Abstract
Overcoming antimicrobial resistance represents a formidable challenge and investigating bacterial growth inhibition by fungal metabolites may yield new strategies. Although the fungal non-ribosomal peptide gliotoxin (GT) is known to exhibit antibacterial activity, the mechanism(s) of action are unknown, although reduced gliotoxin (dithiol gliotoxin; DTG) is a zinc chelator. Furthermore, it has been demonstrated that GT synergises with vancomycin to inhibit growth of Staphylococcus aureus. Here we demonstrate, without precedent, that GT-mediated growth inhibition of both Gram positive and negative bacterial species is reversed by Zn2+ or Cu2+ addition. Both GT, and the known zinc chelator TPEN, mediate growth inhibition of Enterococcus faecalis which is reversed by zinc addition. Moreover, zinc also reverses the synergistic growth inhibition of E. faecalis observed in the presence of both GT and vancomycin (4 µg/ml). As well as zinc chelation, DTG also appears to chelate Cu2+, but not Mn2+ using a 4-(2-pyridylazo)resorcinol assay system and Zn2+ as a positive control. DTG also specifically reacts in Fe3+-containing Siderotec™ assays, most likely by Fe3+ chelation from test reagents. GSH or DTT show no activity in these assays. Confirmatory high resolution mass spectrometry, in negative ion mode, confirmed, for the first time, the presence of both Cu[DTG] and Fe[DTG]2 chelates. Label free quantitative proteomic analysis further revealed major intracellular proteomic remodelling within E. faecalis in response to GT exposure for 30–180 min. Globally, 4.2–7.2% of detectable proteins exhibited evidence of either unique presence/increased abundance or unique absence/decreased abundance (n = 994–1160 total proteins detected), which is the first demonstration that GT affects the bacterial proteome in general, and E. faecalis, specifically. Unique detection of components of the AdcABC and AdcA-II zinc uptake systems was observed, along with apparent ribosomal reprofiling to zinc-free paralogs in the presence of GT. Overall, we hypothesise that GT-mediated bacterial growth inhibition appears to involve intracellular zinc depletion or reduced bioavailability, and based on in vitro chelate formation, may also involve dysregulation of Cu2+ homeostasis.
Subject terms: Antimicrobials, Applied microbiology, Bacteria, Industrial microbiology, Proteomics, Chemical biology, Microbiology
Introduction
Consequent to persistent overuse and abuse of antibiotics over many years, antimicrobial resistance (AMR) has developed in key bacterial pathogens. This has led to the identification of a priority group of ESKAPE bacterial pathogens comprising of Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp., many of which are resistant to multiple classes of antibiotics1. Ultimately, this has led to a reduction in the effectiveness of many established antibiotics, which has consequentially resulted in increased patient mortality. Thus, AMR has been identified as a major global threat2. Although next generation antibiotics, vaccination and bacteriophage therapies have been proposed to overcome AMR, the problem is still growing, which means that additional strategies must be considered to either identify new bacterial drug targets or augment existing antibiotic therapies3–6. Herein work is presented on the effects of the fungal non-ribosomal peptide, gliotoxin (GT), on ESKAPE pathogens and Enterococcus faecalis, selected due to the worrying trend of antibiotic resistant isolates that have been found for this bacterial species, often leading to nosocomial infections7,8.
Gliotoxin (GT) has a molecular mass of 326 Da, its biosynthesis is encoded by the gli biosynthetic gene cluster, and it is most notably produced by Aspergillus fumigatus9,10. It is a cytotoxic epipolythiodioxopiperazine (ETP) which comprises an essential intramolecular disulphide bridge, which can be converted to the dithiol form and exhibit potent Zn2+ chelation properties11,12. In addition to showing potent cytotoxicity against eukaryotic cells, GT has been shown to display broad spectrum antibiotic properties13,14. The accepted hypothesis has been that the oxidation and reduction of the disulphide bridge, which are key to ETP activity, produces cytotoxic reactive oxygen species15.
The growth inhibitory effects and mechanism of action of GT have mainly been studied in animal cells and in fungi16–21. It is now clear that upon entry into animal cells GT is reduced to dithiol gliotoxin (DTG) via cellular GSH, this results in ROS generation, depletion of intracellular GSH and GT efflux from cells13. Cell death ultimately follows. No information is available on the specific and direct impact of DTG in animal cells, although it has been demonstrated to directly inhibit the activity of LTH412. In fungi which produce GT (e.g., A. fumigatus) and related ETPs, a self-protection system involving DTG oxidation to GT via oxidoreductase GliT and efflux via transporter GliA, is present22–25. If levels of GT/DTG increase beyond the capacity of the oxido-efflux system, then DTG is bis-thiomethylated to an inactive form by bis-thiomethyltransferase GtmA26,27. Additionally, many fungi lacking ETP biosynthetic capacity possess a gtmA ortholog, possibly to dissipate environmentally acquired GT or other ETPs18,26. DTG chelates Zn2+ and in A. fumigatus, it is now clear the GT is produced in low Zn2+ environments11,28,29. Although the precise relationship between GT/DTG and Zn2+ in fungi remains to be elucidated, elevated DTG levels can deplete intracellular Zn2+, possibly inhibit intracellular metalloenzymes and inhibit growth11,30. There is some evidence that fungal Zn2+ acquisition systems may be activated under these conditions11,31. Reduced holomycin, a functional dithiolopyrrolone (DTP) analogue of DTG, has been shown to inhibit bacterial metallo-β-lactamases (MBL) and proposed to alter bacterial metal homeostasis32.
Although there is a plethora of publications describing the inhibitory activity of GT and other ETPs against bacteria33,34, to the best of our knowledge, there is limited data on the GT mechanism of inhibitory action against bacterial species. Indeed, apart from a publication inferring interference with GSH levels35, no mechanistic details have been forthcoming about how GT inhibits bacterial growth. Interestingly, it has been demonstrated14 that while GT inhibited growth of a range of bacterial species and interfered with biofilm formation, culture supernatant from A. fumigatus ΔgliG, deficient in GT biosynthesis36, exhibited no antimicrobial activity. It has also been revealed that in addition to inhibiting growth of Staphylococcus aureus, GT augments the activity of vancomycin, thereby revealing synergistic activity, of unknown origin, between both antimicrobial agents37. Thus, although GT and other ETPs have been shown to inhibit bacterial growth, often at low µg/ml concentrations, there is a real dearth of information on how bacterial growth is inhibited.
It is not unreasonable to speculate that upon entry into bacterial cells that GT is reduced by unknown cellular reductants and that it is the reduced (DTG) form of GT which effects growth inhibition38. Given that Zn2+ chelation by DTG has been observed11,12, we speculate that this could also occur in bacteria, along with possible Zn2+ ejection from bacterial enzymes. We further hypothesise that depletion of intracellular Zn2+ levels would be predicted to activate uptake systems and metalloenzyme activity/stability, or abundance, could be adversely affected by GT/DTG-mediated Zn2+-ejection, in part as previously speculated39. This manuscript describes work to explore the effect of the non-ribosomal peptide GT on the Gram-positive bacterial species E. faecalis. Specifically, this work aims to decipher the mechanism of action behind the inhibitory activity of GT against bacteria. It is framed in the context of the reduced form of GT, dithiol gliotoxin (DTG), and its demonstrated ability to chelate zinc (Zn[DTG]).
Results and discussion
GT inhibits Gram positive and negative bacterial growth, which is relieved by zinc addition
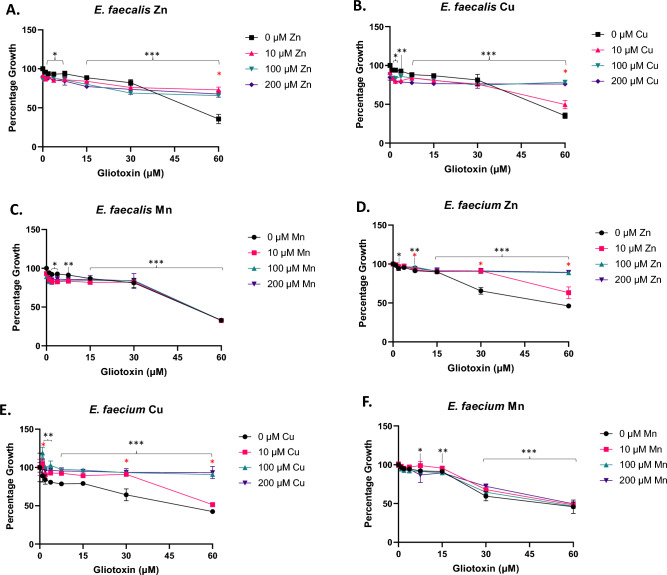
Both Gram positive species were susceptible to growth inhibition in the presence of GT. Specifically, E. faecalis showed significant and dose-dependent growth inhibition when grown in the presence of gliotoxin (0.9375–60 µM; p < 0.05–0.0001). Addition of Zn (10, 100, or 200 µM) relieved most of this inhibition at 10 µM and above Zn supplementation (p < 0.0001) (Fig. 1A). Cu supplementation (10, 100, or 200 µM) relieved some of this inhibition at as little as 10 µM Cu and total relief was seen at 100 and 200 µM Cu supplementation (p < 0.0001) (Fig. 1B). Addition of Mn (10, 100, or 200 µM) did not relieve growth inhibition (Fig. 1C). E. faecium showed significant and dose dependent growth inhibition when grown in the presence of gliotoxin (0.9375–60 µM) (optimally p < 0.0001). Zn (10, 100, or 200 µM) addition showed some relief of inhibition from 10 µM Zn and total relief was seen at 100 and 200 µM Zn supplementation (p < 0.0001) (Fig. 1D). Cu (10, 100, or 200 µM) also relieved growth inhibition at as little as 10 µM Cu. Total relief was seen at 100 (p < 0.005) and 200 µM Cu (p < 0.0001) supplementation (Fig. 1E), however, Mn addition (10, 100, or 200 µM) did not relieve GT-mediated growth inhibition (Fig. 1F). In E. faecalis, we interpret relief of GT-mediated inhibition requiring higher Cu concentration as indicative of differential biosystem dependency on this metal ion. E. faecalis V4932 has previously been shown to be sensitive to A. fumigatus GT-containing culture supernatant33, in accordance with our observations. As far as we can ascertain, there is only a single report of GT-mediated growth inhibition of E. faecium, or associated metal relief of same, in the literature40. Future work will be required to elucidate which Gram positive bacterial systems are subject to metallo-mediated GT inhibition.
Figure 1.
Zinc and copper relieve GT-mediated Gram positive growth inhibition. (A) E. faecalis growth in the presence of gliotoxin (0–60 µM) supplemented with Zn (0, 10, 100, or 200 µM). (B) E. faecalis growth in the presence of gliotoxin (0–60 µM) supplemented with Cu (0, 10, 100, or 200 µM). (C) E. faecalis growth in the presence of gliotoxin (0–60 µM) supplemented with Mn (0, 10, 100, or 200 µM). (D) E. faecium growth in the presence of gliotoxin (0–60 µM) supplemented with Zn (0, 10, 100, or 200 µM). (E) E. faecium growth in the presence of gliotoxin (0–60 µM) supplemented with Cu (0, 10, 100, or 200 µM). (F) E. faecium growth in the presence of gliotoxin (0–60 µM) supplemented with Mn (0, 10, 100, or 200 µM). Culture conditions: GT 15 µM = 5 µg/ml. TSB media, metals added as ZnSO4.7H2O, FeSO4.7H2O, CuCl2 or MnCl2.4H2O and 18 h incubation. The OD600 values equate to the following CFU/ml for E. faecalis: 0.2 OD600 = 1.2 × 108 CFU/ml and 0.1 OD600 = 7 × 107 CFU/ml. Significant inhibition due to GT exposure is indicated with black asterisks (* = P < 0.05, ** = P < 0.01, *** = P < 0.0001). Significant relief of inhibition at any metal concentration is shown with red asterisks (* = P < 0.05).
