Abstract
Background:
The microbiome associations of food protein-induced enterocolitis syndrome (FPIES) are understudied. We sought to prospectively define the clinical features of FPIES in a birth cohort, and investigate for the evidence of gut dysbiosis.
Methods:
We identified children diagnosed with FPIES in the Gastrointestinal Microbiome and Allergic Proctocolitis Study, a healthy infant cohort. Children were assessed and stools were collected at each well child visit. The clinical features of the children with FPIES were summarized. Stool microbiome was analyzed using 16S rRNA sequencing comparing children with and without FPIES.
Results:
Of the 874 children followed up for 3 years, 8 FPIES cases (4 male) were identified, yielding a cumulative incidence of 0.92%. The most common triggers were oat and rice (n = 3, each) followed by milk (n = 2). The children with FPIES were more likely to have family history of food allergy (50% vs. 15.9% among unaffected, p = .03). The average age of disease presentation was 6 months old. During the first 6 months of life, stool from children with FPIES contained significantly less Bifidobacterium adolescentis, but more pathobionts, including Bacteroides spp. (especially Bacteroides fragilis), Holdemania spp., Lachnobacterium spp., and Acinetobacter lwoffii. The short-chain fatty acid (SCFA)-producing Bifidobacterium shunt was expressed significantly less in the stool from FPIES children.
Conclusions:
In this cohort, the cumulative incidence over the 3-year study period was 0.92%. During the first 6 months of life, children with FPIES had evidence of dysbiosis and SCFA production pathway was expressed less in their stool, which may play an important role in the pathogenesis of FPIES.
Keywords: birth cohort study, food allergy, food protein-induced enterocolitis syndrome, FPIES, Stool microbiome
GRAPHICAL ABSTRACT
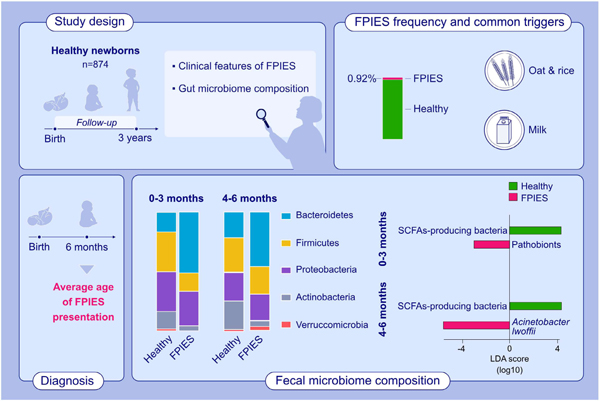
The incidence of FPIES in 3 years in a healthy birth cohort was 0.92%. The average age of disease presentation was 6 months. The most common FPIES triggers are oat and rice, followed by cow’s milk. Dysbiosis, more pathobionts and less commensal bacteria were noted since birth from the stools of FPIES children.
1 |. INTRODUCTION
Food protein-induced enterocolitis syndrome (FPIES) is a non-immunoglobulin E (IgE)-mediated food allergic disease, in which predominant symptoms are delayed profuse vomiting, pallor, lethargy, and diarrhea.1 The disease has been identified worldwide, and its incidence may be increasing.2–5 In different geographic areas and age groups, FPIES triggers vary from cow’s milk, soy milk, grains, to fish and shellfish.3–10 Because symptoms are reproducibly elicited by various specific offending dietary triggers, they are thought to be immune-mediated, but the pathophysiology of reactions is not well understood. From studies of peripheral immune cells, innate immune activation and interleukin-17-predominant inflammatory signatures have been described post-FPIES reaction.11–13 There is relatively little data to shed light on how or why this untoward immune-mediated response develops.14
Variations of the intestinal microbiome, by interacting with our immune system, have been associated with many immune-mediated diseases.14–17 Several studies have investigated the relationship of the intestinal microbiome to IgE-mediated food allergies.18,19 Dysbiosis has been identified in patients with other non-IgE-mediated food allergic disease or mixed IgE and non-IgE-mediated disease, including food protein-induced allergic proctocolitis and eosinophilic esophagitis.20–22 However, the role of the intestinal microbiome in FPIES has not been reported. In this study, we identified FPIES cases prospectively from the Gastrointestinal Microbiome and Allergic Proctocolitis Study (GMAP) cohort23 to better define disease incidence, investigate clinical features, and assess the stool microbiome.
