Abstract
Human Tapasin (hTapasin) is the main chaperone of MHC-I molecules, enabling peptide loading and antigen repertoire optimization across HLA allotypes. However, it is restricted to the endoplasmic reticulum (ER) lumen as part of the protein loading complex (PLC), and therefore is highly unstable when expressed in recombinant form. Additional stabilizing co-factors such as ERp57 are required to catalyze peptide exchange in vitro, limiting uses for the generation of pMHC-I molecules of desired antigen specificities. Here, we show that the chicken Tapasin (chTapasin) ortholog can be expressed recombinantly at high yields in a stable form, independent of co-chaperones. chTapasin can bind the human HLA-B∗37:01 with low micromolar-range affinity to form a stable tertiary complex. Biophysical characterization by methyl-based NMR methods reveals that chTapasin recognizes a conserved β2m epitope on HLA-B∗37:01, consistent with previously solved X-ray structures of hTapasin. Finally, we provide evidence that the B∗37:01/chTapasin complex is peptide-receptive and can be dissociated upon binding of high-affinity peptides. Our results highlight the use of chTapasin as a stable scaffold for protein engineering applications aiming to expand the ligand exchange function on human MHC-I and MHC-like molecules.
Keywords: major histocompatibility complex (MHC), Tapasin, molecular chaperone, protein ortholog, antigen processing and presentation
The display of peptide-loaded major histocompatibility complex class I (pMHC-I) molecules on the cell surface is crucial for T cell and natural killer (NK) cell-mediated immunity. The loading of peptides onto MHC-I within cells relies heavily on the activity of the protein loading complex (PLC), which comprises several molecules including the transporter associated with antigen processing (TAP), Tapasin, calreticulin, ERp57, and the MHC-I-β2m heterodimer (1, 2). The intracellular enzymes endoplasmic reticulum aminopeptidase 1 and 2 (ERAP1/2) optimize the peptide cargos for MHC-I presentation, playing an indirect role in the modulation of adaptive immune responses (3, 4, 5). Tapasin is a transmembrane glycoprotein that acts as a bridge between the TAP transporter and MHC-I to facilitate peptide loading. Its significance in antigen presentation is evident in Tapasin-depleted cells by the reduced levels of surface MHC-I, which affects antigen presentation, CD8+ T cell development, and antiviral immunity (6, 7). Additionally, many tumors evade immune surveillance by targeting components of the PLC, including Tapasin. The available crystal structures of Tapasin–ERp57 complex (8) and a high-resolution structure of Tapasin bound to B∗44:05 (9) have proposed a model where Tapasin interacts with the peptide binding domain of MHC-I via the α2-1 helix and the β-sheet floor, while the membrane-proximal domain engages the MHC-I via the α3 domain and β2m (9). However, understanding the mechanism by which Tapasin performs its peptide loading function has been challenging due to the dynamic flexibility of both the chaperone and the MHC-I molecule. Studies focusing on the Tapasin homolog, TAPBPR, which is not a component of the PLC have provided insights into Tapasin function (10, 11, 12). TAPBPR functions both as a chaperone and a peptide-editor with distinct allelic specificity (13). We have recently characterized a chicken ortholog of TAPBPR (chTAPBPR) that interacts with a broader repertoire of HLAs (human leukocyte antigens; the human MHCs) and has a strong preference for empty HLA-I molecules (14). However, further exploration of Tapasin/MHC-I interactions is necessary to understand chaperone dependencies among allelic variations in HLA molecules.
In this work, we aim to expand the function and applications of chaperones to classical, non-classical, and MHC-like molecules. As a first step, we characterize an ortholog of Tapasin from chicken (Gallus gallus) that has co-evolved with MHC-I to effectively promote peptide-loading (15), and show that it can be stably expressed in recombinant form without the need of co-chaperones. We further identify xeno reactivity with a panel of HLA allotypes using single antigen beads (SABs), and define low micromolar interactions with the disease-relevant HLA-B∗37:01 allotype (16, 17). Using biophysical assays and solution NMR, we show that a stabilized B∗37:01/chTapasin complex is peptide-receptive. Our results highlight the use of chicken Tapasin as a standalone molecular chaperone to stabilize empty, receptive HLA molecules, toward a range of in vitro and in cell applications (18, 19, 20, 21, 22).
