Graphical abstract
Keywords: Msalais wine, Functional microbes, Aromatic compounds, Furaneol, 5-Methyfurfural
Highlights
-
•
At species level, dominant microbes for Msalais wine differ greatly from that for regular wine.
-
•
Saccharomyces cerevisiae, Kazachstania humilis, Lactobacillus plantarum, and Lactobacillus farraginis are dominant species in Msalais.
-
•
Fungi contribute more to aroma of Msalais than bacteria, with the four species as most important ones.
-
•
The four dominant species all produce the key aromatic compounds of Msaiais, especially furaneol and 5-methylfurfural.
Abstract
Msalais is a traditional wine produced from naturally fermented boiled local grape juice in China. It has characteristic dried fruit and caramel odors, mainly attributed to aromatic compounds, such as furaneol and 5-methylfurfural. However, it is unclear how microbes involved in the natural fermentation of Msalais contribute to this characteristic aroma. Here, we analyzed the Msalais-fermenting microbes and aromatic compounds formed during natural Msalais fermentation by using high-throughput sequencing and gas chromatography–mass spectrometry, respectively. The analysis revealed that Saccharomyces cerevisiae, Kazachstania humilis, Lactobacillus plantarum, and Lactobacillus farraginis are the dominant and key functional species that produce high amounts of furaneol and 5-methylfurfural during Msalais fermentation. Of these, K. humilis and L. farraginis are rarely detected during regular wine fermentation. The identified functional species could be used to control typical aromatic characteristics of Msalais.
Introduction
Currently, developments in omics methodologies are rapidly changing information of wine in level and complexity (Sirén, Mak, Fischer, Hansen & Gilbert, 2019). It has been proven that microbes associated with grapes and wine are a decisive factor influencing wine aroma and consumer’s preferences (Belda et al., 2017). Analyses of the associations between the microbial community and volatile wine components indicate that the characteristic flavor compounds are largely determined by the dominant species during fermentation (Di et al., 2020, Liang et al., 2023). Considering wine flavor, bacterial activity provides fewer sensorially active biochemical conversions than fungi during wine fermentation (Dim, Qinglin, Pangzhen, Chen & Howell,2020). Further, yeasts generate a distinctive aromatic profile that is also attributed to interactions among strains (Lappa, Kachrimanidou, Pateraki, Koulougliotis, Eriotou & Kopsahelis, 2020).
Wine microbiome from specific vinicultural regions produces “terroir wine” with a distinctive aromatic profile. In addition to the geographical location, grape variety, soil, climatic conditions, and agronomical practices, the microbial terroir also depends on metabolic interactions that take place during spontaneous fermentation (Belda et al., 2017, Dim et al., 2020, Lappa et al., 2020). For instance, fungal communities play a principal role in shaping wine aroma profile and its regional distinctiveness(Di et al., 2020). Further, the microbial profile of grapes can be used to predict the composition and abundance of certain wine impact metabolites (Belda et al., 2017), and the soil fungal communities in the vineyard are of primary importance for the wine aroma (Dim et al., 2020).
During wine fermentation, the initial microbial species are specific and unique to the grape juice, several species are found in most musts independent of the grape variety or region of origin, while common species are considered as the core of the wine fermentation ecosystem. These species interact and compete with one another: the species with a higher relative fitness will persist longer, and significantly influence the chemical composition and sensorial features of the final product (Bagheri, Bauer, Cardinali and Setati, 2020). Different types of microbial interactions, e.g., mutualism and commensalism or competition and amensalism, may positively or negatively, accordingly, affect yeast populations. These interactions are intimately linked to yeast metabolic activities that influence the wine analytical profile and shape the wine character (Zilelidou & Nisiotou, 2021).
Msalais is a popular traditional alcohol beverage naturally fermented from boiled grape juice in the A’wati Region in Southern Xinjiang (China), currently the only production region of Msalais. The local grape Hotan Tianhong (Vitis vinifera Hotan Tianhong) is used for its production. Because of the unique production technology and the use of local grape, Msalais has brown–red color and typical aroma, without astringency. Previously, we have fully profiled the aromatic characteristics of Msalais as including strong dried fruit, fruit jam, and fruity odors, intermediate strength caramel and baked odors, and weak floral and herbaceous odors (Li-Xia et al., 2021). We have attributed them to 24 key aromatic compounds with odor activity value ≥ 1 or flavor dilution ≥ 4. Specifically, furaneol, methionol, and 5-methylfurfural (5MF) greatly contribute to the dried fruit, fruit jam, and caramel odors, respectively (Li-Xia et al., 2021).
The technology, grape variety, and viticultural region of Msalais fermentation are completely different from those used in the production of regular wine. Hence, the Msalais wine microflora is greatly different from that of other wines. The indigenous Msalais-fermenting microorganisms have a long, over thousands of years, domestication history of natural fermentation and most likely shape the typical aroma of Msalais. Importantly, during the natural fermentation of Msalais, the fermenting substrate is concentrated grape juice and not fresh grape juice. Our original culture-based studies of Msalais yeasts revealed that, unlike fresh grape juice, which contains a high biomass of greatly variable species of microbes mainly derived from the grape ecosystem, the starting fungal community in concentrated grape juice only comprises a low mass of dominant S. cerevisiae (Li-xia et al., 2012), which must adapt to the new environment of concentrated grape juice. A successful start of natural fermentation is thus more challenging than the start of regular wine fermentation by niche microbes already presents in the grape juice. We also showed that the indigenous yeasts associated with Msalais fermentation have two basic features: high adaptability and excellent enological characteristics (Li-Xia, Guan-Qiong, Ju-Lan, Dong-Qi & Chang-Qing, 2017). However, it is currently unclear how the microbes associated with Msalais fermentation, especially the dominant species, impact the Msalais aroma with its distinctive characteristics. The current knowledge of the relationship between the aroma and microflora of regular wine cannot be applied to Msalais wine.
In the current study, we used high-throughput sequencing and gas chromatography–mass spectrometry (GC–MS) to analyze the dominant functional microbes that contribute to the distinctive aroma of Msalais. The findings inform the development of scientific strategies for natural fermentation of Msalais to better protect this traditional wine from the loss of its typical aroma characteristics during production, which is necessary for its industrialization to meet customer demand.
2. Material and methods
2.1. Standards
Absolute ethanol and dichloromethane (GC-grade) were from Honeywell (Marris Township, NJ, USA). Aromatic compound standards, 4-methyl-2-pentanol (internal standard, purity over 97%), and C7–C24 n-alkanes were all purchased from Sigma-Aldrich (St. Louis, MO, USA).
2.2. Sample collection during natural fermentation of Msalais in a winery
Samples were collected in 2017 from a 2000-L fermentation tank (denoted as “9f” in the current study) at Dolang Msalais Co. Ltd. in the A’wati Region (China). Concentrated grape juice newly transferred to the “9f” tank (day 0) was sampled, as well as the fermentation liquid throughout the natural fermentation process. Samples were collected in triplicate. The mean Brix, pH, and turbidity values of the samples are shown in Table S1. For high-throughput sequencing, 150 mL of sample were centrifuged (6149×g, 15 min, 4 °C) and the pellet retained for analysis. For GC–MS analysis, 50 mL of sample were used.
2.3. Headspace solid-phase microextraction (HS-SPME)–GC–MS
The volatile compounds were determined using HS-SPME–GC–MS (Li-Xia et al., 2021), in triplicate. The retention index of each compound was calculated by analyzing C7–C24 n-alkane data under the same chromatographic conditions. The aromatic compounds were identified by comparing their retention indices with those of reference standards, and comparing the experimental mass spectra with those in the standard NIST 11 MS database (National Institute of Science and Technology, Gaithersburg, MD, USA).
2.4. High-throughput sequencing and analysis
After sample centrifugation (section 2.2), DNA was extracted from the pellet using a rapid DNA extraction kit (BioTeke Corporation, Beijing, China), following the manufacturer’s instructions. Polymerase chain reaction was performed according to the 16S Illumina Amplicon Protocol and ITS Illumina Amplicon Protocol available on the Earth Microbiome website (https://earthmicrobiome.org/). The primers 33815F and 806R, and ITS1 and ITS2 were used to amplify fragments of the bacterial 16S rRNA gene and the fugal internal transcribed spacer (ITS) gene, respectively. The amplification products were purified using a QIAquick gel extraction kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions, and quantified using QuantiFluotTM-ST (Promega, Madison, WI, USA). The purified amplicons were sequenced using an Illumina MiSeq platform (Allwegene, Beijing, China), according to standard protocols. Sequencing quality control was performed by image analysis, base calling, and error estimation using the Illumina Analysis Pipeline v.6 (Illumina, Inc., San Diego, CA, USA). The sequences were clustered into operational taxonomic units (OTUs) at a similarity level of 97% using Uparse software (v7.0). The most frequently occurring sequence in each OTU was used as the representative sequence. Bacterial 16 S rRNA genes referred to the Greengenes database (Release 13.8, http:// greengenes.secondgenome.com/). The sequence of fungal ITS selected the UNITE database (Release 8.0, https://unite.ut.ee/), and adopted QIIME2’s classify-sklearn algorithm (https://github.com/QIIME2/q2-feature-classifier) to annotate the species of each OTU representative sequence.
2.5. Microfermentation experiments
Representative strains, i.e., indigenous strains [A1-4d5 (S. cerevisiae), bjkh (Kazachstania humilis), alf1 (Lactobacillus farraginis), alf2 (L. farraginis), and alp1 (L. plantarum)] and commercial EC1118 strain (S. cerevisiae, LALVIN EC-1118™), were used in microfermentation experiments. Concentrated juice from Hotan Tianhong grape was donated by Dolang Msalais Co. Ltd. For the experiment, 30 mL of the concentrated juice were sterilized at 115 °C (0.1 Mpa) for 15 min in a 50-mL Erlenmeyer flask, cooled to room temperature (approximately 25 °C), and then inoculated with 0.6 mL of an overnight yeast culture in YPD broth(1% yeast extract, 2% peptone, 2% dextrose), pre-grown at 28 °C. The fermentation was allowed to proceed at 28 °C until the residual sugar content stabilized (approximately 15 d). The obtained Msalais was aged at room temperature (20–23 °C) for 115 d. Each strain was tested in triplicate. Bacteria were activated in Man- Rogosa-Sharpe broth(Difco Laboratories, Detroit, MI) by culturing at 32 °C overnight. Bacterial fermentations were performed as the yeast fermentations, except that the fermenting temperature was 32 °C. Freshly concentrated grape juice without microbial inoculation was used as the control sample CK1. The sample was treated in the same manner as the inoculated fermentations, for 115 d, to obtain control sample CK2. After 115 d, furaneol and 5MF were analyzed, as previously described, using LC-20AB Shimadzu Series high-performance liquid chromatography (Shimadzu Technologies, Shanghai, China) (Zhu, Zhang, Liu, Shi & Duan, 2019).
The peak area for each compound was divided by the peak area of 4-methyl-2-amyl alcohol (internal standard, 0.9898 μg/L) to calculate the relative content (). of each compound in a sample was standardized to obtain the corresponding , using the formula:
where and are the minimum and maximum relative contents in the same sample, respectively. Based on the obtained matrix, a heat map of the evolution of volatile compounds whose increased with the increasing fermentation time was constructed in Henm 1.1.0.3.3 (https://hem.biocuckoo.org).
2.6. Calculation
The relative abundance matrix of annotated OTUs at the species level and the matrix of volatile compounds were used in correlation analysis of species and aromatic compounds based on redundancy analysis (RDA) using CANOCO 4.5 (Biometris, Wageningen, Netherlands) (Braak and Smilauer, 2002, Jiang et al., 2015). The evolution of top fungal and bacterial species during Msalais fermentation were plotted, and the relationship between the top fungal and bacterial species and aromatic compound increase during fermentation was analyzed by multiple factor analysis (MFA), using the XLSAT 2019 (Addinsoft 2019, Boston, MA, USA, https://www.xlstat.com). The 5MF and furaneol content was plotted using Originpro 8 (OriginLab Corporation, Northampton, MA, USA).
3. Results
3.1. Fungal succession during Msalais fermentation
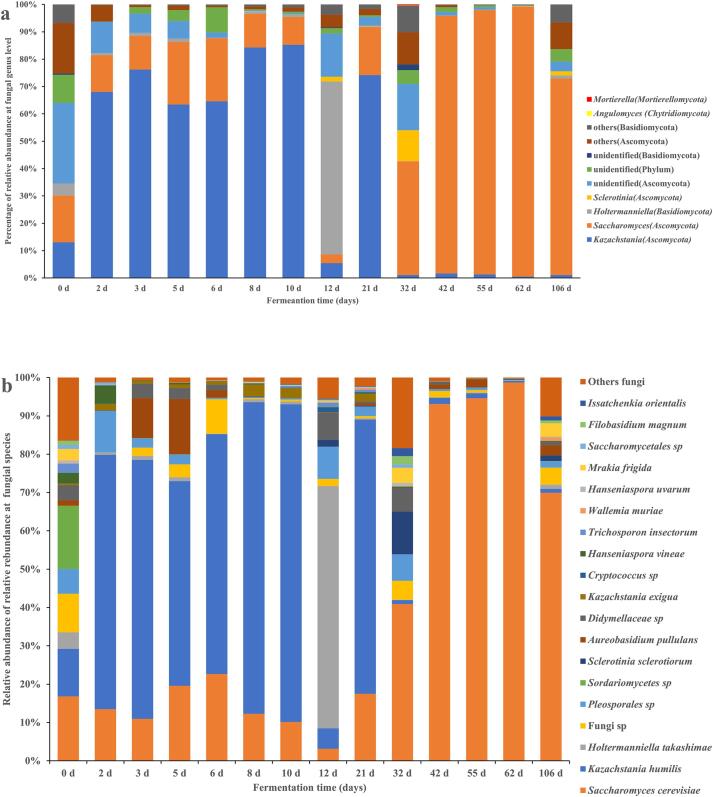
The obtained ITS sequences were annotated to 4 phyla, 92 genera, and 160 species at 97% similarity level (Tables S2-S3). At the phylum level, Ascomycota was dominant during the spontaneous fermentation of Msalais, except on day 12, when Basidiomycota was dominant. The abundance of Ascomycota varied from 3874 to 12,518, and that of Basidiomycota from 21 to 8436. The evolution of the two phyla during fermentation showed contrasting patterns (Table S2). Chytridiomycota and Mortierellomycota were only detected on day 32, with the abundance below 25. Among the 91 annotated genera, we identified 58 Ascomycota genera and 31 Basidiomycota genera. We detected only one Chytridiomycota genus and one Mortierellomycota genus, on day 32 (Table S2). The top fungal genera with an abundance over 1000 were Kazachstania and Saccharomyces. The former was dominant in the first 12 d fermentation, while the latter was dominant after 21 d of fermentation. Holtermanniella (Basidiomycota) was dominant on day 12 (Fig. 1a). The top 19 fungal species with an abundance over 100 and the detection frequency of 6 out of 14 collection time points are shown in Fig. 1b and Table S3. At the beginning of fermentation (day 0), the relative abundance of S. cerevisiae and K. humilis was 16.8% and 12.4%, respectively. During the subsequent fermentation, K. humilis was dominant on days 2–21 (except for day 12), while S. cerevisiae was dominant on days 32–106. S. cerevisiae and K. humilis were detected in all samples. Holtermanniella takashimae was dominant on day 12 (relative abundance of 63.2%). Aureobasidium pullulans was dominant on days 3 and 5, Pleosporales_sp. was dominant on days 2, 12, and 32, and Sclerotinia sclerotiorum was dominant on day 32, all with the abundance of approximately 1000 (Fig. 1b; Table S3).
Fig. 1.
The distribution of top fungal genera (a) and species (b) during natural fermentation of Msalais.
3.2. Bacterial succession during Msalais fermentation
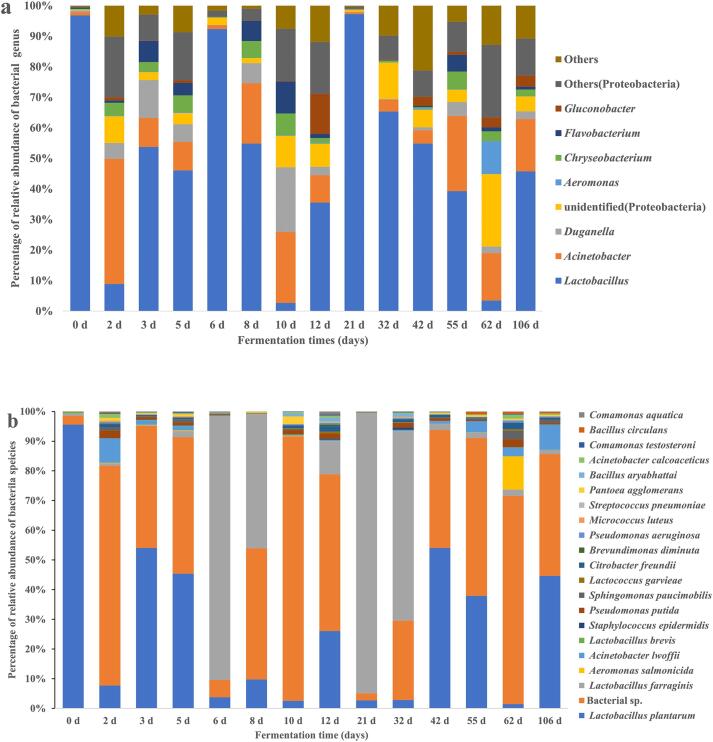
At 97% similarity level, the obtained 16S rRNA sequences were annotated to 11 phyla and 203 genera, but only 48 species (most of them were unidentified at species level, Tables S4–S3). The top 7 bacterial genera with the highest abundance over ≥500 in at least one fermentation sample included Lactobacillus, Acinetobacter, and Duganella (Fig. 2a; Table S4). The top 20 bacterial species with the highest abundance over ≥100 in at least one fermentation sample included L. plantarum and L. farraginis (Fig. 2b; Table S5). L. farraginis and L. plantarum were detected in all fermentation samples. L. plantarum was dominant at the early fermentation stage (days 0–5) and at the mature stage (days 42–106), while L. farraginis was dominant at the middle stage (days 6–32) (Fig. 2b). Some unidentified species were highly abundant on days 2, 10, and 62 (Fig. 2b). Most of them were classed into Acinetobacter and Duganella genera, and Proteobacteria phylum (Fig. 2a; Table S5). A. salmonicida had high relative abundance (11.3%) on day 62. The relative abundance of Acinetobacter lwoffii was high (approximately 8.5%) on days 2 and 106. The remaining 16 species had low relative abundance (<12%).
Fig. 2.
The distribution of fungi genera (a) and species (b) during natural fermentation of Msalais.
3.3. Volatile compounds whose concentration increased during Msalais fermentation
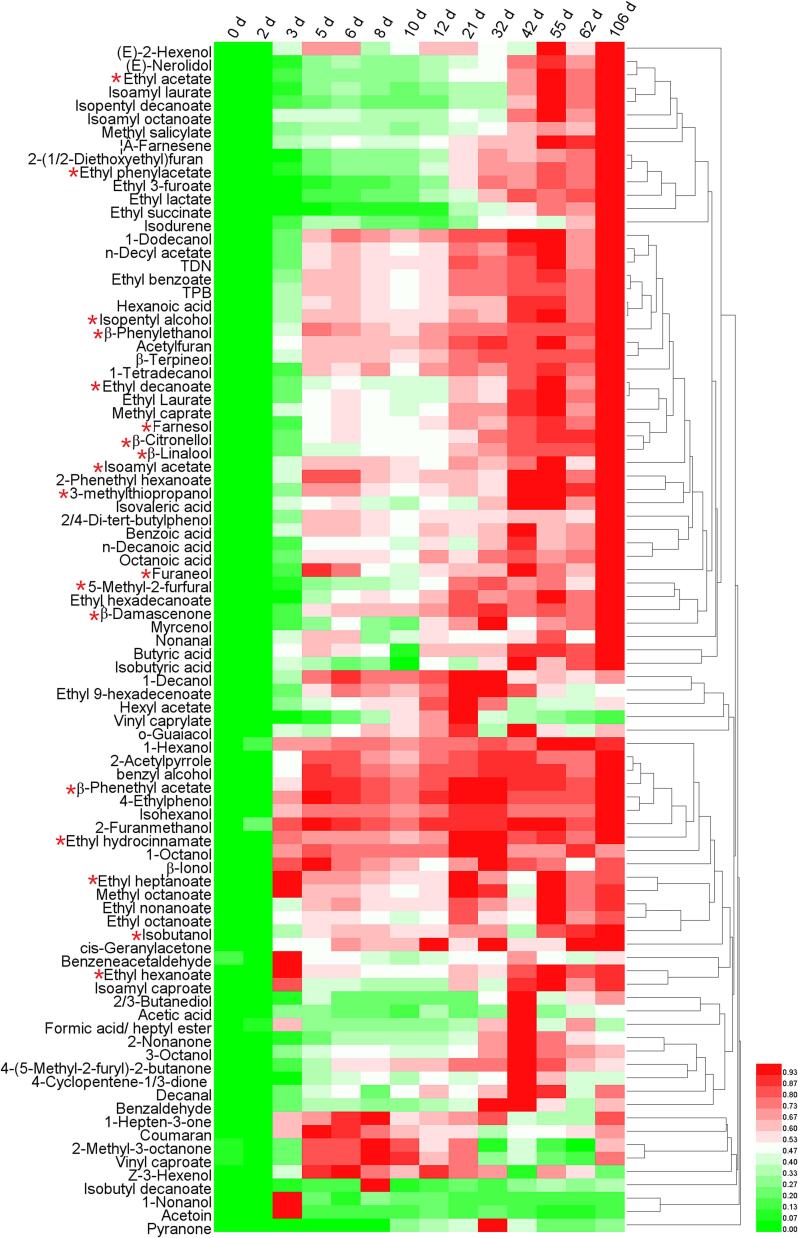
We identified 89 volatile compounds whose relative content increased during Msalais fermentation, compared to that in the concentrated grape juice (day 0 sample) (Fig. 3). These included higher alcohols, esters, terpenes, norisoprene, sulfides, and some furans (Table S6). The levels of most aromatic compounds (alcohols, esters, terpenes, norisoprene, sulfides) continually increased during the 106 d of fermentation. The levels of some furans, e.g., furaneol and 5MF, also increased during Msalais fermentation. The levels of other compounds fluctuated during fermentation, and were high at the middle stage but obviously decreased at the mature stage, e.g., vinyl caprylate, acetic acid, and pyranone. The levels of 18 of 24 key aromatic compounds of Msalais(Li-Xia et al., 2021) increased during Msalais fermentation (Fig. 3; Table S6).
Fig. 3.
Eighty-nine volatile compounds whose relative levels increased during the natural fermentation of Msalais. *, Key aromatic compound of Msalais.
3.4. Analysis of functional species responsible for the characteristic aroma of Msalais
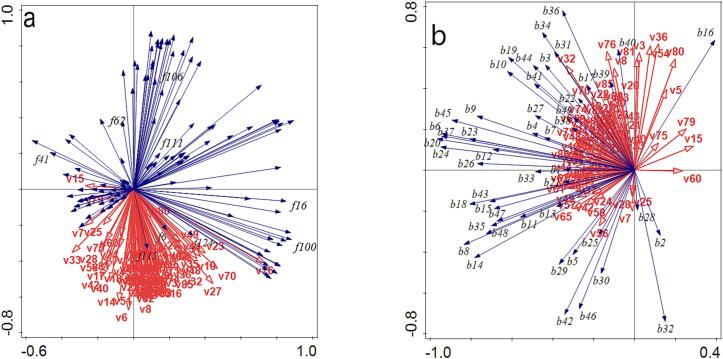
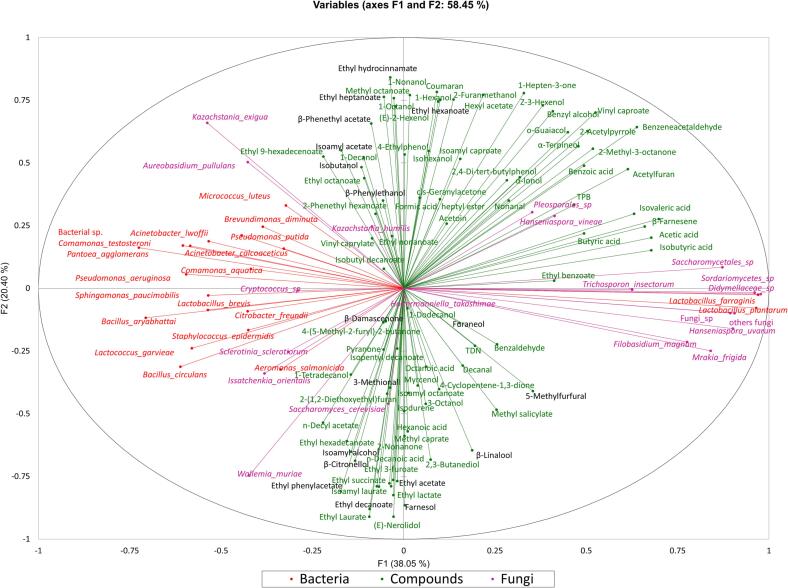
Next, we performed a preliminary identification of functional microbial species that contribute to aromatic compound production in Msalais. Accordingly, we used RDA (Fig. 4a, 4b) to analyze the abundances of the identified 160 fungal species (Table S3) and 48 bacterial species (Table S5), and 89 aromatic compounds whose levels increased during Msalais fermentation(Table S6). S. cerevisiae (f115) clustered with most aromatic compounds, indicating a strong correlation (Fig. 4 a). Although K. humilis (f41) did not cluster with as many aromatic compounds as S. cerevisiae (f115), this species was more closely correlated with most aromatic compounds than A. pullulans (f16), Pleosporales_sp. (f106), Fungi_sp. (f111), and S. sclerotiorum (f62).
Fig. 4.
Correlation analysis of fungi and aroma compounds (a), and bacteria and aroma compounds (b) during natural fermentation of Msalais. Notation: f[number], fungal species[Table S5]; b[number], bacterial species (Table S3); v[number], volatile compound (as in Table S6).
Most bacteria and most aromatic compounds were clustered together, indicating a strong correlation (Fig. 4b). The dominant bacterial species, L. farraginis (b16) and L. plantarum (b32), did not clustered with most aromatic compounds as closely as other bacteria, and hence, their contribution to most aroma compounds was not as strong as that of other bacteria.
Using multiple factor analysis (MFA) (Fig. 5), we next analyzed the correlations between the top microbial species (19 identified fungal species, Table S3, 20 identified bacterial species, Table S5) and 89 compounds during Msalais fermentation (Table S6). The two top fungal species S. cerevisiae and K. humilis clustered with most aromatic compounds. H. takashimae, were positioned close to the center of the MFA plot, which indicated it had somewhat contribution to aroma of Msalais, with the highest relative abundance on day 12 very and low of that during the other Msalais fermentation days. Hanseniaspora vineae and Pleosporales sp. were clustered with some non-key aromatic compounds and, hence, they could contribute to the aromatic characteristics of Msalais. Wallemia muriae, Issatchenkia orientails, Aureobasidium pullulans, and Kazachstania exigua were positioned along the left edge of the aromatic compound distribution in the plot, and hence, they could be closely associated with aromatic compounds located in their vicinity in the plot.
Fig. 5.
Correlation analysis of dominant fungal and bacterial species and aromatic compounds of Msalais.
L. farraginis and L. plantarum were the two top bacterial species clustering with some top fungal species on the right side of the plot. The other top bacterial species clustered together on the left side of the plot, together with the fungus Cyptococcus sp.
Considering the species at the opposite ends of the F1 axis, the ones on the right should contribute to the aromatic compound production more so than the ones on the left, because all (89) aromatic compounds were distributed to the center and right of the F1 axis in the plot.
While S. cerevisiae and K. humilis were also important for key aroma compound production (Fig. 5, black font), L. farraginis and L. plantarum should not be ignored as the dominant bacterial species that contribute to the aromatic characteristics of Msalais. The two key aromatic compounds, furaneol and 5MF, were in the lower-right quadrant of the plot, between S. cerevisiae and the right-hand species cluster including L. plantarum and L. farraginis. Hence, these two key aromatic compounds could be contributed by S. cerevisiae, L. plantarum, and L. farraginis rather than by the other species.
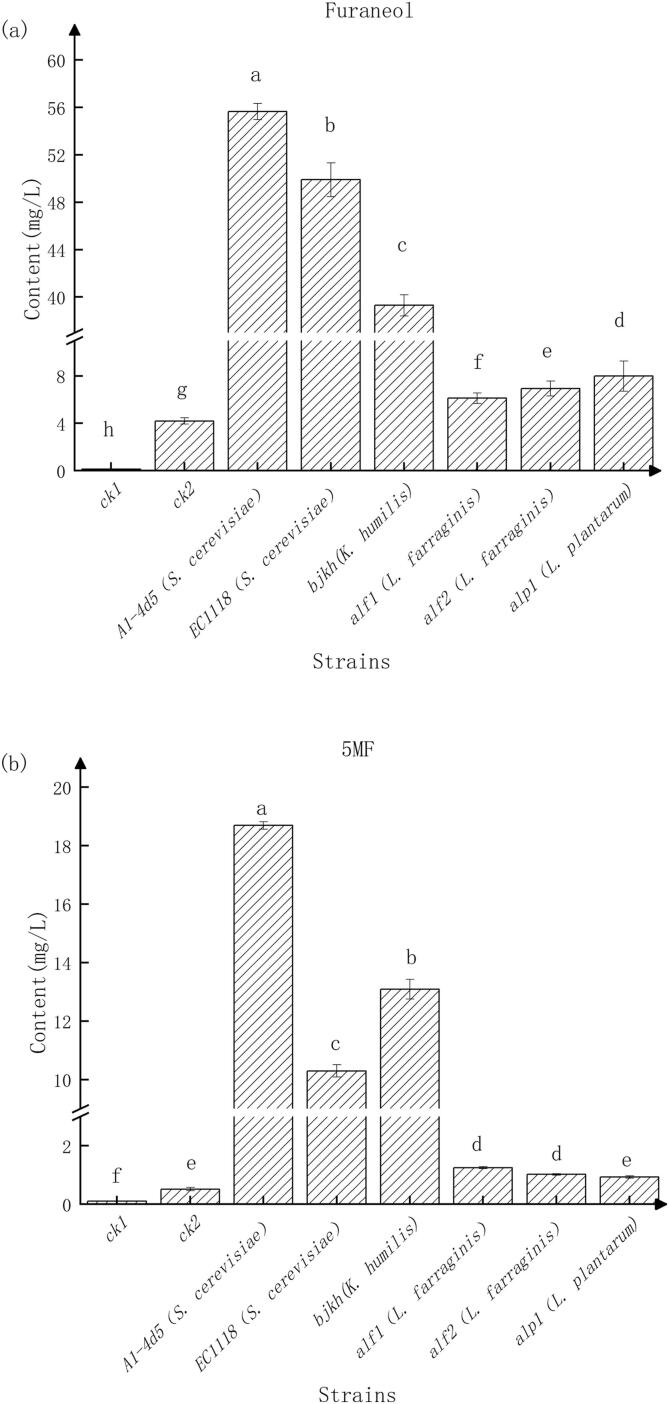
3.5. Functional yeast and bacteria contributing to the caramel aroma
The RDA (Fig. 4a, 4b) and MFA (Fig. 5) revealed that the dominant species are also among the functional microbes that produce furaneol and 5MF, the key compounds responsible for the caramel odor of Msalais. Indeed, Msalais microfermentation experiments with representative strains of the four species confirmed their ability to produce 5MF and furaneol, with the concentrations of these compounds significantly higher in the fermentation samples than in the control samples CK1 and CK2 (Fig. 6). Further, the accumulation of furaneol and 5MF in samples containing the dominant yeast species (S. cerevisiae, A1-4d5 and EC1118; and K. humilis, bjkh) was significantly higher than that in samples with the dominant bacterial species (L. farraginis, alf1 and alf2; and L. plantarum, alp1). Furthermore, the furaneol and 5MF levels in CK2 sample were higher than those in CK1 sample, indicating some non-microbial accumulation of these two compounds, albeit one that was far lower than the microbial-dependent accumulation. S. cerevisiae was the most important functional species during Msalais fermentation: it produced higher quantities of key aromatic compounds (furaneol and 5MF) than the other dominant species, and produced over 70% of aromatic compounds analyzed, i.e., 89 whose concentration increased with fermentation, out of 127 compounds detected during Msalais fermentation (not shown).
Fig. 6.
The furaneol (a) and 5MF content (b) of Msalais fermented by different microbes. The differences between values indicated by different lowercase letters are significant (one-way ANOVA, p < 0.05, Tukey’s test, triplicate per strain; data are presented as the mean ± SD).
4. Discussion
The niche microbes associated with the aromatic characteristics of Msalais have the following main traits: (1) they are the local dominant microbial community involved in natural Msalais fermentation; and (2) they show strong adaptability and have excellent enological characteristics. In the current study, we showed that S. cerevisiae and K. humilis are the dominant fungi, and L. plantarum and L. farraginis are the dominant bacteria during the natural fermentation of Msalais, and that these microbes are important contributors to the caramel odor of Msalais.
In the current study, we showed that L. plantarum, L. farraginis, S. cerevisiae, and K. humilis were the dominant species throughout the entire Msalais fermentation process, which is strikingly different from the microbial community involved in regular wine fermentation, i.e., directly from grape juice. During regular wine fermentation, Aureobacter is the dominant bacterial genus in grape juice, and Lactococcus is the dominant bacterial genus in grape juice and during fermentation (Wei et al., 2018). In one study on the fermentation of Gehenna grape juice, Glueconobacter was dominant throughout the entire fermentation, Hansenula was dominant on day 0 of fermentation, Candida was dominant during the first 10 d of fermentation, while Saccharomyces became dominant after 10 d of fermentation (Portillo & Mas, 2016). In Chardonnay grape juice and during its fermentation, Metschnikowia pulcherrima and Hanseniaspora uvarum are the dominant species (David et al., 2014). Acetic acid bacteria are dominant in grape juice and during fermentation under low SO2, while lactic acid bacteria are dominant in grape juice and its fermentation without SO2 addition. The common bacteria associated wine fermentation are Lactobacillus, Lactococcus, Leuconostoc, and Prococcus (Morgan, Toit & Setati, 2017).
S. cerevisiae is the major dominant species during Msalais fermentation, as determined not only by high-throughput sequencing in the current study, but also by culturing (Li-xia et al., 2012). Regarding Msalais aroma, S. cerevisiae contributes to more than 70% of identified aromatic compounds (89 out of 127) in Msalais, and produces higher amounts of key aromatic compounds, such as furaneol and 5MF, than the other dominant species. In addition, it is highly adaptable, as it grows well on various substrates and under various environmental conditions; it also has excellent enological characteristics, with high β-glucosidase and galacturonidase production (Li-Xia et al., 2017).
In the current study, K. humilis was dominant during Msalais fermentation, with a high production of 5MF and furaneol. K. humilis, also known as Candida humilis and Candida milleri (Tongjie et al., 2018), is a common species involved in the fermentation of floury foods (Gutiérrez et al., 2018, Wittwer et al., 2022). It does not assimilate maltose, and has a stable symbiotic relationship with Lactobacillus, with which it does not compete for carbon and nitrogen nutrients (Wittwer et al., 2022). The CO2 production capacity of K. humilis is greater than that of S. cerevisiae, and the yeast produces 3-methyl-3-butene-1-ol, (E,E)-2,4-decanediol, higher alcohols, esters, acetic acid, butyric acid, octanoic acid, and decanoic acid (Tongjie et al., 2018). Further, it has been detected during the fermentation of cocoa bean (Papalexandratou et al., 2019), soybean paste (Jianxin et al., 2009), cheese (Cardinali et al., 2016), Baijiu (Wang et al., 2019), in fruit alcohol fermentation (Bovo, Nardi, Fontana, Carlot, Giacomini & Corich, 2012; Xavier et al., 2009), and in fermented vegetables (Shang et al., 2022). K. humilis is highly abundant in the yellow liquor during Baijiu fermentation (Lai, Cheng, Lai & Lai, 2019). In fact, it is the most important contributing species to Baijiu, next to Pichia kudriavzevii, with a high yield of 1-propanol, 2-methyl-1-propanol, 2,3-butanediol, and 3-methyl-1-butanol (Liu, Xiong, Wang & Miao, 2017). Further, K. humilis and P. kudriavzevii are frequently isolated during the natural fermentation of orange wine (Hu, Wang, Ji, Liu & Chen, 2018). The active aromatic compounds in orange wine fermented by K. humilis mainly include esters (ethyl caproate, ethyl 3-hydroxyphenylpropionate, isoamyl acetate, and hexyl acetate), higher alcohols (1-pentanol and phenylethanol), and terpenoids (limonene and β-citronellol) (Hu et al., 2018). Finally, K. humilis NRRLY-7245 strain significantly contributes to ketone production during the fermentation of grape juice, malt juice, and apple juice, and to phenol and monoterpene production during the fermentation of grape juice (Gutiérrez et al., 2018).
K. humilis is dominant during the early and middle stages of Baijiu fermentation, while S. cerevisiae is dominant during the later fermentation stage(You, Zhao, Zhou, Tan, Wang & Zheng, 2021). This is similar to the succession pattern of the two species during the fermentation of Msalais observed in the current study. Compared with pure fermentation by S. cerevisiae, a combined fermentation with S. cerevisiae and K. humilis results in a significantly reduced ethanol production, while sequential fermentation with the two species significantly increases the ester content and, at the same time, the content of β-damascenone, during low-temperature fermentation (Ya et al., 2018). In the current study, K. humilis was dominant during the early and middle stages of Msalais fermentation, and its presence was highly correlated with the key aromatic compounds of Msalais. Overall, K. humilis is an important functional species that generates 5MF and furaneol, a new finding in wine aroma research.
Although the contribution of bacteria to the aroma of Msalais is likely smaller than that of yeasts, it should not be ignored. L. plantarum and L. farraginis are dominant during Msalais fermentation, and produce 5MF and furaneol. This constitutes direct evidence that the niche microbial flora contributes to the typical aromatic characteristics of Msalais, and is different from that found in other alcoholic fermented beverages. In the past decade, L. plantarum has become an important industrial starter in the wine industry and plays an important role in wine aroma modification (Natalia et al., 2019). However, the contribution mainly concerns the content of esters, terpenes, benzene, volatile phenolic acids, sulfide, diacetyl, etc., with no previous reports on its effect on 5MF and furaneol. L. plantarum is a good producer of lipase, esterase, acetylesterase, and carboxylesterase (Sestelo, Poza & Villa, 2004); undertakes enzymatic hydrolysis of over-C8 esters (Pérez-Martín, Seseña, Izquierdo, & Palop, 2013); and possesses an enzyme system for the synthesis of ethyl esters (acyl coenzyme A:acyltransferase and reverse esterase) (Costello, Siebert, Solomon & Bartowsky, 2013), β-naphthyl esterase of C2–C10 fatty acids (especially for the conversion of β-naphthyl esterase of butyrate) (Gobbetti, Fox & Stepaniak,1995), and a low-temperature (5 °C) esterase (Esteban-Torres, Mancheño, De Las Rivas, Muñoz, 2014). The levels of other aromatic compounds present in wine, such as butyl acetate, acetaldehyde, caprylic acid, decanoic acid, acetaldehyde, and γ-butyrolactone, are also influenced by L. plantarum (Pozo-bayón et al., 2005). In addition, L. plantarum reduces the alcohol content, and increases the ester content, during the second fermentation of sterilized Pinot Noir wine (Brizuela, Bravo-Ferrada, Pozo-Bayón, Semorile & Elizabeth Tymczyszyn, 2018) Compared to Oenococcus oenolyticus, L. plantarum shows a good malolactic fermentation ability; is characterized by a high leucine arylamidase, α-glucosidase, and β-glucosidase activity during malolactic fermentation; and releases aroma compounds from bonded glycosides, mainly limonene, β-linalool, oxidized linalool, β-myrcene, benzyl alcohol, β-phenylethanol, 1-hexanol, trans-2-hexen-1-ol, etc. (Natalia et al., 2019). L. plantarum also shows a high benzyl alcohol dehydrogenase activity, releasing p-benzyl alcohol, nerol, geraniol, phenylethanol, cinnamyl alcohol, and coniferol (Landete, Rodríguez, Las Rivas & Muñoz, 2008). Finally, it contributes to the phenolic acid, sulfide, diacetyl, and phenol content of wine (Natalia et al., 2019).
L. farraginis has been discovered in 2007 in the lees of sake (Endo & Okada, 2007). Since then, it has been detected during the fermentation of agave (Escalante-Minakata, Blaschek, Barba, Santos & De León-Rodríguez, 2008), but there have been no reports on its involvement in the fermentation of wine prior to the current study. In the current study, we showed that L. plantarum and L. farraginis produce low amounts of characteristic aroma compounds as the dominant bacteria during Msalais fermentation. Based on the published information, L. plantarum may contribute to the accumulation of ester compounds and the release of aroma from bound odorless forms. L. farraginis and other unknown bacteria may contribute to the key aromatic compounds of Msalais to a greater extent than L. plantarum, but this requires further verification.
5. Conclusion
The dominant microbial community during the natural fermentation of Msalais is different from that observed during regular fermentation of wine, with L. plantarum, L. farraginis, S. cerevisiae, and K. humilis as the dominant species. These microbes contribute to the characteristic aroma of Msalais, with S. cerevisiae the most important among them. Furaneol and 5MF, i.e., the typical aromatic compounds attributed to the caramel odor of Msalais, accumulated in S. cerevisiae culture in vitro at a significantly higher level than in L. farraginis, L. plantarum, and L. farraginis cultures. L. farraginis and K. humilis are rarely detected in wine. However, as the dominant species, they produce 5MF and furaneol aroma compounds in Msalais, and the presence of L. farraginis is highly correlated with that of key aromatic compounds of Msalais. The other dominant bacterium, L. plantarum, also produces a small amount of 5MF and furaneol, and could contribute other important aromatic compounds.
In conclusion, we here identified the dominant functional microbes that contribute to the characteristic aroma of Msalais, a unique traditional wine. Identification of the indigenous microbes involved in the natural fermentation of Msalais is a crucial first step toward improving and standardizing Msalais quality by effectively controlling the fermentation not only in small craft breweries but also during large-scale production. The functional dominant species identified herein could inform a new direction of research to control and improve this traditional alcohol beverage, especially its characteristic aroma.
CRediT authorship contribution statement
Li-Xia Zhu: Methodology, Writing – original draft. Hui Wang: Data curation, Software, Visualization. Pei-jie Han: Investigation, Supervision. Yi-Bin Lan: Conceptualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant numbers 32272457, 31660460, 31871777]. The sponsor did not have any role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2023.100778.
Contributor Information
Li-Xia Zhu, Email: 120050068@taru.edu.cn.
Hui Wang, Email: wanghui@taru.edu.cn.
Pei-jie Han, Email: hanpj@im.ac.cn.
Yi-Bin Lan, Email: lanyibin@cau.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
The data that has been used is confidential.
References
- Bagheri B., Bauer F.F., Cardinali G., Setati M.E. Ecological interactions are a primary driver of population dynamics in wine yeast microbiota during fermentation. Scientific Reports. 2020;10(1):1–12. doi: 10.1038/s41598-020-61690-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda I., Ruiz J., Esteban-Fernández A., Navascués E., Marquina D., Santos A., Moreno-Arribas M.V. Microbial contribution to Wine aroma and its intended use for Wine quality improvement. Molecules. 2017;22(2):1–29. doi: 10.3390/molecules22020189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak, C. J. F. ter, & Smilauer, P. (2002). CANOCO Reference Manual and CanoDraw for Windows User’s Guide: Software for Canonical Community Ordination (version 4.5). In www. canoco. com (Microcomputer Power). www.canoco.com. http://edepot.wur.nl/405659.
- Cardinali F., Taccari M., Milanović V., Osimani A., Polverigiani S., Garofalo C.…Aquilanti L. Yeast and mould dynamics in Caciofiore della Sibilla cheese coagulated with an aqueous extract of Carlina acanthifolia All. Yeast. 2016;33(8):403–414. doi: 10.1002/yea.3168. [DOI] [PubMed] [Google Scholar]
- David V., Terrat S., Herzine K., Claisse O., Rousseaux S., Tourdot-Maréchal R.…Alexandre H. High-throughput sequencing of amplicons for monitoring yeast biodiversity in must and during alcoholic fermentation. Journal of Industrial Microbiology and Biotechnology. 2014;41(5):811–821. doi: 10.1007/s10295-014-1427-2. [DOI] [PubMed] [Google Scholar]
- Di L., Legras J.-L., Pangzhen Z., Deli C., Howell K. Diversity and dynamics of fungi during spontaneous fermentations and association with unique aroma profiles in wine. International Journal of Food Microbiology. 2020;108983 doi: 10.1016/j.ijfoodmicro.2020.108983. [DOI] [PubMed] [Google Scholar]
- Dim L., Qinglin C., Pangzhen Z., Chen D., Howell K.S. The fungal microbiome is an important component of vineyard ecosystems and correlates with regional distinctiveness of wine. MSphere. 2020;5(4):1–15. doi: 10.1128/msphere.00534-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A., Okada S. Lactobacillus farraginis sp. nov. and Lactobacillus parafarraginis sp. nov., heterofermentative lactobacilli isolated from a compost of distilled shochu residue. International Journal of Systematic and Evolutionary Microbiology. 2007;57(4):708–712. doi: 10.1099/ijs.0.64618-0. [DOI] [PubMed] [Google Scholar]
- Esteban-Torres M., Mancheño J.M., De Las Rivas B., Muñoz R. Characterization of a cold-active esterase from Lactobacillus plantarum suitable for food fermentations. Journal of Agricultural and Food Chemistry. 2014;62(22):5126–5132. doi: 10.1021/jf501493z. [DOI] [PubMed] [Google Scholar]
- Gobbetti M., Fox P.F., Stepaniak L. Isolation and characterization of a tributyrin esterase from Lactobacillus plantarum 2739. Journal of Dairy Science. 1995;80(12):3099–3106. doi: 10.3168/jds.S0022-0302(97)76280-5. [DOI] [PubMed] [Google Scholar]
- Gutiérrez A., Boekhout T., Gojkovic Z., Katz M. Evaluation of non-Saccharomyces yeasts in the fermentation of wine, beer and cider for the development of new beverages. Journal of the Institute of Brewing. 2018;124(4):389–402. doi: 10.1002/jib.512. [DOI] [Google Scholar]
- Hu L., Wang J., Ji X., Liu R., Chen F. Selection of non- Saccharomyces yeasts for orange wine fermentation based on their enological traits and volatile compounds formation. Journal of Food Science and Technology. 2018 doi: 10.1007/s13197-018-3325-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z.Y., Wang Y.S., Cheng H., Zhang J.D., Fei J. Spatial variation of phytoplankton community structure in Daya Bay, China. Ecotoxicology. 2015;24(7–8):1450–1458. doi: 10.1007/s10646-015-1471-3. [DOI] [PubMed] [Google Scholar]
- Jianxin, Z., Xiaojun, D., Xiaoming, L., Haiqin, C., Jian, T., Hao, Z., & Wei, C. (2009). Original article Changes in microbial community during Chinese traditional soybean paste fermentation. 2526–2530. https://doi.org/10.1111/j.1365-2621.2009.02079.x.
- Lai Y.-T., Cheng K.-C., Lai C.-N., Lai Y.-J. Isolation and identification of aroma producing strain with esterification capacity from yellow water. PLOS ONE. 2019;14(2):e0211356. doi: 10.1371/journal.pone.0211356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landete J.M., Rodríguez H., De Las Rivas B., Muñoz R. Characterization of a benzyl alcohol dehydrogenase from Lactobacillus plantarum WCFS1. Journal of Agricultural and Food Chemistry. 2008;56(12):4497–4503. doi: 10.1021/jf800500v. [DOI] [PubMed] [Google Scholar]
- Lappa I.K., Kachrimanidou V., Pateraki C., Koulougliotis D., Eriotou E., Kopsahelis N. Indigenous yeasts: Emerging trends and challenges in winemaking. Current Opinion in Food Science. 2020;32:133–143. doi: 10.1016/j.cofs.2020.04.004. [DOI] [Google Scholar]
- Li-xia Z., Gong M., Guo D., Christensen M., Xu-jie H., Hong-mei L.…Fen S. Preliminary analysis of yeast communities associated with the spontaneous fermentation of musalais, a traditional alcoholic beverage of Southern Xinjiang, China. South African Journal of Enology and Viticulture. 2012;33(1):95–104. [Google Scholar]
- Li-Xia Z., Guan-Qiong W., Ju-Lan X., Dong-Qi G., Chang-Qing D. Direct stamp of technology or origin on the genotypic and phenotypic variation of indigenous Saccharomyces cerevisiae population in a natural model of boiled grape juice fermentation into traditional Msalais wine in China. FEMS Yeast Research. 2017;17(5):1–32. doi: 10.1093/femsyr/fow108. [DOI] [PubMed] [Google Scholar]
- Li-Xia Z., Meng-Meng Z., Xiao-Feng X., Yi-Bing L., Ying S., Chang-qing D., Rui-Li Z. Aromatic characterization of traditional Chinese wine Msalais by partial least-square regression analysis based on sensory quantitative descriptive and odor active values, aroma extract dilution analysis, and aroma recombination and omission tests. Food Chemistry. 2021;361 doi: 10.1016/j.foodchem.2021.129781. [DOI] [PubMed] [Google Scholar]
- Liang L., Ma Y., Jiang Z., Sam F.E., Peng S., Li M., Wang J. Dynamic analysis of microbial communities and flavor properties in Merlot wines produced from inoculation and spontaneous fermentation. Food Research International. 2023;164(September 2022) doi: 10.1016/j.foodres.2022.112379. [DOI] [PubMed] [Google Scholar]
- Liu, P., Xiong, X., Wang, S., & Miao, L. (2017). Population dynamics and metabolite analysis of yeasts involved in a Chinese miscellaneous-flavor liquor fermentation. 553–565. doi: 10.1007/s13213-017-1286-y.
- Morgan, H. H., Toit, M., & Setati, M. E. (2017). The Grapevine and Wine Microbiome : Insights from High-Throughput Amplicon Sequencing. 8(May). doi: 10.3389/fmicb.2017.00820. [DOI] [PMC free article] [PubMed]
- Natalia B., Tymczyszyn E.E., Semorile L.C., Valdes La Hens D., Delfederico L., Hollmann A., Bravo-Ferrada B. Lactobacillus plantarum as a malolactic starter culture in winemaking: A new (old) player? Electronic Journal of Biotechnology. 2019;38:10–18. doi: 10.1016/j.ejbt.2018.12.002. [DOI] [Google Scholar]
- Papalexandratou Z., Kaasik K., Kauffmann L.V., Skorstengaard A., Bouillon G., Espensen J.L.…Nielsen D.S. Linking cocoa varietals and microbial diversity of Nicaraguan fine cocoa bean fermentations and their impact on final cocoa quality appreciation. International Journal of Food Microbiology. 2019;304(June):106–118. doi: 10.1016/j.ijfoodmicro.2019.05.012. [DOI] [PubMed] [Google Scholar]
- Pérez-Martín F., Seseña S., Izquierdo P.M., Palop M.L. Esterase activity of lactic acid bacteria isolated from malolactic fermentation of red wines. International Journal of Food Microbiology. 2013;163(2–3):153–158. doi: 10.1016/j.ijfoodmicro.2013.02.024. [DOI] [PubMed] [Google Scholar]
- Portillo M. del C., Mas A. Analysis of microbial diversity and dynamics during wine fermentation of Grenache grape variety by high-throughput barcoding sequencing. LWT – Food Science and Technology. 2016;72:317–321. doi: 10.1016/j.lwt.2016.05.009. [DOI] [Google Scholar]
- Pozo-bayón M.Á., Algeria E.G., Polo M.C., Tenorio C., Martin-alvarez P.J., Calvo De La Banda M.T.…Moreno-arribas M.V. Wine volatile and amino acid composition after malolactic fermentation: Effect of Oenococcus oeni and Lactobacillus plantarum Starter cultures. Journal of Agricultural and Food Chemistry. 2005;53:8729–8735. doi: 10.1021/jf050739y. [DOI] [PubMed] [Google Scholar]
- Sestelo, A. B. F., Poza, M., & Villa, T. G. (2004). β-Glucosidase activity in a Lactobacillus plantarum wine strain. 633–637. doi: 10.1023/B:WIBI.0000043195.80695.17.
- Shang Z., Ye Z., Li M., Ren H., Cai S., Hu X., Yi J. Dynamics of microbial communities, flavor, and physicochemical properties of pickled chayote during an industrial-scale natural fermentation: Correlation between microorganisms and metabolites. Food Chemistry. 2022;377(November 2021) doi: 10.1016/j.foodchem.2021.132004. [DOI] [PubMed] [Google Scholar]
- Sirén K., Mak S.S.T., Fischer U., Hansen L.H., Gilbert M.T.P. Multi-omics and potential applications in wine production. Current Opinion in Biotechnology. 2019;56:172–178. doi: 10.1016/j.copbio.2018.11.014. [DOI] [PubMed] [Google Scholar]
- Tongjie L., Yang L., Sadiq F.A., Huanyi Y., Gu J., Lei Y.…Guoqing H. Predominant yeasts in Chinese traditional sourdough and their influence on aroma formation in Chinese steamed bread. Food Chemistry. 2018;242:404–411. doi: 10.1016/j.foodchem.2017.09.081. [DOI] [PubMed] [Google Scholar]
- Wang D., Chen L., Yang F., Wang H., Wang L. Yeasts and their importance to the flavour of traditional Chinese liquor: A review. Journal of the Institute of Brewing. 2019;125(2):214–221. doi: 10.1002/jib.552. [DOI] [Google Scholar]
- Wei Y.J., Wu Y., Yan Y.Z., Zou W., Xue J., Ma W.R.…Wang L.Y. High-throughput sequencing of microbial community diversity in soil, grapes, leaves, grape juice and wine of grapevine from China. PLoS ONE. 2018;13(3):1–17. doi: 10.1371/journal.pone.0193097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer A.E., Sicard D., Howell K.S. Kazachstania humilis. Trends in Microbiology. 2022;xx(xx):10–11. doi: 10.1016/j.tim.2022.05.007. [DOI] [PubMed] [Google Scholar]
- Ya Y., ChangQing D., GuoLiang Y. Effects of mixed fermentation of Candida humilis and Saccharomyces cerevisiae on ethanol content and aroma of wine. Shipin Kexue / Food Science. 2018;39(20):146–154. [Google Scholar]
- You L., Zhao D., Zhou R., Tan Y., Wang T., Zheng J. Distribution and function of dominant yeast species in the fermentation of strong-flavor baijiu. World Journal of Microbiology and Biotechnology. 2021;37(2):1–12. doi: 10.1007/s11274-020-02988-y. [DOI] [PubMed] [Google Scholar]
- Zhu L.-X., Zhang M.-M., Liu Z., Shi Y., Duan C.-Q. Levels of furaneol in Msalais wines: A comprehensive overview of multiple stages and pathways of its formation during Msalais winemaking. Molecules. 2019;24(17):18–21. doi: 10.3390/molecules24173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilelidou E.A., Nisiotou A. Understanding wine through yeast interactions. Microorganisms. 2021;9(8):1–16. doi: 10.3390/microorganisms9081620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.