim, 2020; Wang et al., 2015). People get infected by these pathogens through consumption of contaminated water/seafood or open wound exposure. By expressing multiple virulence factors, such as the MARTX toxin and phospholipase (Kim et al., 2017; Cho et al., 2022), these non-cholera vibrios often cause severe outcomes including necrotizing fasciitis and septic death. In addition to human-infecting vibrios, there are other species of Vibrio (Vibrio harveyi, Vibrio anguillarum and Vibrio ordalii) that can infect marine life, causing a big loss in the aquaculture industry (Plaza et al., 2018).
Based on the Continuous Plankton Recorder survey conducted in the North Atlantic Ocean from 1958 to 2011, it has been reported that the incidence of Vibrio spp. is rising due to the increase in sea surface temperature worldwide (Vezzulli et al., 2016). Especially in Korea, the rate of change of sea surface temperature has been reported to be higher than the global average, suggesting that the incidence of Vibrio-related diseases is increasing (Korea Meteorological Administration, 2020). In this country, about 10% of the food-borne outbreaks from 2007 to 2012 were attributed to V. parahaemolyticus, while the average incidence of V. vulnificus infection from 2003 to 2016 was 0.12 cases per 100,000 people but with a case fatality rate as high as 48.9% (Kim et al., 2022; Moon et al., 2014).
Due to the misuse of antibiotics in veterinary medicine, the development of antibiotic-resistant Vibrio spp. is an alarming problem. A study conducted in the United States in 2006 revealed that V. vulnificus and V. parahaemolyticus are resistant to eight or more antibiotics commonly used to treat Vibrio infections, as well as ampicillin and chloramphenicol (Elmahdi et al., 2016). Meanwhile, Vibrio spp. is a major biofilm forming strain. Nevertheless, antibiotics have shown little effect in controlling biofilm. Bacteria in biofilm can transfer resistance genes more easily than those present as plankton cells, increasing the risk of spreading antibiotic resistance. Therefore, new alternatives to target antibiotic-resistant bacteria and/or biofilm need to be developed and deployed (Abe et al., 2020).
A promising solution to this problem is the use of bacteriophages. Bacteriophages are viruses that can only infect and replicate inside their host bacteria. Among these, Vibrio-infecting phages are viruses that are ubiquitous in seawater and can only infect bacteria from the Vibrionaceae family, being important for the control of the population of Vibrio spp. and maintaining the health of the marine ecosystems. These phages have DNA or RNA as their genetic material, are mostly tailed or filamentous and the grand majority follow a lytic infecting cycle. Lytic Vibrio-infecting phages are attracting attention as a potential treatment for vibriosis. In addition, they have been proposed as control agents to mitigate antibiotic-resistant and biofilm-forming bacteria, which are currently considered to be significant threats to the food industry (Bischoff et al., 2021).
This review focuses on the use of Vibrio-infecting phages as a potential alternative control agent for pathogens causing human infections and losses in aquaculture. Specifically, the current research status of Vibrio-infecting phages and their ability to control biofilms will be covered. In addition, the prospects and future opportunities for the use of bacteriophages as biocontrol agents are explored.
Bacteriophages
Phage life cycle
To infect the host bacteria, phages recognize bacterial surface receptors via the receptor binding proteins (RBPs), which are usually located in the phage tail. After this tight adsorption, the phages inject their genome into the host cell. The subsequent infection strategy is determined by whether the phage follows a lytic (virulent) or lysogenic (temperate) life cycle (Salmond and Fineran, 2015). If the phage is a virulent phage, it hijacks the bacterial gene expression machinery to support phage proliferation and assembly of new virions (Madigan et al., 2020). Consequently, the production of phage enzymes, such as holins and endolysins, lyses the bacterial cell wall, allowing the release of new phage progeny and restarting the cycle (Liu et al., 2022b).
Temperate phages, on the other hand, do not replicate inside or kill the host bacteria. Rather, once inside the bacteria, the phage genome is synchronized within the host chromosome as a prophage. Under normal circumstances, the phage genome is kept silent; however, under certain stress conditions, such as exposure to antibiotics, the prophage undergoes an induction cycle that leads to excision of the phage genome and initiation of a lytic cycle (Salmond and Fineran, 2015).
Phages as biocontrol agents
Phages can be used to control pathogenic and multidrug resistant bacteria in a variety of applications, including the food industry. In the food industry, bacteriophages have several advantages over traditional chemical and physical disinfectants. These include effective elimination of target host bacteria, relatively low cost of discovery/production, and minimal impact over the sensory and quality characteristics of the applied food (Moye et al., 2018; O'Sullivan et al., 2019; Sillankorva et al., 2012). In general, in this industry, bacteriophages have been employed to prevent, control, sanitize and preserve food, by applying it on pre-harvested, post-harvested products, food contact surfaces and ready-to-eat foods. With this, a significant reduction of Salmonella, Campylobacter, Escherichia coli, Listeria monocytogenes has been achieved (Endersen and Coffey, 2020).
Among the food industries, the aquaculture industry is gradually increasing in size and is considered to play an important role as a future food supplier. However, commercial aquaculture has a high incidence of bacterial diseases, especially vibriosis, due to overfeeding, use of high temperatures, lack of water renewal, and improper removal of injured or dead fish (Almeida et al., 2009). For these problems, phages are likely to be a key solution. In fact, Guenther et al. (2009) demonstrated that when phages were applied to food, there was a greater reduction in food-borne pathogens in the aquatic matrix than in the solid matrix. In addition, Culot et al. (2019) demonstrated that when phages were applied to water, the phages could easily and efficiently enter the body of fish through the gills, facilitating the spread of phages. A number of examples have verified the likelihood of phages to control pathogenic bacteria in aquaculture (Higuera et al., 2013; Karunasagar et al., 2007; Nakai and Park, 2002; Nakai et al., 1999; Park and Nakai, 2003; Park et al., 2000; Silva et al., 2014, 2016; Vinod et al., 2006), proving the efficacy of phages in the aquatic environment. Furthermore, due to their ubiquitous nature, the isolation of bacteriophages is more cost-effective than the development of vaccines or antibiotics (Culot et al., 2019). These characteristics make bacteriophages, especially lytic phages, suitable for use as biocontrol agents in aquaculture.