Results
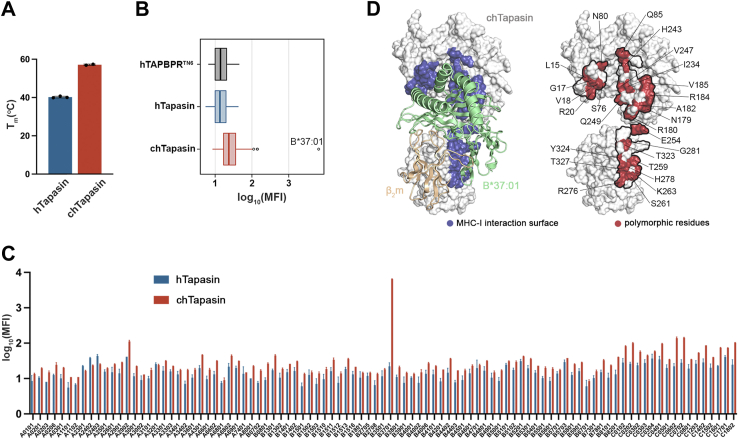
To characterize differences between human Tapasin (hTapasin) and its ortholog from chicken (chTapasin), we expressed both proteins recombinantly in Drosophila S2 cells and purified them by size exclusion chromatography (SEC) (Fig. S1A) (23). Notably, the protein yields from equal volumes of culture were significantly higher for chTapasin (retention time ∼80 min), which also showed enhanced thermal stability with a melting temperature (Tm) of 57.1 °C compared to 40.2 °C for hTapasin, as measured by differential scanning fluorimetry (DSF) (Figs. 1A and S1B). Hence, the observed high-molecular-weight protein aggregates that elute in void volume (retention time 40–50 min) (Fig. S1A) are due to the increased total amount of protein loaded on the column and do not imply a pronounced aggregation of chTapasin. To thoroughly evaluate interactions with different HLA-I allotypes in a high-throughput manner, we created multivalent chaperone tetramers (24) and incubated them with single antigen beads (SABs), as previously described (14, 25). The beads are coated with 97 different HLA-I molecules that are loaded with a variety of peptides derived from EBV-transfected cell lines (26, 27). Using the monoclonal, pan-HLA class I antibody W6/32, we demonstrated comparable mean fluorescence intensity (MFI) levels for all allotypes (Fig. S2A). As a negative control, we used a construct of the homologous chaperone TAPBPR carrying substitutions on important residues that have been shown to disrupt interactions with HLA-A∗02:01 (hTAPBPRTN6: E205L, R207E, Q209S, and Q272S) (23, 28). To evaluate a threshold for statistical significance, we calculated z-scores corresponding to all HLA allotypes on the SABs and identified HLA-C∗17:01 as the point with the highest z-score (+4.18). As expected, staining with hTapasin tetramers revealed low MFI levels of binding, similar to hTAPBPRTN6, indicating the lack of interactions across the tested HLA panel in the absence of co-chaperones. In contrast, chTapasin showed a systematic shift towards higher MFI levels compared to both hTAPBPRTN6 and hTapasin, and a clear interaction with HLA-B∗37:01 (Figs. 1, B and C and S2B). While we cannot distinguish any other allele with significant binding to chTapasin based on the SAB experiments alone (z-score values lower than 4.18), this is still possible to occur for other HLAs in their peptide-deficient form which warrants a more detailed biochemical evaluation, to be performed in future studies.
Figure 1.
The chicken Tapasin ortholog shows enhanced stability and distinct interactions with HLA allotypes.A, comparison of the thermal stabilities of human versus chicken Tapasin by differential scanning fluorimetry (DSF) experiments. Tm, melting temperature in degrees Celsius. The plotted data represent triplicate assays (n = 3). B, a Box plot of the distribution of human and chicken Tapasin MFI levels for 97 different MHC-I allotypes. Human TAPBPR carrying the substitutions E205L, R207E, Q209S, and Q272S (hTAPBPRTN6) was used as a negative control. The left boundary of the box represents the 25th percentile, the line within the box represents the median, and the right boundary of the box represents the 75th percentile. Whiskers extending above and below the box indicate the 10th and 90th percentiles, respectively. Any points appearing above the whiskers represent outliers that fall beyond the 90th percentile. Plotted data are mean from three replicates (n= 3). C, bar graph showing the binding levels of tetramerized hTapasin and chTapasin to 97 different HLA allotypes on the SABs plotted in (B). Data are mean ± SD of n = 3 independent experiments. D, surface representation of the chTapasin bound to HLA-B∗37:01 structure model generated using the BAKER-ROBETTA server (29). The predicted contact surfaces of chTapasin with the heavy chain of B∗37:01 are highlighted in blue (left). Surface representation of chTapasin where the polymorphic residues within the contact surfaces (denoted by the black line) are marked in red (right).
We next examined to what extent these HLA interaction trends can be determined by sequence divergence between hTapasin and chTapasin. For this purpose, we generated a model of chTapasin in complex with HLA-B∗37:01 using the BAKER-Robetta server (29), based on the crystal structure of hTapasin with HLA-B∗44:05T73C (PDB ID: 7TUE) (9). Comparison of the interacting residues with the MHC-I heavy chain within 3.5 Å (Fig. 1D) revealed that the majority are polymorphic between the two orthologs and these variations could enable interactions with distinct HLA allotypes (Figs. 1D and S3). Interestingly, even though multiple chTapasin allomorphs have been described (15), none of the polymorphic sites are located within the interacting surfaces with MHC-I.