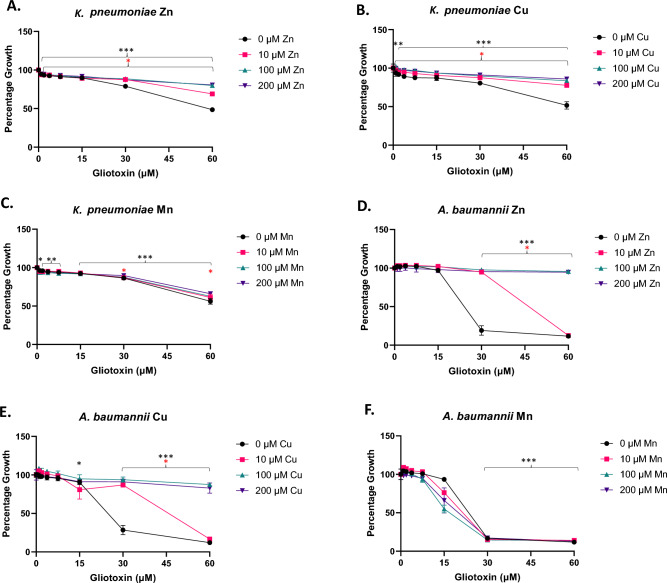
Both Gram negative species were also susceptible to GT-mediated growth inhibition. K. pneumoniae showed significant and dose-dependent growth inhibition at GT concentrations ≥ 30 µM (p < 0.0001). Addition of Zn (10, 100, or 200 µM) relieved most inhibition at as little as 10 µM Zn, with near total relief evident at 100 and 200 µM Zn supplementation (p < 0.05–0.0001) (Fig. 2A). Cu (10, 100, or 200 µM) relieved almost all this inhibition (p < 0.05–0.0005) (Fig. 2B), yet addition of Mn (10, 100, or 200 µM) showed no relief of inhibition (Fig. 2C). Of all bacterial species tested, growth of A. baumannii was most significantly inhibited in a dose-dependent manner in the presence of GT (≥ 30 µM) (p < 0.0001). Addition of Zn at 10 µM relieved almost all this inhibition in the presence of 30 µM GT (p < 0.0001) but provided no relief at 60 µM GT. Near total relief was seen at 100 and 200 µM Zn supplementation (p < 0.0001) (Fig. 2D). Cu at as low as 10 µM also relieved almost all growth inhibition (p < 0.0001), at 30 µM GT, but provided no relief at 60 µM GT. Near total relief was seen at 100 and 200 µM Cu supplementation (p < 0.0001)(Fig. 2E). As with all other species, Mn (10, 100, or 200 µM) did not relieve GT-mediated growth inhibition in A. baumannii (Fig. 2F). Experimentation showed no significant (p < 0.05) relief of GT-induced inhibition with supplemental Fe addition at any concentration to E. faecalis (Supplementary Fig. 1). This leads to the conclusion that either there is sufficient Fe present in the TSB media (5.37 µM)41 to overcome GT inhibition or that intracellular DTG preferentially chelates Zn or Cu. This preferential chelation may be due to the need for two DTG molecules per Fe ion to form the Fe[DTG]2 complex as opposed to one for Zn and Cu. ICP-MS or similar technology could be used to gain insight into the intracellular concentrations of Fe ions when the cells are challenged with GT/DTG or protected from challenge with supplementary Zn or Cu, and this will form the basis of future detailed work.
Figure 2.
Zinc and copper relieve GT-mediated Gram negative growth inhibition. (A) K. pneumoniae growth in the presence of gliotoxin (0–60 µM) supplemented with Zn (0, 10, 100, or 200 µM). (B) K. pneumoniae growth in the presence of gliotoxin (0–60 µM) supplemented with Cu (0, 10, 100, or 200 µM). (C) K. pneumoniae growth in the presence of gliotoxin (0–60 µM) supplemented with Mn (0, 10, 100, or 200 µM). (D) A. baumannii growth in the presence of gliotoxin (0–60 µM) supplemented with Zn (0, 10, 100, or 200 µM). (E) A. baumannii growth in the presence of gliotoxin (0–60 µM) supplemented with Cu (0, 10, 100, or 200 µM). (F) A. baumannii growth in the presence of gliotoxin (0–60 µM) supplemented with Mn (0, 10, 100, or 200 µM). Culture conditions: GT 15 µM = 5 µg/ml. TSB media, metals added as ZnSO4.7H2O, FeSO4.7H2O, CuCl2 or MnCl2.4H2O and 18 h incubation. Significant inhibition due to GT exposure is indicated with black asterisks (* = P < 0.05, ** = P < 0.01, *** = P < 0.0001). Significant relief of inhibition at any metal concentration is shown with red asterisks (* = P < 0.05).
E. faecalis cultures grown in the presence of GT (5 µg/ml) for 1 h contained 12.6% less Zn than MeOH treated counterparts, as measured by zinquin fluorescence. To our knowledge this is the first demonstration that GT/DTG alters the internal metal abundance in bacteria, which suggests that the metal-chelating action of DTG has a negative effect on cellular growth and that GT may not be the actual active agent. This apparent Zn depletion likely plays a significant role in the inhibitory effects of GT/DTG and explains why Zn supplementation ameliorates the effects of GT. However, we observed no significant difference between intracellular Zn levels in the presence or absence of GT at 5, 15 and 30 min GT exposure which suggests that it is the bioavailability of Zn, as opposed to total intracellular Zn, which may mediate the bacterial response to GT (Supplementary Fig. 2). Importantly, the 30 min time-point overlaps with the quantitative proteomic experimentation where increased abundance of the Zn uptake receptor system is observed. This leads us to hypothesise the GT is reduced to DTG upon cellular uptake, which in turn chelates intracellular Zn thereby generating a Zn-limiting environment which activates the Zn uptake system. Future work will be focused on identifying intracellular Zn-DTG chelate and using ICP-MS to absolutely quantify intracellular Zn levels, following chelate formation and purification.
Relevantly, a range of zinc chelators, including TPEN, were evaluated for their ability to resensitize metallo-β-lactamase (MBL)-producing bacteria to antibiotics42. Synergistic activity between TPEN and meropenem was observed against K. pneumoniae, Chryseobacterium indologenes, Elizabethkingia meningoseptica and Stenotrophomonas maltophilia in in vitro experiments. Moreover, using the Galleria mellonella in vivo infection model, chelators TPEN or nitroxoline in combination with meropenem resulted in increased larval survival following infection with either K. pneumoniae, E. meningoseptica or S. maltophilia42. Thus, it is clear that artificial and naturally-occurring zinc chelators have potential to either prevent microbial growth or inactivate MBLs to overcome AMR. The demonstration of GSH-mediated activation of holomycin to reduced holomycin, with zinc-chelating ability, further underscores this biomedical application43.
GT is bactericidal at high concentrations
An antibacterial agent is defined as bactericidal if it reduces the CFU/ml of a culture by more than a 3 log10-fold decrease. At 8 h, all GT concentrations show bactericidal activity against E. faecalis with a 3 log10-fold decrease (Supplementary Fig. 3). At 24 h, a dose-dependent recovery of the bacterial cells was observed where GT (1.875 µM) was no longer sufficient to inhibit growth. Both GT (3.75 and 7.5 µM) maintained their efficacy. GT (3.75 µM) was still effective though it fell slightly below the 3 log10-fold decreased cut off with 6.4 × 107 CFU/ml compared to the control cultures 1.05 × 1010 CFU/ml. However, the highest GT concentration (7.5 µM) maintained bactericidal activity with 2.12 × 105 CFU/ml.
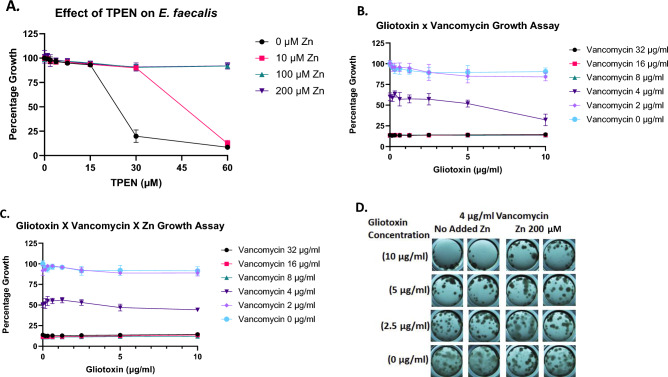
Zinc relieves chelator-mediated (GT and TPEN) growth inhibition of E. faecalis in a dose-dependent manner
To show that metal chelation is a viable mechanism of bacterial growth inhibition, and that Zn supplementation relieves growth inhibition, the known Zn chelator TPEN was used as GT comparator. Indeed, our data shows that both molecular species chelate Zn whereby we determined the GT Zn pKd = 8.72 and TPEN Zn pKd = 9.69, respectively (Supplementary data). GT and TPEN (0–60 µM) were used to treat cultures of E. faecalis with and without Zn supplementation. When E. faecalis was grown in the presence of GT there was insignificant inhibition of growth (< 5%) at low concentrations (< 3.75 µM). The effects of GT increased with the dosage showing minor (6.1%) but significant (p = 0.0001) growth inhibition at 7.5 µM GT. The most significant (p < 0.0001) inhibition was seen at 15–60 µM GT. Inhibition rapidly increased at these concentrations 15 µM (8.0%), 30 µM (10.0%), and 60 µM (65.9%). Supplementation of 60 µM GT treated samples with all Zn concentrations showed significant (p < 0.0001) relief of inhibition with the percentage growth relative to the 0 µM Zn treated samples increasing by 53.3–55.8% (Supplementary Fig. 4). When E. faecalis was grown in the presence of TPEN significant (p = 0.0167) growth inhibition (5.1%) was seen at 7.5 µM TPEN, which became more significant (p = 0.0005) at 15 µM TPEN with slightly greater growth inhibition (6.9%). The most significant (p < 0.0001) growth inhibition was seen at 30 µM (80.1%) and 60 µM (91.5%) TPEN. Zn supplementation showed significant (p < 0.0001) relief of growth inhibition (70.0–71.0%). At 60 µM TPEN, 10 µM Zn was no longer able to provide significant (p > 0.05) relief of growth inhibition. However, 100 and 200 µM Zn supplementation provided significant (p < 0.0001) relief of inhibition, reducing growth inhibition by 83% (Fig. 3A). These data provide strong evidence that GT inhibits growth through intracellular Zn limitation similar to TPEN. Moreover, it is in accordance with previous work that GT/DTG, acts as a zinc chelator and either binds free zinc, or ejects it from metalloenzymes in fungi11,29. TPEN is a well-characterised zinc chelator, and it too appears to inhibit E. faecalis growth in a dose-dependent manner, which is also completely reversible by zinc addition (Fig. 3A). Combined, these data demonstrate, for the first time, a potential mechanism(s) of action, involving disruption of intracellular zinc homeostasis, for GT/DTG-mediated bacterial growth inhibition. Moreover, it complements the work of others who have deployed a range of zinc chelators (e.g., TPEN, reduced holomycin, D-alanyl-D-alanyl-D-alanine methyl ester functionalized N,N,N'-tris(2-pyridylmethyl)-ethylenediamine and tris-picolylamine (TPA) to overcome MBL degradation of target antibiotics32,42,44,45. Conversely, in pioneering work, microarray analysis of E. faecalis exposure to Zn and Cu revealed that two modules were implicated in the microbial response to metal exposure46. So-called module I and II, comprised genes implicated in Zn homeostasis (including the Zur transcription factor) and those involved in stress responses/basal metabolism, respectively. Relevantly, exposure to Zn resulted in downregulation of module I-encoded genes (adcABC/adcA-II) responsible for the Zn uptake system in E. faecalis 46.
Figure 3.
Factors influencing E. faecalis growth and phenotype related to Zn2+ availability. (A) Relief of TPEN-mediated growth inhibition of E. faecalis by Zn2+ addition (0—200 µM). (B) GT (up to 10 µg/ml) augments vancomycin-mediated growth inhibition of E. faecalis in a concentration-dependent manner. E. faecalis OD600 0.2 = 1.2 × 108 CFU/ml. (C) Zn2+ (200 µM) relieves GT augmentation of vancomycin-mediated growth inhibition of E. faecalis. (D) A speckled growth phenotype of E. faecalis unique to the presence of vancomycin (4 µg/ml) is decreased in the presence of gliotoxin. Zn2+ addition (200 µM) reverses the GT dissipation effect and restores the speckled growth phenotype.
GT augments vancomycin-mediated growth inhibition of E. faecalis—which is relieved by Zn2+
Vancomycin has been shown to transiently increase expression of selected Zur regulon genes in Streptomyces coelicolor and also to bind to zinc in vitro47. Moreover, zinc has been demonstrated to induce vancomycin polymerisation thereby leading to enhanced antibiotic activity against E. faecalis48. Of further relevance is that it has been independently shown that GT acts synergistically with vancomycin, fusidic acid and linezolid, to inhibit Staphylococcus aureus growth37, an observation which suggests differential mechanistic actions and strongly underpins the rationale for further investigating that of GT/DTG. Interestingly, this previous work did not suggest a metal-dependent basis of GT-mediated inhibition37. However, herein we have observed that GT/DTG augments the vancomycin-mediated inhibition of E. faecalis growth, and that zinc addition (200 µM) negates the observed combinatorial inhibition (Fig. 3B, C). We show that GT inhibits the growth of E. faecalis in a dose-dependent manner, and that this inhibition is readily relieved by the addition of zinc. This implicates the disruption of microbial zinc homeostasis as a mechanism of GT antimicrobial action and provides an insight into microbial systems targeted by GT. This inhibitory effect was further explored in combination with vancomycin, the antibiotic of last resort. Checkerboard assays using GT and vancomycin showed that the presence of sub-inhibitory concentrations of GT (5 and 10 µg/ml) were sufficient to significantly (p < 0.03) lower the MIC of vancomycin against E. faecalis. This combinatorial work has also shown that GT may inhibit biofilm formation as there were apparent differences in biofilm appearance between shaking and static incubation. A unique speckled phenotype was seen at vancomycin (4 µg/ml) which was diminished with increasing GT. The presence of Zn (200 µM) attenuated the effects of GT on this phenotype (Fig. 3D). Interestingly, although GT (0.15–10 µg/ml) impeded biofilm formation in the presence of vancomycin (4 µg/ml), this inhibition did not reach statistical significance (Supplementary Fig. 5). Thus, alternative approaches will be required to further dissect any combinatorial or synergistic activity of both antimicrobials and the role of zinc in inducing vancomycin polymerisation to augment its antimicrobial activity48.