2 |. METHODS
2.1 |. Study design
The GMAP study is a prospective observational healthy infant cohort in suburban Massachusetts, USA.23 Infants were recruited at their first office visit after birth at the Pediatrics at Newton Wellesley, a private primary care pediatric office, between April 2014 and February 2017. All infants were approached for participation in the study up to 2 months of age. To preserve the assumption of independence of observations, one infant from those families who enrolled more than one child in the GMAP study was randomly selected and the other sibling(s) were excluded in our final analyses. Because more than 95% of FPIES cases were diagnosed before 3 years old,14 children with incomplete followup to 3 years old were excluded. Participants were followed at all routine well-child visits according to the schedule of the American Academy of Pediatrics (1 week, 2 weeks, and 1, 2, 4, 6, 9, 12, 15, 18, 24, and 36 months of age) and at unscheduled sick visits. At the first visit, parents completed a questionnaire about family histories of atopy and food allergies, antibiotics and pet exposure, and their infant’s birth condition. At each well visit, parents completed a questionnaire about their infant’s current feeding, stooling, and sleeping patterns as well as any gastrointestinal or allergic symptoms. Fresh stool samples were collected at each visit, placed in sterile tubes, kept immediately at −20°C, and transported to Massachusetts General Hospital. All stool samples were stored at −80°C. The GMAP study was approved by the Partners Human Research Committee (IRB Protocol No: 2013P002374) and a parent of all enrolled infants gave written informed consent.
During each visit, parents were asked about symptoms of adverse food reactions generally and FPIES diagnosis specifically. Study staff also reviewed contemporaneous documentation by their pediatricians for any signs and symptoms of adverse food reactions. Once children with possible FPIES were noted, the research study staff comprehensively chart reviewed each case of suspected FPIES, using the diagnostic criteria from the international consensus guidelines for the diagnosis and management of food protein-induced enterocolitis syndrome.1 For comparisons of the proportion of children with FPIES versus children without FPIES by demographic and clinical variables, Fisher’s exact test was used.
2.2 |. Stool DNA extraction, 16S sequencing, and microbiome analysis
Stool samples were aliquoted from the frozen vials in a sterile manner. Bacterial DNA was isolated with the DNeasy® PowerSoil® Kit (Qiagen, Venlo, Netherlands). 16S V4 region sequencing was performed at the Microbial ‘Omics Core, Broad Institute of the Harvard University, and the Massachusetts Institute of Technology.
By using the R package, MatchIt (version 3.0.2),24 77 matched controls without FPIES (matched for age, gender, mode of delivery, gestational age, antibiotics exposure, and breastfeeding) were selected from the same cohort, and their demographic data, are similar to that of the FPIES cases. Stool microbiome data from the matched controls were used for comparative analysis. Because infant stool microbiome undergoes dynamic changes during the first year after birth, comparisons were performed within four different age bins: 0–3 months, 4–6 months, 7–9 months, and 10–12 months of age. The alpha diversity, beta diversity, and specific bacterial taxa were analyzed by QIIME2 (version 2019.10).25 The alpha diversity comparison was performed by the Kruskal-Wallis test. The beta diversity comparison was performed by pairwise permutational multivariate analysis of variance test. Multiple tools, including microbiome multivariable association with linear model 2 (MaAsLin2 (version 1.6.0)), linear discriminant analysis effect size (LEfSe (Galaxy version 1.0)), and analysis of composition of microbiomes (ANCOM (version 2019.10.0)) were used to compare the microbiome difference between FPIES cases and controls.26–28 We firstly used a linear model to determine the difference between groups in different taxa levels by using MaAsLin2.26 LEfSe is applied to measure the consistency of differences in normalized relative abundance of taxa in different groups. We considered bacteria taxa reached significance when the p-value was less than .05 and linear discriminant analysis (LDA) score was more than 2.0.27 We also applied a more conservative methodology, ANCOM, to detect differences in microbial mean taxa abundance after a log-ratio transformation. W value generated by ANCOM is a count of a number value rejecting the null hypothesis during the multiple Wilcoxon rank sum tests.28 We used another microbiome tool, phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt (version 1.1.4)), to predict the identified intestinal microbiome functions on the pathogenesis of FPIES.29 The Benjamini-Hochberg procedure was used to adjust the false discovery rate for multiple comparisons, and a priori levels of significance were set at false discovery rate-adjusted p-value (q-value) < .05. A random forest machine learning using the R package, caret (version 6.0–78), was used to reveal the most differential microbes between children with FPIES and matched controls.30
3 |. RESULTS
3.1 |. Characteristics of children with FPIES in the GMAP birth cohort
1003 infants were enrolled initially, and the median age at enrollment was 8 days. 60 children were excluded due to siblings in the study and 69 children were excluded because of incomplete followup. 874 children were completely followed at 3 years old (Figure S1).
Of the 874 children, eight cases were diagnosed with acute FPIES, yielding a cumulative incidence of 0.92% over 3 years. Of the children with FPIES, 50% were female and all 8 were Caucasian (vs. 66.7% among those without FPIES, p = .06, Table 1). Of the eight cases of acute FPIES, four had a family history of IgE food allergy, significantly higher than those without FPIES (50% vs. 15.9%, p = .03). One had a family history of FPIES. Four had eczema, three were diagnosed with food protein-induced allergic proctocolitis, and six had a family history of atopy (all p > .05). The C-section (12.5% vs. 32.1%, p = .45) and initial breastfeeding rates (50% vs. 60.7%, p = .72) in children with FPIES were not significantly different from those without FPIES. There was also no difference in gender, race, preterm delivery rates, or early antibiotic exposure (Table 1).