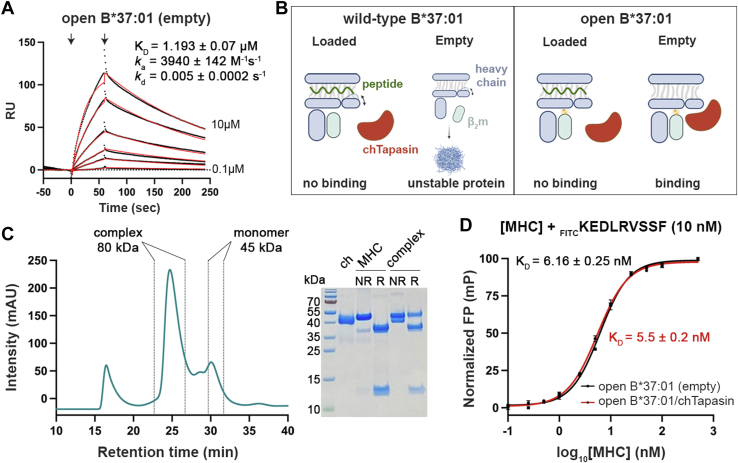
To thoroughly investigate the interaction between chTapasin and HLA-B∗37:01, we used surface plasmon resonance experiments (SPR), where chTapasin was immobilized to the chip surface at approx. 2000 RU. HLA-B∗37:01 refolded with the conditional peptide FEDLRVJSF (photo B37; J = 3-amino-3-(2-nitrophenyl)-propionic acid) that is cleaved upon UV irradiation (30, 31) was flown over the chip at different concentrations. No binding was observed with either peptide-loaded or empty, wild-type (WT) B∗37:01 molecules (Fig. S4, A and B). We hypothesized that the lack of interaction could be due to the instability of empty HLA-B∗37:01 molecules. In a recently proposed, universal platform for the generation of MHC-I molecules with enhanced stability, we engineered an interchain disulfide bond between the light and heavy chains (32). Therefore, we used a similar, disulfide-linked HLA-B∗37:01 molecule, referred to as open B∗37:01, that carried the substitutions H31C and G120C in the light and heavy chains, respectively (32). We found that chTapasin showed low micromolar affinity to the UV-irradiated (empty), open B∗37:01 with a KD of ∼1.2 μM (Fig. 2A), but not with the open, peptide-loaded B∗37:01 (Fig. S4C). The low Rmax observed compared to the conjugated chTapasin indicates that not all the molecules are active or adopt a favorable orientation to bind the analyte. The selective binding to the open MHC-I is likely due to rapid light-chain dissociation from the WT empty molecule, as a result of the known cooperativity between the peptide and β2m for binding to the heavy chain (33, 34). Contrarily, the covalent attachment of the β2m promotes the open state of the peptide-binding groove, facilitating interactions with the chaperone (Fig. 2B).
Figure 2.
The chTapasin ortholog binds to empty HLA-B∗37:01 forming a stable, peptide-receptive complex.A, representative sensorgrams of UV-irradiated HLA-B∗37:01. KD, equilibrium constant; ka, association rate constant; kd, dissociation rate constant; RU, resonance units. Fits from the kinetic analysis are shown in red lines and the concentrations. B, schematic representation of the chTapasin interactions with wild-type versus open HLA-B∗37:01. Created with BioRender.com. C, SEC analysis of the mixture of chTapasin (ch) with open HLA-B∗37:01/FEDLRVJSF (MHC) upon 40 min UV-irradiation. The peak corresponding to the empty complex was collected and all the components were identified by SDS/PAGE under reduced (R) or non-reduced (NR) conditions. D, FP saturation binding curve of FITCKEDLRVSSF with increasing concentrations of open B∗37:01/chTapasin complex. Log10(KD) = 0.74. Results of n = 3 replicates (mean ± SD) are plotted.
To explore further whether the open B∗37:01/chTapasin was receptive to incoming peptides, we purified the empty complex by incubating 1:1.3 ratio of chTapasin and UV-irradiated open B∗37:01, followed by SEC (Fig. 2C). Next, we followed the binding of a fluorescently labeled, high-affinity peptide by real-time fluorescence polarization (FP) (35). Titration of open B∗37:01/FEDLRVJSF, upon 20 min UV irradiation to generate empty monomers, and of purified complex using an optimized concentration of the fluorescently labeled peptide (FITCKEDLRVSSF) (32), showed increasing binding of the FITC-peptide as a function of MHC-I concentration (Fig. S5). After 1 h, FP values reached a plateau with estimated affinity values (KD) at the low nanomolar range for both the monomer and the complex (6.16 vs. 5.5 nM) (Fig. 2D) (35, 36), suggesting that the open B∗37:01/chTapasin complex is peptide-receptive, and that chTapasin does not affect the binding equilibrium of the peptide.
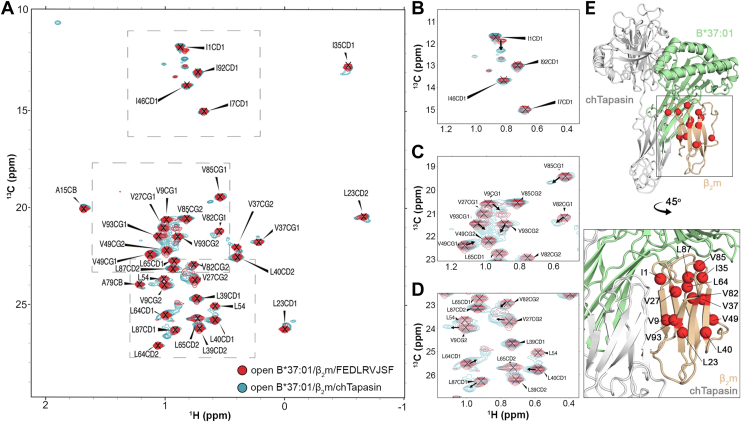
To evaluate the effects of chTapasin binding to the open B∗37:01 complex in a solution environment, we employed selective isotopic labeling and NMR methods established previously by our group to study a range of MHC-I molecules (37, 38). We focused on the alanine, isoleucine, leucine, and valine side chain methyl groups in β2m, which are known to be impacted by Tapasin binding (9), as highly sensitive probes for changes in the local magnetic environment. We first assigned the 2D methyl HMQC spectrum for the light chain of open B∗37:01 loaded with the photo B37 peptide (Fig. 3A). We then incubated the open MHC-I with an excess of chTapasin and UV-irradiated the peptide to form the empty complex. Overlay of the resulting 2D methyl spectra with the apo-open B∗37:01/photo B37 spectra shows significant chemical shift effects upon Tapasin binding, where the interaction between the two proteins is slow on the NMR timescale (Fig. 3, A–D). We observed that several methyl probes appear as two peaks in the chTapasin-bound NMR spectra, corresponding to the apo-open B∗37:01 state and the chTapasin-bound state. Specifically, residues Val9 and Val93 are located on the β-sheet (residues 6–10) and the C-terminus of β2m that interact with chTapasin, as previously reported for hTapasin (9). The methyl groups with affected 2D HMQC resonances corresponding to residues Leu23, Val27, Val37, Leu40, and Val82 are dispersed throughout the β-sheets of the light chain, indicating widespread allosteric effects on the β2m structure induced by binding to chTapasin followed by peptide release (Fig. 3E). Additionally, the observed chemical shift perturbations for residues Ile35, Val85, and Leu87 that are located beneath the peptide binding groove, supporting previous data that indicate a possible mechanism for peptide release by interfering with the β2m/α2 interface (9). These results are consistent with a prolonged lifetime of the open B∗37:01/chTapasin protein complex (>200 msec timescale), which exists as an equilibrium of unbound and bound states in solution.