Currently, there are several phage-related products that have been classified as Generally Recognized as Safe (GRAS) and are commercially available. These products can be used in food products to reduce contamination by some of the major pathogenic and antibiotic resistant bacteria, such as E. coli (Ecolicide PX™) (Vikram et al., 2021), Listeria (ListShield™) (Lang, 2006), and Salmonella (PhageGuard S™) (Parveen et al., 2017; Ye et al., 2022). Currently, there are no commercially available phage products for the treatment of pathogenic Vibrio spp. However, there are multiple ongoing efforts to characterize and evaluate the efficacy of novel bacteriophages that can infect Vibrio spp. in food and aquaculture products (Table 1), resulting in several Vibrio-infecting products that are under development (Hodgson, 2013; Letchumanan et al., 2016; Richards, 2014).
Table 1.
List of recently reported Vibrio spp.-infecting bacteriophages (From 2020 to 2023)
| Phage | Family | Genome size (kb) | Host | Efficacy test | Reference |
|---|---|---|---|---|---|
| VVP001 | Siphoviridae | 76.42 | V. vulnificus | Abalones | (Kim et al., 2021) |
| vB_VpaP_DE10 | Autographiviridae | 42.87 | V. parahaemolyticus | (Ye et al., 2022) | |
| VPT02 | Siphoviridae | 120.55 | V. parahaemolyticus | RTE raw fish flesh slices | (You et al., 2021) |
| VPG01 | Siphoviridae | 120.02 | V. parahaemolyticus | Cutting board and seafood item | (Lee et al., 2023) |
| KIT04 | Demerecviridae | 114.93 | V. parahaemolyticus | (Tu et al., 2023) | |
| vB_VpP_DE17 | Podoviridae | 43.397 | V. parahaemolyticus | (Yang et al., 2022a) | |
| F23s1 | Siphoviridae | 76.648 | V. parahaemolyticus | (Xia et al., 2022) | |
| Vp33 | Podoviridae | V. parahaemolyticus | (Tan et al., 2021a) | ||
| Vp22 | Podoviridae | V. parahaemolyticus | (Tan et al., 2021a) | ||
| Vp21 | Podoviridae | V. parahaemolyticus | (Tan et al., 2021a) | ||
| Vp02 | Podoviridae | V. parahaemolyticus | (Tan et al., 2021a) | ||
| Vp08 | Siphoviridae | V. parahaemolyticus | (Tan et al., 2021a) | ||
| Vp11 | Siphoviridae | V. parahaemolyticus | (Tan et al., 2021a) | ||
| vB_VpaS_PG07 | Siphoviridae | 112.106 | V. parahaemolyticus | (Ding et al., 2020) | |
| phiTY18 | Myoviridae | 191.5 | V. parahaemolyticus | (Liu et al., 2022a) | |
| vB_VpP_WS1 | Microviridae | 5.564 | V. parahaemolyticus | (Xu et al., 2022) | |
| V5 | Inoviridae | 6.658 | V. parahaemolyticus | Shrimp | (Dubey et al., 2021; Tyagi et al., 2022) |
| vB_VpS_BA3 | Siphoviridae | 58.648 | V. parahaemolyticus | (Yang et al., 2020) | |
| vB_VpS_CA8 | Siphoviridae | 58.48 | V. parahaemolyticus | (Yang et al., 2020) | |
| vB_VpaP_FE11 | Podoviridae | 43.397 | V. parahaemolyticus | (Yang et al., 2022b) | |
| 27Ua.3 | Siphoviridae | 76.890 | V. parahaemolyticus | (Stoos et al., 2022) | |
| 29Fa.3 | Siphoviridae | 79.348 | V. parahaemolyticus | (Stoos et al., 2022) | |
| 31Fb.4 | Siphoviridae | 77.620 | V. parahaemolyticus | (Stoos et al., 2022) | |
| 33Fb.4 | Siphoviridae | 77.632 | V. parahaemolyticus | (Stoos et al., 2022) | |
| vB_VpaM_PG19 | Microviridae | 5.572 | V. parahaemolyticus | (Guo et al., 2022) | |
| vB_VpaP_CHI | Queuovirinae | 57.805 | V. parahaemolyticus | (Orozco-Ochoa et al., 2023) | |
| vB_VpaP_ALK | Queuovirinae | 57.805 | V. parahaemolyticus | (Orozco-Ochoa et al., 2023) | |
| vB_VpaP_M3 | Autographiviridae | 43.446 | V. parahaemolyticus | (Orozco-Ochoa et al., 2023) | |
| vB_VpaP_C2 | Autographiviridae | 43.494 | V. parahaemolyticus | (Orozco-Ochoa et al., 2023) | |
| vB_VpaP_M9 | Autographiviridae | 43.268 | V. parahaemolyticus | (Orozco-Ochoa et al., 2023) | |
| vB_VpaP_M83 | Autographiviridae | 43.901 | V. parahaemolyticus | (Orozco-Ochoa et al., 2023) | |
| vB_VpaP_MGD2 | Autographiviridae | 45.105 | V. parahaemolyticus | (Cao et al., 2021) | |
| ϕVP-1 | Myoviridae | 150.764 | V. parahaemolyticus | (Matamp and Bhat, 2020) | |
| vB_VpaP_AL-1 | Autographiviridae | 42.854 | V. parahaemolyticus | (González-Gómez et al., 2022) | |
| vB_VpaS_AL-2 | Siphoviridae | 58.457 | V. parahaemolyticus | (González-Gómez et al., 2022) | |
| vB_VpS_PG28 | Siphoviridae | 82.712 | V. parahaemolyticus | (Tian et al., 2022) | |
| KIT05 | Podoviridae | 50.628 | V. parahaemolyticus | (Anh et al., 2022) | |
| vB_VpaP_GHSM17 | Autographiviridae | 43 | V. parahaemolyticus | (Liang et al., 2022) | |
| vB_VcaS_HC | Siphoviridae | 81.566 | V. campbellii | (Li et al., 2021a) | |
| OPA17 | Siphoviridae | 75.897 | V. campbellii | (Srisangthong et al., 2023) | |
| ΦImVa-1 | Schitoviridae | 77.479 | V. alginolyticus | (Tajuddin et al., 2022) | |
| Φ-5 | Myoviridae | 238.053 | V. alginolyticus | Oyster larvae | (Le et al., 2020) |
| Φ-6 | Myoviridae | V. alginolyticus | Oyster larvae | (Le et al., 2020) | |
| Φ-7 | Myoviridae | V. alginolyticus | Oyster larvae | (Le et al., 2020) | |
| BUCT549 | Siphoviridae | 80.294 | V. alginolyticus | (Li et al., 2021b) | |
| vB_ValP_VA-RY-3 | Podoviridae | 40.271 | V. alginolyticus | (Ren et al., 2022) | |
| VPMCC5 | Zobellviridae | 48.938 | V. harveyi | (Kar et al., 2022) | |
| vB_VhaM_pir03 | Myoviridae | 286.284 | V. harveyi | Brine shrimp | (Misol et al., 2020) |
| V-YDF132 | Siphoviridae | 84.375 | V. harveyi | (Kang et al., 2022) | |
| OY1 | Autographiviridae | 43.479 | V. mimicus | (Gao et al., 2022) | |
| vB_VnaS-L3 | Siphoviridae | 39.99 | V. natriegens | (Li et al., 2022) |
In the meantime, several studies have been conducted to confirm the safety of phage treatment. According to a review from 2008 to 2021 made by Yang et al. (2022b), the administration of phages (which have various bacteria as hosts, including V. parahaemolyticus) to animals and humans did not manifest any major adverse events. In 52 studies investigating various routes of administration, adverse effects were reported in only 7% of the patients treated with phages, and these adverse effects were generally mild and resolvable (Uyttebroek et al., 2022). Based on these reports, the use of phages is considered safe; however, there is still a need to develop an appropriate standard for phage administration.
Biofilm and phages
Vibrio biofilms
Bacteria have developed several mechanisms to withstand external stress. One of these mechanisms is the formation of biofilms. Biofilms are a collection of bacterial communities embedded in a self-produced extracellular polymeric substance (EPS) composed of polysaccharides, proteins, lipids, and extracellular DNA that can adhere to surfaces. Cells within a biofilm are protected from various external stresses (e.g., dehydration, starvation, and predation), are tolerant to antimicrobial agents, and can readily adapt to different environments (Azeredo et al., 2021).
In the case of Vibrio spp., different loci (vps, wcr, syp and cps) control the production of different exopolysaccharides, and thus the production of the exopolysaccharide differs depending on the genes within a particular locus present in the bacteria (Yildiz and Visick, 2009). Notably, the expressions of these Vibrio exopolysaccharide genes are controlled by quorum sensing, a bacterial communication system. Therefore, species specific quorum sensing system determine biofilm formation in Vibrio spp. (Yildiz and Visick, 2009). The flagellum and pili also play an important role in Vibrio biofilm formation. Research has shown that when the bacterium loses the flagellar genes flaA, flrC, flgD or flgE, the attachment ability is significantly reduced and only a weak 3-dimensional biofilm is formed. A similar situation was also observed when the mutations occurred in the main pili (MSHA, ChiRP and PilD) (Yildiz and Visick, 2009).
Although there are not as many studies as with other bacterial strains, Vibrio infecting phages have been used to disrupt the biofilm structure. Yang et al. (2022b) used phage FE11 to control the biofilm produced by V. parahaemolyticus. The results showed that the ability to disrupt the biofilm was directly related to the concentration of the phage, with the highest concentration used being the one that reduced the bacterial load the most. Matamp and Bhat (2020) achieved an 84% reduction of V. parahaemolyticus biofilm after 24 h of phage ϕVP-1 treatment. Furthermore, Tan et al. (2015) applied phage ΦH20 to V. anguillarum biofilm, which resulted in a reduction of the total biofilm area from 41,000 μm2 per mm2 filter to 5000 μm2 per mm2 filter after 6 h of incubation. These are promising results suggesting that phages have multiple mechanisms to overcome biofilm production by Vibrio spp.
Role of phages in control of biofilms
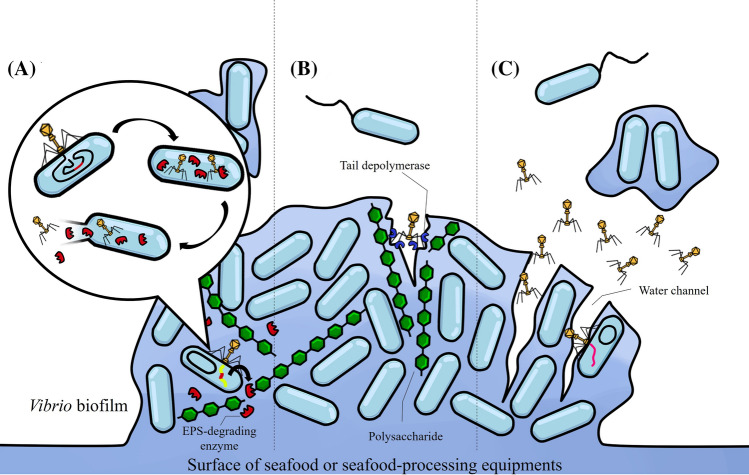
To overcome the EPS barrier, lytic phages have developed several attack strategies (Fig. 1). The first mechanism involves stimulating the host bacteria to produce EPS-degrading enzymes after lytic infection. Once the EPS-degrading enzymes break down the pieces of the biofilm, phages and phage progeny can easily penetrate deep and maintain the infection cycle (Amankwah et al., 2021). For example, Bacillus subtilis produces the hydrolase γ-PGA, which is encoded by the pghP gene of phage ΦNIT. This enzyme breaks down the capsular polysaccharide (poly-γ-glutamate) into tri-, tetra-, and penta-γ-glutamate, facilitating phage movement even through thick biofilms (Geredew et al., 2019; Kimura and Itoh, 2003).
Fig. 1.
Biofilm eradication by bacteriophages. (A) Phages stimulate the target bacteria to produce enzymes (lyases or hydrolases) that degrade the EPS matrix post phage infection. (B) Some phages have enzymes (depolymerases) on their tail spikes that help to degrade the biofilm. (C) Water accessible channels in the mature biofilm facilitate the movement of bacteriophages and increase the chance of infection
Another mechanism that phages can use to degrade the biofilm is to encode tail depolymerases that can digest the polysaccharides, lipids, and proteins that make up the EPS. Matamp and Bhat (2020) discovered that the Vibrio-infecting phage ϕVP-1 can produce a tail tube protein that functions as a polysaccharide-hydrolyzing enzyme that aids in biofilm destruction. A study by Latka and Drulis-Kawa (2020) showed that the K. pneumoniae infecting phage KP34 produces the depolymerase KP34p57 (homologous to a pectin lyase), an enzyme with antibiofilm capacity, achieving a 60% reduction in biofilm mass after 72 h of treatment with 109 PFU/ml. In addition, the bacteriophage JA1 that infects V. cholerae O139 produces a lyase capable of depolymerizing the capsular polysaccharide by β-elimination of a 4-substituted uronic acid residue (Linnerborg et al., 2001).
Although some phages do not encode EPS-degrading enzymes, they can diffuse through the water-accessible channels in biofilm and reach the bacterial cells. Water is the most abundant component in biofilms, making up to 97% of the volume in some cases. This property helps to transport nutrients throughout the matrix. Phages can diffuse through these water channels and initially infect the bacteria at the edges, penetrating the inner layers of the biofilm by increasing the population through active replication (Azeredo et al., 2021). The results of Vilas Boas et al. (2016) confirmed this observation, as their study showed that during the initial stages of phage infections (specifically using the phage phiIBB-PAA2 with P. aeruginosa and vB_AbaP_CEB1 with Acinetobacter), the infected cells were primarily located in the outer layer of the biofilm. However, as the infection progressed, cells located at the deepest depths of the biofilm became susceptible to phage infection.
Phages vs. antibiotics to control biofilm in the food industry
One of the main problems with antibiotic treatment is that antibiotics target not only the problematic bacteria, but also the bacteria that make up the normal microbiota, leading to microbiota imbalances. Moreover, due to the slow diffusion of antibiotics within a biofilm, bacteria have a chance to develop resistance mechanisms against the antibiotics, making biofilms difficult to eradicate with such chemicals. In contrast, lytic phages are highly specific and lyse only the target bacteria, likely preserving the normal bacterial flora in the local environment. Although bacteria can also develop resistance mechanisms against phages, phages are living creatures that are constantly evolving and developing new mechanisms to overcome bacterial resistance (Samson et al., 2013). Phages also produce endolysins, enzymes that lyse the peptidoglycan layer, to which bacteria are less likely to develop resistance mechanisms since these enzymes target a highly essential and thus conserved area of the bacterial cell wall (Schmelcher et al., 2012). However, the high specificity of phages to target host bacteria can be a drawback for biofilm treatment, as the biofilm-forming bacteria need to be identified in order to select the appropriate phages. Biofilms are usually composed of different species in nature. Tan et al. (2021b) discovered that V. parahaemolyticus and Shewanella putrefaciens interact with each other in a synergistic manner to produce a mixed biofilm, increasing the cell viability, EPS content and biomass of the biofilms compared to the corresponding mono-species biofilms. Therefore, to efficiently target the biofilm microbiome, a mixture of different phages should be considered (Sulakvelidze et al., 2001).
The effectiveness of antibiotics depends on the amount supplied. However, sometimes the required amount is above the minimum toxic concentration for humans, and thus antibiotics cannot be supplied in sufficient quantities to eliminate the biofilm due to health risks. On the other hand, the toxicity of phage-treated food has not yet been clearly analyzed. However, Plaut and Stibitz (2019) stated that if the phages are administered following Good Manufacturing Practices (GMP), the reported adverse effects are none or relatively small. Following this, the toxicity of medical phage-treatment in humans has demonstrated that phages are not toxic even when applied in concentrations as high as 1011 PFU/mL to treat a P. aeruginosa infection (Suh et al., 2022).
Because biofilms are composed of EPS, which has hydrophobic or hydrophilic properties, the diffusion efficiency of antibiotics is reduced; thus, the delivery of the antibiotic to the bottom of the biofilm is not guaranteed (Azeredo et al., 2021). Nevertheless, there are studies that have shown that phages use the water channels in biofilm to migrate, reach, and infect the innermost cells in a biofilm (Vilas Boas et al., 2016). In addition, phages can self-replicate within the infected bacterial cell, multiplying in large numbers and spreading the infection to the surrounding cells (Azeredo et al., 2021).
When it comes to eliminating persister cells in biofilms, antibiotics are not an option because they can only affect metabolically active cells. Phages also have a limited effect on this type of cells; however, the advantage of phages is that they can infect non-living bacterial cells, remain dormant inside them, and reactivate once the cells become metabolically active (Harper et al., 2014). As bacterial lysis proceeds, the process releases nutrients into the matrix that help restore normal growth in nearby dormant bacterial cells, allowing the phages to infect them. However, one limitation common to both antibiotic and phage treatment of biofilms is that their effectiveness decreases as the thickness, age, and diversity of the biofilm increases (Azeredo et al., 2021).
Future directions
The threat of vibriosis and Vibrio biofilms to the food industry is a significant concern, especially under the growing antibiotic resistance problem. Bacteriophages have emerged as a promising alternative to antibiotics, as they possess a high degree of specificity for their host bacteria and thus, are less likely to harm beneficial bacteria. In addition, phages have demonstrated the ability to eradicate biofilm matrices, thereby eliminating pathogens in the process. While it is true that bacteria can develop resistant mechanisms to block phage infection, phages can evolve on their own and counteract this resistance by developing adaptive systems, such as the anti-CRISPR system.
To expand the usage of bacteriophages, however, further investigation is needed to increase their efficiency and/or efficacy. Research related to the use of phage cocktails or the co-treatment of phages with other antibacterial molecules, such as essential oils (bergamot, lemongrass oil, etc.), free-fatty acids (palmitic, stearic acid, etc.), and natural chelating agents (EDTA, nitrilotriacetic acid, etc.), should be explored to widen the range of target pathogens. These components have a synergistic effect, since they can permeabilize the cell membrane, and thus, increase the effectivity of bacteriophages. Phage engineering should also be utilized to increase host range and burst size while reducing the eclipse and latent periods, as these characteristics could improve phage efficiency. Furthermore, phage enzymes (endolysin and depolymerase) could be engineered to enhance their recognition and degradation ability to various substrates, thus increasing their capacity to eradicate diverse bacterial cell walls and multi-species biofilms. Finally, a regulatory framework to standardize the dosage of phages in food products should be developed.
Overall, the evidence suggests that bacteriophages have the potential to be an effective alternative for controlling pathogens and biofilms, particularly with respect to Vibrio spp. This presents a promising breakthrough for the challenges faced by the seafood industry.
Acknowledgements
This work was supported by the National Research Foundation of Korea funded by the Ministry of Science and ICT (NRF-2020R1F1A1070168 and NRF- 2022R1F1A1074305 to B.S.K.).
Declarations
Conflict of interest
There are no conflicts of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ana Cevallos-Urena and Jeong Yeon Kim contributed equally to this article.
Contributor Information
Ana Cevallos-Urena, Email: belen_cevallos_@hotmail.com.
Jeong Yeon Kim, Email: kimjy0311@ewhain.net.
Byoung Sik Kim, Email: b.kim@ewha.ac.kr.
References
- Abe K, Nomura N, Suzuki S. Biofilms: hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. Federation of European Microbiological Societies. 2020;96:fiaa031. doi: 10.1093/femsec/fiaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida A, Cunha Â, Gomes NC, Alves E, Costa L, Faustino MA. Phage therapy and photodynamic therapy: low environmental impact approaches to inactivate microorganisms in fish farming plants. Marine Drugs. 2009;7:268–313. doi: 10.3390/md7030268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amankwah S, Abdella K, Kassa T. Bacterial biofilm destruction: A focused review on the recent use of phage-based strategies with other antibiofilm agents. Science and Applications: Nanotechnology; 2021. pp. 161–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anh VTT, Pham-Khanh NH, Han NS, Sunahara H, Kamei K. Characterization and Complete Genomic Analysis of Vibrio parahaemolyticus-Infecting Phage KIT05. Current Microbiology. 2022;79:221. doi: 10.1007/s00284-022-02907-4. [DOI] [PubMed] [Google Scholar]
- Azeredo J, García P, Drulis-Kawa Z. Targeting biofilms using phages and their enzymes. Current Opinion in Biotechnology. 2021;68:251–261. doi: 10.1016/j.copbio.2021.02.002. [DOI] [PubMed] [Google Scholar]
- Bischoff V, Zucker F, Moraru C. Marine Bacteriophages. Encyclopedia of Virology. 2021;4:322–341. doi: 10.1016/B978-0-12-809633-8.20988-6. [DOI] [Google Scholar]
- Cao Y, Zhang Y, Lan W, Sun X. Characterization of vB_VpaP_MGD2, a newly isolated bacteriophage with biocontrol potential against multidrug-resistant Vibrio parahaemolyticus. Archives of Virology. 2021;166:413–426. doi: 10.1007/s00705-020-04887-x. [DOI] [PubMed] [Google Scholar]
- CDC. Cholera - Vibrio cholerae infection. CDC. Available from: https://www.cdc.gov/cholera/general/index.html#one. Feb. 20, 2022.
- Cho C, Choi S, Kim MH, Kim BS. Vibrio vulnificus PlpA facilitates necrotic host cell death induced by the pore forming MARTX toxin. Journal of Microbiology. 2022;60(2):224–233. doi: 10.1007/s12275-022-1448-x. [DOI] [PubMed] [Google Scholar]
- Culot A, Grosset N, Gautier M. Overcoming the challenges of phage therapy for industrial aquaculture: A review. Aquaculture. 2019;513:734423. doi: 10.1016/j.aquaculture.2019.734423. [DOI] [Google Scholar]
- Ding T, Sun H, Pan Q, Zhao F, Zhang Z, Ren H. Isolation and characterization of Vibrio parahaemolyticus bacteriophage vB_VpaS_PG07. Virus Research. 2020;286:198080. doi: 10.1016/j.virusres.2020.198080. [DOI] [PubMed] [Google Scholar]
- Dubey S, Singh A, Kumar BN, Singh NK, Tyagi A. Isolation and characterization of bacteriophages from inland saline aquaculture environments to control Vibrio parahaemolyticus contamination in shrimp. Indian Journal of Microbiology. 2021;61:212–217. doi: 10.1007/s12088-021-00934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmahdi S, DaSilva LV, Parveen S. Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: a review. Food Microbiology. 2016;57:128–134. doi: 10.1016/j.fm.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Endersen L, Coffey A. The use of bacteriophages for food safety. Current Opinion in Food Science. 2020;36:1–8. doi: 10.1016/j.cofs.2020.10.006. [DOI] [Google Scholar]
- Gao L, Ouyang M, Li Y, Zhang H, Zheng X-F, Li H-X, Rao S-Q, Yang Z-Q, Gao S. Isolation and Characterization of a Lytic Vibriophage OY1 and Its Biocontrol Effects Against Vibrio spp. Frontiers in Microbiology. 2022 doi: 10.3389/fmicb.2022.830692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geredew K, Legesse M, James G, Speck P. Mini-review: efficacy of lytic bacteriophages on multispecies biofilms. Biofouling. 2019;35:472–481. doi: 10.1080/08927014.2019.1613525. [DOI] [PubMed] [Google Scholar]
- González-Gómez JP, López-Cuevas O, Castro-del Campo N, González-López I, Martínez-Rodríguez CI, Gomez-Gil B, Chaidez C. Genomic and biological characterization of the novel phages vB_VpaP_AL-1 and vB_VpaS_AL-2 infecting Vibrio parahaemolyticus associated with acute hepatopancreatic necrosis disease (AHPND) Virus Research. 2022;312:198719. doi: 10.1016/j.virusres.2022.198719. [DOI] [PubMed] [Google Scholar]
- Guenther S, Huwyler D, Richard S, Loessner MJ. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Applied and Environmental Microbiology. 2009;75:93–100. doi: 10.1128/AEM.01711-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Zheng K, Luo L, Liu Y, Shao H, Guo C, He H, Wang H, Sung YY, Mok WJ. Characterization and Genomic Analysis of ssDNA Vibriophage vB_VpaM_PG19 within Microviridae, Representing a Novel Viral Genus. Microbiology Spectrum. 2022;10:e00585–e1522. doi: 10.1128/spectrum.00585-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper DR, Parracho HM, Walker J, Sharp R, Hughes G, Werthén M, Lehman S, Morales S. Bacteriophages and Biofilms. Antibiotics. 2014;3:270–284. doi: 10.3390/antibiotics3030270. [DOI] [Google Scholar]
- Higuera G, Bastías R, Tsertsvadze G, Romero J, Espejo RT. Recently discovered Vibrio anguillarum phages can protect against experimentally induced vibriosis in Atlantic salmon, Salmo Salar. Aquaculture. 2013;392:128–133. doi: 10.1016/j.aquaculture.2013.02.013. [DOI] [Google Scholar]
- Hodgson K. Bacteriophage Therapy. Microbiology Australia. 2013;34:28–31. doi: 10.1071/MA13009. [DOI] [Google Scholar]
- Kang S, Zhang L, Liao J, Zhang D, Wu S, Zhang X, Qin Q, Wei J. Isolation and Characterization of a Newly Discovered Phage, V-YDF132, for Lysing Vibrio harveyi. Viruses. 2022;14:1802. doi: 10.3390/v14081802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar P, Das TK, Ghosh S, Pradhan S, Chakrabarti S, Mondal KC, Ghosh K. Characterization of a Vibrio-infecting bacteriophage, VPMCC5, and proposal of its incorporation as a new genus in the Zobellviridae family. Virus Research. 2022;321:198904. doi: 10.1016/j.virusres.2022.198904. [DOI] [PubMed] [Google Scholar]
- Karunasagar I, Shivu M, Girisha S, Krohne G, Karunasagar I. Biocontrol of pathogens in shrimp hatcheries using bacteriophages. Aquaculture. 2007;268:288–292. doi: 10.1016/j.aquaculture.2007.04.049. [DOI] [Google Scholar]
- Kim BS. Spatiotemporal regulation of Vibrio exotoxins by HlyU and other transcriptional regulators. Toxins. 2020;12:544. doi: 10.3390/toxins12090544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BS, Gavin HE, Satchell KJF. Variable virulence of biotype 3 Vibrio vulnificus due to MARTX toxin effector domain composition. mSphere. 2017;2:e00272–e317. doi: 10.1128/mSphereDirect.00272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-J, Kim Y-T, Kim HB, Choi SH, Lee J-H. Characterization of bacteriophage VVP001 and its application for the inhibition of Vibrio vulnificus causing seafood-borne diseases. Food Microbiology. 2021;94:103630. doi: 10.1016/j.fm.2020.103630. [DOI] [PubMed] [Google Scholar]
- Kim JS, Lee EG, Chun BC. Epidemiologic characteristics and case fatality rate of Vibrio vulnificus Infection: analysis of 761 cases from 2003 to 2016 in Korea. Journal of Korean Medical Science. 2022;37:9. doi: 10.3346/jkms.2022.37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Itoh Y. Characterization of poly-γ-glutamate hydrolase encoded by a bacteriophage genome: possible role in phage infection of Bacillus subtilis encapsulated with poly-γ-glutamate. Applied and Environmental Microbiology. 2003;69:2491–2497. doi: 10.1128/AEM.69.5.2491-2497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korea Meteorological Administration. Korean Climate Change Assessment Report 2020. Available from: http://www.climate.go.kr/home/cc_data/2020/Korean_Climate_Change_Assessment_Report_2020_2_eng_summary.pdf. Feb. 20, 2020.
- Latka A, Drulis-Kawa Z. Advantages and limitations of microtiter biofilm assays in the model of antibiofilm activity of Klebsiella phage KP34 and its depolymerase. Scientific Reports. 2020;10:1–12. doi: 10.1038/s41598-020-77198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TS, Southgate PC, O’Connor W, Vu SV, Kurtböke Dİ. Application of bacteriophages to control Vibrio alginolyticus contamination in oyster (Saccostrea glomerata) larvae. Antibiotics. 2020;9:415. doi: 10.3390/antibiotics9070415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Oh M, Kim BS. Phage biocontrol of zoonotic food-borne pathogen Vibrio parahaemolyticus for seafood safety. Food Control. 2023;144:109334. doi: 10.1016/j.foodcont.2022.109334. [DOI] [Google Scholar]
- Letchumanan V, Chan K-G, Pusparajah P, Saokaew S, Duangjai A, Goh B-H, Ab Mutalib N-S, Lee L-H. Insights into bacteriophage application in controlling Vibrio species. Frontiers in Microbiology. 2016;7:1114. doi: 10.3389/fmicb.2016.01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang Z, Zhao J, Wang L, Xie G, Huang J, Zhang Y. A novel vibriophage vB_VcaS_HC containing lysogeny-related gene has strong lytic ability against pathogenic bacteria. Virologica Sinica. 2021;36:281–290. doi: 10.1007/s12250-020-00271-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tian F, Hu Y, Lin W, Liu Y, Zhao F, Ren H, Pan Q, Shi T, Tong Y. Characterization and genomic analysis of BUCT549, a novel bacteriophage infecting Vibrio alginolyticus with flagella as receptor. Frontiers in Microbiology. 2021;12:668319. doi: 10.3389/fmicb.2021.668319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liang Y, Wang Z, Yao Y, Chen X, Shao A, Lu L, Dang H. Isolation and Characterization of a Novel Vibrio natriegens—Infecting Phage and Its Potential Therapeutic Application in Abalone Aquaculture. Biology. 2022;11:1670. doi: 10.3390/biology11111670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Wang Y, Hong B, Li Y, Ma Y, Wang J. Isolation and Characterization of a Lytic Vibrio parahaemolyticus Phage vB_VpaP_GHSM17 from Sewage Samples. Viruses. 2022;14:1601. doi: 10.3390/v14081601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnerborg M, Weintraub A, Albert MJ, Widmalm G. Depolymerization of the capsular polysaccharide from Vibrio cholerae O139 by a lyase associated with the bacteriophage JA1. Carbohydrate Research. 2001;333:263–269. doi: 10.1016/S0008-6215(01)00159-8. [DOI] [PubMed] [Google Scholar]
- Liu B, Zheng T, Quan R, Jiang X, Tong G, Wei X, Lin M. Biological characteristics and genomic analysis of a novel Vibrio parahaemolyticus phage phiTY18 isolated from the coastal water of Xiamen China. Frontiers in Cellular and Infection Microbiology. 2022 doi: 10.3389/fcimb.2022.1035364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Han G, Li Z, Cun S, Hao B, Zhang J, Liu X. Bacteriophage therapy in aquaculture: Current status and future challenges. Folia Microbiologica. 2022;67:573–590. doi: 10.1007/s12223-022-00965-6. [DOI] [PubMed] [Google Scholar]
- Madigan MT, Clark DP, Stahl D, Martinko JM. Brock biology of microorganisms. 16. USA: Pearson; 2020. pp. 191–195. [Google Scholar]
- Matamp N, Bhat SG. Genome characterization of novel lytic Myoviridae bacteriophage ϕVP-1 enhances its applicability against MDR-biofilm-forming Vibrio parahaemolyticus. Archives of Virology. 2020;165:387–396. doi: 10.1007/s00705-019-04493-6. [DOI] [PubMed] [Google Scholar]
- Misol GN, Jr, Kokkari C, Katharios P. Biological and genomic characterization of a novel jumbo bacteriophage, vB_VhaM_pir03 with broad host lytic activity against Vibrio harveyi. Pathogens. 2020;9:1051. doi: 10.3390/pathogens9121051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S, Sohn I-W, Hong Y, Lee H, Park J-H, Kwon G-Y, Lee S, Youn S-K. Emerging pathogens and vehicles of food-and water-borne disease outbreaks in Korea, 2007–2012. Osong Public Health and Research Perspectives. 2014;5:34–39. doi: 10.1016/j.phrp.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye ZD, Woolston J, Sulakvelidze A. Bacteriophage applications for food production and processing. Viruses. 2018;10:205. doi: 10.3390/v10040205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai T, Park SC. Bacteriophage therapy of infectious diseases in aquaculture. Research in Microbiology. 2002;153:13–18. doi: 10.1016/S0923-2508(01)01280-3. [DOI] [PubMed] [Google Scholar]
- Nakai T, Sugimoto R, Park K-H, Matsuoka S, Mori K-i, Nishioka T, Maruyama K. Protective effects of bacteriophage on experimental Lactococcus garvieae infection in yellowtail. Diseases of Aquatic Organisms. 1999;37:33–41. doi: 10.3354/dao037033. [DOI] [PubMed] [Google Scholar]
- O'Sullivan L, Bolton D, McAuliffe O, Coffey A. Bacteriophages in food applications: from foe to friend. Annual Review of Food Science and Technology. 2019;10:151–172. doi: 10.1146/annurev-food-032818-121747. [DOI] [PubMed] [Google Scholar]
- Baker-Austin Craig, Oliver James D, Alam Munirul, Ali Afsar, Waldor Matthew K, Qadri Firdausi, Martinez-Urtaza Jaime. Vibrio spp infections. Nature Reviews Disease Primers. 2018;4:1–19. doi: 10.1038/s41572-018-0005-8. [DOI] [PubMed] [Google Scholar]
- Orozco-Ochoa AK, González-Gómez JP, Castro-del Campo N, Lira-Morales JD, Martínez-Rodríguez CI, Gomez-Gil B, Chaidez C. Characterization and genome analysis of six novel Vibrio parahaemolyticus phages associated with acute hepatopancreatic necrosis disease (AHPND) Virus Research. 2023;323:198973. doi: 10.1016/j.virusres.2022.198973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SC, Nakai T. Bacteriophage control of Pseudomonas plecoglossicida infection in ayu Plecoglossus altivelis. Diseases of Aquatic Organisms. 2003;53:33–39. doi: 10.3354/dao053033. [DOI] [PubMed] [Google Scholar]
- Park SC, Shimamura I, Fukunaga M, Mori K-I, Nakai T. Isolation of bacteriophages specific to a fish pathogen, Pseudomonas plecoglossicida, as a candidate for disease control. Applied and Environmental Microbiology. 2000;66:1416–1422. doi: 10.1128/AEM.66.4.1416-1422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parveen S, Schwarz J, Hashem F, Vimini B. Reduction of Salmonella in ground chicken using a bacteriophage. Poultry Science. 2017;96:2845–2852. doi: 10.3382/ps/pex062. [DOI] [PubMed] [Google Scholar]
- Plaut RD, Stibitz S. Phage Therapy: A Practical Approach. Cham: Springer; 2019. Regulatory considerations for bacteriophage therapy products; pp. 337–349. [Google Scholar]
- Plaza N, Castillo D, Pérez-Reytor D, Higuera G, García K, Bastías R. Bacteriophages in the control of pathogenic vibrios. Electronic Journal of Biotechnology. 2018;31:24–33. doi: 10.1016/j.ejbt.2017.10.012. [DOI] [Google Scholar]
- Ren Y, Wang L, Chen R, Li X, Li S, Li J, Li Q, Wang Z, Xu Y. Isolation and characterization of a novel phage vB_ValP_VA-RY-3 infecting Vibrio alginolyticus. Virus Research. 2022;322:198945. doi: 10.1016/j.virusres.2022.198945. [DOI] [PubMed] [Google Scholar]
- Richards GP. Bacteriophage remediation of bacterial pathogens in aquaculture: a review of the technology. Bacteriophage. 2014;4:e975540. doi: 10.4161/21597081.2014.975540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmond GP, Fineran PC. A century of the phage: past, present and future. Nature Reviews Microbiology. 2015;13:777–786. doi: 10.1038/nrmicro3564. [DOI] [PubMed] [Google Scholar]
- Samson JE, Magadán AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nature Reviews Microbiology. 2013;11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- Schmelcher M, Donovan DM, Loessner MJ. Bacteriophage endolysins as novel antimicrobials. Future Microbiology. 2012;7:1147–1171. doi: 10.2217/fmb.12.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillankorva SM, Oliveira H, Azeredo J. Bacteriophages and their role in food safety. International Journal of Microbiology. 2012 doi: 10.1155/2012/863945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva YJ, Costa L, Pereira C, Mateus C, Cunha A, Calado R, Gomes NC, Pardo MA, Hernandez I, Almeida A. Phage therapy as an approach to prevent Vibrio anguillarum infections in fish larvae production. PLoS ONE. 2014;9:e114197. doi: 10.1371/journal.pone.0114197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva YJ, Moreirinha C, Pereira C, Costa L, Rocha RJ, Cunha Â, Gomes NC, Calado R, Almeida A. Biological control of Aeromonas salmonicida infection in juvenile Senegalese sole (Solea senegalensis) with Phage AS-A. Aquaculture. 2016;450:225–233. doi: 10.1016/j.aquaculture.2015.07.025. [DOI] [Google Scholar]
- Srisangthong I, Sangseedum C, Chaichanit N, Surachat K, Suanyuk N, Mittraparp-Arthorn P. Characterization and Genome Analysis of Vibrio campbellii Lytic Bacteriophage OPA17. Microbiology Spectrume. 2023 doi: 10.1128/spectrum.01623-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoos KAB, Ren J, Shields-Cutler RR, Sams KL, Caldwell S, Ho MB, Rivara G, Whistler CA, Jones SH, Wiedmann M. Coastal water bacteriophages infect various sets of Vibrio parahaemolyticus sequence types. Frontiers in Microbiology. 2022 doi: 10.3389/fmicb.2022.1041942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh GA, Lodise TP, Tamma PD, Knisely JM, Alexander J, Aslam S, Barton KD, Bizzell E, Totten KM, Campbell JL. Considerations for the use of phage therapy in clinical practice. Antimicrobial Agents and Chemotherapy. 2022;66(3):e02071–e12021. doi: 10.1128/aac.02071-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulakvelidze A, Alavidze Z, Morris JG., Jr Bacteriophage Therapy. Antimicrobial Agents and Chemotherapy. 2001;45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajuddin S, Khan AM, Chong LC, Wong CL, Tan JS, Ina-Salwany MY, Lau HY, Ho KL, Mariatulqabtiah AR, Tan WS. Genomic analysis and biological characterization of a novel Schitoviridae phage infecting Vibrio alginolyticus. Applied Microbiology and Biotechnology. 2022;107(2–3):749–768. doi: 10.1007/s00253-022-12312-3. [DOI] [PubMed] [Google Scholar]
- Tan CW, Rukayadi Y, Hasan H, Abdul-Mutalib N-A, Jambari NN, Hara H, Thung TY, Lee E, Radu S. Isolation and characterization of six Vibrio parahaemolyticus lytic bacteriophages from seafood samples. Frontiers in Microbiology. 2021;12:616548. doi: 10.3389/fmicb.2021.616548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Dahl A, Middelboe M. Vibriophages differentially influence biofilm formation by Vibrio anguillarum strains. Applied and Environmental Microbiology. 2015;81:4489–4497. doi: 10.1128/AEM.00518-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Li H, Chen B, Huang J, Li Y, Zheng H, Liu H, Zhao Y, Wang JJ. Dual-species biofilms formation of Vibrio parahaemolyticus and Shewanella putrefaciens and their tolerance to photodynamic inactivation. Food Control. 2021;125:107983. doi: 10.1016/j.foodcont.2021.107983. [DOI] [Google Scholar]
- Tian F, Li J, Hu Y, Zhao F, Ren H, Pan Q, Nazir A, Li F, Tong Y. Characterization and complete genome sequence analysis of a newly isolatedphage against Vibrio parahaemolyticus from sick shrimp in Qingdao, China. Plos ONE. 2022;17:e0266683. doi: 10.1371/journal.pone.0266683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu AVT, Pham-Khanh NH, Nguyen SH, Sunahara H, Xuan TDT, Kamei K. Isolation, characterization, and complete genome sequence of vibrio phage KIT04, a novel lytic phage of the subfamily ermolyevavirinae. Virology. 2023 doi: 10.1016/j.virol.2023.01.008. [DOI] [PubMed] [Google Scholar]
- Tyagi A, Dubey S, Sharma C, Sudan P, Rai S, Kumar BN, Chandra M, Arora A. Complete genome sequencing and characterization of single-stranded DNA Vibrio parahaemolyticus phage from inland saline aquaculture environment. Virus Genes. 2022;58:483–487. doi: 10.1007/s11262-022-01913-9. [DOI] [PubMed] [Google Scholar]
- Uyttebroek S, Chen B, Onsea J, Ruythooren F, Debaveye Y, Devolder D, Spriet I, Depypere M, Wagemans J, Lavigne R. Safety and efficacy of phage therapy in difficult-to-treat infections: a systematic review. The Lancet Infectious Diseases. 2022 doi: 10.1016/S1473-3099(21)00612-5. [DOI] [PubMed] [Google Scholar]
- Vezzulli L, Grande C, Reid PC, Hélaouët P, Edwards M, Höfle MG, Brettar I, Colwell RR, Pruzzo C. Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proceedings of the National Academy of Sciences. 2016;113:E5062–E5071. doi: 10.1073/pnas.1609157113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikram A, Woolston J, Sulakvelidze A. Phage biocontrol applications in food production and processing. Current Issues in Molecular Biology. 2021;40:267–302. doi: 10.21775/cimb.040.267. [DOI] [PubMed] [Google Scholar]
- Vilas Boas D, Almeida C, Sillankorva S, Nicolau A, Azeredo J, Azevedo NF. Discrimination of bacteriophage infected cells using locked nucleic acid fluorescent in situ hybridization (LNA-FISH) Biofouling. 2016;32:179–190. doi: 10.1080/08927014.2015.1131821. [DOI] [PubMed] [Google Scholar]
- Vinod M, Shivu M, Umesha K, Rajeeva B, Krohne G, Karunasagar I, Karunasagar I. Isolation of Vibrio harveyi bacteriophage with a potential for biocontrol of luminous vibriosis in hatchery environments. Aquaculture. 2006;255:117–124. doi: 10.1016/j.aquaculture.2005.12.003. [DOI] [Google Scholar]
- Wang R, Zhong Y, Gu X, Yuan J, Saeed AF, Wang S. The pathogenesis, detection, and prevention of Vibrio parahaemolyticus. Frontiers in Microbiology. 2015 doi: 10.3389/fmicb.2015.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Yang H, Yan N, Hou W, Wang H, Wang X, Wang H, Zhou M. Bacteriostatic effects of phage F23s1 and its endolysin on Vibrio parahaemolyticus. Journal of Basic Microbiology. 2022;62:963–974. doi: 10.1002/jobm.202200056. [DOI] [PubMed] [Google Scholar]
- Xu W, Xuan G, Lin H, Wang J. Complete genome analysis of the newly isolated Vibrio phage vB_VpP_WS1 of the family Microviridae. Archives of Virology. 2022;167:1311–1316. doi: 10.1007/s00705-022-05413-x. [DOI] [PubMed] [Google Scholar]
- Yang M, Chen H, Guo S, Tan S, Xie Z, Zhang J, Wu Q, Tan Z. Characterization and genome analysis of a novel Vibrio parahaemolyticus phage vB_VpP_DE17. Virus Research. 2022;307:198580. doi: 10.1016/j.virusres.2021.198580. [DOI] [PubMed] [Google Scholar]
- Yang M, Chen H, Huang Q, Xie Z, Liu Z, Zhang J, Ding Y, Chen M, Xue L, Wu Q. Characterization of the Novel Phage vB_VpaP_FE11 and Its Potential Role in Controlling Vibrio parahaemolyticus Biofilms. Viruses. 2022;14:264. doi: 10.3390/v14020264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Liang Y, Huang S, Zhang J, Wang J, Chen H, Ye Y, Gao X, Wu Q, Tan Z. Isolation and Characterization of the Novel Phages vB_VpS_BA3 and vB_VpS_CA8 for Lysing Vibrio parahaemolyticus. Frontiers in Microbiology. 2020;11:259. doi: 10.3389/fmicb.2020.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Chen H, Huang Q, Huang S, He J, Zhang J, Wu Q, Li X, Hu W, Yang M. Characterization and Genomic Analysis of Novel Vibrio parahaemolyticus Phage vB_VpaP_DE10. Viruses. 2022;14:1609. doi: 10.3390/v14081609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends in Microbiology. 2009;17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You HJ, Lee JH, Oh M, Hong SY, Kim D, Noh J, Kim M, Kim BS. Tackling Vibrio parahaemolyticus in ready-to-eat raw fish flesh slices using lytic phage VPT02 isolated from market oyster. Food Research International. 2021;150:110779. doi: 10.1016/j.foodres.2021.110779. [DOI] [PubMed] [Google Scholar]