TABLE 1.
The demographic comparison between GMAP children with and without FPIES
| Characteristics | Children with FPIES (n = 8) | Children without FPIES (n = 866) | p-Value (Children with vs. without FPIES) | Controls (n = 77) | p-Value (Children with FPIES vs. controls) |
|---|---|---|---|---|---|
| Gender, male | 4 (50.0%) | 465 (53.7%) | .99 | 45 (58.4%) | .72 |
| Race, white | 8 (100%) | 578 (66.7%) | .06 | 51 (66.2%) | .10 |
| Hispanic or Latino | 0 (0%) | 41 (4.7%) | .99 | 2 (2.6%) | .99 |
| C-section | 1 (12.5%) | 278 (32.1%) | .45 | 29 (37.7%) | .25 |
| Preterm (<37 wk) | 1 (12.5%) | 97 (11.2%) | .99 | 4 (5.2%) | .40 |
| Abx during delivery | 3 (37.5%) | 431 (49.8%) | .73 | 43 (55.8%) | .46 |
| Infant perinatal Abx | 1 (12.5%) | 75 (8.7%) | .52 | 8 (10.4%) | .99 |
| Initial exclusively BF | 4 (50.0%) | 526 (60.7%) | .72 | 49 (63.6%) | .47 |
| IgE-mediated food allergy | 0 (0%) | 58 (6.7%) | .99 | 0 (0%) | a |
| Eczema | 4 (50.0%) | 360 (41.6%) | .73 | 36 (46.8%) | .99 |
| Allergic proctocolitis | 3 (37.5%) | 144 (16.6%) | .14 | 0 (0%) | <.01 |
| Family history of atopy | 6 (75.0%) | 401 (46.3%) | .16 | 40 (51.9%) | .28 |
| Family history of IgE-mediated food allergy | 4 (50.0%) | 138 (15.9%) | .03 | 12 (15.6%) | .04 |
| Pet at home | 5 (62.5%) | 336 (38.8%) | .27 | 23 (29.9%) | .11 |
Abbreviations: Abx, antibiotics; BF, breastfeeding; IgE, immunoglobulin E.
Fisher’s exact test is not applicable when two cells in a row are zero.
The average age of disease presentation was 6 months old (interquartile range: 3, 6.1). The median age at diagnosis of acute FPIES was 7 months old (interquartile range: 6, 11). The most common FPIES triggers in this cohort were oat and rice (n = 3 each), followed by milk (n = 2). The remaining trigger foods were sweet potato, shrimp, avocado, and legume (each n = 1). Four cases were triggered by two kinds of foods and the other four cases only had a single food trigger. Two children had initially presented with chronic FPIES due to milk and had subsequent acute FPIES reactions, triggered by different foods (legume and rice). Consistent with diagnostic guidelines, all of them experienced repeated delayed-onset vomiting after ingesting trigger foods. Seven of the acute FPIES cases developed lethargy during the reaction, four were seen in the emergency department, and three received intravenous fluid rehydration. Clinical features are summarized in Table 2.
TABLE 2.
Clinical features of GMAP cases with FPIES
| Case | Food | Age at symptom onset |
Age of diagnosis | Mode of delivery | Breast-feeding ever (feeding length in months) | Solid food introduction timing (months old) | Eczema | Allergic proctocolitis | Clinical presentation of FPIES |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Milk (Chronic) | Solid food (Acute) | Delayed vomiting | Repeated episodes | To different foods | Lethargy | Pale | ED visit | Need IV fluid | Diarrhea | ||||||||
| 1 | Sweet potato | – | 4 months | 6 months | Vaginal | Yes (4) | 4 | No | No | Yes | Yes | – | Yes | – | Yes | – | – |
| 2 | Oat | – | 6 months | 9 months | Vaginal | Yes (1) | 5 | Yes | Yes | Yes | Yes | – | Yes | Yes | Yes | Yes | Yes |
| 3 | Oat, rice | – | 6.5 months | 7 months | Vaginal | Yes (6) | 5 | No | No | Yes | Yes | Yes | Yes | – | Yes | Yes | – |
| 4 | Milk, legume | 4 days | 11 months | 14 months | Vaginal | Yes (1) | 4 | Yes | Yes | Yes | Yes | Yes | Yes | – | – | – | Yes |
| 5 | Shrimp | – | 16 months | 18 months | Vaginal | Yes (6) | 6 | No | No | Yes | Yes | – | Yes | – | – | – | – |
| 6 | Milk, rice | 5 days | 6 months | 7 months | C-section | No | 5 | No | No | Yes | Yes | Yes | – | – | – | – | – |
| 7 | Avocado, Oat | – | 6 months | 6 months | Vaginal | Yes (6) | 5 | Yes | Yes | Yes | Yes | Yes | Yes | – | – | – | – |
| 8 | Rice | – | 6 months | 6 months | Vaginal | Yes (6) | 5 | Yes | No | Yes | Yes | – | Yes | – | Yes | Yes | – |
Abbreviations: ED, emergency department; IV: intravenous fluid.