DTG chelates zinc, copper and iron
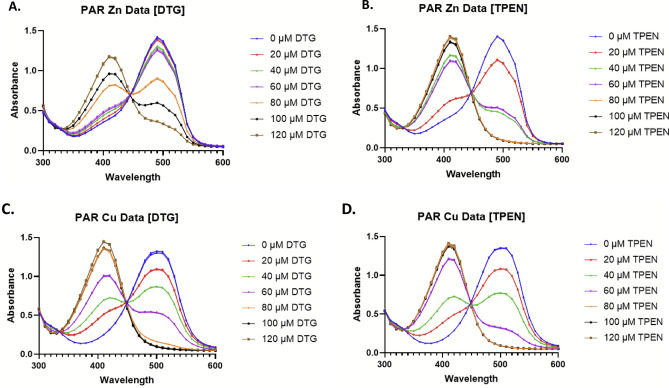
Using the colorimetric metal chelator 4-(2-pyridylazo)resorcinol (PAR), it was shown that DTG exhibits copper chelation properties (Fig. 4A–D). Specifically, DTG-mediated liberation of Zn (positive control;11) and Cu from respective PAR complexes was determined using a decrease in absorbance at 495 nm and 513 nm, respectively (Fig. 4A,C). A dose-dependent decrease in Zn(PAR)2 and Cu(PAR)2 complexes corresponding to an increase in DTG concentration was observed, indicating that DTG competitively liberates both Zn and Cu from PAR (Fig. 4A,C). Under test conditions, DTG (120 µM) dissociated most, but not all, Zn from Zn(PAR)2 (Fig. 4A). In contrast, TPEN removed all Zn from Zn(PAR)2 at concentrations ≥ 80 µM (Fig. 4B). Interestingly, DTG was more effective at dissociating Cu from Cu(PAR)2 with the vast majority dissociated at 80 µM and complete dissociation observed at ≥ 100 µM (Fig. 4C). DTG was only slightly less effective than TPEN at dissociating Cu from PAR, with ≥ 80 µM TPEN resulting in dissociation of all Cu from Cu(PAR)2 (Fig. 4D). DTG does not remove Mn from PAR chelates or Mn does not form chelates with PAR (data not shown), in accordance with previous observations regarding absence of Sporidesmin A:Mn chelates49. It is important to note that the formation of DTG:Zn and DTG:Cu chelates is in accordance with the observation that GT-mediated bacterial growth inhibition is reversed by these metals and not Mn, which does not appear to form a metal chelate with DTG.
Figure 4.
DTG ejects specific metal cations (M2+) from 4-(2-Pyridylazo)resorcinol (PAR). DTG removes Zn from PAR:Zn. (A) The effect of increasing DTG concentration on the PAR:Zn complex (495 nm). As the concentration of DTG increases the 495 nm peak decreases accompanied by an increased peak at 410 nm representing unbound PAR. (B) The metal chelator TPEN requires a lower concentration to remove the same amount of metal. (C) DTG removes Cu from PAR:Cu. The effect of increasing DTG concentration on the PAR:Cu complex (513 nm). Interestingly, DTG was able to remove Cu from PAR at lower concentrations than when removing Zn. (D) When this is compared to the efficacy of the metal chelator TPEN there is little difference between the two.
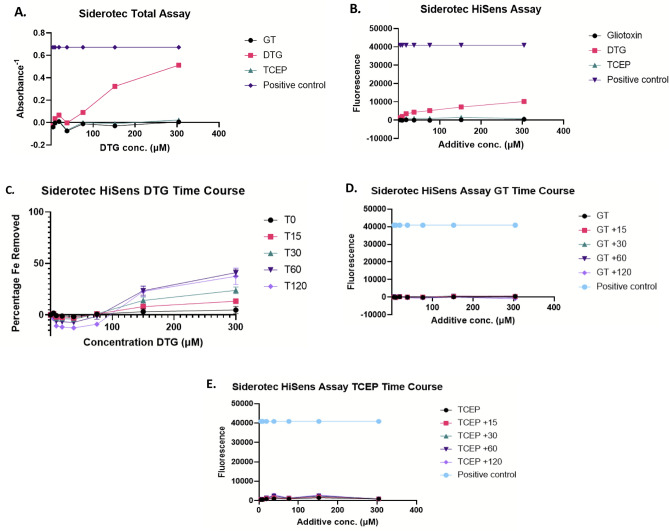
The colorimetric Siderotec Total assay contains a Fe3+ chromophore. DTG (4.7–304 µM) appeared to dissociate Fe from the chromophore in a dose-dependent manner culminating in 76% Fe removal at DTG (304 µM) after 10 min (Fig. 5A). Relevantly, neither GT or TCEP revealed any ability to dissociate Fe from the chromophore in the Siderotec Total™ assay (Fig. 5A). Furthermore, the ability of DTG to interact with Fe was also assessed using a fluorescence-based Siderotec HiSens™ assay, which confirmed a dose-dependent removal of Fe from the constituent fluorophore with increasing DTG (4.7–304 µM) (Fig. 5B). In this assay only 25% Fe was removed at 304 µM DTG, however, a temporal effect was apparent with DTG-mediated dissociation of increased amounts of Fe over time (41% Fe dissociation after 60 min, decreasing to 37.6% after 120 min at 300 µM DTG) (Fig. 5C). Again, neither GT or TCEP (4.7–300 µM) showed any notable impact on the assay at any time point, though TCEP did show slightly higher fluorescence signal than GT (Fig. 5D,E). Moreover, compared to DTG, neither GSH or DTT showed any impact on fluorescent chelate-Fe3+ stability in the Siderotec-HiSens™ assay (Supplementary Fig. 8), indicating the specificity of DTG for Fe3+ displacement from fluorescent chelates.
Figure 5.
DTG specifically reacts with colorimetric and fluorescent iron chelates, possibly via Fe3+ reduction. DTG specifically removes Fe3+ from colorimetric or fluorescent Siderotec™ reagents. (A) The impact of gliotoxin, DTG, and TCEP on the colorimetric Fe3+ assay. The control (purple) indicates the signal at total Fe3+ removal from the chromophore. DTG (red) is the only test compound which also removes Fe3+. (B) The impact of gliotoxin, DTG, and TCEP on the fluorimetric Siderotec HiSens assay. The control (purple) indicates the signal due to Fe3+ removal from the fluorophore. DTG (red) is the only test compound which also removes Fe3+. (C) Temporal effect of DTG on the high sensitivity assay where individual samples were normalized to a percentage relative to the control sample (100%) at each timepoint. Peak fluorescence occurs at T60 min (41% of Fe3+ removed) which then begins to drop slightly by T120 min. (D) Neither gliotoxin or (E) TCEP affect the fluorometric Siderotec HiSens assay. The control (blue) indicates the signal at total Fe3+ removal from the fluorophore.
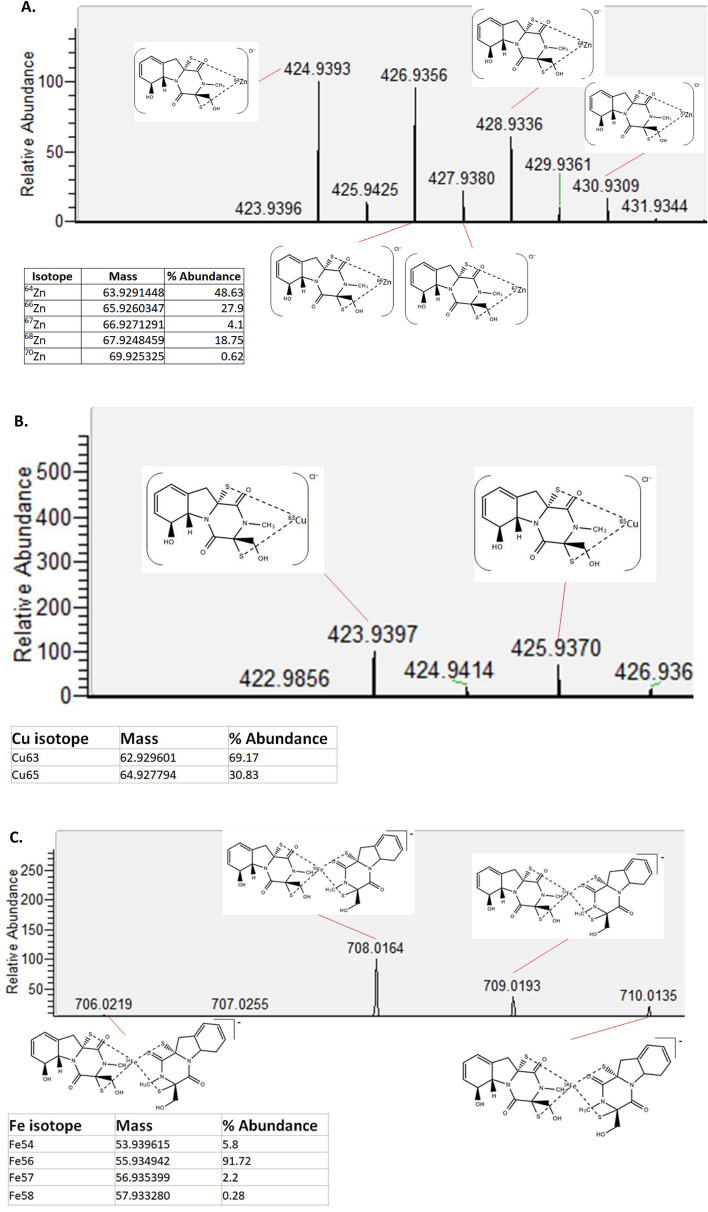
High resolution mass spectrometry in negative mode confirmed the presence of DTG:Zn (positive control), DTG:Cu, and (DTG)2:Fe chelates, and the resultant spectra compared to those from theoretical chelate structures (Fig. 6). The DTG:Zn chelate stoichiometry was known from previous work11 who found the metal adduct had an m/z = 424.93851, which corresponded to a 1:1 complex of DTG:Zn with a Cl− ion. This structure was herein identified to exhibit m/z = 424.9393 ([DTG(Zn)—2H + Cl]−)(Fig. 6A), in accordance with previous data. The DTG:Cu chelate revealed m/z = 423.9397, corresponding to a 1:1 DTG:Cu chelate with Cl- present ([DTG(Cu)—2H + Cl]−)(Fig. 6B) and the (DTG)2:Fe chelate showed an m/z = 708.0164 which corresponds to a 2:1 complex of DTG:Fe with no Cl- adduct present (Fig. 6C), but with an overall single negative charge suggesting Fe3+ as the iron cation ([DTG2(Fe)—2H]−).
Figure 6.
Mass spectrometric confirmation of DTG:metal ion chelates. (A) Identification of DTG:Zn chelate structures detected by negative mode mass spectrometry. Structures are shown with Cl- ion and the relevant Zn isotope. (B) The DTG:Cu chelate structures detected by negative mode mass spectrometry. Structures are shown with Cl- ion and the relevant Cu isotope. (C) The DTG:Fe chelate structures detected by negative mode mass spectrometry. Predicted structures are shown with the two DTG molecular structures and the relevant Fe isotope.