Figure 3.
Conformational changes in open B∗37:01/β2m induced by interaction with chTapasin.A, 2D 1H-13C SOFAST HMQC spectra of U-[15N, 2H], 13CH3-labeled alanine, isoleucine, leucine, and valine residues of human β2m (H31C) in the B∗37:01 (G120C)-FEDLRVJSF bound state (in red). The sample was subsequently incubated with chicken Tapasin and UV irradiated for 40 min prior to collecting additional 1H-13C SOFAST HMQC spectra (in cyan). Both spectra were collected at 600 MHz 1H magnetic field, at 25 °C. Due to lack of stereospecific assignments, the CD1 and CD2 methyl groups of Leu54 are not indicated. B–D, Zoomed regions of the 1H-13C SOFAST HMQC spectra with arrows highlighting methyl probes experiencing minor chemical shift changes upon chTapasin binding, indicated by a slow-exchanging minor state peak. E, Methyl probes that show a detectable slow-exchanging minor peak upon chTapasin binding are shown as red spheres on the predicted B∗37:01/β2m/chTapasin model by BAKER-ROBETTA (29).
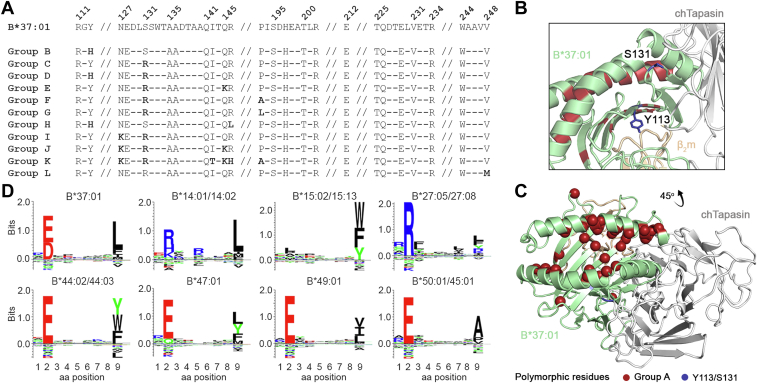
To investigate further the selective binding of chTapasin to B∗37:01, we extracted the interacting residues on MHC-I (9) from all the HLA allotypes present on our SAB panel. We distinguished alleles with identical interacting residues as B∗37:01 (Table S1, group A), and 11 distinct categories of HLA allotypes with polymorphisms in 8 out of the 24 Tapasin-contacting residues (Fig. 4A). These results suggest that the presence of Tyr at position 113 and Ser at position 131 located within 6 Å from the interface with Tapasin, positively correlate with binding to the chaperone consistently with a canonical binding mode that is further supported by our low-resolution NMR mapping experiments (Fig. 4B). However, these are not the only determinants of the interaction, since several allotypes that contain these polymorphisms do not show an interaction with Tapasin in their peptide-loaded form according to our SAB assay. Recognizing that the peptide repertoire and propensity to generate stable, empty molecules is likely to influence interactions with chaperones (10, 11, 23, 25, 39), we aligned the sequences of allotypes in group A and identified 32 polymorphic positions, from which 31 are located in the α1 and α2 helices and the groove floor, and one in the α3 domain (Fig. 4C). Comparing the peptide binding motifs between allotypes in group A using NetMHCpan4.0 (40) we observed that most can accommodate positively charged P2 side chains within a negatively charged B-pocket, and bulky/hydrophobic P9 side chains within the F-pocket. In contrast, B∗37:01 has a positively charged B-pocket and a preference for small hydrophobic side chains in the F-pocket (Fig. 4D). This suggests that B∗37:01 may have a higher propensity to generate empty molecules in solution due to a destabilized interaction with the P9 peptide anchor in the F-pocket, which would be consistent with the observation of a stable complex with chTapasin. Taken together, these results suggest that a combination of polymorphisms in the peptide binding groove, removed from the MHC-I/Tapasin interacting surface, determine the overall peptide exchange and conformational dynamics of HLA-B∗37:01 to favor a selective interaction with chTapasin.
Figure 4.
Polymorphisms in the HLA peptide binding groove govern interactions with chicken Tapasin.A, alignment of the possible chaperone interacting residues from all HLA allotypes present on the SAB panel shown in Fig. 1C. Representation of (B) Y133 and S131 that positively correlate with chaperone binding (blue), and (C) the polymorphic positions within allotypes of group A (red), on the B∗37:01/chTapasin complex. D, the sequence logos of the HLA allotypes in group A, visualized in Seq2Logo from NetMHCpan4.0 (40).