3.2 |. Fecal microbiome features of FPIES patients
Seventy-seven matched healthy children from the same cohort were used for comparative analysis. The stool microbiome diversity was lower in children with FPIES cases at 4–6 months old (Shannon index, q = .04). Children with FPIES had a distinct stool microbiome pattern compared with controls at 0–3 and 4–6 months of age (either by Bray–Curtis dissimilarity or Jaccard index, all q < .05).
At the phylum level, the stools from the children with FPIES were significantly enriched with Bateroidetes phylum at both the 0–3 and 4–6 months age bins (both q < .05). In contrast, Actinobacteria phylum at 0–3 and 4–6 months of age and Firmicutes phylum at 0–3 months of age were less abundant among samples from FPIES cases (all q < .05) (Figure 1A).
FIGURE 1.
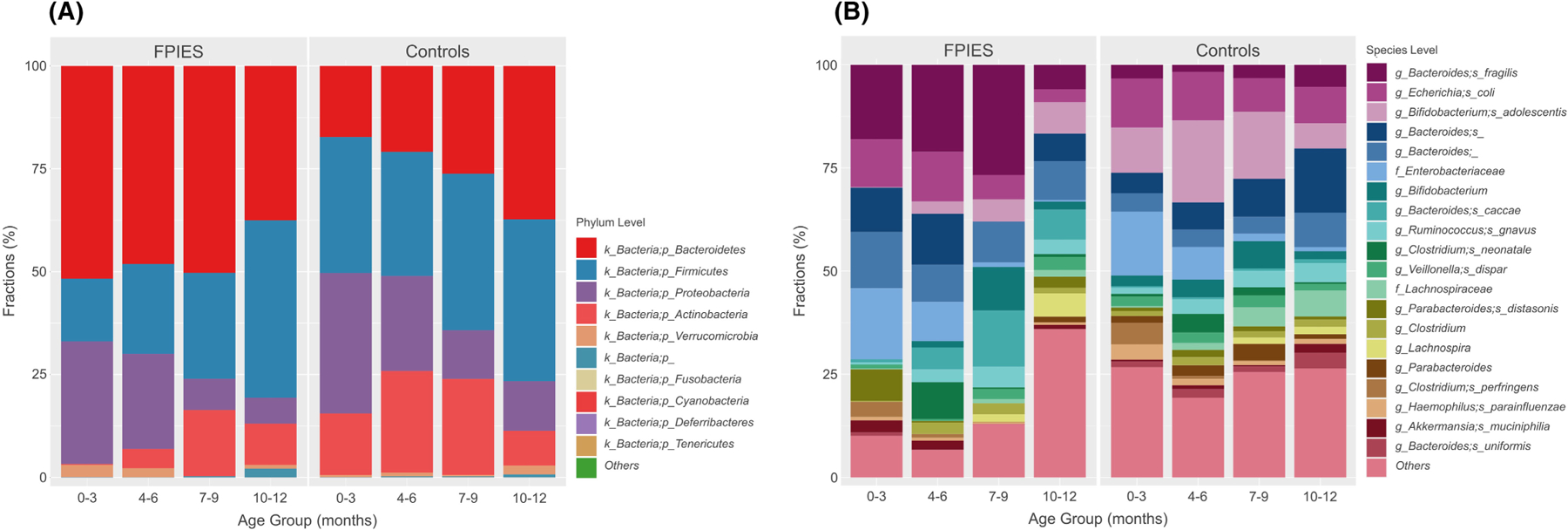
The stacked bar plots demonstrate the difference between FPIES and controls in different age groups at the (A) phylum level and (B) species level. The case numbers provide stools for final analysis at each age groups: FPIES N = 8 (0–3 months), 7 (4–6 months), 4 (7–9 months), 6 (10–12 months); Controls N = 77 (0–3 months), 67 (4–6 months), 48 ((7–9 months), 35 (10–12 months).
At the species level, Bacteroides fragilis, Ruminococcus bromii, Parabacteroides distasonis, and Clostridium baratii were all more abundant in children with FPIES at 0–3 months of age (all q < .05, Figure 1B). At 4–6 months of age, Bacteroides fragilis also trended toward higher abundance in FPIES cases (q = .06, p = 2.92 × 10−4). In contrast, Bifidobacterium spp., especially Bifidobacterium adolescentis, were less abundant in stool from children with FPIES at both 0–3 months and 4–6 months of age (q < .05, Figure 1B).
LEfSe was used to determine taxa that consistently characterize the different groups. This approach identified a higher abundance of pathobionts in the stools from FPIES children, including: Clostridium perfringens, Bacteroides caccae, Ruminococcus bromii, Gemmiger formicilis, and Bacteroides ovatus at 0–3 months of age and Acinetobacter lwoffii at 4–6 months of age (LDA score >2, Figure 2).
