Abstract
The 16S rRNA sequences of enterococcal species E. faecium, E. faecalis, E. gallinarum, E. casseliflavus/flavescens, E. dispar, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. mundtii, E. avium, E. durans, E. columbae, and E. solitarius are presented herein. These data were utilized to confirm the species identification of two nonmotile E. gallinarum isolates which had been previously phenotypically identified as E. faecium. The implications of this finding are discussed.
We previously reported detection of the vanC-1 vancomycin resistance-associated gene in two enterococcal isolates (isolates 41 and 88), which by phenotypic analysis appeared to be Enterococcus faecium (19). Because the vanC-1 gene is believed to be an intrinsic characteristic of Enterococcus gallinarum, our findings raised the question as to whether the species identification of these two isolates, which was performed by conventional phenotypic methodologies, was erroneous (1, 6, 15, 18). To answer this question, we performed small-subunit rRNA (16S rRNA) sequencing of these isolates as well as a collection of enterococci.
To date, molecular analyses of the 16S rRNA sequences of enterococci have only been partially performed (32). Objective, clean 16S rRNA sequencing data for enterococci are important to determine the relationship of clinically relevant enterococcal species, especially in situations such as that delineated above. Herein we present the 16S rRNA sequences of multiple isolates of the enterococcal species E. faecium, E. faecalis, E. gallinarum, and E. casseliflavus/flavescens and single isolates of E. dispar, E. mundtii, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. avium, E. durans, E. columbae, and E. solitarius and use these findings to demonstrate that isolates 41 and 88 are nonmotile E. gallinarum isolates.
Thirty-four clinical isolates of vancomycin-resistant enterococci (VRE) or vanC gene-carrying enterococci (E. faecium, E. faecalis, E. gallinarum, E. casseliflavus/flavescens, and isolates 41 and 88) as well as one isolate of E. avium and one isolate of E. raffinosus identified by the Mayo Clinic microbiology laboratory were identified as previously reported (19). We previously reported detection of the vanC-1 gene in isolates 41 and 88, which initially had appeared to be E. faecalis and E. faecium, respectively (19). Subsequent analysis suggests that isolate 41, referred to in our previous publication as E. faecalis (19), is a mixture of E. faecium and E. faecalis and that the E. faecium component (nonmotile; arginine, mannitol, arabinose, raffinose, sucrose, and bile esculin positive; and sorbitol, sorbose, tellurite, and pyruvate negative) has the vanC-1 gene (19). Therefore, both isolates 41 and 88 appear phenotypically to be E. faecium.
In addition to the 36 aforementioned isolates, the following American Type Culture Collection strains were studied: E. mundtii ATCC 43186, E. dispar ATCC 51266, E. pseudoavium ATCC 49372, E. sulfureus ATCC 49903, E. malodoratus ATCC 43197, E. cecorum ATCC 43198, E. hirae ATCC 8043, E. saccharolyticus ATCC 43076, E. seriolicida ATCC 49156, E. durans ATCC 59607, E. columbae ATCC 51263, and E. solitarius ATCC 49428.
16S rRNA PCR amplification and sequencing were performed with previously described cycling conditions and primers (10) and previously described PCR mixtures (19, 20). The sequence data were analyzed with Sequencher 3.0 (Gene Codes Corporation, Ann Arbor, Mich.).
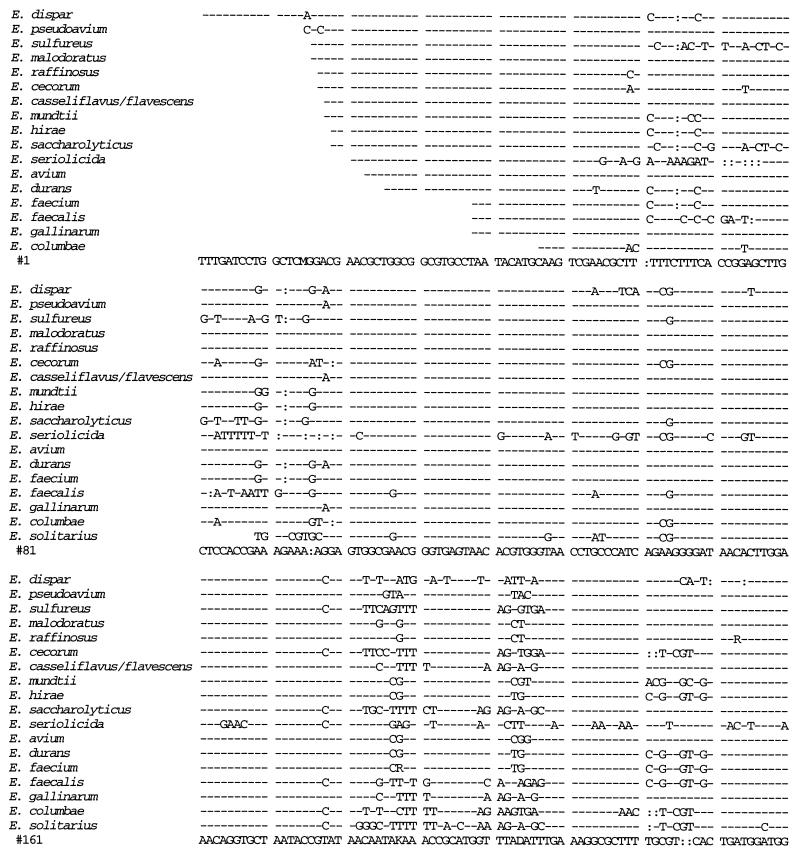
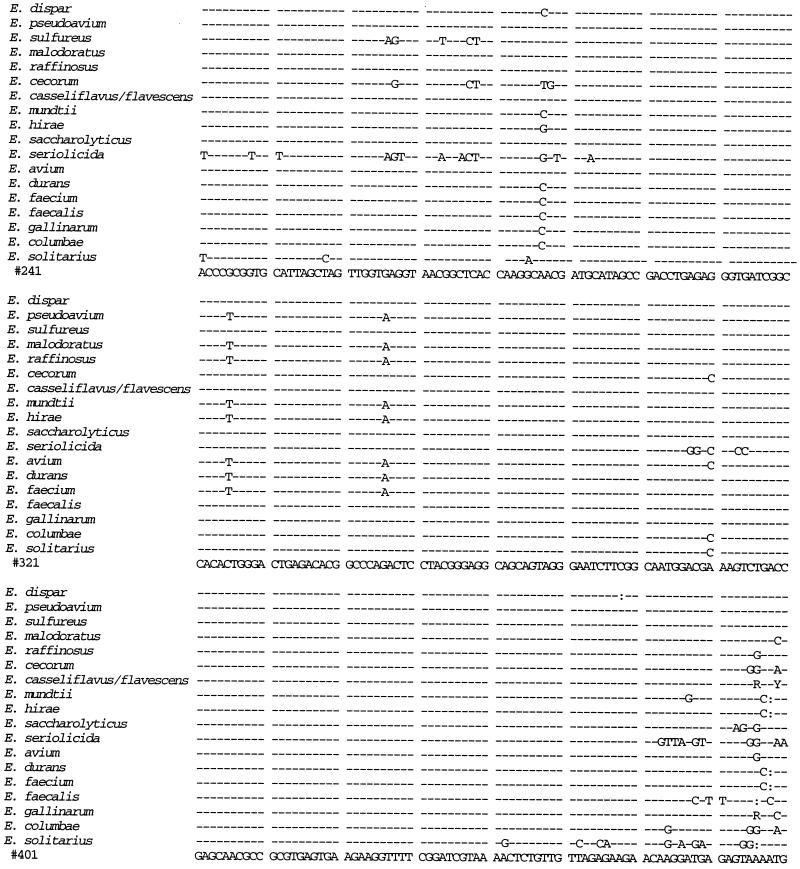
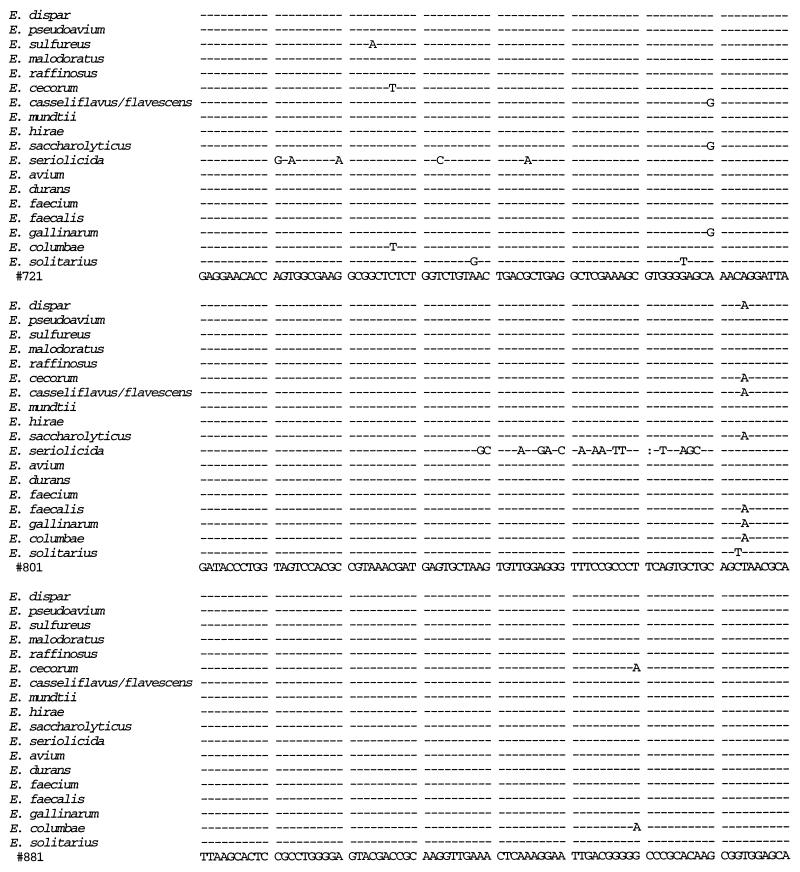
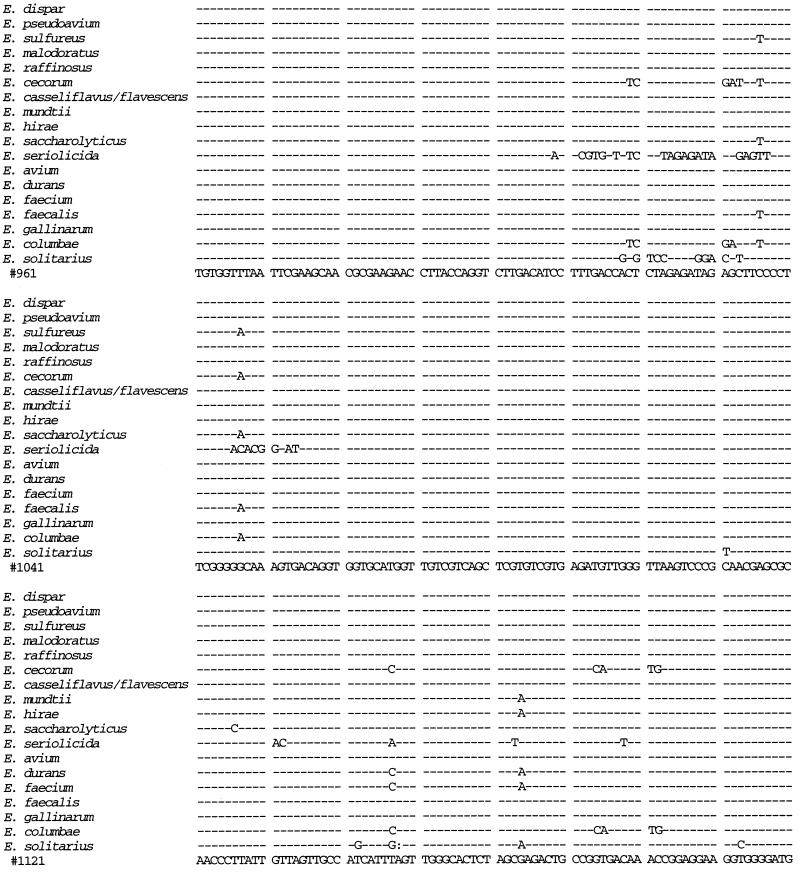
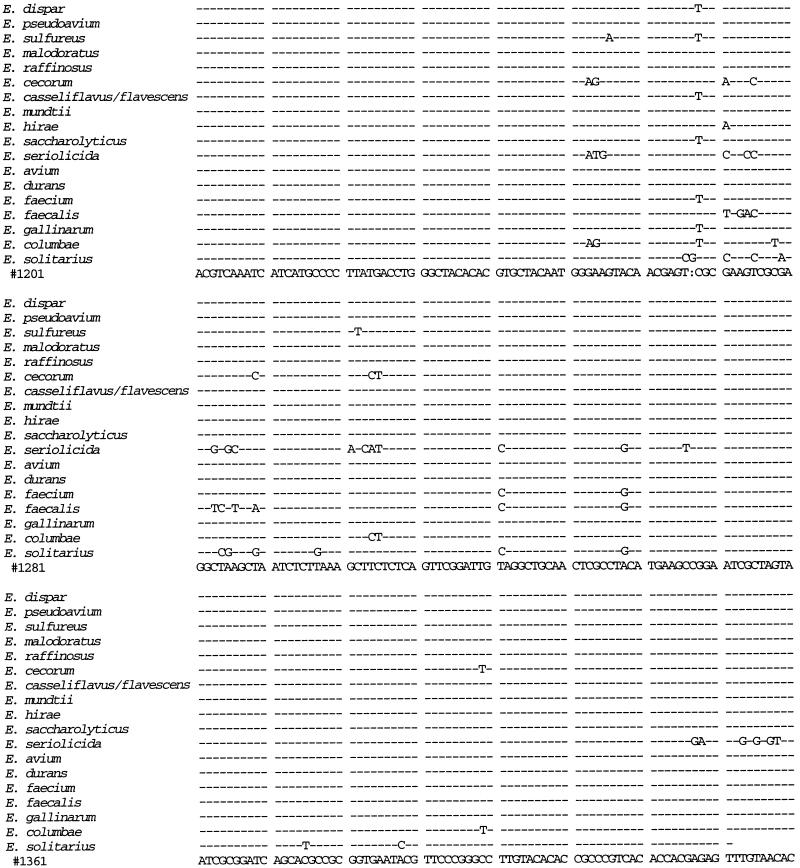
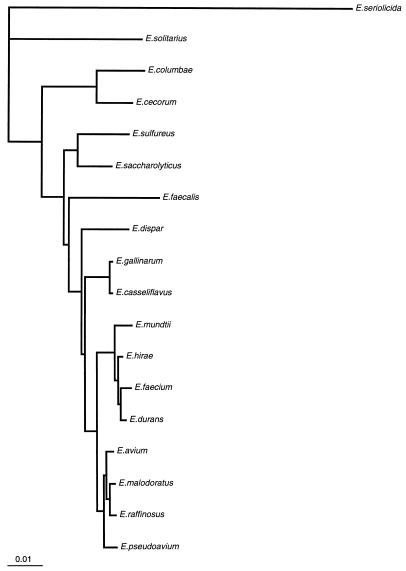
We sequenced the 16S rRNA gene from isolates 41 and 88 and compared the sequences to those from 14 E. faecium isolates, 3 E. faecalis isolates, 10 E. gallinarum isolates, 4 E. casseliflavus isolates, and 1 isolate each of E. flavescens, E. mundtii, E. dispar, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. avium, E. durans, E. columbae, and E. solitarius (Fig. 1). A distance matrix tree constructed from the 16S rRNA gene sequences is presented in Fig. 2, and homology values are presented in Table 1. All three E. faecalis isolates had identical sequences, as did all 14 E. faecium isolates. All 15 of the E. gallinarum, E. casseliflavus, and E. flavescens isolates were identical to each other except as follows. At position 287 (as shown in Fig. 1), the E. casseliflavus and E. flavescens isolates had an adenine and the E. gallinarum isolates had a cytosine. At positions 476 and 479 (as shown in Fig. 1), all of the E. gallinarum isolates except one and one of the E. casseliflavus isolates had a guanosine and a cytosine, one of the E. gallinarum isolates had an adenine and a cytosine, and all of the E. casseliflavus isolates except one and the one E. flavescens isolate had an adenine and a thymidine. Although E. flavescens differs from E. casseliflavus in only one biochemical reaction (21), both possess the vanC-2 gene (5). In three of four isolates of E. casseliflavus, the 16S rRNA sequences were identical to each other and to that of E. flavescens. The 16S rRNA of the fourth E. casseliflavus isolate differed from the rest by 2 bp. This suggests that E. casseliflavus and E. flavescens comprise a single species.
FIG. 1.
16S rRNA sequences of E. faecium, E. faecalis, E. gallinarum, the E. casseliflavus/flavescens group, E. dispar, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. mundtii, E. avium, E. durans, E. columbae, and E. solitarius. Position 99 corresponds to position 100 of the Escherichia coli 16S rRNA gene. R = A or G; Y = C or T.
FIG. 2.
Distance matrix tree of Enterococcus spp. derived from sequence homology determinations of 16S rRNA. The tree was constructed by the neighbor-joining method (23). The tree was rooted by using E. seriolicida as an outgroup. The scale bar represents a 1% difference in nucleotide sequence, as determined by taking the sum of all of the horizontal lines connecting two species.
TABLE 1.
Homology values derived from 16S rRNA sequencesa
| Species | % Homology with:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. solitarius | E. cecorum | E. malodoratus | E. columbae | E. dispar | E. gallinarum | E. hirae | E. mundtii | E. avium | E. casseliflavus | E. pseudoavium | E. sulfureus | E. faecalis | E. raffinosus | E. saccharolyticus | E. durans | E. faecium | |
| E. seriolicida | 86.1 | 88.2 | 88.6 | 88.2 | 88.5 | 88.2 | 88.5 | 88.5 | 88.6 | 88.2 | 88.4 | 88.4 | 87.9 | 88.6 | 88.2 | 88.4 | 88.6 |
| E. solitarius | 93.0 | 93.7 | 93.0 | 93.6 | 94.5 | 93.7 | 93.6 | 93.8 | 94.7 | 93.8 | 93.8 | 94.1 | 93.8 | 94.8 | 93.7 | 93.8 | |
| E. cecorum | 95.9 | 97.9 | 95.8 | 96.6 | 95.8 | 95.7 | 95.9 | 96.6 | 95.8 | 96.8 | 95.4 | 95.9 | 96.6 | 95.8 | 95.7 | ||
| E. malodoratus | 96.0 | 97.8 | 98.7 | 99.4 | 99.3 | 99.5 | 98.8 | 99.4 | 97.3 | 96.7 | 99.8 | 97.8 | 99.3 | 99.1 | |||
| E. columbae | 96.3 | 96.5 | 95.8 | 95.8 | 96.0 | 96.4 | 95.7 | 95.5 | 95.2 | 96.0 | 96.7 | 95.9 | 95.8 | ||||
| E. dispar | 97.9 | 97.7 | 97.6 | 97.6 | 97.9 | 97.7 | 96.6 | 96.9 | 98.0 | 97.7 | 97.7 | 97.7 | |||||
| E. gallinarum | 98.5 | 98.4 | 98.4 | 99.9 | 98.4 | 97.6 | 97.3 | 98.6 | 98.8 | 98.5 | 98.4 | ||||||
| E. hirae | 99.5 | 99.5 | 98.5 | 99.3 | 97.1 | 97.0 | 99.5 | 97.7 | 99.8 | 99.7 | |||||||
| E. mundtii | 99.5 | 98.4 | 99.1 | 97.1 | 96.9 | 99.4 | 97.6 | 99.5 | 99.4 | ||||||||
| E. avium | 98.5 | 99.5 | 97.3 | 96.6 | 99.7 | 97.7 | 99.4 | 99.2 | |||||||||
| E. casseliflavus | 98.6 | 97.7 | 97.3 | 98.8 | 99.0 | 98.4 | 98.3 | ||||||||||
| E. pseudoavium | 97.3 | 97.0 | 99.5 | 97.9 | 99.2 | 99.1 | |||||||||||
| E. sulfureus | 96.4 | 97.3 | 97.9 | 97.0 | 96.9 | ||||||||||||
| E. faecalis | 96.8 | 97.4 | 97.0 | 97.3 | |||||||||||||
| E. raffinosus | 97.9 | 99.4 | 99.2 | ||||||||||||||
| E. saccharolyticus | 97.7 | 97.5 | |||||||||||||||
| E. durans | 99.8 | ||||||||||||||||
Values were based on the data shown in Fig. 1. A total of 1,322 nucleotides were used for this comparison.
Isolates 41 and 88 had 16S rRNA sequences identical to those of E. gallinarum (19). These isolates were previously misidentified as E. faecium because they were not motile, despite being raffinose positive. The vancomycin MICs for these two isolates were 4 and 8 μg/ml, respectively. This finding has important implications for the clinical microbiology laboratory. E. gallinarum is pathogenic for humans (21, 22); in a recent study, enterococci with vanC-associated vancomycin resistance were isolated from nonstool specimens of 9 of 538 patients (1.7%), including two patients with bacteremia who subsequently died (27). Furthermore, vancomycin treatment failure has been associated with intrinsic low-level vancomycin resistance both in humans and in animal models of experimental endocarditis (8, 12, 15). There therefore exists a need to accurately and quickly differentiate enterococci with vanC-associated vancomycin resistance from vancomycin-susceptible enterococci. On the other hand, it has been suggested that vanC-associated vancomycin resistance is not a concern for infection control, because no nosocomial transmission of the involved organisms has been reported (27–29). Costly and cumbersome infection control precautions, such as those recommended for enterococci with vanA- and vanB-associated vancomycin resistance, may therefore not be necessary in the management of patients with vanC enterococci (9, 11, 13, 16). There also exists a need to accurately and quickly differentiate enterococci with vanC-associated vancomycin resistance from VanA or VanB VRE. Stool screening for VRE commonly involves the use of media supplemented with vancomycin at concentrations as low as 6 μg/ml and may also not accurately differentiate E. gallinarum from VanA or VanB VRE or from vancomycin-susceptible enterococci (14, 24, 28).
Unfortunately, both conventional species identification, including commercial test systems, and vancomycin susceptibility tests, including the classical disk diffusion method, are unreliable at detecting E. gallinarum (and E. casseliflavus/flavescens) (25, 26, 31). E. gallinarum can be difficult to differentiate from other enterococci, particularly E. faecium, with commercial biochemical test systems, which may not even include this organism in their databases (1). The motility test is not totally reliable for E. gallinarum (or E. casseliflavus) (4, 18, 27, 30). The MIC breakpoint of vancomycin for one of our alleged vanC-1 E. faecium isolates was low (4 μg/ml), which would have been categorized as susceptible according to current guidelines (17, 18). Recently, the use of a methyl-α-d-glucopyranoside reagent has shown promising results and may prove valuable in the species identification of E. gallinarum; our isolates 41 and 88 produced acid from methyl-α-d-glucopyranoside (2). As an alternative, 16S rRNA sequencing may be helpful in selected situations.
Finally, our findings regarding the 16S rRNA sequences of E. seriolicida (11.4 to 11.8% different from the other Enterococcus spp.) and E. solitarius (5.2 to 7.0% different from the other Enterococcus spp.) are consistent with the findings of previous investigators, who have suggested that these proposed species should not be included in the genus Enterococcus (3, 7, 32). E. seriolicida has been renamed as Lactococcus garvieae (6a).
In summary, we have reported the 16S rRNA sequences of E. faecium, E. faecalis, E. gallinarum, E. casseliflavus/flavescens, E. dispar, E. mundtii, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. avium, E. durans, E. columbae, and E. solitarius and used these sequences to confirm the species identification of two nonmotile E. gallinarum isolates.
Nucleotide sequence accession number.
Representative nucleotide sequences have been submitted to GenBank and have accession no. AF039898 to AF039903 and AF061000 to AF061013.
REFERENCES
- 1.Cartwright C P, Stock F, Fahle G A, Gill V J. Comparison of pigment production and motility tests with PCR for reliable identification of intrinsically vancomycin-resistant enterococci. J Clin Microbiol. 1995;33:1931–1933. doi: 10.1128/jcm.33.7.1931-1933.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Devriese L A, Pot B, Kersters K, Lauwers S, Haesebrouck F. Acidification of methyl-α-d-glucopyranoside: a useful test to differentiate Enterococcus casseliflavus and Enterococcus gallinarum from Enterococcus faecium species group and from Enterococcus faecalis. J Clin Microbiol. 1996;34:2607–2608. doi: 10.1128/jcm.34.10.2607-2608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domenech A, Prieta J, Fernandez-Garayzabal J F, Collins M D, Jones D, Dominguez L. Phenotypic and phylogenetic evidence for a close relationship between Lactococcus garvieae and Enterococcus seriolicida. Microbiologia. 1993;9:63–68. [PubMed] [Google Scholar]
- 4.Donabedian S, Chow J W, Shlaes D M, Green M, Zervos M J. DNA hybridization and contour-clamped homogeneous electric field electrophoresis for identification of enterococci to the species level. J Clin Microbiol. 1995;33:141–145. doi: 10.1128/jcm.33.1.141-145.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutka-Malen S, Molinas C, Arthur M, Courvalin P. Sequence of the vanC gene of Enterococcus gallinarum BM4174 encoding a D-alanine:D-alanine ligase-related protein necessary for vancomycin resistance. Gene. 1992;112:53–58. doi: 10.1016/0378-1119(92)90302-6. [DOI] [PubMed] [Google Scholar]
- 6a.Eldar A, et al. Enterococcus seriolicida is a junior synonym of Lactococcus garvieae, a causative agent of septicemia and meningoencephalitis in fish. Curr Microbiol. 1996;32:85–88. doi: 10.1007/s002849900015. [DOI] [PubMed] [Google Scholar]
- 7.Elliott J A, Collins M D, Pigott N E, Facklam R R. Differentiation of Lactococcus lactis and Lactococcus garvieae from humans by comparison of whole-cell protein patterns. J Clin Microbiol. 1991;29:2731–2734. doi: 10.1128/jcm.29.12.2731-2734.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fantin B, Leclercq R, Arthur M, Duval J, Carbon C. Influence of low-level resistance to vancomycin on efficacy of teicoplanin and vancomycin for treatment of experimental endocarditis due to Enterococcus faecium. Antimicrob Agents Chemother. 1991;35:1570–1575. doi: 10.1128/aac.35.8.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handwerger S, Raucher B, Altarac D, Monka J, Marchione S, Singh K V, Murray B E, Wolff J, Walters B. Nosocomial outbreak due to Enterococcus faecium highly resistant to vancomycin, penicillin, and gentamicin. Clin Infect Dis. 1993;16:750–755. doi: 10.1093/clind/16.6.750. [DOI] [PubMed] [Google Scholar]
- 10.Johnson J L. Similarity analysis of rRNAs. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 683–700. [Google Scholar]
- 11.Jordens J Z, Bates J, Griffiths D T. Faecal carriage and nosocomial spread of vancomycin-resistant Enterococcus faecium. J Antimicrob Chemother. 1994;34:515–528. doi: 10.1093/jac/34.4.515. [DOI] [PubMed] [Google Scholar]
- 12.Kaplan A H, Gilligan P H, Facklam R R. Recovery of resistant enterococci during vancomycin prophylaxis. J Clin Microbiol. 1988;26:1216–1218. doi: 10.1128/jcm.26.6.1216-1218.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karanfil L V, Murphy M, Josephson A, Gaynes R, Mandel L, Hill B C. A cluster of vancomycin-resistant Enterococcus faecium in an intensive care unit. Infect Control Hosp Epidemiol. 1992;13:195–200. doi: 10.1086/646509. [DOI] [PubMed] [Google Scholar]
- 14.Kohner P C, Patel R, Uhl J R, Garin K M, Hopkins M K, Wegener L T, Cockerill F R., III Comparison of agar dilution, broth microdilution, E-test, disk diffusion, and automated Vitek methods for testing susceptibilities of Enterococcus spp. to vancomycin. J Clin Microbiol. 1997;35:3258–3263. doi: 10.1128/jcm.35.12.3258-3263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leclercq R, Dutka-Malen S, Duval J, Courvalin P. Vancomycin resistance gene vanC is specific to Enterococcus gallinarum. Antimicrob Agents Chemother. 1992;36:2005–2008. doi: 10.1128/aac.36.9.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montecalvo M A, Horowitz H, Gedris C, Carbonaro C, Tenover F C, Issah A, Cook P, Wormser G P. Outbreak of vancomycin-, ampicillin-, and aminoglycoside-resistant Enterococcus faecium bacteremia in an adult oncology unit. Antimicrob Agents Chemother. 1994;38:1363–1367. doi: 10.1128/aac.38.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing: eighth informational supplement M100-S8. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 18.Navarro F, Courvalin P. Analysis of genes encoding d-alanine–d-alanine ligase-related enzymes in Enterococcus casseliflavus and Enterococcus flavescens. Antimicrob Agents Chemother. 1994;38:1788–1793. doi: 10.1128/aac.38.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel R, Uhl J R, Kohner P, Hopkins M K, Cockerill F R., III Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J Clin Microbiol. 1997;35:703–707. doi: 10.1128/jcm.35.3.703-707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel R, Uhl J R, Kohner P, Hopkins M K, Steckelberg J M, Kline B, Cockerill F R., III DNA sequence variation within vanA, vanB, vanC-1, and vanC-2/3 genes of clinical Enterococcus isolates. Antimicrob Agents Chemother. 1998;42:202–205. doi: 10.1128/aac.42.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pompei R, Lampis G, Berlutti F, Thaller M C. Characterization of yellow-pigmented enterococci from severe human infections. J Clin Microbiol. 1991;29:2884–2886. doi: 10.1128/jcm.29.12.2884-2886.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruoff K L, de la Maza L, Murtagh M J, Spargo J D, Ferraro M J. Species identities of enterococci isolated from clinical specimens. J Clin Microbiol. 1990;28:435–437. doi: 10.1128/jcm.28.3.435-437.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Swenson J M, Clark N C, Ferraro M J, Sahm D F, Doern G, Pfaller M A, Reller L B, Weinstein M P, Zabransky R J, Tenover F C. Development of a standardized screening method for detection of vancomycin-resistant enterococci. J Clin Microbiol. 1994;32:1700–1704. doi: 10.1128/jcm.32.7.1700-1704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swenson J M, Ferraro M J, Sahm D F, Charache P, Tenover F C The National Committee for Clinical Laboratory Standards Working Group on Enterococci. New vancomycin disk diffusion breakpoints for enterococci. J Clin Microbiol. 1992;30:2525–2528. doi: 10.1128/jcm.30.10.2525-2528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover F C, Tokars J, Swenson J, Paul S, Spitalny K, Jarvis W. Ability of clinical laboratories to detect antimicrobial agent-resistant enterococci. J Clin Microbiol. 1993;31:1695–1699. doi: 10.1128/jcm.31.7.1695-1699.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toye B, Shymanski J, Bobrowska M, Woods W, Ramotar K. Clinical and epidemiologic significance of enterococci intrinsically resistant to vancomycin (possessing the vanC genotype) J Clin Microbiol. 1997;35:3166–3170. doi: 10.1128/jcm.35.12.3166-3170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Horn K G, Gedris C A, Rodney K M. Selective isolation of vancomycin-resistant enterococci. J Clin Microbiol. 1996;34:924–927. doi: 10.1128/jcm.34.4.924-927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Horn K G, Gedris C A, Rodney K M, Mitchell J B. Evaluation of commercial vancomycin agar screen plates for detection of vancomycin-resistant enterococci. J Clin Microbiol. 1996;34:2042–2044. doi: 10.1128/jcm.34.8.2042-2044.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vincent S, Knight R G, Green M, Sahm D F, Shlaes D M. Vancomycin susceptibility and identification of motile enterococci. J Clin Microbiol. 1991;29:2335–2337. doi: 10.1128/jcm.29.10.2335-2337.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willey B M, Kreiswirth B N, Simor A E, Faur Y, Patel M, Williams G, Low D E. Identification and characterization of multiple species of vancomycin-resistant enterococci, including an evaluation of Vitek software version 7.1. J Clin Microbiol. 1993;31:2777–2779. doi: 10.1128/jcm.31.10.2777-2779.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams A, Rodrigues U, Collins M. Intrageneric relationships of Enterococci as determined by reverse transcriptase sequencing of small-subunit rRNA. Res Microbiol. 1991;142:67–74. doi: 10.1016/0923-2508(91)90098-u. [DOI] [PubMed] [Google Scholar]