Discussion
Cells detect foreign or aberrant antigens displayed by MHC-I molecules via T cell receptors (TCRs), initiating the first crucial stage in the development of immunity against viruses and tumors (41). Considering the fundamental importance of T cell responses, fast, high-throughput, and cost-effective methods, including peptide exchange technologies for pMHC-multimers generation, are emerging to study pMHC:TCR antigen recognition. The molecular chaperones Tapasin and TAPBPR determine to a high extent the resulting peptide repertoire (19, 23, 28, 42, 43, 44). We have previously described a robust method to prepare biologically relevant, stable pMHC-I molecules loaded with peptides of choice by exploiting hTAPBPR (20) that can function independent of the PLC, albeit this approach is limited to specific allotypes that can be recognized by the chaperone (25). Aiming to expand the HLA-I repertoire susceptible to TAPBPR-assisted peptide exchange, we characterized a TAPBPR ortholog from chicken (14). While the predicted structure of chTAPBPR exhibits remarkable similarity to hTAPBPR (13), their sequence variability conferred an expanded interaction pattern to include representatives from six additional supertypes (14, 45, 46). These studies underscore the importance of studying chaperones from different species as a means to expand peptide exchange applications on therapeutically relevant HLAs.
Human Tapasin is an indispensable part of the PLC and serves as the primary chaperone for MHC-I molecules, facilitating the loading of peptides and optimizing the antigen repertoire across various allotypes (47). However, Tapasin expression is restricted to the PLC and therefore is highly unstable when produced in recombinant form. To address this limitation, we explore chicken Tapasin, which is encoded by multiple allomorphs, each closely associated with the expression of the dominant MHC molecules (15). We show that chTapasin can function in the absense of co-chaperones and interacts tightly with an engineered, stabilized HLA-B∗37:01 (32), suggesting that structural polymorphisms between Tapasin orthologs can lead to distinct MHC-I interactions. Importantly, deep mutational scanning of hTAPBPR surfaces revealed two prominent regions which can drive these interactions to allow for peptide exchange on desired HLA allotypes (14). In an analogous manner, performing a single site-saturation mutagenesis (SSM) library on different Tapasin orthologs (14) would provide insights into important residues that could be substituted to guide the HLA allelic specificity landscape. Notably, HLA-B∗37:01 is a low hTapasin-dependent allotype (47) that has been associated with diseases like multiple sclerosis and flu (16, 17), underscoring the importance of uncovering new tools to study the immunopeptidome. Overall, our results highlight the potential use of chTapasin as an orthologous chaperone and provide a readily available framework for future protein engineering applications aimed at broadening the ligand exchange function on human MHC I molecules.
Experimental procedures
Sequence analysis
The sequences used in this study are: hTAPBPR (Q9BX59), chTAPBPR (NP_001382952.1), hTapasin (O15533), and chTapasin (A4F5A9). Alignments were performed in ClustalOmega (48) and processed in ESPript (49).
Recombinant protein expression, refolding, and purification
The luminal domains of human and chicken Tapasin, and hTAPBPRTN6 encoding the BirA substrate peptide (BSP; LHHILDAQKMVWNHR) and 6-His tag were stably expressed in the Drosophila melanogaster S2 cells and purified as previously described (23).
The wild-type MHC-I heavy chain tagged with BSP and the β2m light chain were provided by the NIH (Emory university) or synthesized and cloned into pET-22b(+) vector (Genscript). For the open constructs, substitutions in positions H31C (β2m) and G120C (heavy chain) were generated using site-directed mutagenesis (32). Upon Escherichia coli BL21 (DE3) transformation (New England Biolabs), expressed proteins were isolated from inclusion bodies. For pMHC-I generation, we performed in vitro refolding as previously described (30).
Purification of empty MHC-I/Tapasin complex
chTapasin was incubated with open B∗37:01/FEDLRVJSF at 1:1.3 M ratio for 1 h at room temperature (RT), followed by 40-min UV irradiation at 365 nm and 30 min additional RT incubation (14). The empty complex was purified by SEC using a Superdex 200 pg Increase 10/300 Gl. To identify all components, the eluted peaks were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
Peptides
Peptide sequences are given as single-letter codes. Photolabile peptides were purchased from Biopeptek Inc (Malvern) using J as 3-amino-3-(2-nitrophenyl)-propionic acid. FITC-labeled peptides were synthesized as previously described (32). Peptides were solubilized in distilled water and centrifuged at 14,000 rpm for 15 min before measuring their concentration at 205 nm using the respective extinction coefficient.
Differential scanning fluorimetry
The thermal stability of Tapasin was measured by incubating 7 μM of protein with 10X SYPRO Orange dye in PBS buffer in 20 μl. Samples were loaded in a MicroAmp Optical 384-well plate in triplicates and analyzed on a QuantiStudio 5 real-time PCR machine. Excitation and emission wavelengths were set to 470 and 569 nm, and temperature increased at a rate of 1 °C/min between 25 to 95 °C. Data analysis and fitting were performed in GraphPad Prism v9.
Biotinylation and tetramer formation
BSP-tagged proteins were biotinylated using the BirA biotin-ligase bulk reaction kit (Avidity), according to the manufacturer’s instructions. Streptavidin-PE (Agilent Technologies, Inc) at 4:1 monomer:streptavidin molar ratio was added in the dark, every 10 min at RT over 10-time intervals (20).
Single antigen bead assay
Tetramerized Tapasin orthologs and hTAPBPRTN6 (7 μM) were mixed with 4 μl of LABScreen SABs (OneLambda Inc, CA, USA) and incubated for 1 h, 550 rpm at RT. After four washes with the provided buffer to remove the excess of tetramers, beads were resuspended in PBS buffer. The levels of peptide-loaded MHC-I on the beads were tested using the PE-conjugated W6/32 antibody (Biolegend, 311,406). Binding levels were measured in Luminex 100 Liquid Analyzer System, and the results were analyzed in GraphPad Prism v9.