FIGURE 2.
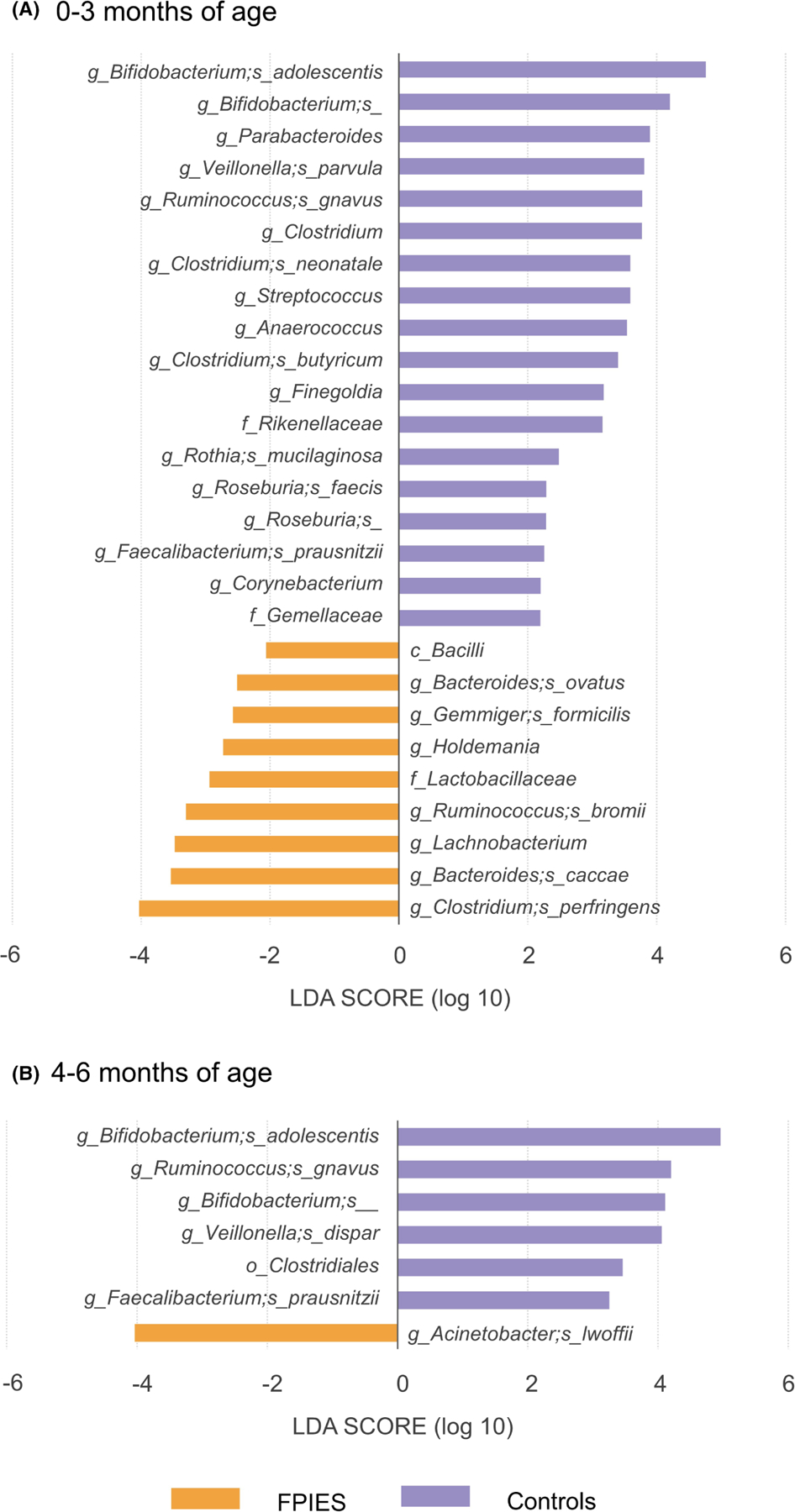
Distinct gut microbiome composition between FPIES cases and controls. The linear discriminant analysis (LDA) scores were calculated by LEfSe to demonstrate the different abundance of taxa at (A) 0–3 months of age and (B) 4–6 months of age.
Short-chain fatty acid (SCFA) producing bacteria and commensal bacteria were found to be less abundant in the stools from children with FPIES. For example, at 0–3 months of age, FPIES children had less Bifidobacterium adolescentis, Veillonella parvula, Ruminococcus gnavus, Clostridium neonatale, Clostridium butyricum, Rothia mucilaginosa, Roseburia faecis, and Faecalibacterium prausnitzii in their stool (LDA score >2, Figure 2A). And at 4–6 months of age, Bifidobacterium adolescentis, Veillonella dispar, Ruminococcus gnavus, and Faecalibacterium prausnitzii were significantly less abundant (LDA score >2, Figure 2B).