Overall, the revelation that DTG can chelate Cu and Fe is of significant biological interest, and to our knowledge has not previously been observed. It is notable that DTG:Mn chelates were not observed and suggests an element of specificity regarding the formation of DTG:Zn, DTG:Cu and (DTG)2:Fe chelates. Although outside the scope of the present work, the potential interaction between DTG and Fe may be of particular relevance in A. fumigatus and other fungi which biosynthesise ETP-type biomolecules as it suggests an additional rationale for controlling intracellular levels of DTG to avoid Fe3+ reduction and disrupted redox homeostasis. In E. faecalis, GT exposure at T = 60 min resulted in the loss of detection of ferrous transport protein A which indicates metal ion remodeling may occur due to hitherto unidentified effects of GT/DTG in bacteria. It is notable that gliT expression in A. fumigatus was reduced upon deletion of SreA, a transcriptional repressor of siderophore biosynthetic enzymes transporters to avoid excess iron uptake and resultant toxicity under iron-replete conditions50. This observation, in combination with our demonstration of (DTG)2:Fe chelate formation, suggests that GliT may potentially contribute to Fe2+ homeostasis in A. fumigatus wild-type. However, further work is undoubtedly required to fully dissect the biological significance of these observations. When a nucleophile donates lone-pair (n) electron density into the empty π* orbital of a nearby carbonyl group, it is referred to as an n → π* interaction51. In ETPs, the n → π* interactions can decrease the disulphide reduction potential, which may confer biologically-relevant stability on the disulphide bond in physiological environments. Indeed, it has been proposed that the two strong n → π* interactions in an ETP, like GT, can nearly completely compensate for the molecular instability caused by the strained conformation of the disulphide bond52. Moreover, intramolecular stability may also be affected by the hydrophobicity of the environment in which the molecule is located. Thus, we speculate that the intracellular bacterial environment is compatible with GT reduction and consequent metal ion chelation.
GT causes significant remodelling of the E. faecalis proteome
Extensive alterations, including protein unique presence and increased abundance as well as unique absence and decreased abundance were observed in the E. faecalis proteome following GT addition (5 µg/ml) for 30, 60 and 180 min respectively (Tables 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12). Overall, between T = 30–180 min GT exposure, 4.2–7.2% of detectable proteins exhibited evidence of either unique presence/increased abundance or unique absence/decreased abundance, where total proteins detected ranged from 994 to 1160 across all analyses. To our knowledge, this is the first demonstration that GT affects the bacterial proteome in general, and E. faecalis, specifically.
Table 1.
Proteins which are uniquely present in E. faecalis when grown in the presence of gliotoxin (5 µg/ml), compared to identical cultures grown without gliotoxin (5 µg/ml). Grown using tryptic soy broth (TSB) media for 30 min in the log phase (0.3–0.4 OD600).
| Protein description | Fold Change (log2) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| Pyridine nucleotide-disulphide oxidoreductase | Unique | N/A | 12 | 36.1 | H7C719 |
| 50S ribosomal protein L33 4 | Unique | N/A | 2 | 40.8 | P59629 |
| Cobalamin synthesis protein/P47K family protein | Unique | N/A | 6 | 25.6 | Q82Z69 |
| 30S ribosomal protein S14 1 | Unique | N/A | 1 | 13.5 | Q8KU58 |
| Protein EbsA | Unique | N/A | 3 | 19 | P36920 |
| 3-dehydroquinate dehydratase | Unique | N/A | 3 | 17.4 | P36923 |
| Protein of unknown function (DUF3114) | Unique | N/A | 2 | 7.3 | Q82YY0 |
| Uncharacterized protein | Unique | N/A | 2 | 18.3 | Q82ZK3 |
| Transcriptional antiterminator, bglG family | Unique | N/A | 3 | 8.8 | Q82ZT1 |
| Xanthine/uracil permease family protein | Unique | N/A | 2 | 8 | Q82ZW2 |
| tRNA(Met) cytidine acetate ligase | Unique | N/A | 3 | 13.3 | Q830C2 |
| PTS system, beta-glucoside-specific IIABC component | Unique | N/A | 3 | 6.9 | Q831B4 |
| Uncharacterized protein | Unique | N/A | 2 | 55.9 | Q834Y7 |
| Histidine kinase | Unique | N/A | 3 | 9.7 | Q837B6 |
| Abhydrolase_3 domain-containing protein | Unique | N/A | 3 | 14.3 | Q838Q5 |
| Glyoxylase family protein* | Unique | N/A | 3 | 11.2 | Q834I3 |
*This protein exhibited overall increased abundance > 1.5-fold and was only detectable in 1 of 3 untreated samples.
Table 2.
Proteins which are increased in abundance in E. faecalis when grown in the presence of gliotoxin (5 µg/ml), compared to identical cultures grown without gliotoxin (5 µg/ml). Grown using tryptic soy broth (TSB) media for 30 min in the log phase (0.3–0.4 OD600).
| Protein description | Fold Change (log2) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| Adhesion lipoprotein (adcA-II) | 7.28739 | 0.000462756 | 15 | 39.3 | Q82Z67 |
| ABC transporter, ATP-binding protein | 4.10372 | 0.00277729 | 6 | 35.7 | Q839U4 |
| Adhesion lipoprotein (adcABC) | 2.41358 | 0.00528037 | 9 | 45.1 | Q839U5 |
| Transcriptional regulator, Fur family | 1.6883 | 0.00314565 | 4 | 27.8 | Q831T2 |
| Acyl carrier protein | 1.27213 | 0.00210808 | 2 | 44.3 | Q82ZE9 |
| Copper-exporting P-type ATPase | 0.938649 | 0.0166494 | 17 | 29.1 | Q838Y5 |
| Spermidine/putrescine ABC transporter, ATP-binding protein | 0.935763 | 0.0242974 | 2 | 6.9 | Q835Z8 |
| Cold shock protein CspC | 0.826335 | 0.0414736 | 4 | 80.3 | Q833G3 |
| Cellulose biosynthesis cyclic di-GMP-binding regulatory protein BcsB | 0.809572 | 0.0170368 | 12 | 23 | Q837F4 |
| DUF2187 domain-containing protein | 0.75339 | 0.0432523 | 3 | 63.8 | Q835R0 |
| Segregation and condensation protein A | 0.692021 | 0.0318054 | 2 | 9.1 | Q834U4 |
| 30S ribosomal protein S15 | 0.690959 | 0.00618528 | 3 | 39.3 | Q82ZJ1 |
| Thioredoxin | 0.690918 | 0.0330177 | 5 | 79.8 | Q835H2 |
| Uncharacterized protein | 0.688846 | 0.0210995 | 3 | 47.7 | Q830I5 |
| Guanosine monophosphate reductase | 0.656947 | 0.030067 | 8 | 35.1 | Q831S1 |
Table 3.
Proteins which are uniquely absent in E. faecalis when grown in the presence of gliotoxin (5 µg/ml), compared to identical cultures grown without gliotoxin (5 µg/ml). Grown using tryptic soy broth (TSB) media for 30 min in the log phase (0.3–0.4 OD600).
| Protein description | Fold Change (log2) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| Endonuclea_NS_2 domain-containing protein | Absent | N/A | 2 | 17.4 | Q835Q6 |
| ABC transporter, ATP-binding/permease protein | Absent | N/A | 5 | 12.3 | Q837A1 |
| Uncharacterized protein | Absent | N/A | 3 | 41.7 | Q82YV6 |
| Lipase, putative | Absent | N/A | 4 | 15.1 | Q82Z80 |
| ATP-dependent DNA helicase RecG | Absent | N/A | 4 | 8.6 | Q82ZE7 |
| YitT family protein | Absent | N/A | 3 | 12.4 | Q82ZG6 |
| Beta-hydroxyacyl-ACP dehydratase | Absent | N/A | 2 | 27.3 | Q833N7 |
| Phosphate transport system permease protein PstA | Absent | N/A | 2 | 7.1 | Q834B2 |
| ABC transporter, ATP-binding protein | Absent | N/A | 3 | 23.2 | Q835Q5 |
| PTS system, IIA component | Absent | N/A | 3 | 50 | Q836T9 |
| DUF2200 domain-containing protein* | Absent | N/A | 4 | 49.1 | Q833J9 |
| YlbF family regulator* | Absent | N/A | 2 | 20.1 | Q831P6 |
*This protein exhibited overall decreased abundance > 1.5-fold and was only detectable in 1 of 3 treated samples.
Table 4.
Proteins which are decreased in abundance in E. faecalis when grown in the presence of gliotoxin (5 µg/ml), compared to identical cultures grown without gliotoxin (5 µg/ml). Grown using tryptic soy broth (TSB) media for 30 min in the log phase (0.3–0.4 OD600).
| Protein description | Fold Change (log2) | P-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| 30S ribosomal protein S12 | − 2.08302 | 0.0128374 | 2 | 25.5 | Q839H1 |
| Sodium/dicarboxylate symporter family protein | − 0.929064 | 0.0460545 | 11 | 23.5 | Q837T6 |
| Cysteine synthase B, putative | − 0.843203 | 0.0200332 | 10 | 49.8 | Q838Z3 |
| Cyclopropane-fatty-acyl-phospholipid synthase | − 0.711893 | 0.0158166 | 15 | 54.9 | Q839G6 |
| Magnesium-transporting ATPase, P-type 1 | − 0.63794 | 0.0195624 | 16 | 23.7 | Q835M5 |
Table 5.
Proteins which are uniquely present in E. faecalis when grown in the presence of gliotoxin (5 µg/ml), compared to identical cultures grown without gliotoxin (5 µg/ml). Grown using tryptic soy broth (TSB) media for 60 min in the log phase (0.3–0.4 OD600).
| Protein description | Fold Change (log2) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| Pyridine nucleotide-disulphide oxidoreductase | Unique | N/A | 11 | 33.2 | H7C719 |
| 50S ribosomal protein L33 3 | Unique | N/A | 2 | 26.5 | P59628 |
| 50S ribosomal protein L33 4 | Unique | N/A | 3 | 44.9 | P59629 |
| Cobalamin synthesis protein/P47K family protein | Unique | N/A | 6 | 25.6 | Q82Z69 |
| 50S ribosomal protein L28 | Unique | N/A | 4 | 48.4 | Q82ZE4 |
| UPF0637 protein EF_3078 | Unique | N/A | 3 | 26.6 | Q82ZH9 |
| Glycosyl transferase, group 2 family protein | Unique | N/A | 4 | 18.7 | Q832N3 |
| Na+/H+ antiporter, putative | Unique | N/A | 7 | 11.4 | Q834R5 |
| MazG domain-containing protein | Unique | N/A | 3 | 28.3 | Q836X0 |
| Tributyrin esterase, putative | Unique | N/A | 2 | 8 | Q837P7 |
| Uncharacterized protein | Unique | N/A | 7 | 71.9 | H7C700 |
| Transcriptional regulator MraZ | Unique | N/A | 4 | 27.3 | O07103 |
| Glycerol kinase | Unique | N/A | 24 | 72.1 | O34154 |
| Gluconate kinase, putative | Unique | N/A | 6 | 17 | Q82Z43 |
| 30S ribosomal protein S14 1 | Unique | N/A | 1 | 13.5 | Q8KU58 |
| OsmC/Ohr family protein | Unique | N/A | 2 | 19.2 | Q82Z71 |
| ATP-dependent DNA helicase RecG | Unique | N/A | 7 | 14.2 | Q82ZE7 |
| Serine/threonine transporter SstT | Unique | N/A | 4 | 11 | Q82ZN5 |
| Lipoprotein, putative | Unique | N/A | 3 | 13.5 | Q82ZP7 |
| Hydrolase, haloacid dehalogenase-like family | Unique | N/A | 5 | 31.3 | Q82ZY1 |
| Hydroxyethylthiazole kinase | Unique | N/A | 3 | 13.6 | Q830K4 |
| Transcriptional regulator | Unique | N/A | 6 | 37.1 | Q830S0 |
| Acetyltransferase, GNAT family | Unique | N/A | 4 | 27.6 | Q830S1 |
| DUF2188 domain-containing protein | Unique | N/A | 5 | 33.3 | Q833X9 |
| Lipoprotein signal peptidase | Unique | N/A | 2 | 16.1 | Q834D8 |
| Sucrose operon repressor ScrR | Unique | N/A | 5 | 30.1 | Q834N9 |
| Phosphoenolpyruvate–glycerone phosphotransferase | Unique | N/A | 3 | 18.2 | Q835L8 |
| Uncharacterized protein | Unique | N/A | 1 | 22 | Q835Q1 |
| ABC transporter, ATP-binding protein | Unique | N/A | 4 | 29.2 | Q835Q5 |
| Transcriptional regulator, Cro/CI family | Unique | N/A | 8 | 33.5 | Q835Q9 |
| Amino acid ABC transporter, amino acid-binding protein | Unique | N/A | 5 | 28.4 | Q836J2 |
| 50S ribosomal protein L32-3 | Unique | N/A | 4 | 37.3 | Q836R0 |
| ABC transporter permease | Unique | N/A | 3 | 13.4 | Q837A0 |
| Isopentenyl-diphosphate delta-isomerase | Unique | N/A | 4 | 15 | Q837E2 |
| 50S ribosomal protein L36* | Unique | N/A | 2 | 60.5 | Q839E1 |
| ABC transporter, ATP-binding protein* | Unique | N/A | 7 | 47 | Q839U4 |
| Dihydropteroate synthase* | Unique | N/A | 4 | 20.9 | Q82Z14 |
| CBS domain protein* | Unique | N/A | 8 | 54.7 | Q830P4 |
| 4-hydroxy-tetrahydrodipicolinate synthase* | Unique | N/A | 10 | 54.5 | Q836D1 |
*This protein exhibited overall increased abundance > 1.5-fold and was only detectable in 1 of 3 untreated samples.