Surface plasmon resonance
SPR experiments were performed in a BiaCore X100 instrument (Cytiva) in SPR buffer (150 mM NaCl, 20 mM sodium phosphate pH 7.4, 0.1% Tween-20). Approximately 2000 resonance units (RU) of biotinylated chTapasin were conjugated on a streptavidin-coated chip (Cytiva) at 10 μl/min. Various concentrations of pMHC-I were flown over the chip for 60 s at 30 μl/min followed by a buffer wash with 180 s dissociation time, at 25°C. SPR sensorgrams, association/dissociation rate constants (ka, kd), and equilibrium dissociation constant KD values were analyzed in BiaCore X100 evaluation software (Cytiva) using kinetic analysis settings of 1:1 binding. SPR sensorgrams and affinity-fitted curves were prepared in GraphPad Prism v9.
Fluorescence polarization
FP was used to monitor the kinetic association of fluorescently labeled peptides and MHC-I. Various concentrations of open B∗37:01/FEDLRVJSF upon 20 min UV-irradiation (empty), and open B∗37:01/chTapasin complex were incubated with an optimized concentration of FITCKEDLRVSSF (10 nM) in FP buffer (PBS and 0.05% Tween-20). Excitation and emission values were 475 and 525 nm, respectively.
NMR spectroscopy
NMR samples of open HLA-B∗37:01/β2m/FEDLRVJSF was prepared by in vitro refolding, by selectively labeling the light chain hβ2m with a {U-[15N,2H]; Ala, Ile, Leu, Val-[13CH3]}-labeled isotope-selective labeling scheme using established protocols and reagents (11, 50). The purified sample was prepared in standard NMR buffer (150 mM NaCl, 20 mM sodium phosphate pH 7.2, 0.001 M sodium azide, 5% D2O) in the presence of 2-fold molar excess of peptide. 2D SOFAST methyl 1H-13C HMQC and 2D 1H-15N TROSY spectra were collected. Methyl resonance assignments for the light chain were obtained using 3D SOFAST Cm-CmHm NOESY experiments recorded with 150 ms mixing time (51). The complex was then incubated with chTapasin and UV-irradiated for 40 min to cleave the photo B37 peptide. Approximately 70 μM of complex was prepared in identical buffer conditions with excess of chTapasin to collect 2D SOFAST methyl 1H-13C HMQC and 2D 1H-15N TROSY spectra for the complex. All NMR data were recorded at 600 MHz and 298 K, processed with NMRPipe (52), and analyzed using POKY (53).
Data availability
All other data are contained within this article and in the supporting information. Final methyl assignments were deposited in the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu) under accession number 51999.
Supporting information
This article contains supporting information (9, 48, 49, 54).
Conflict of interest
N. G. S and G. F. P. are co-inventors in provisional patent applications related to this work.
Acknowledgments
We acknowledge Dr Monos and the CHOP Immunogenetics Laboratory for discussions and access to equipment and reagents.
Author contributions
N. G. S. conceptualization; N. G. S. investigation; G. F. P., M. C. Y., T. J. W. and G. M. B. resources; G. F. P. and C. H. W. formal analysis; G. F. P. visualization; N. G. S. and G. F. P writing–original draft; N. G. S., G. F. P., C. H. W., M. C. Y, T. J. W. and G. M. B. writing–review & editing; N. G. S. supervision; N. G. S. funding acquisition.
Funding and additional information
This research was supported through grants by NIAID (5R01AI143997), NIGMS (5R35GM125034), and NIDDK (5U01DK112217) to N. G. S. This work was delivered as part of the NexTGen team supported by the Cancer Grand Challenges partnership funded by Cancer Research UK (CGCATF-2021/100002) and the National Cancer Institute (CA278687-01) and the Mark Foundation.
Reviewed by members of the JBC Editorial Board. Edited by Clare E. Bryant
Supporting information
References
- 1.Blum J.S., Wearsch P.A., Cresswell P. Pathways of antigen processing. Annu. Rev. Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cresswell P., Bangia N., Dick T., Diedrich G. The nature of the MHC class I peptide loading complex. Immunol. Rev. 1999;172:21–28. doi: 10.1111/j.1600-065x.1999.tb01353.x. [DOI] [PubMed] [Google Scholar]
- 3.De Castro J.A.L., Stratikos E. Intracellular antigen processing by ERAP2: molecular mechanism and roles in health and disease. Hum. Immunol. 2019;80:310–317. doi: 10.1016/j.humimm.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Mpakali A., Maben Z., Stern L.J., Stratikos E. Molecular pathways for antigenic peptide generation by ER aminopeptidase 1. Mol. Immunol. 2019;113:50–57. doi: 10.1016/j.molimm.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martín-Esteban A., Rodriguez J.C., Peske D., Lopez De Castro J.A., Shastri N., Sadegh-Nasseri S. The ER Aminopeptidases, ERAP1 and ERAP2, synergize to self-modulate their respective activities. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1066483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garbi N., Tiwari N., Momburg F., Hämmerling G.J. A major role for tapasin as a stabilizer of the TAP peptide transporter and consequences for MHC class I expression. Eur. J. Immunol. 2003;33:264–273. doi: 10.1002/immu.200390029. [DOI] [PubMed] [Google Scholar]
- 7.Grandea A.G., Van Kaer L. Tapasin: an ER chaperone that controls MHC class I assembly with peptide. Trends Immunol. 2001;22:194–199. doi: 10.1016/s1471-4906(01)01861-0. [DOI] [PubMed] [Google Scholar]
- 8.Müller I.K., Winter C., Thomas C., Spaapen R.M., Trowitzsch S., Tampé R. Structure of an MHC I–tapasin–ERp57 editing complex defines chaperone promiscuity. Nat. Commun. 2022;13:5383. doi: 10.1038/s41467-022-32841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang J., Taylor D.K., Kim E.J., Boyd L.F., Ahmad J., Mage M.G., et al. Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation. Nat. Commun. 2022;13:5470. doi: 10.1038/s41467-022-33153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McShan A.C., Natarajan K., Kumirov V.K., Flores-Solis D., Jiang J., Badstübner M., et al. Peptide exchange on MHC-I by TAPBPR is driven by a negative allostery release cycle. Nat. Chem. Biol. 2018;14:811–820. doi: 10.1038/s41589-018-0096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McShan A.C., Devlin C.A., Overall S.A., Park J., Toor J.S., Moschidi D., et al. Molecular determinants of chaperone interactions on MHC-I for folding and antigen repertoire selection. Proc. Natl. Acad. Sci. U. S. A. 2019;116:25602–25613. doi: 10.1073/pnas.1915562116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truong H.V., Sgourakis N.G. Dynamics of MHC-I molecules in the antigen processing and presentation pathway. Curr. Opin. Immunol. 2021;70:122–128. doi: 10.1016/j.coi.2021.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margulies D.H., Taylor D.K., Jiang J., Boyd L.F., Ahmad J., Mage M.G., et al. Chaperones and catalysts: how antigen presentation pathways cope with biological necessity. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.859782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y., Papadaki G.F., Devlin C.A., Danon J.N., Young M.C., Winters T.J., et al. Xeno interactions between MHC-I proteins and molecular chaperones enable ligand exchange on a broad repertoire of HLA allotypes. Sci. Adv. 2023;9 doi: 10.1126/sciadv.ade7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Hateren A., Carter R., Bailey A., Kontouli N., Williams A.P., Kaufman J., et al. A mechanistic basis for the Co-evolution of chicken tapasin and major histocompatibility complex class I (MHC I) proteins. J. Biol. Chem. 2013;288:32797–32808. doi: 10.1074/jbc.M113.474031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant E.J., Josephs T.M., Loh L., Clemens E.B., Sant S., Bharadwaj M., et al. Broad CD8+ T cell cross-recognition of distinct influenza A strains in humans. Nat. Commun. 2018;9:5427. doi: 10.1038/s41467-018-07815-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.James L.M., Georgopoulos A.P. Immunogenetic epidemiology of multiple sclerosis in 14 continental western European countries. J. Immunol. Sci. 2021;5:40–46. [Google Scholar]
- 18.Ilca F.T., Neerincx A., Wills M.R., de la Roche M., Boyle L.H. Utilizing TAPBPR to promote exogenous peptide loading onto cell surface MHC I molecules. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E9353–E9361. doi: 10.1073/pnas.1809465115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan H., Abualrous E.T., Sticht J., Fernandez L.M.A., Werk T., Weise C., et al. Exchange catalysis by tapasin exploits conserved and allele-specific features of MHC-I molecules. Nat. Commun. 2021;12:4236. doi: 10.1038/s41467-021-24401-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Overall S.A., Toor J.S., Hao S., Yarmarkovich M., O’Rourke S.M., Morozov G.I., et al. High throughput pMHC-I tetramer library production using chaperone-mediated peptide exchange. Nat. Commun. 2020;11:1909. doi: 10.1038/s41467-020-15710-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Rourke S.M., Morozov G.I., Roberts J.T., Barb A.W., Sgourakis N.G. Production of soluble pMHC-I molecules in mammalian cells using the molecular chaperone TAPBPR. Protein Eng. Des. Select. 2019;32:525–532. doi: 10.1093/protein/gzaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sengupta S., Zhang J., Reed M.C., Yu J., Kim A., Boronina T.N., et al. A cell-free antigen processing system informs HIV-1 epitope selection and vaccine design. J. Exp. Med. 2023;220 doi: 10.1084/jem.20221654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morozov G.I., Zhao H., Mage M.G., Boyd L.F., Jiang J., Dolan M.A., et al. Interaction of TAPBPR, a tapasin homolog, with MHC-I molecules promotes peptide editing. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E1006–E1015. doi: 10.1073/pnas.1519894113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altman J.D., Moss P.A.H., Goulder P.J.R., Barouch D.H., McHeyzer-Williams M.G., Bell J.I., et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 25.Ilca F.T., Drexhage L.Z., Brewin G., Peacock S., Boyle L.H. Distinct polymorphisms in HLA class I molecules govern their susceptibility to peptide editing by TAPBPR. Cell Rep. 2019;29:1621–1632.e3. doi: 10.1016/j.celrep.2019.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei R., Lee J., Shih N.-J., Chen M., Terasaki P.I. Single human leukocyte antigen flow cytometry beads for accurate identification of human leukocyte antigen antibody specificities. Transplantation. 2003;75:43–49. doi: 10.1097/00007890-200301150-00008. [DOI] [PubMed] [Google Scholar]