In an effort to corroborate and increase confidence in these findings, we also applied ANCOM, a more conservative microbiome method, to evaluate for any differences between groups. The decrease of Bifidobacterium adolescentis in the stool of FPIES children at 0–3 and 4–6 months of age was again demonstrated by ANCOM (W value = 270). Furthermore, metabolic pathway analysis by PICRUSt also revealed that the Bifidobacterium shunt, a metabolic pathway responsible for producing SCFA from oligosaccharides,31 was expressed significantly less in the stool from FPIES patients. The significantly different bacterial genera and species by MaAsLin2 linear model, ANCOM, and LEfSe are summarized in Table 3. Finally, we applied a random forest machine learning model to differentiate FPIES from controls. Bifidobacterium spp., Bifidobacterium adolescentis, Roseburia spp., and Bacteroides fragilis were the top four bacteria that differentiated children with FPIES from controls (Figure 3).
TABLE 3.
Microbiome predominant in FPIES versus controls at different age bins
| Age bin | Group | MaAsLin2 | ANCOM | LEfSe |
|---|---|---|---|---|
| 0–3 months | FPIES |
Bacteroides fragilis Ruminococcus bromii Parabacteroides distasonis Clostridium baratii Bacteroides spp. Holdemania spp. Lachnobacterium spp. Sutterella spp. Phascolarctobacterium spp. Anaerotruncus spp. |
None |
Clostridium perfringens Bacteroides caccae Ruminococcus bromii Gemmiger formicilis Bacteroides ovatus Holdemania spp. Lachnobacterium spp. |
| Controls | Bifidobacterium spp. | Bifidobacterium adolescentis |
Bifidobacterium adolescentis Veillonella parvula Ruminococcus gnavus Clostridium neonatale Clostridium butyricum Rothia mucilaginosa Roseburia faecis Faecalibacterium prausnitzii Bifidobacterium spp. Parabacteroides spp. Clostridium spp. Streptococcus spp. Anaerococcus spp. Finegoldia spp. Roseburia spp. Corynebacterium spp. |
|
| 4–6 months | FPIES | Bacteroides fragilis * | Acinetobacter lwoffii | |
| Controls | None | Bifidobacterium adolescentis |
Bifidobacterium adolescentis Ruminococcus gnavus Veillonella dispar Faecalibacterium prausnitzii Bifidobacterium spp. |
Note: The q-value of all bacteria <.05 except the one marked with asterisk.
p = 2.92 × 10−4, q = .06.
FIGURE 3.
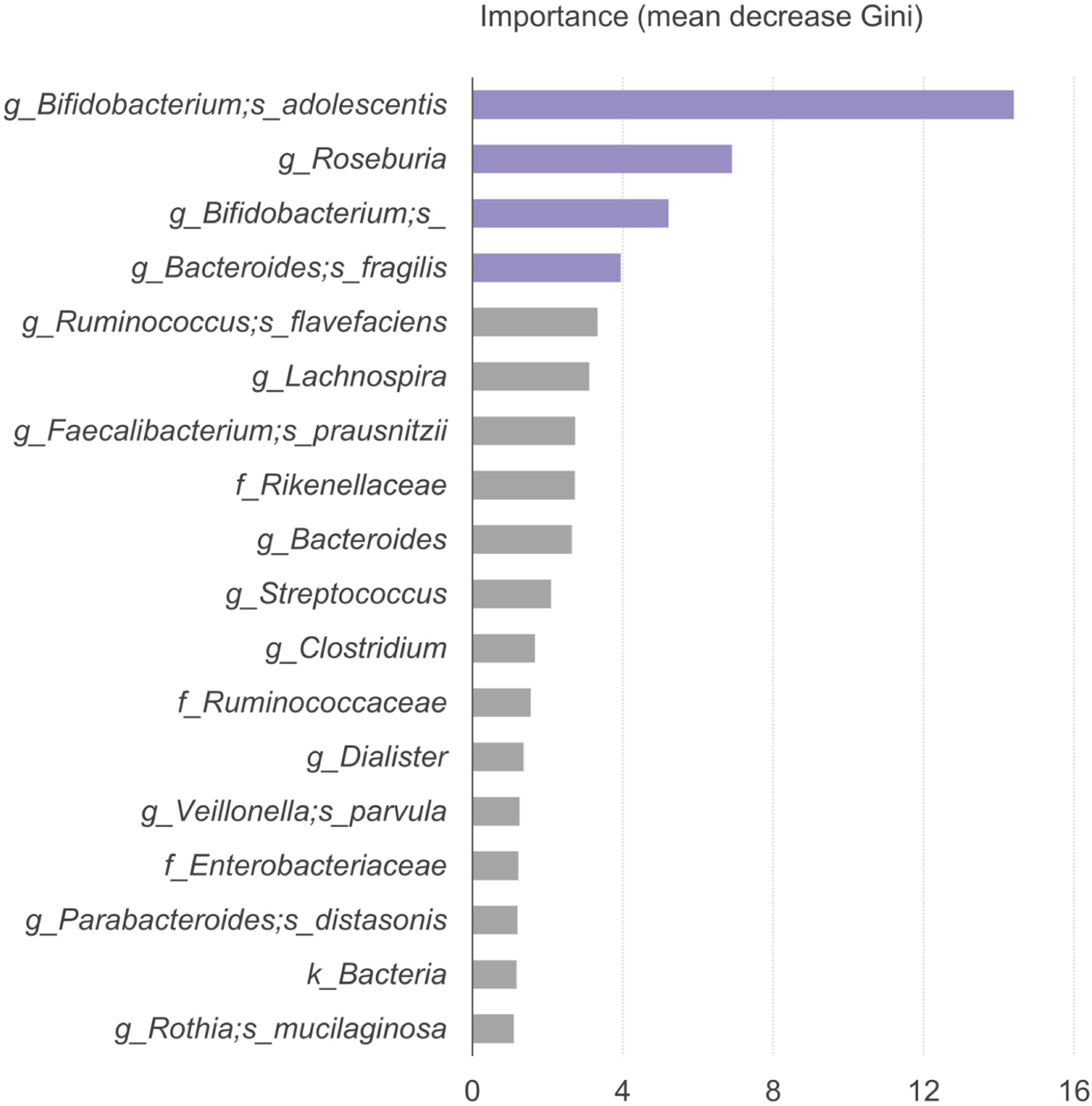
The bacteria predicted by machine learning random forest model to differentiate FPIES from controls. The microbe with higher mean decreases in the Gini index indicates higher importance in differentiating children with FPIES from healthy controls.
4 |. DISCUSSION
FPIES is regarded as a rare non-IgE-mediated food allergic disease. Using a population-based study design, Mehr et al. estimated FPIES annual incidence during the years 2012–2014 in Australian children under 2 years old to be 0.0154%.3 By comparison, Katz et al. reported the accumulated incidence of cow’s milk-triggered FPIES during the first 2 years was 0.34% in one Israel hospital.2 Nowak-Wegrzyn et al. reported that by a telephone survey in the United States, the estimated prevalence was 0.51% in children and 0.22% in adults.4 In a prospective unselected birth cohort, Alonso et al. identified 8 FPIES of 1142 Spanish children—a cumulative incidence of 0.7% in 3 years,5 very similar to the 0.92% prospectively identified here.