Table 6.
Proteins which are increased in abundance in E. faecalis when grown in the presence of gliotoxin (5 µg/ml), compared to identical cultures grown without gliotoxin (5 µg/ml). Grown using tryptic soy broth (TSB) media for 60 min in the log phase (0.3–0.4 OD600).
| Protein description | Fold Change (log2) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| Adhesion lipoprotein (adcA-II) | 8.49828 | 0.0020639 | 17 | 47.2 | Q82Z67 |
| Adhesion lipoprotein (adcABC) | 2.61741 | 0.00538351 | 8 | 44.8 | Q839U5 |
| Peptide ABC transporter, ATP-binding protein | 1.28554 | 0.00946214 | 12 | 70.7 | Q82ZF0 |
| 5'-nucleotidase family protein | 0.948218 | 0.011422 | 23 | 26.9 | Q839U0 |
| LemA family protein | 0.664767 | 0.00432472 | 6 | 40.7 | Q838I0 |
| Phage portal protein | 0.64703 | 0.0486542 | 16 | 56 | Q835K9 |
| ABC transporter, ATP-binding protein | 0.643841 | 0.0456337 | 4 | 35.8 | Q833S0 |
| Xanthine phosphoribosyltransferase | 0.614899 | 0.00209845 | 12 | 84.5 | Q831Y0 |
| Adenine phosphoribosyltransferase | 0.575857 | 0.0317383 | 10 | 71.2 | Q834G6 |
| Peptide methionine sulfoxide reductase MsrB* | 1.6171 | N/A | 5 | 51.7 | P0DM32 |
*This protein was found in 2/3 GT treated samples and 1/3 untreated controls which prevented calculation of a P-value.
Table 7.
Proteins which are uniquely absent in E. faecalis when grown in the presence of gliotoxin (5 µg/ml), compared to identical cultures grown without gliotoxin (5 µg/ml). Grown using tryptic soy broth (TSB) media for 60 min in the log phase (0.3–0.4 OD600).
| Protein description | Fold Change (log2) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| Glycosyl hydrolase, family 1 | Absent | N/A | 9 | 22.8 | Q836T7 |
| Thymidine kinase | Absent | N/A | 5 | 29.9 | Q831F5 |
| Probable transcriptional regulatory protein EF_0663 | Absent | N/A | 3 | 22.3 | Q838A9 |
| Transcriptional regulator, MerR family | Absent | N/A | 4 | 15.3 | Q832K1 |
| DNA-binding response regulator | Absent | N/A | 5 | 25.3 | Q833S2 |
| Ferrous iron transport protein A | Absent | N/A | 3 | 22.9 | Q838H4 |
| Cell division protein FtsL | Absent | N/A | 3 | 40 | H7C6Z7 |
| Uncharacterized protein | Absent | N/A | 2 | 19.1 | Q833H3 |
| MazG domain-containing protein | Absent | N/A | 3 | 33.6 | Q834S7 |
| PTS system, IIA component | Absent | N/A | 3 | 22.6 | Q831R2 |
| PC4 domain-containing protein | Absent | N/A | 2 | 48.6 | Q82ZD2 |
Table 8.
Proteins which are decreased in abundance in E. faecalis when grown in the presence of gliotoxin (5 µg/ml), compared to identical cultures grown without gliotoxin (5 µg/ml). Grown using tryptic soy broth (TSB) media for 60 min in the log phase (0.3–0.4 OD600).
| Protein description | Fold Change (log2) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| Sodium/dicarboxylate symporter family protein | − 0.6585 | 0.008642 | 11 | 26.8 | Q837T6 |
| Mn2+ /Fe2+ transporter, NRAMP family | − 0.61846 | 0.026495 | 2 | 5.6 | Q836Q1 |
| PTS system mannose-specific EIIAB component | − 0.60834 | 0.007484 | 22 | 78.8 | Q839X9 |
| PTS system, mannose-specific IIC component | − 0.6067 | 0.018615 | 2 | 12.4 | Q839X8 |
Table 9.
Proteins which are uniquely present in E. faecalis when grown in the presence of gliotoxin (5 µg/ml), compared to identical cultures grown without gliotoxin (5 µg/ml). Grown using tryptic soy broth (TSB) media for 180 min in the log phase (0.3–0.4 OD600).
| Protein description | Fold Change (log2) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| ABC transporter, ATP-binding protein | Unique | N/A | 4 | 24.1 | Q833B4 |
| UPF0316 protein EF_1609 | Unique | N/A | 4 | 22.4 | Q834N5 |
| Uracil-DNA glycosylase | Unique | N/A | 7 | 42.9 | Q836Z5 |
| ABC transporter, ATP-binding protein | Unique | N/A | 7 | 38.7 | Q839U4 |
| 50S ribosomal protein L33 4 | Unique | N/A | 2 | 40.8 | P59629 |
| 30S ribosomal protein S14 2 | Unique | N/A | 3 | 39.3 | Q82Z70 |
| Lipoprotein, putative | Unique | N/A | 3 | 19.6 | Q82ZP7 |
| DUF2829 domain-containing protein | Unique | N/A | 4 | 57.1 | Q833Z9 |
| MutT/nudix family protein | Unique | N/A | 4 | 23.7 | Q834Q4 |
| UPF0291 protein EF_1580 | Unique | N/A | 4 | 37.5 | Q834Q9 |
| Histidine kinase | Unique | N/A | 6 | 12.7 | Q835W1 |
| Amino acid ABC transporter, ATP-binding protein | Unique | N/A | 7 | 41.2 | Q837N1 |
| Transcriptional regulator, ArsR family | Unique | N/A | 3 | 26.1 | Q839Q2 |
| Adhesion lipoprotein (AdcABC)* | Unique | N/A | 9 | 44.8 | Q839U5 |
| Uncharacterized protein* | Unique | N/A | 2 | 32.2 | Q835Q1 |
| Adhesion lipoprotein (AdcA-II)* | Unique | N/A | 22 | 51.9 | Q82Z67 |
*This protein exhibited overall increased abundance > 1.5-fold and was only detectable in 1 of 3 untreated samples.
Table 10.
Proteins which are increased in abundance in E. faecalis when grown in the presence of gliotoxin (5 µg/ml), compared to identical cultures grown without gliotoxin (5 µg/ml). Grown using tryptic soy broth (TSB) media for 180 min in the log phase (0.3–0.4 OD600).
| Protein description | Fold Change (log2) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| Pyridoxal phosphate homeostasis protein | 1.84387 | 0.0140765 | 8 | 64 | Q836V5 |
| Transcriptional regulator, GntR family | 0.915665 | 0.0140919 | 17 | 68.2 | Q833B3 |
| Uncharacterized protein | 0.854289 | 0.0111249 | 7 | 85.6 | Q833L4 |
| Methylglyoxal synthase | 0.778122 | 0.0358049 | 5 | 52.1 | Q837A4 |
| Cysteine synthase | 0.748689 | 0.0245636 | 18 | 77.1 | Q834Q6 |
| D-isomer specific 2-hydroxyacid dehydrogenase family protein | 0.690603 | 0.00288734 | 8 | 39.1 | Q82ZZ6 |
| Oxidoreductase, short chain dehydrogenase/reductase family | 0.64496 | 0.0218784 | 13 | 47.8 | Q839T0 |
| Ribosomal RNA small subunit methyltransferase I | 0.588148 | 0.0470489 | 10 | 38.3 | Q830M1 |
Table 11.
Proteins which are uniquely absent in E. faecalis when grown in the presence of gliotoxin (5 µg/ml), compared to identical cultures grown without gliotoxin (5 µg/ml). Grown using tryptic soy broth (TSB) media for 180 min in the log phase (0.3–0.4 OD600).
| Protein description | Fold Change (log2) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| 50S ribosomal protein L33 3 | Absent | N/A | 2 | 26.5 | P59628 |
| PTS system, beta-glucoside-specific IIABC component | Absent | N/A | 5 | 15.2 | Q831B4 |
| 6-aminohexanoate-cyclic-dimer hydrolase, putative | Absent | N/A | 10 | 19.1 | Q836S5 |
| Prephenate dehydrogenase | Absent | N/A | 7 | 23.4 | H7C6X1 |
| Mga domain-containing protein | Absent | N/A | 6 | 15.6 | Q82ZN7 |
| Protease synthase and sporulation negative regulatory protein pai 1 | Absent | N/A | 2 | 11.7 | Q82ZP9 |
| Transcriptional antiterminator, bglG family | Absent | N/A | 3 | 7 | Q82ZT1 |
| FolC family protein | Absent | N/A | 6 | 19.3 | Q82ZW9 |
| DNA polymerase III, delta prime subunit | Absent | N/A | 3 | 8.9 | Q830L8 |
| Acetyltransferase, GNAT family | Absent | N/A | 3 | 21.3 | Q830S1 |
| Diacylglycerol kinase catalytic domain protein | Absent | N/A | 3 | 12.9 | Q830V8 |
| ABC transporter, permease protein | Absent | N/A | 3 | 16.7 | Q831K7 |
| ComE operon protein 2, putative | Absent | N/A | 4 | 29.3 | Q831Q2 |
| Uncharacterized protein | Absent | N/A | 1 | 25.5 | Q831U5 |
| YxeA family protein | Absent | N/A | 3 | 26.5 | Q832L5 |
| tRNA-specific adenosine deaminase | Absent | N/A | 2 | 16.2 | Q832M0 |
| Probable dual-specificity RNA methyltransferase RlmN | Absent | N/A | 9 | 34.5 | Q833B6 |
| Putative 3-methyladenine DNA glycosylase | Absent | N/A | 4 | 19.2 | Q833H5 |
| YitT family protein | Absent | N/A | 5 | 21.2 | Q833I6 |
| Uncharacterized protein | Absent | N/A | 3 | 63.3 | Q833K1 |
| Glycerol uptake facilitator protein | Absent | N/A | 2 | 9.8 | Q833L8 |
| Phosphate transport system permease protein PstA | Absent | N/A | 2 | 7.1 | Q834B2 |
| Bacteriocin-protection, YdeI or OmpD-Associated | Absent | N/A | 2 | 10.5 | Q834C8 |
| Cold-shock domain family protein | Absent | N/A | 2 | 63.2 | Q834D5 |
| Metal-independent alpha-mannosidase | Absent | N/A | 10 | 33.8 | Q834E7 |
| Glyoxylase family protein | Absent | N/A | 5 | 18.8 | Q834I3 |
| V-type ATP synthase subunit D | Absent | N/A | 4 | 32.7 | Q834X7 |
| Exodeoxyribonuclease 7 small subunit | Absent | N/A | 2 | 46.1 | Q836W5 |
| Pyrroline-5-carboxylate reductase | Absent | N/A | 3 | 14.8 | Q836Y3 |
| 3-demethylubiquinone-9 3-methyltransferase | Absent | N/A | 2 | 12.1 | Q837B9 |
| UPF0178 protein EF_0842 | Absent | N/A | 5 | 45.3 | Q837J5 |
| DUF1189 domain-containing protein | Absent | N/A | 2 | 8.2 | Q838I5 |
| Putative N-acetylmannosamine-6-phosphate 2-epimerase | Absent | N/A | 3 | 18.1 | Q839T3 |
| 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase | Absent | N/A | 4 | 15.2 | Q839U9 |
| PTS system, IIB component* | Absent | N/A | 8 | 50 | Q82ZC7 |
*This protein exhibited overall decreased abundance > 1.5-fold and was only detectable in 1 of 3 treated samples.
Table 12.
Proteins which are decreased in abundance in E. faecalis when grown in the presence of gliotoxin (5 µg/ml), compared to identical cultures grown without gliotoxin (5 µg/ml). Grown using tryptic soy broth (TSB) media for 180 min in the log phase (0.3–0.4 OD600).
| Protein description | Fold Change (log2) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| Glycosyl hydrolase, family 1 | − 3.88117 | 0.0106769 | 13 | 34.1 | Q831B5 |
| Glycerol kinase | − 2.9383 | 0.00501984 | 24 | 64.1 | O34154 |
| Alpha-glycerophosphate oxidase | − 2.89753 | 0.0117899 | 33 | 68.1 | Q833L7 |
| Glycosyl hydrolase, family 1 | − 2.76907 | 0.012451 | 15 | 39.9 | Q839A6 |
| PTS system, IIBC components | − 2.73649 | 0.0463181 | 7 | 16.2 | Q832L3 |
| ABC transporter, substrate-binding protein | − 1.82212 | 0.0241417 | 19 | 49.8 | Q832K5 |
| Ornithine carbamoyltransferase, catabolic | − 1.73602 | 0.00170647 | 17 | 67.6 | Q839Q5 |
| 30S ribosomal protein S15 | − 1.64109 | 0.0088054 | 6 | 56.2 | Q82ZJ1 |
| Hydrolase, haloacid dehalogenase-like family | − 1.4352 | 0.0409972 | 6 | 48.6 | Q82ZA8 |
| Acyl carrier protein | − 1.34991 | 0.0196974 | 3 | 88 | Q830B0 |
| Biotin carboxyl carrier protein of acetyl-CoA carboxylase | − 1.32096 | 0.0104951 | 4 | 32.1 | Q830B2 |
| Acyl carrier protein | − 1.16376 | 0.0267896 | 3 | 55.7 | Q82ZE9 |
| Arginine deiminase | − 1.09113 | 0.00625365 | 25 | 74.3 | Q93K67 |
| Mn2+/Fe2+ transporter, NRAMP family | − 1.04732 | 0.0290044 | 3 | 11.1 | Q836Q1 |
| Carbamate kinase 1 | − 1.02868 | 0.0133442 | 14 | 66.8 | P0A2X7 |
| Citrate lyase subunit beta | − 0.941302 | 0.017623 | 8 | 26.4 | Q82YW2 |
| Sodium/dicarboxylate symporter family protein | − 0.814864 | 0.0266616 | 11 | 29.2 | Q837T6 |
Three ribosomal proteins are either uniquely present or increased in abundance following GT addition, and two Zn2+ uptake membrane proteins (AdcA and AdcC of the Gram positive AdcABC system), out of 32 proteins in this dataset, were significantly increased in abundance (log2-fold 2.4–7.28) at 30 min (Tables 1, 2; Supplementary Fig. 9). This is the first demonstration that GT affects bacterial ribosomal protein presence, as has been predicted39. Increased Zn2+ uptake transporter abundance clearly suggests that intracellular Zn2+ may be complexed by GT/DTG resulting in a limiting intracellular environment. Our observations complement previous work which revealed significantly increased adcABC and adcA-II gene expression following exposure to 5 µM TPEN46. Moreover, it is in complete accordance with our data which shows that Zn2+ relieves GT-mediated growth inhibition and that GT/DTG is a Zn2+ chelator. It is further conceivable that DTG may eject Zn2+ from intracellular proteins resulting in inactivity and possibly denaturation. The increased abundance of the Zn2+ uptake membrane proteins was also evident at T = 60 and they were uniquely detected at T = 180 min post-GT addition (Tables 5, 6, 9, 10).
Our hypothesis that GT-mediated zinc depletion (reduced bioavailability) was primarily responsible for quantitative proteomic alterations, was investigated by assessment if Zn co-addition (200 µM) for 30 min with GT would reverse the observed GT-induced proteomic alterations. This chemo-complementation approach revealed that 1 of 16 proteins uniquely detected by GT addition only exhibited reversed abundance in the presence of GT and Zn, of the 12 proteins uniquely absent with GT, 3 were decreased in abundance in the presence of Zn while 1 (Q834B2) was uniquely detected in the Zn supplemented samples (Supplementary Tables 4 and 5). After 30 min exposure to GT (5 µg/ml) and Zn (200 µM), there was a minor and insignificant increase in AdcA, AdcC, and AdcA-II when compared to GT (5 µg/ml) only. This data indicates that 30 min may be too short a time frame to see a complete disappearance of the AdcABC system proteins. After 60 min, AdcA was decreased in abundance (− 1.5 log2 fold change), AdcC also exhibited decreased abundance (− 1.3 log2 fold change) (although not statistically significant (p < 0.5)) and AdcA-II was also insignificantly decreased in abundance (p < 0.2; − 1.3 log2 fold change) in the presence of GT (5 µg/ml) and Zn (400 µM), when compared to GT (5 µg/ml) only (Supplementary Table 6). In combination, these data indicate that addition of Zn (400 µM) relieves some of the effects of GT (5 µg/ml), but was either insufficient to result in a rapid decrease in the requirement for the Zur-regulated zinc uptake system or 60 min incubation was sub-optimal for this comparative study. Hence, future work will focus on evaluating and defining the optimal conditions of zinc reversal of significantly elevated AdcABC and Adc-II uptake systems in E. faecalis.
However, the increased abundance or unique presence of E. faecalis zinc import proteins (AdcA and C) in the presence of GT at all timepoints is in complete accordance with the independent demonstration that disruption of orthologous genes encoding AdcA or AdcB and AdcC, components of the zinc uptake system in Streptococcus mutans, caused severe growth inability under zinc-deplete conditions53. Furthermore, it was demonstrated that the S. mutans ΔadcBC mutant exhibited a severe colonisation defect in a rat infection model system53. In a further study in E. faecalis, it has been elegantly shown that AdcABC and adhesion lipoprotein AdcA-II function in a cooperative manner to ensure Zn homeostasis. Deletion of system components resulted in zinc-associated growth defects and increased sensitivity to antibiotics which target the bacterial cell wall54. These authors also showed that bacterial virulence was attenuated in zinc transporter deletion mutants and propose interference with high affinity zinc importers as a potential therapeutic strategy to combat E. faecalis infection54. In combination, these deletion studies and associated identification of attenuated virulence in the absence of zinc uptake systems underpin the strategy and utility of using GT/DTG couple as a means of identifying new antibacterial drug targets38. Interestingly, an E. faecalis pyridine nucleotide-disulphide oxidoreductase (PNDO)(H7C719) was uniquely detected in the presence of GT (Table 1 and 5) at T = 30 and 60 min exposure. To our knowledge, this is a protein of unknown function in E. faecalis and its presence is suggestive of altered redox homeostasis upon GT exposure. It is also relevant that the self-protection enzyme gliotoxin oxidoreductase GliT in A. fumigatus converts DTG to GT22,23 and is also classified as a PNDO-type enzyme. Future work will involve targeting this bacterial PNDO for deletion and mutant characterisation.
The unique or elevated abundance of specific ribosomal proteins ((T30 = 50S ribosomal protein L33 4 (P59629), 30S ribosomal protein S14 1 (Q8KU58), and 30S ribosomal protein S15 (Q82ZJ1); T60 = 50S ribosomal protein L33 4 (P59629), 50S ribosomal protein L28 (Q82ZE4), 30S ribosomal protein S14 1 (Q8KU58); T180 = 50S ribosomal protein L33 4 (P59629), 30S ribosomal protein S14 2 (Q82Z70); (plus uniquely absent 50S ribosomal protein L33 3 (P59628)) was also observed (Supplementary Fig. 10 and Supplemental Table 1). These changes in ribosomal protein abundance were indicative of a shift towards non-zinc binding paralogs upon GT addition, which is in accordance with the proposal of Danchin that the prokaryotic ribosome may act as a zinc store39. Thus, when GT is added to E. faecalis, we hypothesise that resultant zinc depletion would lead to zinc release from ribosomes and consequent increased presence of selected zinc-free ribosomal protein paralogs. Future RNAseq work will further investigate this GT-induced phenomenon in E. faecalis, as has been deployed in Mycobacterium smegmatis55. In the work of Dow et al., non-zinc-binding paralogs were identified as functional replacements for Zn2+-dependent paralogs and were involved in the transcriptomic response to Zn2+-limitation.
GT uptake and predicted conversion to DTG by intracellular reduction by GSH, L-Cys or other reductants would likely lead to dissipation of these metabolites and increased biosynthesis of same. In accordance with this prediction, a cysteine synthase (Q834Q6) was detected as significantly increased in abundance (p < 0.025) at T = 180 min, with high confidence given that 77% sequence coverage was evident (Tables 9,10). This enzyme may result in either L-Cys or ultimately GSH formation in E. faecalis. Importantly, this proteomic observation aligns with the detection of higher (+ 38%) total intracellular thiol concentration (0.156 nmol per mg cells) following GT addition to E. faecalis. Detection of a putative cysteine synthase B (Q838Z3) as significantly decreased in abundance (p < 0.02) at T = 30 min requires future analysis (Tables 3, 4).
Detection of a cobalamin synthesis protein (Q82Z69) at T = 30 and 60 only post-GT addition (Tables 1, 2, 5, 6) suggests that vitamin B12 biosynthesis is increased in E. faecalis due to GT presence. Allied to this is the increased abundance of Met sulphoxide reductase MsrB (P0DM32) which was identified in 2/3 GT treated samples versus 1/3 controls therefore precluding the calculation of a p-value (Tables 5, 6). This enzyme has been shown to contribute to E. faecalis response to oxidative stress, by L-Met regeneration, and virulence56. The detection of both these enzymes, along with significantly increased abundance (p < 0.03) of thioredoxin (Table 1, 2) suggests that the GT effect on E. faecalis also involves disrupted L-Met oxidation, redox homeostasis and possibly Methyl/Met cycle due to SAM requirements. Future work will investigate the altered abundance of enzymes involved in L-Met metabolism by targeted deletion studies, since few data is currently available on these phenotypes.
Our quantitative proteomic analyses also revealed absence or significantly reduced abundance of components of the E. faecalis PTS (phosphoenolpyruvate (PEP):carbohydrate phosphotransferase system), mainly PTS system: IIA component, mannose-specific EIIAB and IIC components between 30 and 180 min post-GT addition, though one protein in this system, PTS System, beta-glycoside-specific IIABC component was uniquely present with GT (Q831B4) (Tables 1, 3, 4, 7, 8, 11, 12). This system has recently been shown to repress ribosomal biosynthesis and to increase the sensitivity of E. faecalis to gentamycin. Overexpression of PTS also increased the sensitivity of E. faecalis to the antibiotic, daptomycin57. Interestingly, alkaline stress caused reduced abundance of PTS system components in E. faecalis V583 as determined by proteomic analysis58. Our observation of decreased abundance of PTS system components, in response to GT presence is in accordance with these studies which suggests that deactivation of the PTS system reduces the impact of antibacterial compounds (e.g., daptomycin, GT or gentamycin) or conditions (alkaline stress).
Through proteomic analysis we show that GT may affect intracellular metal ion homeostasis, possibly by interference with intracellular zinc pools in E. faecalis. Proteins associated with key pathways, several of which are controlled by the zinc-dependent Zur transcription factor46, were altered in abundance when E. faecalis was exposed to GT. This metallo-centric model implicates DTG as the active form of GT and supplants the conventional perspective of altered redox homeostasis due to GT presence as the sole factor responsible for the antimicrobial activity of GT/DTG. In conclusion, this work illustrates the GT/DTG couple potential for providing new ways to combat the growing antibiotic resistance threat as well as yielding new mechanistic approaches for treating antibiotic resistant infections38.
In summary, DTG causes Zn2+ and Cu2+ to specifically dissociate from PAR in a concentration-dependent manner. DTG can specifically strip or remove Fe3+ from low and high sensitivity Fe3+ chelates in a concentration-dependent manner. Notably, this interaction is also time-dependent. The reducing agent DTT and antioxidant GSH had no effect on fluorescent chelate:Fe3+ stability, implicating chelation as the mechanism of Fe3+ removal from this chelate by DTG. When exposed to GT (5 µg/ml) E. faecalis showed significant and specific proteome alterations, many of which are linked to the occurrence Zn2+-limiting or bioavailability conditions; and for the first time imply that GT/DTG induce zinc starvation within E. faecalis. DTG may chelate free Zn2+ and eject it from intracellular metalloproteins, thereby causing growth inhibition. During manuscript revision a report emerged which also addressed the role of gliotoxin as an anti-bacterial agent59.
Methods
Bacterial growth inhibition assays
Single colonies of E. faecalis (ATCC 19433), E. faecium (ATCC 19434), K. pneumoniae (ATCC 70063), and A. baumannii (ATCC 19606) were used to inoculate tryptic soy broth (TSB) (Sigma-Aldrich, cat no. 22092-500G). These cultures were incubated overnight (37°C; 170 rpm). The OD600 nm of the cultures were determined and aliquots of each culture were diluted to OD600 nm = 0.2 using TSB. Cultures were added to wells (100 µl/well) of 96-well plates, in triplicate, containing 100 µl TSB supplemented with GT (0–120 µM) and Zn, Cu, or Mn (0, 20, 200, or 400 µM), incubated (37°C; Static; 18 h) and OD600 nm determined using a BioTek Synergy HT 96-well plate reader. Following GT exposure (GT: 0–60 µM), iron supplementation (0, 10, 100 and 200 µM) was carried out for E. faecalis only. TSB has been shown via ICP-AES41 to basally contain: Zn: 0.6 ppm = 9.177 µM; Cu: < 0.1 ppm = < 1.5737 µM and Fe: 0.3 ppm = 5.372 µM. Data analysis was performed using GraphPad Prism 9.5.0 via two-way ANOVA.