- 27.Wittenbrink N., Herrmann S., Blazquez-Navarro A., Bauer C., Lindberg E., Wolk K., et al. A novel approach reveals that HLA class 1 single antigen bead-signatures provide a means of high-accuracy pre-transplant risk assessment of acute cellular rejection in renal transplantation. BMC Immunol. 2019;20:11. doi: 10.1186/s12865-019-0291-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermann C., Strittmatter L.M., Deane J.E., Boyle L.H. The binding of TAPBPR and tapasin to MHC class I is mutually exclusive. J. Immunol. 2013;191:5743–5750. doi: 10.4049/jimmunol.1300929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song Y., DiMaio F., Wang R.Y.-R., Kim D., Miles C., Brunette T., et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garboczi D.N., Hung D.T., Wiley D.C. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Eschericlhia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. U. S. A. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodenko B., Toebes M., Hadrup S.R., van Esch W.J.E., Molenaar A.M., Schumacher T.N.M., et al. Generation of peptide–MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc. 2006;1:1120–1132. doi: 10.1038/nprot.2006.121. [DOI] [PubMed] [Google Scholar]
- 32.Sun Y., Young M.C., Woodward C.H., Danon J.N., Truong H.V., Gupta S., et al. Universal open MHC-I molecules for rapid peptide loading and enhanced complex stability across HLA allotypes. Proc. Natl. Acad. Sci. U. S. A. 2023;120 doi: 10.1073/pnas.2304055120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Achour A., Michaëlsson J., Harris R.A., Ljunggren H.-G., Kärre K., Schneider G., et al. Structural basis of the differential stability and receptor specificity of H-2Db in complex with murine versus human β2-microglobulin. J. Mol. Biol. 2006;356:382–396. doi: 10.1016/j.jmb.2005.11.068. [DOI] [PubMed] [Google Scholar]
- 34.Gakamsky D.M., Boyd L.F., Margulies D.H., Davis D.M., Strominger J.L., Pecht I. An allosteric mechanism controls antigen presentation by the H-2K b complex. Biochemistry. 1999;38:12165–12173. doi: 10.1021/bi9905821. [DOI] [PubMed] [Google Scholar]
- 35.Buchli R., VanGundy R.S., Hickman-Miller H.D., Giberson C.F., Bardet W., Hildebrand W.H. Real-time measurement of in Vitro peptide binding to soluble HLA-A∗0201 by fluorescence polarization. Biochemistry. 2004;43:14852–14863. doi: 10.1021/bi048580q. [DOI] [PubMed] [Google Scholar]
- 36.Rossi A.M., Taylor C.W. Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 2011;6:365–387. doi: 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sgourakis N.G., May N.A., Boyd L.F., Ying J., Bax A., Margulies D.H. A novel MHC-I surface targeted for binding by the MCMV m06 immunoevasin revealed by solution NMR. J. Biol. Chem. 2015;290:28857–28868. doi: 10.1074/jbc.M115.689661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardner K.H., Kay L.E. Production and incorporation of 15 N, 13 C, 2 H (1 H-δ1 methyl) isoleucine into proteins for multidimensional NMR studies. J. Am. Chem. Soc. 1997;119:7599–7600. [Google Scholar]
- 39.Rizvi S.M., Salam N., Geng J., Qi Y., Bream J.H., Duggal P., et al. Distinct assembly profiles of HLA-B molecules. J. Immunol. 2014;192:4967–4976. doi: 10.4049/jimmunol.1301670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucl. Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Germain R.N., Margulies D.H. The biochemistry and cell biology of antigen processing and presentation. Annu. Rev. Immunol. 1993;11:403–450. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- 42.Boyle L.H., Hermann C., Boname J.M., Porter K.M., Patel P.A., Burr M.L., et al. Tapasin-related protein TAPBPR is an additional component of the MHC class I presentation pathway. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3465–3470. doi: 10.1073/pnas.1222342110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen M., Bouvier M. Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. EMBO J. 2007;26:1681–1690. doi: 10.1038/sj.emboj.7601624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teng M.S., Stephens R., Pasquier L.D., Freeman T., Lindquist J.A., Trowsdale J. A humanTAPBP (TAPASIN)-related gene,TAPBP-R. Eur. J. Immunol. 2002;32:1059–1068. doi: 10.1002/1521-4141(200204)32:4<1059::AID-IMMU1059>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 45.Sette A., Sidney J. HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Curr. Opin. Immunol. 1998;10:478–482. doi: 10.1016/s0952-7915(98)80124-6. [DOI] [PubMed] [Google Scholar]
- 46.Sidney J., Peters B., Frahm N., Brander C., Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bashirova A.A., Viard M., Naranbhai V., Grifoni A., Garcia-Beltran W., Akdag M., et al. HLA tapasin independence: broader peptide repertoire and HIV control. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28232–28238. doi: 10.1073/pnas.2013554117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucl. Acids Res. 2014;42:W320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tugarinov V., Kanelis V., Kay L.E. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 51.Rossi P., Xia Y., Khanra N., Veglia G., Kalodimos C.G. 15N and 13C- SOFAST-HMQC editing enhances 3D-NOESY sensitivity in highly deuterated, selectively [1H,13C]-labeled proteins. J. Biomol. NMR. 2016;66:259–271. doi: 10.1007/s10858-016-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995 doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 53.Lee W., Rahimi M., Lee Y., Chiu A. POKY: a software suite for multidimensional NMR and 3D structure calculation of biomolecules. Bioinformatics. 2021;37:3041–3042. doi: 10.1093/bioinformatics/btab180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang J., Natarajan K., Boyd L.F., Morozov G.I., Mage M.G., Margulies D.H. Crystal structure of a TAPBPR–MHC I complex reveals the mechanism of peptide editing in antigen presentation. Science. 2017;358:1064–1068. doi: 10.1126/science.aao5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other data are contained within this article and in the supporting information. Final methyl assignments were deposited in the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu) under accession number 51999.