Unlike early studies in the United States and elsewhere,8,32 solid foods, such as oat and rice, are more commonly reported as FPIES triggers in recent studies. Blackman et al. in Texas revealed that oat (53%) and rice (35%) became the leading cause in their study.6 In the Boston area, our group also previously reported from a retrospective review of more than 5 million medical records identifying 203 FPIES cases, that oat (34.5%) and rice (29.6%) were the most common triggers in acute FPIES children.7 This prospective cohort study has similar findings. Oat and rice (each with three cases and one case triggered by both) were the most common foods inducing acute FPIES, while two children had cow’s milk FPIES. There are several possible explanations, including the awareness of solid food-triggered FPIES, the change of habits of solid food introduction, and selection bias.6
Early life gut microbiome changes have been associated with food allergies and other respiratory allergies in different studies.18,19,33 The decreased production of SCFA is one of the common mechanistic explanations for the association between dysbiosis and allergic diseases.33 Taking advantage of the serial collection of stool samples in a prospective study design, we revealed that the bacterial diversity between FPIES and controls was different during early infancy. The stool microbiome diversity was lower in FPIES than in controls at 4–6 months of age. The stool microbiome in children with FPIES had a distinct pattern from birth to 6 months old. Commensal Bifidobacterium and Clostridium species were less abundant among FPIES children from 0 to 6 months of age, consistent with potentially lower SCFA production. FPIES infants had more pathobiont species detected in their fecal microbiome, even before the onset of FPIES symptoms, consistent with a propensity for inflammation. Because the percentage of probiotics supplements in FPIES infants was higher than in controls (25% in FPIES vs. 13% in controls), the increase of Bifidobacterium species observed in the control group was unlikely due to the supplement of probiotics at the first 6 months of age.
The gut microbiome interacts with many aspects of our immune system that may increase the risk of food allergy, including FPIES.14,18,19 Regulatory T cells, an important immune cell for oral tolerance development, are known to be decreased when SCFA produced by the commensal microbes are lower.34 When commensal anaerobic bacteria, such as Bifidobacterium and Clostridium, are decreased, food allergens have been shown to more easily penetrate the intestinal barrier, leading to more food sensitization.35 The relative deficiency of commensal Bifidobacterium and Clostridium species, including Bifidobacterium adolescentis, Clostridium neonatale, and C. butyricum, observed among these children with FPIES may be an important factor causing barrier dysfunction.35 And, barrier dysfunction is likely an important factor causing FPIES.14
Interleukin-17-dominant inflammation has been observed following FPIES reactions.13 Henrick et al.36 have demonstrated that a lack of Bifidobacteria leads to T helper 17 and T helper 2-dominant intestinal inflammation. Bifidobacterium shunt, a metabolic pathway responsible for producing SCFA from oligosaccharides,31 was expressed significantly less in the stool from FPIES patients. Therefore, the dysbiosis we have documented in these patients from birth may play an important role in the inflammatory pathogenesis of FPIES.
After literature review, we identified one article and one abstract reporting on fecal microbiome associations with FPIES. Caparrós et al. compared the stool microbiome in 17 FPIES children (mean age, 7.5 ± 3.2 years) with 12 age-matched controls. They demonstrated stools from patients with FPIES were enriched with Lachnospiraceae species and had fewer Ruminococcaceae, Lactobacillaceae, and Leuconostocaceae species.37 In an abstract, Boyer et al. reported in a study of 41 infants with FPIES and 34 controls, that significantly more infants with FPIES had abundant (defined as >4%) of Gammaproteobacteria (primarily Escherich-Shigella and Balneatrix) and Porphyromonadaceae (primarily Parabacteroides) taxa while significantly more allergy-free infants had abundant levels of Prevotella (p < .05).38 The findings of the two researches were comparable to ours. Nevertheless, we demonstrated that dysbiosis appeared in early infancy before the FPIES symptoms onset.
This study has several important limitations. First is the relatively small number of FPIES cases. Multi-centered nationwide or international birth cohorts will be necessary to provide more cases to perform more detailed serial microbiome analyses. Another limitation is that FPIES in this study was not diagnosed by oral food challenge. According to the current FPIES diagnostic guideline, oral food challenge is not necessary when the clinical presentation is typical.1 The diagnosis of FPIES in this study was reviewed and confirmed by study investigators with experience according to the consensus guideline.1 The major strength of this study is the prospective birth cohort study design. Even though such design is labor-intensive, it provides detailed demographic data and serial stool samples before and after the onset of FPIES.
In conclusion, in a prospective birth cohort, we report the cumulative incidence of FPIES was 0.92% over 3 years, slightly higher than other retrospective and prospective studies. The most common trigger foods in this cohort were oat and rice, followed by cow’s milk, similar to our recent retrospective chart-review results.7 The average age of disease presentation was 6 months old. Over the first 6 months of life, children with FPIES have more potentially pathogenic bacteria in their stools and significantly less Bifidobacterium spp. compared with controls. We hypothesize that the resultant decreased SCFA production may play an important role in the pathogenesis of FPIES. Validation of these findings in additional cohorts and elucidation of the detailed pathophysiologic mechanisms warrant further study.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Gerber Foundation, the Demarest Lloyd Jr. Foundation (230465), and the Food Allergy Science Initiative (229711). K.W.S. was supported by a grant from Chang Gung Memorial Hospital (CMRPG2K0332). VMM and YVV were supported by grants from the National Institute of Allergy and Infectious Diseases of the US National Institutes of Health (1K23AI151555-01A1 and K23AI130408 respectively). R.I.S. was supported by NIH/NIDDK grant P30 DK040561. We acknowledged Hera Vlamakis, Ph.D., Bahar Sayoldin, M.S., Thomas Cleland B.S., and other researchers at the Microbial Omics ‘Core of the Broad Institute for their technical expertise in performing stool DNA extraction, library preparation, and sequencing. We sincerely thank all the participants and their families in our GMAP birth cohort study and the entire staff at the Pediatrics of Newton Wellesley for their support of the study.
Funding information
Chang Gung Medical Foundation, Grant/Award Number: CMRPG2K0332; Demarest Lloyd Jr. Foundation, Grant/Award Number: 230465; Food Allergy Science Initiative, Grant/Award Number: 229711; Gerber Foundation; National Institute of Allergy and Infectious Diseases of the US National Institutes of Health, Grant/Award Number: 1K23AI151555-01A1 and K23AI130408; NIH/NIDDK, Grant/Award Number: P30 DK040561
Abbreviations:
- FPIES
food protein-induced enterocolitis syndrome
- LDA
linear discriminant analysis
- SCFAs
short-chain fatty acid
Footnotes
CONFLICT OF INTEREST
All authors have no relevant conflict of interest.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Nowak-Wegrzyn A, Chehade M, Groetch ME, et al. International consensus guidelines for the diagnosis and management of food protein-induced enterocolitis syndrome: executive summary-workgroup report of the adverse reactions to foods committee, American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol 2017;139(4):1111–1126.e4. [DOI] [PubMed] [Google Scholar]
- 2.Katz Y, Goldberg MR, Rajuan N, Cohen A, Leshno M. The prevalence and natural course of food protein-induced enterocolitis syndrome to cow’s milk: a large-scale, prospective population-based study. J Allergy Clin Immunol 2011;127(3):647–653.e1–3. [DOI] [PubMed] [Google Scholar]
- 3.Mehr S, Frith K, Barnes EH, