Abstract
Biofilms are a major concern within the food industry since they have the potential to reduce productivity in situ (within the field), impact food stability and storage, and cause downstream food poisoning. Within this review, predatory bacteria as potential biofilm control and eradication agents are discussed, with a particular emphasis on the intraperiplasmic Bdellovibrio-and-like organism (BALO) grouping. After providing a brief overview of predatory bacteria and their activities, focus is given to how BALOs fulfill four attributes that are essential for biocontrol agents to be successful in the food industry: (1) Broad spectrum activity against pathogens, both plant and human; (2) Activity against biofilms; (3) Safety towards humans and animals; and (4) Compatibility with food. As predatory bacteria possess all of these characteristics, they represent a novel form of biofilm biocontrol that is ripe for use within the food industry.
Keywords: Biofilms, Pathogens, Predatory bacteria, Bdellovibrio, Food
Introduction
The food industry is a broad business network covering the various aspects of food production, food processing, and distribution. In this industry, bacterial contamination can cause serious problems at each stage, such as impacting the food production process, thereby lowering productivity or quality, leading to food spoilage during processing and distribution and potentially causing food poisoning within the consumers. There is also an economic cost. For instance, in New Zealand, a country of barely 4 million people, the cost associated with food-borne diseases was estimated to be $88.8 million NZD (Scott et al., 2000), while the global costs associated with biofilms incurred in all sectors in 2019 were reported to be more than $5 trillion USD annually, with $324 billion USD in food and agriculture alone (Cámara et al., 2022). To prevent bacterial contamination, therefore, the food industry needs to make every effort to introduce good agricultural and manufacturing practices, including managing hygiene and adopting various methods for microbial inactivation, such as thermal or radiation treatments, or the use of chemicals, such as hypochlorous acid. Despite these efforts, however, the problem of bacterial contamination persists, causing significant financial loss and deaths each year. Among the various factors that make it difficult to prevent contamination, one major concern is bacterial biofilms (Galie et al., 2018; Srey et al., 2013; Van Houdt and Michiels, 2010).
Biofilms, which represent the form in which most bacteria in nature are present and are defined as three-dimensional structures created by microbes within them as they gather and grow (Flemming and Wingender, 2010; Tolker-Nielsen et al., 2015). Biofilms are often found on surfaces and tend to be highly resistant to common bacterial eradication methods due to their structural and physiological properties, which are defined by extracellular polymeric substances (EPS). The bacterial EPS is a complex structure of macromolecules, such as polysaccharides (i.e., poly-N-acetylglucosamine), proteins and DNA, that not only surrounds the bacterial cells and hold them fast, but also limits the penetration of antibiotics and other antibacterials into the biofilm and, as a result, allows the bacteria within to survive treatment (Flemming and Wingender, 2010). Furthermore, the bacterial cells inside experience stress conditions such as limited oxygen and nutrient availabilities, quorum sensing mechanisms, etc., that forces them to enter a less active state, thereby increasing their resistance by not responding to drugs and treatments (Kim et al., 2009; Borriello et al., 2004; Nguyen et al., 2011; Zhao et al., 2020). As a result of both factors, i.e., EPS and low activity, bacteria present in biofilms are reported to be 10- to 1000-times more resistant to antibiotics (Mah and O'Toole, 2001), making the removal of the bacteria located within more difficult.
Biofilms are found in all stages of the food industry, and their incomplete sterilization in food processing plants may contaminate food or lead to serious cases of food poisoning. In response, methods for effective biofilm removal are a topic of constant discussion (Carrascosa et al., 2021; Galie et al., 2018). In this review, therefore, recent work related to predatory bacteria and their applications as novel biofilm biocontrol agents is discussed, highlighting some of the clear benefits found with these unique microorganisms.
Predatory bacteria and their life cycles
As a group of microbes, predatory bacteria have a particular lifestyle in that they grow by preying on other microbes. Although many people, including microbiologists, are not familiar with them, these predators exist ubiquitously throughout the environment and have been found in diverse locales, including rivers (Fry and Staples, 1976; Jang et al., 2022b; Pineiro et al., 2013), soil (Jurkevitch et al., 2000; Klein and Casida Jr, 1967; Oyedara et al., 2016), the ocean (Baer et al., 2004; Williams and Li, 2018), wastewater treatment plants (Cohen et al., 2021; Feng et al., 2016; Jurkevitch, 2020; Mun et al., 2022) and even within animals (Guo et al., 2017; Kelley and Williams, 1992; Schwudke et al., 2001). Moreover, various predatory “families” and their different lifestyles have been identified and studied. Predatory bacteria are commonly sub-categorized into four distinct groups by their predatory lifestyles (Table 1)—wolf-pack, epibiotic, intraperiplasmic and cytoplasmic—which are defined by the location of the predator in relation to its prey. Each of these is briefly described below.
Table 1.
Classification of predatory bacteria according to their lifestyle
| Subclass | Predatory strain | References |
|---|---|---|
| ‘Wolf-pack’ predators | Lysobacter spp. | Seccareccia et al. (2015) |
| Myxobacteria; Corallococcus spp., Myxococcus spp., Pyxidicoccus spp. | Livingstone et al. (2017) | |
| Epibiotic predators | Pseudobdellovibrio exovorus | Koval et al. (2013), Pasternak et al. (2013) |
| Micavibrio aeruginosavorus | Pasternak et al. (2013), Wang et al. (2011) | |
| Vampirococcus (Candidate Phyla Radiation) | Esteve et al. (1983), Moreira et al. (2021) | |
| Vampirovibrio chlorellavorus | Coder and Starr (1978), Hovde et al. (2019) | |
| Intraperiplasmic predators | Bacteriovorax stolpii | Seideler et al. (1972) |
| Bdellovibrio bacteriovorus | Im et al. (2019), Sathyamoorthy et al. (2019), Shilo (1969) | |
| Halobacteriovorax spp. | Baer et al. (2004) | |
| Peredibacter starrii | Seideler et al. (1972) | |
| Pseudobacteriovorax antillogorgiicola | McCauley et al. (2015) | |
| Cytoplasmic predators | Daptobacter | Guerrero et al. (1986) |
The first group is ‘wolf-pack’ predators. These are generally opportunistic predators, capable of growth axenically but also possess the ability to predate on other microbes. Wolf-pack predators, of which myxobacteria and Lysobacter are examples (Livingstone et al., 2017; Seccareccia et al., 2015), use social gliding motility to encounter prey. While it is not clear if wolf-pack predators need to physically touch their prey as they secrete membrane vesicles and secondary metabolites that can kill the prey from a distance (Evans et al., 2012; Xiao et al., 2011), this form of predation is often viewed as being social since a minimum number of predators are needed to secrete sufficient enzymes and metabolites necessary to ensure prey lysis (McBride and Zusman, 1996). This perspective, however, has been called into question as microscopic evidence shows individual myxobacterial cells can also lyse their prey (Berleman and Kirby, 2009). A key benefit of wolf-pack predators that arises since these predators neither attach to nor invade their prey, but rather secrete antibacterial enzymes and substances, is they prey on a wide range of bacterial species (Livingstone et al., 2017). On the other hand, they are not obligate predators, and under starvation conditions also form multicellular biofilms called fruiting bodies (Muñoz-Dorado et al., 2016; Shimkets, 1999).
In contrast with wolf-pack predators, those located within the next two groupings all tend to be obligate predators, requiring prey bacteria for their growth and survival. The first is epibiotic predators, which include many different of bacterial species, including Micavibrio aeruginosavorus (Pasternak et al., 2013; Wang et al., 2011), Bdellovibrio exovorus (Koval et al., 2013; Pasternak et al., 2013), Vampirococcus (Candidate Phyla Radiation) (Esteve et al., 1983; Moreira et al., 2021) and Vampirovibrio chlorellavorus (Coder and Starr, 1978; Hovde et al., 2019). As a group, these microbes predate others by attaching to the surface of susceptible prey and consuming them while remaining outside of the prey cell. This is achieved in four stages, namely, (1) attachment, where they recognize and adhere to the surface of the prey, (2) pore formation, where the cell wall of the prey bacterium is dissolved, (3) degradation and utilization of the prey’s macromolecules, and (4) septation, during which the elongated predator septates and releases progeny via binary fission (Koval et al., 2013).
Within the second obligate predatory grouping, i.e., intraperiplasmic, the most studied is Bdellovibrio bacteriovorus, a Gram-negative bacterium belonging to the Deltaproteobacteria that a very motile due to a single polar flagellum (Im et al., 2019; Sathyamoorthy et al., 2019; Shilo, 1969). In contrast with wolf-pack and epibiotic predators, intraperiplasmic predation is quite complex and consists of several different stages [attachment, invasion, bdelloplast formation, elongation, septation via segmentation (not binary fission) and, finally, lysis of the prey and release of the predatory progeny (Fenton et al., 2010; Rotem et al., 2015)] (Fig. 1). Among the characterized predatory bacteria, various genus’ have similar intraperiplasmic lifestyles, including Peredibacter (Seideler et al., 1972), Bacteriovorax (Seideler et al., 1972), Halobacteriovorax (Baer et al., 2004), and Pseudobacteriovorax (Koval et al., 2013).
Fig. 1.
The complex lifecycle of intraperiplasmic predatory bacteria, showing the different stages involved
The final grouping, cytoplasmic predators, is represented by only a single predator, i.e., Daptobacter, and is characterized by this microbe penetrating through both membranes and entering into the cytoplasm of its prey, members of the Chromatiaceae (Guerrero et al., 1986). Much like the wolf-pack predators, Daptobacter is also a facultative predator, capable of growing axenically in the absence of prey, and also divides by binary fission (Guerrero et al., 1986).
Despite the diversity of predatory bacteria, both taxonomically and based on their predatory activities, the one common feature shared by all is their ability to kill other bacteria and hydrolyze their macromolecules. This they achieve using the veritable arsenal of proteases, nuclease and other hydrolytic enzymes that are encoded within their genomes (Inoue et al., 2022a; Oyedara et al., 2018; Pasternak et al., 2012; Rendulic et al., 2004; Williams et al., 2019). It should come as no surprise, therefore, that predatory bacteria and their activities have garnered recent attention, with applications in diverse scientific fields being explored, including biofilm removal (Bratanis et al., 2020; Dwidar et al., 2012b; Kadouri and O'Toole, 2005), prevention of membrane biofouling (Kim et al., 2013, 2014), bioplastic recovery (Martínez et al., 2016, 2013) and even sludge treatment (Feng et al., 2017; Yan et al., 2022). However, a majority of the application-based studies that have been published, particularly for the intraperiplasmic Bdellovibrio-and–like organisms (BALOs), focus heavily on their use as potential biocontrol agents (Choi et al., 2017; Cloeckaert et al., 2013; Pérez et al., 2020). Stemming from these previous works, this review will discuss the potential application of predatory bacteria within the food industry (production, process, distribution) and, in particular, as a biofilm eradication agent through the lens of previous studies.
Potential of predatory bacteria as biofilm control and eradication agents for the food industry
Within food processing industries, microbial contamination can occur at multiple stages throughout production, processing, and distribution. When this occurs, it not only leads to declines in productivity but also poses a threat to public health due to increased potential for the development and spread of pathogens associated with food poisoning. Consequently, effective methods for preventing bacterial contamination and spread are paramount in the food industry, with key aspects of these methods including (1) broad activity against pathogens, (2) effectiveness at biofilm removal, (3) safety and (4) compatibility with foods.
While conventional treatments to control bacterial contamination, such as heat, chlorine, ozone or UV, are widely used, these come with some limitations as they may not be very effective at eradicating biofilms (de Carvalho, 2017), may impact the quality of the food being processed (Sert et al., 2020), can negatively affect the equipment being used (Greene et al., 1999) or offer limited scope and applicability due to other reasons, such as toxicity (Baggio et al., 2020). In the following sections, therefore, the importance of each criterion and how predatory bacteria address each is discussed.
Predatory bacteria consume diverse food pathogens
Treatments used to reduce bacterial contamination in the food industry typically have broad-spectrum bactericidal capabilities. This is necessary because food-associated pathogens are diverse (Table 2). For example, Salmonella or E. coli O157:H7, two bacterial strains that cause food poisoning, are present in the breeding farms and enter the food processing pipeline via the animals and meat in a process coined “farm-to-fork” (Collineau et al., 2020; Rothrock et al., 2021; Wilson et al., 2018; Zhang et al., 2022). Once present within the meat processing facilities, these bacteria can grow on the surface of the meat and exposed equipment, form a biofilm in a matter of hours and eventually spread to other foods (Dourou et al., 2011; Silagyi et al., 2009; Wang et al., 2013), potentially causing downstream problems such as food poisoning. Similarly, Pectobacteria spp. and Dickeya spp., are both crop pathogens (Czajkowski et al., 2009; Davidsson et al., 2013). While both of these bacterial species may infect crops in the field and reduce the overall harvest yields, they can also cause serious problems in already harvested produce during its storage (Blancard, 2012; Czajkowski et al., 2011), leading to additional losses in food production and distribution.
Table 2.
Food-associated pathogens and their susceptibility to predation
| Pathogen | Source | Disease target | Predation in liquid culturesa | Biofilm removal by predationb | References |
|---|---|---|---|---|---|
| Gram-negative | |||||
| Acidovorax avenae | Fruits, vegetables | Crop; seedling blight, bacterial fruit blotch (BFB) | +++ | NDc | McNeely et al. (2017), Schaad et al. (2003) |
| Aeromonas spp. | Seafood | Aqua farming; crustaceans | ++ |
Bdellovibrio + |
Chu and Zhu (2010), Dashiff et al. (2011), Hoel et al. (2019) |
| Agrobacterium tumefaciens | Fruits, vegetables | Crop; crown gall disease | ± | ND | Escobar and Dandekar (2003), McNeely et al. (2017) |
| Burkholderia cepacia complex | Raw milk | Human | ++ |
Micavibrio + |
Dashiff et al. (2011), McNeely et al. (2017), Moore et al. (2001) |
| Campylobacter | Vegetables, poultry, eggs, dairy | Human and Livestock | ± | ND | Humphrey et al. (2007), Markelova (2010b) |
| Erwinia carotovora | Vegetables | Crop; soft rot | ++ | ND | Toth and Birch (2005), Yair et al. (2003) |
| Escherichia coli O157:H7 | Vegetables, meat, dairy, and seafood | Human and Livestock | ++ |
Bdellovibrio + |
Fratamico and Cooke (1996) |
| Escherichia coli STEC (non O157:H7) | Vegetables, meat, dairy, seafood | Human and Livestock | ++ | ND | Baker et al. (2016), Ottaviani et al. (2019) |
| Pseudomonas spp. | Fruits, vegetables, meat, dairy | Crop, Human and Livestock | +++ |
Bdellovibrio + Micavibrio + |
Dashiff et al. (2011), González‐Rivas et al. (2018) |
| Salmonella enterica serovar Typhimurium | Fruits, vegetables, poultry, eggs | Human and Livestock; gastroenteritis, septicemia | +++ |
Bdellovibrio + |
Atterbury et al. (2011), Fratamico and Cooke (1996), Nguyen et al. (2014), Im et al. (2018) |
| Serratia spp. | Vegetables, meat, dairy | Human and Livestock | ± | ND | Dashiff et al. (2011), Kurz et al. (2003) |
| Shigella spp. | Fecal contaminated food and water | Human; diarrhea | +++ |
Micavibrio + |
Dashiff et al. (2011), Lima et al. (2015), Willis et al. (2016) |
| Yersinia enterocolitica | Meat, dairy | Human and Livestock | +++ | ND | Monnappa et al. (2014) |
| Vibrio cholerae | Seafood | Human; cholera | ++ |
Bdellovibrio + |
Cao et al. (2015), Reidl and Klose (2002), Wucher et al. (2021) |
| Vibrio parahemolyticus | Seafood | Human; gastroenteritis | +++ |
Bdellovibrio + |
Dashiff et al. (2011), Kongrueng et al. (2017), Letchumanan et al. (2014), Richards et al. (2012) |
| Vibrio vulnificus | Seafood | Human; cellulitis, septicemia | +++ | ND | Diaz (2014), Richards et al. (2012) |
| Xanthomonas spp. | Vegetables | Crop; spots, blights | ++ | ND | Boch and Bonas (2010), Odooli et al. (2021) |
| Gram-positive | |||||
| Clostridium botulinum | Honey, canned food | Human; botulism | ND | ND | Schneider et al. (2011) |
| Listeria monocytogenes | Vegetables, meat, dairy | Human; listeriosis | ND | ND | Genigeorgis et al. (1991) |
| Staphylococcus aureus | Meat, dairy | Human | – |
Bdellovibrio + |
Im et al. (2018), Monnappa et al. (2014) |
aActivity: (–)—no activity, (±)—weak to no activity, ( +)—weakly positive activity, (++)—good activity, (+++)—strong activity
bActivity ( +) Removal of the biofilm was observed by the listed predator based on a qualitative evaluation
cND—Not done or evaluated
Studies on the prey spectrum and predation ability of predatory bacteria have been most actively conducted with B. bacteriovorus. According to Dashiff et al. (2011), one predatory strain, B. bacteriovorus 109J, attacked and reduced the viabilities (1- to 7-log) of numerous pathogens, including major food-associated pathogens, such as Salmonella, Aeromonas, Escherichia, Enterobacter, Citrobacter, Shigella, Vibrio, Yersinia and Serratia. While Campylobacter, another pathogen commonly associated with food poisoning, was not preyed upon by B. bacteriovorus 109J, Markelova (2010a) reported both Campylobacter jejuni and Helicobacter pylori were susceptible to another predatory strain, B. bacteriovorus 100NCJB. Similar to B. bacteriovorus, the epibiotic predator M. aeruginosavorus also has broad spectrum activities against many bacterial species and is capable of reducing Citrobacter, Enterobacter, Escherichia, Shigella and Yersinia populations all by as much as 2-log (Dashiff and Kadouri, 2011).
One limitation of the above predators, however, is their inability to kill Gram-positive bacterial strains, such as Enterococcus faecalis and Staphylococcus aureus (Dashiff et al., 2011; Im et al., 2018; Monnappa et al., 2014). This is not true for all predators as wolf-pack predators are capable of attacking and consuming Gram-positive strains. In a recent study, Inoue et al. (2022b) isolated two novel predatory strains, Bacteriovorax stolpii HI3 and Myxococcus sp. MH1, from a freshwater pond and compared their activities against a diverse assortment of bacterial species, including Gram-positive strains (e.g., S. aureus and Bacillus spp.). While B. stolpii HI3 was active against many of the Gram-negative strains tested (26 out of 45), it did not predate any of the eight Gram-positive bacterial strains. In contrast, Myxococcus sp. MH1 preyed on all 53 strains tested, both Gram-positive and Gram-negative (Inoue et al., 2022b). While Myxococcus sp. MH1 clearly offers a broader spectrum activity than B. stolpii HI3, one caveat associated with using Myxococcus sp. MH1 is the time required—a week was needed for predation to occur with several of the prey tested, limiting its potential as a biocontrol agent within food industries.
In addition to possessing broad spectrum activities against many different pathogens, one other clear benefit of predatory bacteria is their ability to also mitigate antibiotic resistant populations. This was demonstrated by several different groups where CDC-priority resistance markers (colistin- and carbapenem-resistance) and multidrug-resistant pathogens were employed (Dharani et al., 2018; Jang et al., 2022a; Sun et al., 2017). One of these studies, Jang et al. (2022a), delved deeper into the predation process and demonstrated that not only does predation kill the pathogens, E. coli and K. pneumoniae in their study, it also significantly removed the antibiotic resistance gene pools, i.e., mcr-1, blaKPC-2 and blaOXA-51, by as much as 99.3%.