Zn determination by zinquin method
E. faecalis (ATCC 19433) was grown overnight in TSB (37 °C 170 rpm). These cultures were used to inoculate fresh TSB to an OD600 of 0.05 until reaching an OD600 of 0.3–0.4. The cultures were divided into even aliquots and spiked with GT (5 µg/ml final) or MeOH. The cultures were returned to the incubator (37 °C 170 rpm) and 15 ml aliquots were taken after 5, 15, and 30 min. The assay was also performed for 60 min. Aliquots were centrifuged 4500 g at 4 °C for 10 min and the supernatants discarded. Pellets were washed extensively in PBS and transferred to 1.5 ml tubes. Samples were centrifuged 15,000 g for 10 min at 4 °C. After weighing pellets, 400 µl PBS (0.03% (w/v) albumin from chicken egg white) was added to each sample and vortexed into solution followed by sonication on ice (6 times MS72 probe, power ≤ 20%, cycle 6) and transfer to black 96 well plates (200 µl per well) for addition of 3.5 µl Zinquin in DMSO (12 mM stock). After pipette-mixing and incubation (37 °C in the dark for 40 min.), Zn levels were calculated by comparison to a Zn standard curve. All dilutions were done in PBS (0.03% albumin from chicken egg white). Fluorescence detection: excitation: 360/40 nm, emission: 460/40 nm, Gain: 65.
GT versus TPEN bacterial growth Inhibition assays
E. faecalis was added to wells (100 µl/well) of 96-well plates, in triplicate, containing 100 µl TSB supplemented with GT (0–120 µM) or TPEN (0–120 µM) and ZnSO4 (0, 20, 200, or 400 µM), incubated (37 °C; static; 18 h) and OD600 nm determined using a BioTek Synergy HT 96-well plate reader. Data analysis was performed using GraphPad Prism 9.5.0 via two-way ANOVA.
GT-augmented vancomycin bacterial growth inhibition assays, with and without Zn
E. faecalis was added to wells (100 µl/well) of 96-well plates, in triplicate, containing 100 µl TSB supplemented with GT (0–20 µg/ml) and vancomycin (0–64 µg/ml) in varied concentrations. The wells were then inoculated with 100 µl E. faecalis (0.2 OD 600 nm). Identical plates were then prepared using wells supplemented with ZnSO4 (200 µM final concentration). The plates were incubated (37 °C; static; 18 h). The wells were imaged using a camera attached to a microscope (Olympus SZX16 microscope with Olympus SDF PLAPO 2XPFC camera attachment) using cellSens Standard software. The wells were homogenised using a multichannel pipette OD600 nm determined using a BioTek Synergy HT 96-well plate reader.
Gliotoxin x vancomycin biofilm evaluation by crystal violet assay
E. faecalis cultures (0.2 OD600) were added to 96-well plates, 100 µl/well, containing 100 µl TSB supplemented with GT (0–10 µg/ml) and vancomycin (4 µg/ml) followed by incubation (37 °C; Static), 18 h and OD600 determined using a BioTek Synergy HT 96-well plate reader. The media was removed from the wells, washed with deionised H2O then dried. Crystal violet (0.1% (w/v)) was added to each well, incubated for 10 min at room temperature, discarded, the wells washed with deionised H2O and then dried. Acetic acid (30% (v/v)) was added to each well and allowed to incubate for 10 min at room temperature. The wells were pipette-mixed then read at 595 nm using a BioTek Synergy HT 96-well plate reader.
GT time-kill assay
Three colonies of E. faecalis were used to inoculate TSB followed by incubation overnight (37 °C; 170 rpm). The OD600 of the cultures were measured and an aliquot of each culture was taken and diluted to 5 × 105 CFU/ml using TSB. These cultures were divided into aliquots and spiked with GT in MeOH (0, 1.875, 3.75 and 7.5 µM) and further divided into aliquots for each time point in falcon tubes. After incubation (37 °C; 170 rpm), culture growth was analysed at 0, 2, 4, 8, and 24 h whereby at each time point the cultures were serially diluted in TSB and streaked on TSB agar. The plates were grown (37 °C; Static) and counted after 24 h.
Assessment of DTG metal chelation capacity by 4-(2-pyridylazo)resorcinol (PAR) assay
A master stock of 4-(2-pyridylazo)resorcinol (PAR) (1.5 mM)60 was prepared in PBS pH 7.4. A working stock of PAR was made up in PBS pH 7.4 (22.22 µM). PAR (22.22 µM; 180 µl) was transferred to the wells of a 96-well plate. 10 µl of 200 µM metal (Zn, Cu) was added to the appropriate wells. The plate was allowed to incubate in the dark for 2 min and 10 µl of DTG or N,N,N′,N′-Tetrakis(2-pyridylmethyl)ethylenediamine (TPEN) (0–120 µM final) was added to the appropriate wells and read using a BioTek Synergy HT 96-well plate reader with spectrum scan 300–600 nm.
Determination of DTG and TPEN Zn pKd
This methodology was adapted from that of Kocyła et al., with the 10 h incubation being done in plastic cuvettes instead of glass60.
Assessment of DTG and related thiol compound reductant activity by Siderotec™ assays
The Siderotec™ total and Siderotec-HiSens™ tests were performed as per manufacturer’s instructions (Accuplex Diagnostics Limited, Ireland). In brief, these tests deploy chromophoric and fluorimetric compounds, respectively, to detect Fe3+ chelator or reductant activity61, Supplemental information and Supplementary Figs. 6 and 7.
High resolution mass spectrometry analysis of GT metal chelates
Tris(2-carboxyethyl)phosphine hydrochloride (TCEP)-reduced GT (DTG; 304.5 μM final) in 50%(v/v) MeOH was spiked with ZnSO4·7H2O, CuCl2, or FeSO4·7H2O respectively to achieve 1 and threefold molar equivalents of metals to DTG (DTG; 300 μM final). Formic acid was added to the samples (0.1%(v/v) final). Samples were filtered (0.22 μm spin filters) and directly injected onto a Thermo Q-Exactive Mass spectrometer (5 μl/min). Assessment of DTG-metal complex formation was evaluated by both positive and negative ESI mode at 70,000 resolution with full MS scan (m/z 66.7–1000.00). The following settings were used in the analysis: spray voltage (negative mode: 3.6 kV, positive mode 4 kV), capillary temperature 320 °C.
Quantitative proteomic analysis of E. faecalis response to GT exposure
Three biological replicate E. faecalis cultures were grown overnight in TSB. These cultures were then used to inoculate warm TSB to an OD600 nm 0.05 and grown (37°C; 170 rpm) until log phase (OD600 nm 0.3). At this stage, each culture was divided into two equal aliquots, one of each was spiked with GT (5 µg/ml final) while the other was spiked with the same volume of MeOH (solvent control). The cultures were returned to the incubator (37°C; 170 rpm) with aliquots taken at T = 30, 60 and 180 min post spiking. Cell pellets were collected and stored at -20°C until required. Pellets were prepared using a modified FASP protocol62 as follows: pellets were lysed via sonication whereby 25 mg bacteria was resuspended in 150 μl of 1% (w/v) SDS/500 mM ammonium bicarbonate pH 8.3. Lysed samples were heated at 95°C for 5 min. The samples were then centrifuged at 16,000 g for 5 min, supernatants were collected, and protein content quantified using Qubit protein assay. A 20 µg protein sample was taken from each and transferred to 30 kDa molecular weight cut-off (MWCO) centrifugal filters (Sartorius, cat no. VN01H22). Each sample was brought up to 200 µl with 8 M urea, 500 mM ammonium bicarbonate. The samples were vortexed then centrifuged at 14,000 g for 20 min. 500 mM ammonium bicarbonate, 8 M urea pH 8.3 (200 µl) was added to each filter. The samples were vortexed, then centrifuged at 14,000 g for 20 min and 100 μl 5 mM TCEP in 8M urea, 500 mM ammonium bicarbonate pH 8.3 was added to each sample filter. The samples were vortexed and incubated at room temperature for 20 min. 500 mM IAA (3 µl) was added to each sample (15 mM final) and samples were vortexed and incubated in the dark at room temperature for 20 min. The samples were centrifuged at 14,000 g for 20 min. 100 μl of 8 M urea, 500 mM ammonium bicarbonate pH 8.3 was added to each filter. The samples were vortexed then centrifuged at 14,000 g for 20 min. Another 100 μl of 8 M urea, 500 mM ammonium bicarbonate was added to each filter. The samples were vortexed then centrifuged at 14,000 g for 20 min. Two sequential washes with 100 μl of 500 mM ammonium bicarbonate pH 8.3 was carried out by addition to each filter, samples vortexed, centrifuged at 14,000 g for 20 min. The filters were then transferred to fresh tubes. 120 μl of 500 mM ammonium bicarbonate, 1 µl ProteaseMax (1% w/v) and 5 μl trypsin (400 ng/μl) was added to each filter followed by a brief vortex. The samples were wrapped in parafilm and placed in a humid chamber at 37°C for 18 h followed by centrifugation 14,000 g for 20 min. 500 mM ammonium bicarbonate pH 8.3 (50 μl) was added to each filter, samples were then centrifuged 14,000 g for 20 min and a 60 μl aliquot was taken from each filtrate. 12 μl of resuspension buffer (2% (v/v) TFA, 20% (v/v) in deionised H2O) was added to each aliquot, samples were vortexed and stored at − 20 °C until mass spectrometry analysis. Peptide samples were analysed using a Thermo Fisher Q-Exactive mass spectrometer coupled with a Dionex RSLCnano. LC gradients ran from 4 to 40% B (0.1%(v/v) trifluoroacetic acid in acetonitrile) over 2 h 13 min, and data was collected using a Top15 method for MS/MS scans. Comparative proteome abundance and data analysis was performed using MaxQuant software, with Andromeda used for database searching and Perseus used to organise the data11,31.
Supplementary Information
Acknowledgements
This project was funded by Leeds-Beckett University and Maynooth University. Funding for protein mass spectrometry facilities was provided by a Science Foundation Ireland Infrastructure Award [12/RI/2346 (3)] (Principal Awardee: Professor Sean Doyle). Dr Amber Dorey is a Post-Doctoral Research Fellow in the laboratory of SFI Professor J.P. Dalton, University of Galway (Recipient of an SFI Research Professorship grant 17/RP/5368). Funding for the SideroTec-HiSensTM assay was provided by H2020-FNR-11-2020: SECRETED, grant number 101000794.
Author contributions
S.G.D., R.A.O., A.L.D., K.W. and D.A.F. performed experimentation. R.A.O. conceptualised the work. S.G.D. prepared draft manuscript text. S.D. and G.W.J. conceptualised and directed the work, wrote the main manuscript text and obtained funding. S.G.D, D.A.F. and R.A.O prepared all Figures. All authors reviewed and approved the final manuscript.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files). The proteomic datasets generated during and analysed during the current study are available in the MassIVE repository, https://doi.org/doi:10.25345/C5Z892R21. All remaining datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gary W. Jones, Email: gary.jones@leedsbeckett.ac.uk
Sean Doyle, Email: sean.doyle@mu.ie.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-43300-w.
References
- 1.De Oliveira DMP, et al. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020 doi: 10.1128/cmr.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mancuso G, Midiri A, Gerace E, Biondo C. Bacterial antibiotic resistance: The most critical pathogens. Pathogens. 2021 doi: 10.3390/pathogens10101310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Y, et al. Phage products for fighting antimicrobial resistance. Microorganisms. 2022 doi: 10.3390/microorganisms10071324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costanzo V, Roviello GN. The potential role of vaccines in preventing antimicrobial resistance (AMR): An update and future perspectives. Vaccines (Basel) 2023 doi: 10.3390/vaccines11020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narendrakumar L, Chakraborty M, Kumari S, Paul D, Das B. β-Lactam potentiators to re-sensitize resistant pathogens: Discovery, development, clinical use and the way forward. Front. Microbiol. 2022;13:1092556. doi: 10.3389/fmicb.2022.1092556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Lababidi RM, Rizk JG. Cefiderocol: A siderophore cephalosporin. Ann. Pharmacother. 2020;54:1215–1231. doi: 10.1177/1060028020929988. [DOI] [PubMed] [Google Scholar]
- 7.Ikuta KS, et al. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2023;400:2221–2248. doi: 10.1016/s0140-6736(22)02185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ali IAA, Cheung GSP, Neelakantan P. Transition metals and Enterococcus faecalis: Homeostasis, virulence and perspectives. Mol. Oral Microbiol. 2022;37:276–291. doi: 10.1111/omi.12391. [DOI] [PubMed] [Google Scholar]
- 9.Dolan SK, O'Keeffe G, Jones GW, Doyle S. Resistance is not futile: Gliotoxin biosynthesis, functionality and utility. Trends Microbiol. 2015;23:419–428. doi: 10.1016/j.tim.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Huber EM. Epipolythiodioxopiperazine-based natural products: Building blocks, biosynthesis and biological activities. Chembiochem. 2022 doi: 10.1002/cbic.202200341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saleh AA, et al. Systems impact of zinc chelation by the epipolythiodioxopiperazine dithiol gliotoxin in Aspergillus fumigatus: A new direction in natural product functionality. Metallomics. 2018;10:854–866. doi: 10.1039/c8mt00052b. [DOI] [PubMed] [Google Scholar]
- 12.König S, et al. Gliotoxin from Aspergillus fumigatus abrogates leukotriene B(4) formation through inhibition of leukotriene A(4) hydrolase. Cell Chem. Biol. 2019;26:524–534.e525. doi: 10.1016/j.chembiol.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Bernardo PH, Brasch N, Chai CL, Waring P. A novel redox mechanism for the glutathione-dependent reversible uptake of a fungal toxin in cells. J. Biol. Chem. 2003;278:46549–46555. doi: 10.1074/jbc.M304825200. [DOI] [PubMed] [Google Scholar]
- 14.Reece E, Doyle S, Greally P, Renwick J, McClean S. Aspergillus fumigatus inhibits Pseudomonas aeruginosa in co-culture: Implications of a mutually antagonistic relationship on virulence and inflammation in the CF airway. Front. Microbiol. 2018;9:1205. doi: 10.3389/fmicb.2018.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waring P, Sjaarda A, Lin QH. Gliotoxin inactivates alcohol dehydrogenase by either covalent modification or free radical damage mediated by redox cycling. Biochem. Pharmacol. 1995;49:1195–1201. doi: 10.1016/0006-2952(95)00039-3. [DOI] [PubMed] [Google Scholar]
- 16.Coleman JJ, Ghosh S, Okoli I, Mylonakis E. Antifungal activity of microbial secondary metabolites. PLoS One. 2011;6:e25321. doi: 10.1371/journal.pone.0025321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carberry S, et al. Gliotoxin effects on fungal growth: Mechanisms and exploitation. Fungal Genet. Biol. 2012;49:302–312. doi: 10.1016/j.fgb.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Manzanares-Miralles L, et al. Quantitative proteomics reveals the mechanism and consequence of gliotoxin-mediated dysregulation of the methionine cycle in Aspergillus niger. J. Proteomics. 2016;131:149–162. doi: 10.1016/j.jprot.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 19.Cook KM, et al. Epidithiodiketopiperazines block the interaction between hypoxia-inducible factor-1alpha (HIF-1alpha) and p300 by a zinc ejection mechanism. J. Biol. Chem. 2009;284:26831–26838. doi: 10.1074/jbc.M109.009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reece KM, et al. Epidithiodiketopiperazines (ETPs) exhibit in vitro antiangiogenic and in vivo antitumor activity by disrupting the HIF-1alpha/p300 complex in a preclinical model of prostate cancer. Mol. Cancer. 2014;13:91. doi: 10.1186/1476-4598-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chamilos G, Lewis RE, Lamaris GA, Albert ND, Kontoyiannis DP. Genomewide screening for genes associated with gliotoxin resistance and sensitivity in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 2008;52:1325–1329. doi: 10.1128/AAC.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scharf DH, et al. Transannular disulfide formation in gliotoxin biosynthesis and its role in self-resistance of the human pathogen Aspergillus fumigatus. J. Am. Chem. Soc. 2010;132:10136–10141. doi: 10.1021/ja103262m. [DOI] [PubMed] [Google Scholar]
- 23.Schrettl M, et al. Self-protection against gliotoxin-a component of the gliotoxin biosynthetic cluster, gliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathogens. 2010 doi: 10.1371/journal.ppat.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang DN, et al. GliA in Aspergillus fumigatus is required for its tolerance to gliotoxin and affects the amount of extracellular and intracellular gliotoxin. Med. Mycol. 2014;52:506–518. doi: 10.1093/mmy/myu007. [DOI] [PubMed] [Google Scholar]
- 25.Owens RA, et al. Interplay between gliotoxin resistance, secretion, and the methyl/methionine cycle in Aspergillus fumigatus. Eukaryot. Cell. 2015;14:941–957. doi: 10.1128/ec.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolan SK, et al. Regulation of nonribosomal peptide synthesis: Bis-thiomethylation attenuates gliotoxin biosynthesis in Aspergillus fumigatus. Chem. Biol. 2014;21:999–1012. doi: 10.1016/j.chembiol.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Scharf DH, Habel A, Heinekamp T, Brakhage AA, Hertweck C. Opposed effects of enzymatic gliotoxin N- and S-methylations. J. Am. Chem. Soc. 2014;136:11674–11679. doi: 10.1021/ja5033106. [DOI] [PubMed] [Google Scholar]
- 28.Seo H, Kang S, Park YS, Yun CW. The role of zinc in gliotoxin biosynthesis of Aspergillus fumigatus. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20246192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Traynor AM, et al. At the metal-metabolite interface in Aspergillus fumigatus: Towards untangling the intersecting roles of zinc and gliotoxin. Microbiology (Reading) 2021 doi: 10.1099/mic.0.001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Traynor AM, Sheridan KJ, Jones GW, Calera JA, Doyle S. Involvement of sulfur in the biosynthesis of essential metabolites in pathogenic fungi of animals, particularly Aspergillus spp.: Molecular and therapeutic implications. Front. Microbiol. 2019;10:2859. doi: 10.3389/fmicb.2019.02859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doyle S, Jones GW, Dolan SK. Dysregulated gliotoxin biosynthesis attenuates the production of unrelated biosynthetic gene cluster-encoded metabolites in Aspergillus fumigatus. Fungal Biol. 2018;122:214–221. doi: 10.1016/j.funbio.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Chan AN, et al. Role for dithiolopyrrolones in disrupting bacterial metal homeostasis. Proc. Natl. Acad. Sci. U S A. 2017;114:2717–2722. doi: 10.1073/pnas.1612810114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svahn KS, et al. Antimicrobial activity of filamentous fungi isolated from highly antibiotic-contaminated river sediment. Infect. Ecol. Epidemiol. 2012 doi: 10.3402/iee.v2i0.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo X, et al. Antituberculosis compounds from a deep-sea-derived fungus Aspergillus sp. SCSIO Ind09F01. Nat. Prod. Res. 2017;31:1958–1962. doi: 10.1080/14786419.2016.1266353. [DOI] [PubMed] [Google Scholar]
- 35.Jones RW, Hancock JG. Mechanism of gliotoxin action and factors mediating gliotoxin sensitivity. Microbiology. 1988;134:2067–2075. doi: 10.1099/00221287-134-7-2067. [DOI] [Google Scholar]
- 36.Davis C, et al. The role of glutathione S-transferase GliG in gliotoxin biosynthesis in Aspergillus fumigatus. Chem. Biol. 2011;18:542–552. doi: 10.1016/j.chembiol.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 37.Esteban P, et al. In vitro and in vivo antibacterial activity of gliotoxin alone and in combination with antibiotics against Staphylococcus aureus. Toxins (Basel) 2021 doi: 10.3390/toxins13020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Downes SG, Doyle S, Jones GW, Owens RA. Gliotoxin and related metabolites as zinc chelators: Implications and exploitation to overcome antimicrobial resistance. Essays Biochem. 2023 doi: 10.1042/ebc20220222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danchin A. Zinc, an unexpected integrator of metabolism? Microb. Biotechnol. 2020;13:895–898. doi: 10.1111/1751-7915.13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng H, et al. Gliotoxin is antibacterial to drug-resistant piscine pathogens. nps. 2018;24:225–228. doi: 10.20307/nps.2018.24.4.225. [DOI] [Google Scholar]
- 41.Akhidime ID, et al. The antimicrobial effect of metal substrates on food pathogens. Food Bioprod. Process. 2019;113:68–76. doi: 10.1016/j.fbp.2018.09.003. [DOI] [Google Scholar]
- 42.Principe L, et al. Zinc chelators as carbapenem adjuvants for metallo-β-lactamase-producing bacteria. In vitro and in vivo evaluation. Microb. Drug Resist. 2020;26:1133–1143. doi: 10.1089/mdr.2020.0037. [DOI] [PubMed] [Google Scholar]
- 43.Albini F, Bormann S, Gerschel P, Ludwig VA, Neumann W. Dithiolopyrrolones are prochelators that are activated by glutathione. Chemistry. 2023;29:e202202567. doi: 10.1002/chem.202202567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.La Piana L, et al. Polypyridine ligands as potential metallo-β-lactamase inhibitors. J. Inorg. Biochem. 2021;215:111315. doi: 10.1016/j.jinorgbio.2020.111315. [DOI] [PubMed] [Google Scholar]
- 45.Schnaars C, et al. Synthesis and preclinical evaluation of TPA-based zinc chelators as metallo-β-lactamase inhibitors. ACS Infect. Dis. 2018;4:1407–1422. doi: 10.1021/acsinfecdis.8b00137. [DOI] [PubMed] [Google Scholar]
- 46.Latorre M, et al. Interplay between copper and zinc homeostasis through the transcriptional regulator Zur in Enterococcus faecalis. Metallomics. 2015;7:1137–1145. doi: 10.1039/c5mt00043b. [DOI] [PubMed] [Google Scholar]
- 47.Zarkan A, Macklyne HR, Truman AW, Hesketh AR, Hong HJ. The frontline antibiotic vancomycin induces a zinc starvation response in bacteria by binding to Zn(II) Sci. Rep. 2016;6:19602. doi: 10.1038/srep19602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zarkan A, et al. Zn(II) mediates vancomycin polymerization and potentiates its antibiotic activity against resistant bacteria. Sci. Rep. 2017;7:4893. doi: 10.1038/s41598-017-04868-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodcock JC, Henderson W, Miles CO. Metal complexes of the mycotoxins sporidesmin A and gliotoxin, investigated by electrospray ionisation mass spectrometry. J. Inorg. Biochem. 2001;85:187–199. doi: 10.1016/S0162-0134(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 50.Schrettl M, et al. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 2008;70:27–43. doi: 10.1111/j.1365-2958.2008.06376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newberry RW, Raines RT. The n→π* Interaction. Acc. Chem. Res. 2017;50:1838–1846. doi: 10.1021/acs.accounts.7b00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kilgore HR, Olsson CR, D'Angelo KA, Movassaghi M, Raines RT. n→π* Interactions modulate the disulfide reduction potential of epidithiodiketopiperazines. J. Am. Chem. Soc. 2020;142:15107–15115. doi: 10.1021/jacs.0c06477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ganguly T, Peterson AM, Kajfasz JK, Abranches J, Lemos JA. Zinc import mediated by AdcABC is critical for colonization of the dental biofilm by Streptococcus mutans in an animal model. Mol. Oral Microbiol. 2021;36:214–224. doi: 10.1111/omi.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lam LN, Brunson DN, Molina JJ, Flores-Mireles AL, Lemos JA. The AdcACB/AdcAII system is essential for zinc homeostasis and an important contributor of Enterococcus faecalis virulence. Virulence. 2022;13:592–608. doi: 10.1080/21505594.2022.2056965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dow A, Burger A, Marcantonio E, Prisic S. Multi-omics profiling specifies involvement of alternative ribosomal proteins in response to zinc limitation in Mycobacterium smegmatis. Front. Microbiol. 2022;13:811774. doi: 10.3389/fmicb.2022.811774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao C, et al. Role of methionine sulfoxide reductases A and B of Enterococcus faecalis in oxidative stress and virulence. Infect. Immun. 2010;78:3889–3897. doi: 10.1128/iai.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei L, et al. Phosphate transport system mediates the resistance of Enterococcus faecalis to multidrug. Microbiol. Res. 2021;249:126772. doi: 10.1016/j.micres.2021.126772. [DOI] [PubMed] [Google Scholar]
- 58.Cathro P, McCarthy P, Hoffmann P, Kidd S, Zilm P. Enterococcus faecalis V583 cell membrane protein expression to alkaline stress. FEMS Microbiol. Lett. 2022 doi: 10.1093/femsle/fnac082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vasilchenko AS, et al. Exploring the antibacterial action of gliotoxin: Does it induce oxidative stress or protein damage? Biochimie. 2023;214:86–95. doi: 10.1016/j.biochi.2023.06.009. [DOI] [PubMed] [Google Scholar]
- 60.Kocyła A, Pomorski A, Krężel A. Molar absorption coefficients and stability constants of metal complexes of 4-(2-pyridylazo)resorcinol (PAR): Revisiting common chelating probe for the study of metalloproteins. J. Inorg. Biochem. 2015;152:82–92. doi: 10.1016/j.jinorgbio.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 61.Were E, Schöne J, Viljoen A, Rasche F. De ovo synthesis of ferrichrome by Fusarium oxysporum f. sp. cubense TR4 in response to iron starvation. Fungal Biol. 2022;126:521–527. doi: 10.1016/j.funbio.2022.05.005. [DOI] [PubMed] [Google Scholar]
- 62.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files). The proteomic datasets generated during and analysed during the current study are available in the MassIVE repository, https://doi.org/doi:10.25345/C5Z892R21. All remaining datasets generated and analysed during the current study are available from the corresponding author on reasonable request.