Predatory bacteria effectively remove bacterial biofilms
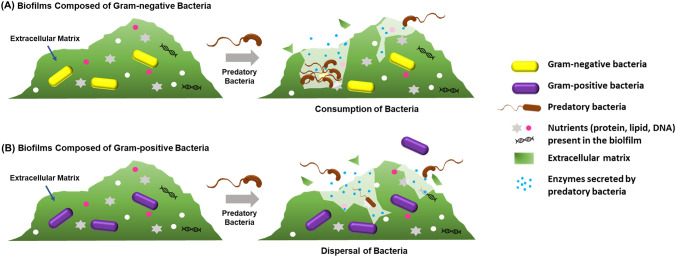
Current estimates place approximately 80% of all bacteria throughout nature are present within biofilms (Flemming and Wuertz, 2019). Within food industries, biofilms can be found on the surfaces of the food, stainless steel worktops or conveyor belts used in processing (Galie et al., 2018). While these biofilms may be resistant to many forms of treatment, including antibiotic/chemical treatments, based on studies conducted, they likely are quite susceptible to predatory bacteria and their activities. The first example of biofilm predation reported was by Kadouri and O'Toole (2005), where B. bacteriovorus 109J was capable of significantly removing E. coli and Pseudomonas fluorescens biofilms in either static or flow cell environments. The same group expanded on these results with a different predatory strain, M. aeruginosavorus, and its activities against biofilms of several different pathogenic bacterial species, including K. pneumoniae and Pseudomonas aeruginosa (Kadouri et al., 2007). In subsequent studies, Im et al. (2018) demonstrated BALOs also dismantle Salmonella enterica biofilms, a concern particularly in poultry industries (Joseph et al., 2001; Merino et al., 2019), while Chanyi and Koval (2014) explored the role of the predator’s pili in biofilm predation efficiencies, finding the loss of one pili gene (pilT1) had no observable effect but the other (pilT2) abolished the ability of their predator, B. bacteriovorus 109JA, to effectively remove E. coli biofilms (Fig. 2a). Table 2 lists many of the studies where predation was used to reduce biofilms of food-borne Gram-negative pathogens. However, the published results often employed qualitative, rather than quantitative, assessments to determine if predation was effective against these pathogens.
Fig. 2.
Removal of biofilms by intraperiplasmic predatory bacteria. (A) Susceptible Gram-negative bacterial biofilms are consumed by the predator, reducing the viability of the prey present within. (B) In contrast, Gram-positive bacteria are not predated on, meaning their viabilities do not decrease when the predator encounters their biofilms but are rather dispersed as the predator hydrolyzes the extracellular polymeric substances composing the extracellular matrix
While Gram-positive bacterial species, such as Staphylococcus spp., Streptococcus spp., Listeria spp., and Enterococcus spp., are not predated on by intraperiplasmic predatory strains (Dashiff et al., 2011; Im et al., 2017, 2018), their biofilms are dispersed by both the predators and their secreted hydrolytic enzymes (Im et al., 2018; Monnappa et al., 2014) (Fig. 2b). In fact, the study by Im et al. (2018) found this occurs even when the predator, B. bacteriovorus HD100, was washed to remove any free proteases that may be present in the media. Through transcriptomic analyses, they demonstrated the predator utilizing the biofilm’s EPS as a source of amino acids and secrete proteases in response (Im et al., 2018). Aside from these, other studies considered additional predator-pathogenic bacterial biofilm combinations, including the removal of Stenotrophomonas maltophilia biofilms by B. exovorus (Chanyi et al., 2016) and Staphylococcus epidermidis biofilms by Lysobacter gummosus, a wolf-pack predator (Gökçen et al., 2014).
While many of the above studies used polystyrene as the substrate for their biofilms, predatory strains are also effective against biofilms on other surfaces, including stainless steel (Fratamico and Cooke, 1996), silicon (Dwidar et al., 2012a), membrane filters (Kim et al., 2013) and even eukaryotic cells (Dwidar et al., 2013), all of which should be considered as they are used in the food industry. While the first three illustrate predators can be used to remove biofilms from a variety of abiotic surfaces, including the removal of E. coli O157:H7 and Salmonella spp. from stainless steel surfaces like those found in food processing plants (Fratamico and Cooke, 1996), the study by Dwidar et al. (2013) proved bacterial biofilms present on the surfaces of eukaryotic cells are also effectively removed while the underlying eukaryotic cells are not harmed by the predators or their activities. This is important as it suggests predators like B. bacteriovorus can be used within the food industry to reduce the presence of pathogenic species and their biofilms on the surfaces of meat, as was demonstrated recently (Ottaviani et al., 2019), without impacting the quality of the food product, or causing harm to the workers. This latter idea is discussed further later in this review.
Co-applications of predatory bacteria with other antimicrobials have synergetic effects
Whereas many studies report on the ability of different predators and their enzymes to remove bacterial biofilms, one limitation is that this removal is never complete as some of the biofilm, and its bacteria, survive. In response, researchers have combined the activities of predators with other antimicrobials with the goal of making them more effective than when used alone. For instance, the study by Dwidar et al. (2012a) used carbon dioxide (CO2) particles alongside predation to remove biofilms of E. coli. Not only did they achieve much better removal when these two methods were used together, but the predator itself reduced the E. coli viabilities by 50,000-fold when used alone. In contrast, use of CO2 aerosols alone to disperse the biofilm saw only a 50% reduction in the E. coli viabilities (Singh et al., 2015), meaning the number of bacteria being dispersed without predation was much higher, effectively increasing the chances for secondary exposures to occur. Another group found the use of either DNase I or DspB (a poly-N-acetylglucosaminidase) improved the ability of B. bacteriovorus 109J to remove biofilms (Dashiff and Kadouri, 2011). In contrast, co-applications with proteinase K reduced the effectiveness of this predator. The authors shared that while the exact reasons for proteinase K inhibition are not clear (prey-associated and/or predator-associated changes), the use of trypsin did not have the same negative impacts, suggesting the detrimental effects seen are specific for proteinase K and its activities.
In addition to using physical (CO2 aerosols) and enzymatic treatments, other groups also explored the use of predatory bacteria alongside antibiotics. Although the focus was not on biofilms, one such study considered the co-application of B. bacteriovorus HD100 with violacein (Im et al., 2017). As noted above, B. bacteriovorus strains only predate on Gram-negative bacterial species, meaning Gram-positive bacteria are not targeted. To circumvent this, Im et al. (2017) selected violacein, a hydrophobic secondary metabolite produced by various bacterial species that has antibacterial activities against Gram-positive bacterial species as it attacks their membranes (Aruldass et al., 2015; Cauz et al., 2019; Choi et al., 2015, 2021). Im et al. (2017) found that when used against a mixed population consisting of both Gram-positive and Gram-negative bacterial pathogens (S. aureus, Acinetobacter baumannii, Bacillus cereus and K. pneumoniae), violacein or the predator alone reduced the total pathogen viabilities by 19% and 68%, respectively, but led to a 99.98% reduction when used together, illustrating the potential use of predators alongside chemical antimicrobials. A similar concept was employed by Chanyi et al. (2016) in their study where they evaluated the potential co-application of B. exovorus with either ciprofloxacin or kanamycin in the removal of S. maltophilia biofilms. They reported that, whereas kanamycin inhibited predation of the biofilms, ciprofloxacin tended to have no observable impact, positive or negative, on the activities of B. exovorus. The reported effects of kanamycin are not too surprising given other translation inhibitors, i.e., streptomycin, chloramphenicol and puromycin, were all shown previously to allow the predator to attach to their prey but prevented predation from occurring (Varon and Shilo, 1968).
Safety of predatory bacteria shown in in vitro and in vivo tests
A third area that requires consideration is the safety of predatory bacteria. While examples of safety were indirectly reported several decades ago, such as in the study by Lenz and Hespell (1978) where it was shown BALOs do not grow within eukaryotic cells (rabbit ova), the last two decades have seen a plethora of studies exploring this issue, with heavy emphasis once more given to the BALO predators. Examples of this are two recent studies where the impact of several different BALO strains, including B. bacteriovorus, M. aeruginosavorus and a natural isolate of Bacteriovorax stolpii, on numerous human and mouse cell lines were evaluated (Gupta et al., 2016; Monnappa et al., 2016). Both studies concluded none of the predatory strains evaluated cause any loss in viability and that they led to only mild cellular responses (i.e., induced cytokine production levels), even when added at very high densities (1,000 predators per human or mouse cell).
In addition to the above in vitro studies, several groups explored the safety of BALOs within various animal models (Table 3). As discussed in the next section, predatory bacteria are present in and help protect shellfish. They have also been found in and isolated from other animals (Guo et al., 2017; Kelley and Williams, 1992; Schwudke et al., 2001), but their potential impact when provided artificially and at higher doses was not known. One of the first studies to explore this was conducted within chicks and explored the ability of the predator (B. bacteriovorus HD100) to also mitigate Salmonella enterica (Atterbury et al., 2011). They found, when the predator was added alone, it had no adverse effects on the growth and well-being of the chicks. Moreover, its addition after pre-dosing the chicks with S. enterica had beneficial impacts on the birds, significantly reducing both pathogen numbers in the gut cecal contents as well as abnormal cecal morphologies. Other in vivo studies have also been performed, including looking at the impacts of BALOs in the gut (Johnke et al., 2020; Shatzkes et al., 2017), on wound healing skin (Liu et al., 2022; Tajabadi et al., 2022), on corneal wound healing (Romanowski et al., 2016), when introduced into the lungs (Shatzkes et al., 2015, 2016) or in the bloodstream (Shatzkes et al., 2015). In every case, the predator had no ill effects on the animal host and, when tested alongside a bacterial pathogen, was often capable of significantly reducing the population of the latter.
Table 3.
Safety tests performed evaluating predatory bacteria
| Animal host | Tested cells (organ or organelle) | Loss in vitality? | References |
|---|---|---|---|
| Bovine | Madin–Darby bovine kidney (MDBK) cells | –a | Boileau et al. (2011) |
| Chick | Gut (Oral gavage) | – | Atterbury et al. (2011) |
| Human | HaCaT (Keratinocytes) | – | Gupta et al. (2016), Monnappa et al. (2016) |
| HepG2 (Liver epithelial) | |||
| HK-2 (Kidney epithelial) | |||
| MD (Spleen monocytes) | |||
| THP-1 (Blood monocytes) | |||
| T84, Caco2 (Colon carcinoma) | |||
| NuLi-1 (Alveolar epithelial) | |||
| Mice | Blood vessel (Intravenous) | – | Shatzkes et al. (2015) |
| Lung (Intranasal) | – | Shatzkes et al. (2015) | |
| Skin (Full-thickness skin wound) | – | Liu et al. (2022), Tajabadi et al. (2022) | |
| Raw 264.7 (Murine monocytes) | |||
| Rabbit | Eye (Ocular surface) | – | Romanowski et al. (2016) |
| Rat | Gut (Intrarectal) | – | Shatzkes et al. (2017) |
| Lung (Intranasal) | – | Shatzkes et al. (2016) | |
| Zebrafish | Larval microinjection (Hindbrain, Tail muscle, Caudal vein) | – | Willis et al. (2016) |
aActivity (–)—no loss in viability observed
BALOs have low immunogenic potential
Several potential reasons for the gentle nature of BALOs towards animal hosts are already known. For instance, Gram-negative outer membranes contain lipopolysaccharides (LPS), endotoxins that are recognized by toll-like receptor (TLR) proteins, leading to induced inflammatory responses and cytokine production levels within our cells (Akira and Hemmi, 2003). While BALOs are themselves Gram-negative bacteria, and also possess LPS within their outer membrane, their LPS has an altered structure as the phosphate groups have been replaced with mannose, effectively making them neutral in charge and much less immunogenic (Schwudke et al., 2003). Similarly, another component present within bacteria that induces immunomodulatory responses is flagellin, the sub-unit protein found within the flagellum (Hajam et al., 2017). BALOs once more also possess a flagellum but this is sheathed by the outer membrane (Thomashow and Rittenberg, 1985), which acts to mask the flagellin from host TLR proteins and immune response.
Methods to remove BALOs after treatment
While the above in vitro and in vivo studies all clearly illustrate the safe nature of BALOs, if the need to control their presence does arise, a simple application of soap (surfactant) will suffice based on the studies by Cho et al. (2019) and Jang et al. (2022b), As discussed in both of studies, BALOs are highly sensitive to detergents, with concentrations of sodium dodecyl sulfate (SDS) as low as 0.02% causing complete or near complete and instantaneous killing of the predator. As such, one can envision using BALOs to remove pathogens from fruits or vegetables and, with a quick wash in a detergent solution and rinse, effectively eliminate most or all of the predators before consuming the produce. One other option to reduce the viability of predatory bacteria on treated produce is to use radiation. This was demonstrated in the study by Olanya et al. (2020) where low doses of gamma irradiation were selected since they do not harm the produce (Niemira and Fan, 2014) but were able to kill B. bacteriovorus 109 present on butter lettuce (Lactuca sativa), with a 0.5 kGy treatment reducing the predator viabilities by 3- to 5-log.
Predatory bacteria are compatible with foods
To prevent food spoilage or food poisoning, excessive contamination of raw materials and equipment with bacteria needs to be prevented. More than just controlling the presence of pathogens, the antibacterial agents used should also not harm or alter the raw materials or the equipment but, rather, be compatible with both. As illustrated in the above sections, predatory bacteria do not harm the underlying substrate and, as such, can be used on many different surfaces, including stainless steel.
As it currently stands, only a few studies used predatory bacteria with the aim of protecting, or removing pathogens associated with, foodstuffs (Table 4). One of the earliest of these explored the presence of Vibrio parahaemolyticus and Vibrio vulnificus, two pathogenic bacteria that infect shellfish, within the Eastern oyster, Crassostrea virginica (Richards et al., 2012). The authors found predatory bacteria (Bdellovibrio, Bacteriovorax and Micavibrio) naturally present in the water and in the oysters acted to suppress and reduce both pathogens. Their study also validated the earlier work by Kelley et al. (1997) who found predatory bacterial strains preferred to associate with surfaces, particularly the shells of oysters. Similar results were reported in another shellfish (black tiger shrimp; Penaeus monodon), where the addition of a predatory isolate (Bdellovibrionales bacterium BDHSH06) led to better survival and pathogen reductions of between 1.3- and 4.5-log in both the water and the intestines of the shrimp (Li et al., 2014).
Table 4.
Case studies of predatory bacteria being applied to protect food
| Food | Predatory strain | Target pathogen | References |
|---|---|---|---|
| Crops | |||
| Cucumber | Corallococcus sp. | Fusarium oxysporum f. sp. cucumerinum (FOC) | Ye et al. (2020) |
| Mushroom | B. bacteriovorus | Pseudomonas tolaasii | Saxon et al. (2014) |
| Potato | B. bacteriovorus | Pectobacterium | Sason et al. (2022), Youdkes et al. (2020), Epton et al. (1989) |
| Dickeya solani | |||
| Erwinia carotovora | |||
| Rice | B. bacteriovorus | Xanthomonas oryzae | Uematsu (1980) |
| Soybean | B. bacteriovorus | Pseudomonas glycinea | Scherff (1973) |
| Seafood | |||
| Oyster | B. bacteriovorus | Vibrio spp. | Richards et al. (2012) |
| B. stolpii | |||
| Shrimp | Bdellovibrio sp. | Vibrio spp. | Cao et al. (2019), Kongrueng et al. (2017), Lu et al. (2022) |
| Bacteriovorax sp. | |||
In addition to seafood, researchers have also tested the ability of BALOs and other predatory bacteria to protect produce. Examples include protecting mushrooms (Agaricus bisporus) from Pseudomonas tolaasii (Saxon et al., 2014) and potatoes (Solanum tuberosum) from the phytopathogens (Pectobacterium and Dickeya) (Sason et al., 2022; Youdkes et al., 2020). In both cases, the actions of the predators used reduced the formation of lesion (in the case of the mushrooms) and macerations on the potato slices, illustrating the potential benefits BALOs may offer in protecting and preserving produce. At least one study also explored their effects in situ. Ye et al. (2020) found, over a two-year study, that myxobacterium Corallococcus sp. strain EGB was capable of protecting cucumber (Cucumis sativus L.) plants from Fusarium wilt caused by Fusarium oxysporum f. sp. cucumerinum in both greenhouse and field studies. After application of Corallococcus sp. strain EGB to the soil, this predator not only reduced the Fusarium oxysporum f. sp. cucumerinum abundance but also migrated to the roots of the plants, where it modulated the soil microbiome, leading the authors to conclude this predator has the potential to be used as a biocontrol agent to prevent Fusarium wilt in situ. These results highlight several simple facts—predatory bacteria already exist ubiquitously in nature alongside food products (i.e., shellfish or crops) and their presence does not affect food quality but rather can protect these products from bacterial contamination and poisoning. Therefore, predatory bacteria could be useful to use for the food industry.
Predatory bacteria: potential biocontrol agents in the food industry
As noted above, bacterial contamination and biofilms are major concerns within the food industry as they reduce productivity and storage stability, and can cause downstream food poisoning when the contaminated food is consumed. Current estimates indicate global food and agricultural costs incurred due to unwanted bacterial biofilms exceed $320 billion USD each year, illustrating the extent of this problem. Work from diverse groups around the globe show predatory bacteria may be used as biocontrol agents to help mitigate this, with their broad spectrum activities against many pathogens and their biofilms, as well as their demonstrated safety towards plants and animals, including humans. One caveat, though, is this field, i.e., the use of predatory bacteria within the food industry to control bacterial contaminants and pathogens, is still relatively young and populated with only a small handful of studies. Considering the demonstrated ability of predatory bacteria to dismantle biofilms, however, as well as the description of new and novel predatory strains each year, this area clearly has immense potential for growth and development.
Acknowledgements
Funding was sponsored by the National Research Foundation of Korea under the Mid-Career Project (Grant No. 2020R1A2C2012158) and the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (Grant Nos. 2022R1I1A1A01066417 and 2021R1A6A3A01087091). The authors appreciate the support.
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wonsik Mun, Email: wmun@unist.ac.kr.
Seong Yeol Choi, Email: asterafe@gmail.com.
Sumudu Upatissa, Email: sumudunipuni@gmail.com.
Robert J. Mitchell, Email: esgott@unist.ac.kr
References
- Akira S, Hemmi H. Recognition of pathogen-associated molecular patterns by TLR family. Immunology Letters. 2003;85:85–95. doi: 10.1016/S0165-2478(02)00228-6. [DOI] [PubMed] [Google Scholar]
- Aruldass CA, Rubiyatno R, Venil CK, Ahmad WA. Violet pigment production from liquid pineapple waste by Chromobacterium violaceum UTM5 and evaluation of its bioactivity. RSC Advances. 2015;5:51524–51536. doi: 10.1039/C5RA05765E. [DOI] [Google Scholar]
- Atterbury RJ, Hobley L, Till R, Lambert C, Capeness MJ, Lerner TR, Fenton AK, Barrow P, Sockett RE. Effects of orally administered Bdellovibrio bacteriovorus on the well-being and Salmonella colonization of young chicks. Applied and Environmental Microbiology. 2011;77:5794–5803. doi: 10.1128/AEM.00426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer ML, Ravel J, Piñeiro SA, Guether-Borg D, Williams HN. Reclassification of salt-water Bdellovibrio sp. as Bacteriovorax marinus sp. nov. and Bacteriovorax litoralis sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2004;54:1011–1016. doi: 10.1099/ijs.0.02458-0. [DOI] [PubMed] [Google Scholar]
- Baggio A, Marino M, Innocente N, Celotto M, Maifreni M. Antimicrobial effect of oxidative technologies in food processing: an overview. European Food Research and Technology. 2020;246:669–692. doi: 10.1007/s00217-020-03447-6. [DOI] [Google Scholar]
- Baker CA, Rubinelli PM, Park SH, Carbonero F, Ricke SC. Shiga toxin-producing Escherichia coli in food: incidence, ecology, and detection strategies. Food Control. 2016;59:407–419. doi: 10.1016/j.foodcont.2015.06.011. [DOI] [Google Scholar]
- Berleman JE, Kirby JR. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiology Reviews. 2009;33:942–957. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancard D. Diagnosis of parasitic and nonparasitic diseases. pp. 35–411. In: Tomato Diseases. (2012)
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annual Review of Phytopathology. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- Boileau MJ, Clinkenbeard KD, Iandolo JJ. Assessment of Bdellovibrio bacteriovorus 109J killing of Moraxella bovis in an in vitro model of infectious bovine keratoconjunctivitis. Canadian Journal of Veterinary Research. 2011;75:285–291. [PMC free article] [PubMed] [Google Scholar]
- Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrobial Agents and Chemotherapy. 2004;48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratanis E, Andersson T, Lood R, Bukowska-Faniband E. Biotechnological potential of Bdellovibrio and like organisms and their secreted enzymes. Frontiers in Microbiology. 2020;11:662. doi: 10.3389/fmicb.2020.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cámara M, Green W, MacPhee CE, Rakowska PD, Raval R, Richardson MC, Slater-Jefferies J, Steventon K, Webb JS. Economic significance of biofilms: a multidisciplinary and cross-sectoral challenge. NPJ Biofilms and Microbiomes. 2022;8:42. doi: 10.1038/s41522-022-00306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, An J, Zheng W, He S. Vibrio cholerae pathogen from the freshwater-cultured whiteleg shrimp Penaeus vannamei and control with Bdellovibrio bacteriovorus. Journal of Invertebrate Pathology. 2015;130:13–20. doi: 10.1016/j.jip.2015.06.002. [DOI] [PubMed] [Google Scholar]
- Cao H, Wang H, Yu J, An J, Chen J. Encapsulated Bdellovibrio powder as a potential bio-disinfectant against whiteleg shrimp-pathogenic Vibrios. Microorganisms. 2019;7:244. doi: 10.3390/microorganisms7080244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrascosa C, Raheem D, Ramos F, Saraiva A, Raposo A. Microbial biofilms in the food industry—a comprehensive review. International Journal of Environmental Research and Public Health. 2021;18:2014. doi: 10.3390/ijerph18042014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauz ACG, Carretero GPB, Saraiva GKV, Park P, Mortara L, Cuccovia IM, Brocchi M, Gueiros-Filho FJ. Violacein targets the cytoplasmic membrane of bacteria. ACS Infectious Diseases. 2019;5:539–549. doi: 10.1021/acsinfecdis.8b00245. [DOI] [PubMed] [Google Scholar]
- Chanyi RM, Koval SF. Role of type IV pili in predation by Bdellovibrio bacteriovorus. PLoS ONE. 2014;9:e113404. doi: 10.1371/journal.pone.0113404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanyi RM, Koval SF, Brooke JS. Stenotrophomonas maltophilia biofilm reduction by Bdellovibrio exovorus. Environmental Microbiology Reports. 2016;8:343–351. doi: 10.1111/1758-2229.12384. [DOI] [PubMed] [Google Scholar]
- Cho G, Kwon J, Soh SM, Jang H, Mitchell RJ. Sensitivity of predatory bacteria to different surfactants and their application to check bacterial predation. Applied Microbiology and Biotechnology. 2019;103:8169–8178. doi: 10.1007/s00253-019-10069-w. [DOI] [PubMed] [Google Scholar]
- Choi SY, Kim S, Lyuck S, Kim SB, Mitchell RJ. High-level production of violacein by the newly isolated Duganella violaceinigra str. NI28 and its impact on Staphylococcus aureus. Scientific Reports. 2015;5:15598. doi: 10.1038/srep15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SY, Im H, Mitchell RJ. Violacein and bacterial predation: promising alternatives for priority multidrug resistant human pathogens. Future Microbiology. 2017;12:835–838. doi: 10.2217/fmb-2017-0090. [DOI] [PubMed] [Google Scholar]
- Choi SY, Lim S, Yoon K-h, Lee JI, Mitchell RJ. Biotechnological activities and applications of bacterial pigments violacein and prodigiosin. Journal of Biological Engineering. 2021;15:10. doi: 10.1186/s13036-021-00262-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WH, Zhu W. Isolation of Bdellovibrio as biological therapeutic agents used for the treatment of Aeromonas hydrophila infection in fish. Zoonoses and Public Health. 2010;57:258–264. doi: 10.1111/j.1863-2378.2008.01224.x. [DOI] [PubMed] [Google Scholar]
- Coder DM, Starr MP. Antagonistic association of the chlorellavorus bacterium (“Bdellovibrio” chlorellavorus) with Chlorella vulgaris. Current Microbiology. 1978;1:59–64. doi: 10.1007/BF02601710. [DOI] [Google Scholar]
- Cohen Y, Pasternak Z, Müller S, Hübschmann T, Schattenberg F, Sivakala KK, Abed-Rabbo A, Chatzinotas A, Jurkevitch E. Community and single cell analyses reveal complex predatory interactions between bacteria in high diversity systems. Nature Communications. 2021;12:5481. doi: 10.1038/s41467-021-25824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collineau L, Chapman B, Bao X, Sivapathasundaram B, Carson CA, Fazil A, Reid-Smith RJ, Smith BA. A farm-to-fork quantitative risk assessment model for Salmonella Heidelberg resistant to third-generation cephalosporins in broiler chickens in Canada. International Journal of Food Microbiology. 2020;330:108559. doi: 10.1016/j.ijfoodmicro.2020.108559. [DOI] [PubMed] [Google Scholar]
- Czajkowski R, Grabe GJ, van der Wolf JM. Distribution of Dickeya spp. and Pectobacterium carotovorum subsp. carotovorum in naturally infected seed potatoes. European Journal of Plant Pathology. 2009;125:263–275. doi: 10.1007/s10658-009-9480-9. [DOI] [Google Scholar]
- Czajkowski R, Pérombelon MCM, van Veen JA, van der Wolf JM. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: a review. Plant Pathology. 2011;60:999–1013. doi: 10.1111/j.1365-3059.2011.02470.x. [DOI] [Google Scholar]
- Dashiff A, Kadouri D. Predation of oral pathogens by Bdellovibrio bacteriovorus 109J. Molecular Oral Microbiology. 2011;26:19–34. doi: 10.1111/j.2041-1014.2010.00592.x. [DOI] [PubMed] [Google Scholar]
- Dashiff A, Junka R, Libera M, Kadouri D. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. Journal of Applied Microbiology. 2011;110:431–444. doi: 10.1111/j.1365-2672.2010.04900.x. [DOI] [PubMed] [Google Scholar]
- Davidsson PR, Kariola T, Niemi O, Palva ET. Pathogenicity of and plant immunity to soft rot pectobacteria. Frontiers in Plant Science. 2013;4:191. doi: 10.3389/fpls.2013.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho CCCR. Biofilms: microbial strategies for surviving UV exposure. In: Ahmad SI, editor. Ultraviolet Light in Human Health, Diseases and Environment. Cham: Springer; 2017. pp. 233–239. [DOI] [PubMed] [Google Scholar]
- Dharani S, Kim DH, Doi Y, Shanks RM, Kadouri DE. Susceptibility of colistin-resistant pathogens to predatory bacteria. Research in Microbiology. 2018;169:52–55. doi: 10.1016/j.resmic.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz JH. Skin and soft tissue infections following marine injuries and exposures in travelers. Journal of Travel Medicine. 2014;21:207–213. doi: 10.1111/jtm.12115. [DOI] [PubMed] [Google Scholar]
- Dourou D, Beauchamp CS, Yoon Y, Geornaras I, Belk KE, Smith GC, Nychas G-JE, Sofos JN. Attachment and biofilm formation by Escherichia coli O157:H7 at different temperatures, on various food-contact surfaces encountered in beef processing. International Journal of Food Microbiology. 2011;149:262–268. doi: 10.1016/j.ijfoodmicro.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Dwidar M, Hong S, Cha M, Jang J, Mitchell RJ. Combined application of bacterial predation and carbon dioxide aerosols to effectively remove biofilms. Biofouling. 2012;28:671–680. doi: 10.1080/08927014.2012.701286. [DOI] [PubMed] [Google Scholar]
- Dwidar M, Monnappa AK, Mitchell RJ. The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Reports. 2012;45:71–78. doi: 10.5483/BMBRep.2012.45.2.71. [DOI] [PubMed] [Google Scholar]
- Dwidar M, Leung BM, Yaguchi T, Takayama S, Mitchell RJ. Patterning bacterial communities on epithelial cells. PLoS ONE. 2013;8:e67165. doi: 10.1371/journal.pone.0067165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epton H, Walker N, Sigee D. Bdellovibrio: a potential control agent for soft rot and black leg of potato. Plant pathogenic bacteria: part A. Akademia Kiado, Budapest, Hungary. pp. 207-212 (1989)
- Escobar MA, Dandekar AM. Agrobacterium tumefaciens as an agent of disease. Trends in Plant Science. 2003;8:380–386. doi: 10.1016/S1360-1385(03)00162-6. [DOI] [PubMed] [Google Scholar]
- Esteve I, Guerrero R, Montesinos E, Abellà C. Electron microscope study of the interaction of epibiontic bacteria with Chromatium minus in natural habitats. Microbial Ecology. 1983;9:57–64. doi: 10.1007/BF02011580. [DOI] [PubMed] [Google Scholar]
- Evans AGL, Davey HM, Cookson A, Currinn H, Cooke-Fox G, Stanczyk PJ, Whitworth DE. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology. 2012;158:2742–2752. doi: 10.1099/mic.0.060343-0. [DOI] [PubMed] [Google Scholar]
- Feng S, Tan CH, Cohen Y, Rice SA. Isolation ofBdellovibrio bacteriovorusfrom a tropical wastewater treatment plant and predation of mixed species biofilms assembled by the native community members. Environmental Microbiology. 2016;18:3923–3931. doi: 10.1111/1462-2920.13384. [DOI] [PubMed] [Google Scholar]
- Feng S, Tan CH, Constancias F, Kohli GS, Cohen Y, Rice SA. Predation by Bdellovibrio bacteriovorus significantly reduces viability and alters the microbial community composition of activated sludge flocs and granules. FEMS Microbiology Ecology. 2017 doi: 10.1093/femsec/fix020. [DOI] [PubMed] [Google Scholar]
- Fenton A, Kanna M, Woods R, Aizawa S-I, Sockett R. Shadowing the actions of a predator: backlit fluorescent microscopy reveals synchronous nonbinary septation of predatory Bdellovibrio inside prey and exit through discrete bdelloplast pores. Journal of Bacteriology. 2010;192:6329–6335. doi: 10.1128/JB.00914-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H-C, Wingender J. The biofilm matrix. Nature Reviews Microbiology. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Flemming H-C, Wuertz S. Bacteria and archaea on Earth and their abundance in biofilms. Nature Reviews Microbiology. 2019;17:247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- Fratamico PM, Cooke PH. Isolation of Bdellovibrios that prey on Escherichia coli O157:H7 and Salmonella species and application for removal of prey from stainless steel surfaces. Journal of Food Safety. 1996;16:161–173. doi: 10.1111/j.1745-4565.1996.tb00157.x. [DOI] [Google Scholar]
- Fry JC, Staples DG. Distribution of Bdellovibrio bacteriovorus in sewage works, river water, and sediments. Applied and Environmental Microbiology. 1976;31:469–474. doi: 10.1128/aem.31.4.469-474.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galie S, García-Gutiérrez C, Miguélez EM, Villar CJ, Lombó F. Biofilms in the food industry: health aspects and control methods. Frontiers in Microbiology. 2018;9:898. doi: 10.3389/fmicb.2018.00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genigeorgis C, Carniciu M, Dutulescu D, Farver TB. Growth and survival of Listeria monocytogenes in market cheeses stored at 4 to 30 °C. Journal of Food Protection. 1991;54:662–668. doi: 10.4315/0362-028X-54.9.662. [DOI] [PubMed] [Google Scholar]
- Gökçen A, Vilcinskas A, Wiesner J. Biofilm-degrading enzymes from Lysobacter gummosus. Virulence. 2014;5:378–387. doi: 10.4161/viru.27919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Rivas F, Ripolles-Avila C, Fontecha-Umaña F, Ríos-Castillo AG, Rodríguez-Jerez JJ. Biofilms in the spotlight: detection, quantification, and removal methods. Comprehensive Reviews in Food Science and Food Safety. 2018;17:1261–1276. doi: 10.1111/1541-4337.12378. [DOI] [PubMed] [Google Scholar]
- Greene AK, Smith GW, Knight CS. Ozone in dairy chilling water systems: effect on metal materials. International Journal of Dairy Technology. 1999;52:126–128. doi: 10.1111/j.1471-0307.1999.tb02853.x. [DOI] [Google Scholar]
- Guerrero R, Pedrós-Alió C, Esteve I, Mas J, Chase D, Margulis L. Predatory prokaryotes: predation and primary consumption evolved in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:2138–2142. doi: 10.1073/pnas.83.7.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Pan Q, Yan S, Chen Y, Li M, Chen D, Han H, Wu B, Cai J. Bdellovibrio and like organisms promoted growth and survival of juvenile abalone Haliotis discus hannai Ino and modulated bacterial community structures in its gut. Aquaculture International. 2017;25:1625–1643. doi: 10.1007/s10499-017-0138-x. [DOI] [Google Scholar]
- Gupta S, Tang C, Tran M, Kadouri DE. Effect of predatory bacteria on human cell lines. PLoS ONE. 2016;11:e0161242. doi: 10.1371/journal.pone.0161242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajam IA, Dar PA, Shahnawaz I, Jaume JC, Lee JH. Bacterial flagellin—a potent immunomodulatory agent. Experimental & Molecular Medicine. 2017;49:e373–e373. doi: 10.1038/emm.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoel S, Vadstein O, Jakobsen AN. The significance of mesophilic Aeromonas spp in minimally processed ready-to-eat seafood. Microorganisms. 2019;7:91. doi: 10.3390/microorganisms7030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovde BT, Steichen SA, Starkenburg SR, Brown JK. Vampirovibrio chlorellavorus draft genome sequence, annotation, and preliminary characterization of pathogenicity determinants. Phycological Research. 2019;68:23–29. doi: 10.1111/pre.12392. [DOI] [Google Scholar]
- Humphrey T, O'Brien S, Madsen M. Campylobacters as zoonotic pathogens: a food production perspective. International Journal of Food Microbiology. 2007;117:237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Im H, Choi SY, Son S, Mitchell RJ. Combined application of bacterial predation and violacein to kill polymicrobial pathogenic communities. Scientific Reports. 2017;7:14415. doi: 10.1038/s41598-017-14567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H, Dwidar M, Mitchell RJ. Bdellovibrio bacteriovorus HD100, a predator of gram-negative bacteria, benefits energetically from Staphylococcus aureus biofilms without predation. The ISME Journal. 2018;12:2090–2095. doi: 10.1038/s41396-018-0154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H, Kwon H, Cho G, Kwon J, Choi SY, Mitchell RJ. Viscosity has dichotomous effects on Bdellovibrio bacteriovorus HD100 predation. Environmental Microbiology. 2019;21:4675–4684. doi: 10.1111/1462-2920.14799. [DOI] [PubMed] [Google Scholar]
- Inoue D, Hiroshima N, Ishizawa H, Dohra H, Ike M, Maresca JA. Complete genome sequences of two predatory bacterial strains, Bacteriovorax sp. HI3 and Myxococcus sp. MH1, isolated from a freshwater pond. Microbiology Resource Announcements. 2022;11:e0114622. doi: 10.1128/mra.01146-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Hiroshima N, Nakamura S, Ishizawa H, Ike M. Characterization of two novel predatory bacteria, Bacteriovorax stolpii HI3 and Myxococcus sp. MH1, isolated from a freshwater pond: prey range, and predatory dynamics and efficiency. Microorganisms. 2022;10:1816. doi: 10.3390/microorganisms10091816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Choi SY, Mun W, Jeong SH, Mitchell RJ. Predation of colistin-and carbapenem-resistant bacterial pathogenic populations and their antibiotic resistance genes in simulated microgravity. Microbiological Research. 2022;255:126941. doi: 10.1016/j.micres.2021.126941. [DOI] [PubMed] [Google Scholar]
- Jang H, Mun W, Choi SY, Mitchell RJ. Use of resazurin to rapidly enumerate Bdellovibrio and like organisms and evaluate their activities. Microbiology Spectrum. 2022;10:e00825–e922. doi: 10.1128/spectrum.00825-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnke J, Fraune S, Bosch TC, Hentschel U, Schulenburg H. Bdellovibrio and like organisms are predictors of microbiome diversity in distinct host groups. Microbial Ecology. 2020;79:252–257. doi: 10.1007/s00248-019-01395-7. [DOI] [PubMed] [Google Scholar]
- Joseph B, Otta SK, Karunasagar I, Karunasagar I. Biofilm formation by Salmonella spp. on food contact surfaces and their sensitivity to sanitizers. International Journal of Food Microbiology. 2001;64:367–372. doi: 10.1016/S0168-1605(00)00466-9. [DOI] [PubMed] [Google Scholar]
- Jurkevitch E. The ecology of Bdellovibrio and like organisms in wastewater treatment plants. In: Mitchell RJ, editor. The Ecology of Predation at the Microscale. Cham: Springer; 2020. pp. 37–64. [Google Scholar]
- Jurkevitch E, Minz D, Ramati B, Barel G. Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Applied and Environmental Microbiology. 2000;66:2365–2371. doi: 10.1128/AEM.66.6.2365-2371.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadouri D, O'Toole GA. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Applied and Environmental Microbiology. 2005;71:4044–4051. doi: 10.1128/AEM.71.7.4044-4051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadouri D, Venzon NC, O'Toole GA. Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Applied and Environmental Microbiology. 2007;73:605–614. doi: 10.1128/AEM.01893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadouri DE, To K, Shanks RMQ, Doi Y. Predatory bacteria: a potential ally against multidrug-resistant gram-negative pathogens. PLoS ONE. 2013;8:e63397. doi: 10.1371/journal.pone.0063397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JI, Williams HN. Bdellovibrios in Callinectus sapidus, the blue crab. Applied and Environmental Microbiology. 1992;58:1408–1410. doi: 10.1128/aem.58.4.1408-1410.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JI, Turng B, Williams HN, Baer ML. Effects of temperature, salinity, and substrate on the colonization of surfaces in situ by aquatic Bdellovibrios. Applied and Environmental Microbiology. 1997;63:84–90. doi: 10.1128/aem.63.1.84-90.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hahn J-S, Franklin MJ, Stewart PS, Yoon J. Tolerance of dormant and active cells in Pseudomonas aeruginosa PA01 biofilm to antimicrobial agents. Journal of Antimicrobial Chemotherapy. 2009;63:129–135. doi: 10.1093/jac/dkn462. [DOI] [PubMed] [Google Scholar]
- Kim E-H, Dwidar M, Mitchell RJ, Kwon Y-N. Assessing the effects of bacterial predation on membrane biofouling. Water Research. 2013;47:6024–6032. doi: 10.1016/j.watres.2013.07.023. [DOI] [PubMed] [Google Scholar]
- Kim E-H, Dwidar M, Kwon Y-N, Mitchell RJ. Pretreatment with alum or powdered activated carbon reduces bacterial predation-associated irreversible fouling of membranes. Biofouling. 2014;30:1225–1233. doi: 10.1080/08927014.2014.970538. [DOI] [PubMed] [Google Scholar]
- Klein DA, Casida LE., Jr Occurrence and enumeration of Bdellovibrio bacteriovorus in soil capable of parasitizing Escherichia coli and indigenous soil bacteria. Canadian Journal of Microbiology. 1967;13:1235–1241. doi: 10.1139/m67-168. [DOI] [PubMed] [Google Scholar]
- Kongrueng J, Pimonsri Mitraparp-arthorn P, Bangpanwimon K, Robins W, Vuddhakul V, Mekalanos J. Isolation of Bdellovibrio and like organisms and potential to reduce acute hepatopancreatic necrosis disease caused by Vibrio parahaemolyticus. Diseases of Aquatic Organisms. 2017;124:223–232. doi: 10.3354/dao03120. [DOI] [PubMed] [Google Scholar]
- Koval SF, Hynes SH, Flannagan RS, Pasternak Z, Davidov Y, Jurkevitch E. Bdellovibrio exovorus sp. nov., a novel predator of Caulobacter crescentus. International Journal of Systematic and Evolutionary Microbiology. 2013;63:146–151. doi: 10.1099/ijs.0.039701-0. [DOI] [PubMed] [Google Scholar]
- Kurz CL, Chauvet S, Andrès E, Aurouze M, Vallet I, Michel GP, Uh M, Celli J, Filloux A, De Bentzmann S. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. The EMBO Journal. 2003;22:1451–1460. doi: 10.1093/emboj/cdg159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz RW, Hespell RB. Attempts to grow Bdellovibrios micurgically-injected into animal cells. Archives of Microbiology. 1978;119:245–248. doi: 10.1007/BF00405402. [DOI] [Google Scholar]
- Letchumanan V, Chan K-G, Lee L-H. Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Frontiers in Microbiology. 2014;5:705. doi: 10.3389/fmicb.2014.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chen C, Sun Q, Liu R, Cai J. Bdellovibrio and like organisms enhanced growth and survival of Penaeus monodon and altered bacterial community structures in its rearing water. Applied and Environmental Microbiology. 2014;80:6346–6354. doi: 10.1128/AEM.01737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima IF, Havt A, Lima AA. Update on molecular epidemiology of Shigella infection. Current Opinion in Gastroenterology. 2015;31:30–37. doi: 10.1097/MOG.0000000000000136. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhuang B, Yuan B, Zhang H, Li J, Wang W, Li R, Du L, Ding P, Jin Y. Predatory bacterial hydrogels for topical treatment of infected wounds. Acta Pharmaceutica Sinica B. 2022;13:315–326. doi: 10.1016/j.apsb.2022.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone PG, Morphew RM, Whitworth DE. Myxobacteria are able to prey broadly upon clinically-relevant pathogens, exhibiting a prey range which cannot be explained by phylogeny. Frontiers in Microbiology. 2017;8:1593. doi: 10.3389/fmicb.2017.01593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Li X, Qiu Q, Chen J, Xiong J. Gut interkingdom predator-prey interactions are key determinants of shrimp health. Aquaculture. 2022;546:737304. doi: 10.1016/j.aquaculture.2021.737304. [DOI] [Google Scholar]
- Mah T-FC, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiology. 2001;9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Markelova NY. Interaction of Bdellovibrio bacteriovorus with bacteria Campylobacter jejuni and Helicobacter pylori. Microbiology. 2010;79:777–779. doi: 10.1134/S0026261710060093. [DOI] [PubMed] [Google Scholar]
- Markelova NY. Predacious bacteria, Bdellovibrio with potential for biocontrol. International Journal of Hygiene and Environmental Health. 2010;213:428–431. doi: 10.1016/j.ijheh.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Martínez V, Jurkevitch E, García JL, Prieto MA. Reward forBdellovibrio bacteriovorusfor preying on a polyhydroxyalkanoate producer. Environmental Microbiology. 2013;15:1204–1215. doi: 10.1111/1462-2920.12047. [DOI] [PubMed] [Google Scholar]
- Martínez V, Herencias C, Jurkevitch E, Prieto MA. Engineering a predatory bacterium as a proficient killer agent for intracellular bio-products recovery: the case of the polyhydroxyalkanoates. Scientific Reports. 2016;6:24381. doi: 10.1038/srep24381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride MJ, Zusman DR. Behavioral analysis of single cells of Myxococcus xanthus in response to prey cells of Escherichia coli. FEMS Microbiology Letters. 1996;137:227–231. doi: 10.1111/j.1574-6968.1996.tb08110.x. [DOI] [PubMed] [Google Scholar]
- McCauley X, Haltli Z, Kerr T. Description of Pseudobacteriovorax antillogorgiicola gen. nov., sp. nov., a bacterium isolated from the gorgonian octocoral Antillogorgia elisabethae, belonging to the family Pseudobacteriovoracaceae fam. nov., within the order Bdellovibrionales. International Journal of Systematic Evolution and Microbiology. 2015;65:522–530. doi: 10.1099/ijs.0.066266-0. [DOI] [PubMed] [Google Scholar]
- McNeely D, Chanyi RM, Dooley JS, Moore JE, Koval SF. Biocontrol of Burkholderia cepacia complex bacteria and bacterial phytopathogens by Bdellovibrio bacteriovorus. Canadian Journal of Microbiology. 2017;63:350–358. doi: 10.1139/cjm-2016-0612. [DOI] [PubMed] [Google Scholar]
- Merino L, Procura F, Trejo FM, Bueno DJ, Golowczyc MA. Biofilm formation by Salmonella sp. in the poultry industry: detection, control and eradication strategies. Food Research International. 2019;119:530–540. doi: 10.1016/j.foodres.2017.11.024. [DOI] [PubMed] [Google Scholar]
- Monnappa AK, Dwidar M, Seo JK, Hur J-H, Mitchell RJ. Bdellovibrio bacteriovorus inhibits Staphylococcus aureus biofilm formation and invasion into human epithelial cells. Scientific Reports. 2014;4:1–8. doi: 10.1038/srep03811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnappa AK, Bari W, Choi SY, Mitchell RJ. Investigating the responses of human epithelial cells to predatory bacteria. Scientific Reports. 2016;6:1–14. doi: 10.1038/srep33485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JE, McILHATTON B, Shaw A, Murphy PG, Elborn JS. Occurrence of Burkholderia cepacia in foods and waters: clinical implications for patients with cystic fibrosis. Journal of Food Protection. 2001;64:1076–1078. doi: 10.4315/0362-028X-64.7.1076. [DOI] [PubMed] [Google Scholar]
- Moreira D, Zivanovic Y, López-Archilla AI, Iniesto M, López-García P. Reductive evolution and unique predatory mode in the CPR bacterium Vampirococcus lugosii. Nature Communications. 2021;12:2454. doi: 10.1038/s41467-021-22762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun W, Upatissa S, Lim S, Dwidar M, Mitchell RJ, Kim M. Outer membrane porin F in E. coli is critical for effective predation by Bdellovibrio. Microbiology Spectrum. 2022;10:e0309422. doi: 10.1128/spectrum.03094-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Dorado J, Marcos-Torres FJ, García-Bravo E, Moraleda-Muñoz A, Pérez J. Myxobacteria: moving, killing, feeding, and surviving together. Frontiers in Microbiology. 2016;7:781. doi: 10.3389/fmicb.2016.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H, Yang Y, Yuk H. Biofilm formation of Salmonella Typhimurium on stainless steel and acrylic surfaces as affected by temperature and pH level. LWT. 2014;55:383–388. doi: 10.1016/j.lwt.2013.09.022. [DOI] [Google Scholar]
- Niemira BA, Fan X. Fruits and Vegetables|Advances in processing technologies to preserve and enhance the safety of fresh and fresh-cut fruits and vegetables. Encyclopedia of Food Microbiology. pp. 983–991. (2014)
- Odooli S, Roghanian R, Ghasemi Y, Mohkam M, Emtiazi G. Predatory and biocontrol potency of Bdellovibrio bacteriovorus toward phytopathogenic strains of Pantoea sp. and Xanthomonas campestris in the presence of exo-biopolymers in vitro and in vivo assessments. International Microbiology. 2021;24:399–413. doi: 10.1007/s10123-021-00177-x. [DOI] [PubMed] [Google Scholar]
- Olanya OM, Niemira BA, Cassidy JM, Boyd G, Uknalis J. Pathogen reduction by predatory bacteria and survival of Bdellovibrio bacteriovorus and Escherichia coli on produce and buffer treated with low-dose gamma radiation. LWT. 2020;130:109630. doi: 10.1016/j.lwt.2020.109630. [DOI] [Google Scholar]
- Ottaviani D, Pieralisi S, Angelico G, Mosca F, Tiscar PG, Rocchegiani E, Scuota S, Petruzzelli A, Fisichella S, Blasi G, DiRaimo E, Leoni F, Latini M, Altissimi S, Haouet N. Bdellovibrio bacteriovorus to control Escherichia coli on meat matrices. International Journal of Food Science & Technology. 2019;55:988–994. doi: 10.1111/ijfs.14355. [DOI] [Google Scholar]
- Oyedara OO, De Luna-Santillana EdJ, Olguin-Rodriguez O, Guo X, Mendoza-Villa MA, Menchaca-Arredondo JL, Elufisan TO, Garza-Hernandez JA, Garcia Leon I, Rodriguez-Perez MA. Isolation of Bdellovibrio sp. from soil samples in Mexico and their potential applications in control of pathogens. MicrobiologyOpen. 2016;5:992–1002. doi: 10.1002/mbo3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyedara OO, Segura-Cabrera A, Guo X, Elufisan TO, Cantú González RA, Rodríguez Pérez MA. Whole-genome sequencing and comparative genome analysis provided insight into the predatory features and genetic diversity of two Bdellovibrio species isolated from soil. International Journal of Genomics. 2018;2018:1–10. doi: 10.1155/2018/9402073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak Z, Pietrokovski S, Rotem O, Gophna U, Lurie-Weinberger MN, Jurkevitch E. By their genes ye shall know them: genomic signatures of predatory bacteria. The ISME Journal. 2012;7:756–769. doi: 10.1038/ismej.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak Z, Njagi M, Shani Y, Chanyi R, Rotem O, Lurie-Weinberger MN, Koval S, Pietrokovski S, Gophna U, Jurkevitch E. In and out: an analysis of epibiotic vs periplasmic bacterial predators. The ISME Journal. 2013;8:625–635. doi: 10.1038/ismej.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez J, Contreras-Moreno FJ, Marcos-Torres FJ, Moraleda-Muñoz A, Muñoz-Dorado J. The antibiotic crisis: how bacterial predators can help. Computational and Structural Biotechnology Journal. 2020;18:2547–2555. doi: 10.1016/j.csbj.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro S, Chauhan A, Berhane T-k, Athar R, Zheng G, Wang C, Dickerson T, Liang X, Lymperopoulou DS, Chen H, Christman M, Louime C, Babiker W, Stine OC, Williams HN. Niche partition of Bacteriovorax operational taxonomic units along salinity and temporal gradients in the Chesapeake bay reveals distinct estuarine strains. Microbial Ecology. 2013;65:652–660. doi: 10.1007/s00248-013-0186-3. [DOI] [PubMed] [Google Scholar]
- Reidl J, Klose KE. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiology Reviews. 2002;26:125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, Meyer F, Sockett RE, Schuster SC. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- Richards GP, Fay JP, Dickens KA, Parent MA, Soroka DS, Boyd EF. Predatory bacteria as natural modulators of Vibrio parahaemolyticus and Vibrio vulnificus in seawater and oysters. Applied and Environmental Microbiology. 2012;78:7455–7466. doi: 10.1128/AEM.01594-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski EG, Stella NA, Brothers KM, Yates KA, Funderburgh ML, Funderburgh JL, Gupta S, Dharani S, Kadouri DE, Shanks RM. Predatory bacteria are nontoxic to the rabbit ocular surface. Scientific Reports. 2016;6:1–9. doi: 10.1038/srep30987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem O, Pasternak Z, Shimoni E, Belausov E, Porat Z, Pietrokovski S, Jurkevitch E. Cell-cycle progress in obligate predatory bacteria is dependent upon sequential sensing of prey recognition and prey quality cues. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E6028–E6037. doi: 10.1073/pnas.1515749112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothrock MJ, Guard JY, Oladeinde A. Salmonella diversity along the farm-to-fork continuum of pastured poultry flocks in the Southeastern United States. Frontiers in Animal Science. 2021;2:52. doi: 10.3389/fanim.2021.761930. [DOI] [Google Scholar]
- Sason G, Jurkevitch E, Nussinovitch A. Biological control of soft rot in potato by κ-carrageenan carriers encapsulated microbial predators. Applied Microbiology and Biotechnology. 2022;107:81–96. doi: 10.1007/s00253-022-12294-2. [DOI] [PubMed] [Google Scholar]
- Sathyamoorthy R, Maoz A, Pasternak Z, Im H, Huppert A, Kadouri D, Jurkevitch E. Bacterial predation under changing viscosities. Environmental Microbiology. 2019;21:2997–3010. doi: 10.1111/1462-2920.14696. [DOI] [PubMed] [Google Scholar]
- Saxon EB, Jackson RW, Bhumbra S, Smith T, Sockett RE. Bdellovibrio bacteriovorus HD100 guards against Pseudomonas tolaasii brown-blotch lesions on the surface of post-harvest Agaricus bisporus supermarket mushrooms. BMC Microbiology. 2014;14:1–12. doi: 10.1186/1471-2180-14-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad N, Postnikova E, Randhawa P, et al. Emergence of Acidovorax avenae subsp. citrulli as a crop threatening disease of watermelon and melon. In: Iacobellis NS, et al., editors. Pseudomonas syringae and Related Pathogens. Cham: Springer; 2003. pp. 573–581. [Google Scholar]
- Scherff R. Control of bacterial blight of soybean by Bdellovibrio bacteriovorus. Phytopathology. 1973;328:400–402. doi: 10.1094/Phyto-63-400. [DOI] [Google Scholar]
- Schneider KR, Schneider RMG, Kurdmongkoltham P, Bertoldi B. Preventing foodborne illness: Clostridium botulinum. Food Science and Human Nutrition. 2011;2017(1):1–7. [Google Scholar]
- Schwudke D, Strauch E, Krueger M, Appel B. Taxonomic studies of predatory Bdellovibrios based on 16S rRNA analysis, ribotyping and the hit locus and characterization of isolates from the gut of animals. Systematic and Applied Microbiology. 2001;24:385–394. doi: 10.1078/0723-2020-00042. [DOI] [PubMed] [Google Scholar]
- Schwudke D, Linscheid M, Strauch E, Appel B, Zähringer U, Moll H, Müller M, Brecker L, Gronow S, Lindner B. The obligate predatory Bdellovibrio bacteriovorus possesses a neutral lipid a containing α-D-mannoses that replace phosphate residues. Journal of Biological Chemistry. 2003;278:27502–27512. doi: 10.1074/jbc.M303012200. [DOI] [PubMed] [Google Scholar]
- Scott WG, Scott HM, Lake RJ, Baker MG. Economic cost to New Zealand of foodborne infectious disease. The New Zealand Medical Journal. 2000;113:281–284. [PubMed] [Google Scholar]
- Seccareccia I, Kost C, Nett M, Schottel JL. Quantitative analysis of Lysobacter predation. Applied and Environmental Microbiology. 2015;81:7098–7105. doi: 10.1128/AEM.01781-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seideler RJ, Mandel M, Baptist JN. Molecular heterogeneity of the Bdellovibrios: evidence of two new species. Journal of Bacteriology. 1972;109:209–217. doi: 10.1128/jb.109.1.209-217.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sert D, Mercan E, Kara Ü. Butter production from ozone-treated cream: effects on characteristics of physicochemical, microbiological, thermal and oxidative stability. LWT. 2020;131:109722. doi: 10.1016/j.lwt.2020.109722. [DOI] [Google Scholar]
- Shatzkes K, Chae R, Tang C, Ramirez GC, Mukherjee S, Tsenova L, Connell ND, Kadouri DE. Examining the safety of respiratory and intravenous inoculation of Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus in a mouse model. Scientific Reports. 2015;5:1–12. doi: 10.1038/srep12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatzkes K, Singleton E, Tang C, Zuena M, Shukla S, Gupta S, Dharani S, Onyile O, Rinaggio J, Connell ND. Predatory bacteria attenuate Klebsiella pneumoniae burden in rat lungs. Mbio. 2016;7:e01847–e1916. doi: 10.1128/mBio.01847-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatzkes K, Tang C, Singleton E, Shukla S, Zuena M, Gupta S, Dharani S, Rinaggio J, Connell ND, Kadouri DE. Effect of predatory bacteria on the gut bacterial microbiota in rats. Scientific Reports. 2017;7:1–12. doi: 10.1038/srep43483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilo M. Morphological and physiological aspects of the interaction of Bdellovibrio with host bacteria. Current Topics in Microbiology and Immunology. 1969;50:174–204. doi: 10.1007/978-3-642-46169-9_6. [DOI] [PubMed] [Google Scholar]
- Shimkets LJ. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annual Review of Microbiology. 1999;53:525–549. doi: 10.1146/annurev.micro.53.1.525. [DOI] [PubMed] [Google Scholar]
- Silagyi K, Kim S-H, Martin Lo Y, Wei C-I. Production of biofilm and quorum sensing by Escherichia coli O157:H7 and its transfer from contact surfaces to meat, poultry, ready-to-eat deli, and produce products. Food Microbiology. 2009;26:514–519. doi: 10.1016/j.fm.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Singh R, Monnappa AK, Hong S, Mitchell RJ, Jang J. Effects of carbon dioxide aerosols on the viability of Escherichia coli during biofilm dispersal. Scientific Reports. 2015;5:13766. doi: 10.1038/srep13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srey S, Jahid IK, Ha S-D. Biofilm formation in food industries: a food safety concern. Food Control. 2013;31:572–585. doi: 10.1016/j.foodcont.2012.12.001. [DOI] [Google Scholar]
- Sun Y, Ye J, Hou Y, Chen H, Cao J, Zhou T. Predation efficacy of Bdellovibrio bacteriovorus on multidrug-resistant clinical pathogens and their corresponding biofilms. Japanese Journal of Infectious Diseases. 2017;70:485–489. doi: 10.7883/yoken.JJID.2016.405. [DOI] [PubMed] [Google Scholar]
- Tajabadi FH, Karimian SM, Mohsenipour Z, Mohammadi S, Salehi M, Sattarzadeh M, Fakhari S, Momeni M, Dahmardehei M, Feizabadi MM. Biocontrol treatment: application of Bdellovibrio bacteriovorus HD100 against burn wound infection caused by Pseudomonas aeroginosa in mice. Burns. 2022 doi: 10.1016/j.burns.2022.08.020. [DOI] [PubMed] [Google Scholar]
- Thomashow LS, Rittenberg SC. Isolation and composition of sheathed flagella from Bdellovibrio bacteriovorus 109J. Journal of Bacteriology. 1985;163:1047–1054. doi: 10.1128/jb.163.3.1047-1054.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolker-Nielsen T. Biofilm development. Microbiology Spectrum. 2015 doi: 10.1128/microbiolspec.MB-0001-2014. [DOI] [PubMed] [Google Scholar]
- Toth IK, Birch PR. Rotting softly and stealthily. Current Opinion in Plant Biology. 2005;8:424–429. doi: 10.1016/j.pbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Uematsu T. Ecology of Bdellovibrio parasitic to rice bacterial leaf blight pathogen, Xanthomonas oryzae. Review Plant Protection Research. 1980;13:12–26. [Google Scholar]
- Van Houdt R, Michiels CW. Biofilm formation and the food industry, a focus on the bacterial outer surface. Journal of Applied Microbiology. 2010;109:1117–1131. doi: 10.1111/j.1365-2672.2010.04756.x. [DOI] [PubMed] [Google Scholar]
- Varon M, Shilo M. Interaction of Bdellovibrio bacteriovorus and host bacteria I. Kinetic studies of attachment and invasion of Escherichia coli B by Bdellovibrio bacteriovorus. Journal of Bacteriology. 1968;95:744–753. doi: 10.1128/jb.95.3.744-753.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kadouri DE, Wu M. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics. 2011;12:453. doi: 10.1186/1471-2164-12-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ding S, Dong Y, Ye K, Xu X, Zhou G. Biofilm formation of Salmonella serotypes in simulated meat processing environments and its relationship to cell characteristics. Journal of Food Protection. 2013;76:1784–1789. doi: 10.4315/0362-028X.JFP-13-093. [DOI] [PubMed] [Google Scholar]
- Williams HN, Li N. Data report exploring the presence of Bdellovibrio and like organisms in deep-sea sediment by culture-independent and culture-dependent methods. International Ocean Discovery Program. 2018 doi: 10.14379/iodp.proc.349.202.2018. [DOI] [Google Scholar]
- Williams LE, Cullen N, DeGiorgis JA, Martinez KJ, Mellone J, Oser M, Wang J, Zhang Y. Variation in genome content and predatory phenotypes between Bdellovibrio sp. NC01 isolated from soil and B. bacteriovorus type strain HD100. Microbiology. 2019;165:1315–1330. doi: 10.1099/mic.0.000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis AR, Moore C, Mazon-Moya M, Krokowski S, Lambert C, Till R, Mostowy S, Sockett RE. Injections of predatory bacteria work alongside host immune cells to treat Shigella infection in zebrafish larvae. Current Biology. 2016;26:3343–3351. doi: 10.1016/j.cub.2016.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D, Dolan G, Aird H, Sorrell S, Dallman TJ, Jenkins C, Robertson L, Gorton R. Farm-to-fork investigation of an outbreak of Shiga toxin-producing Escherichia coli O157. Microbial Genomics. 2018;4:e000160. doi: 10.1099/mgen.0.000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucher BR, Elsayed M, Adelman JS, Kadouri DE, Nadell CD. Bacterial predation transforms the landscape and community assembly of biofilms. Current Biology. 2021;31:2643–265.e13. doi: 10.1016/j.cub.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Wei X, Ebright R, Wall D. Antibiotic production by Myxobacteria plays a role in predation. Journal of Bacteriology. 2011;193:4626–4633. doi: 10.1128/JB.05052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yair S, Yaacov D, Susan K, Jurkevitch E. Small eats big: ecology and diversity of Bdellovibrio and like organisms, and their dynamics in predator-prey interactions. Agronomie. 2003;23:433–439. doi: 10.1051/agro:2003026. [DOI] [Google Scholar]
- Yan C, Zhan M, Xv K, Zhang S, Liang T, Yu R. Sludge dewaterability enhancement under low temperature condition with cold-tolerant Bdellovibrio sp. CLL13. Science of the Total Environment. 2022;820:153269. doi: 10.1016/j.scitotenv.2022.153269. [DOI] [PubMed] [Google Scholar]
- Ye X, Li Z, Luo X, Wang W, Li Y, Li R, Zhang B, Qiao Y, Zhou J, Fan J. A predatory myxobacterium controls cucumber Fusarium wilt by regulating the soil microbial community. Microbiome. 2020;8:1–17. doi: 10.1186/s40168-020-00824-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youdkes D, Helman Y, Burdman S, Matan O, Jurkevitch E. Potential control of potato soft rot disease by the obligate predators Bdellovibrio and like organisms. Applied and Environmental Microbiology. 2020;86:e02543–e2619. doi: 10.1128/AEM.02543-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Schmidt JW, Arthur TM, Wheeler TL, Zhang Q, Wang B. A farm-to-fork quantitative microbial exposure assessment of β-lactam-resistant Escherichia coli among U.S. beef consumers. Microorganisms. 2022;10:661. doi: 10.3390/microorganisms10030661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu Z, Ding T. Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms. 2020;8:425. doi: 10.3390/microorganisms8030425. [DOI] [PMC free article] [PubMed] [Google Scholar]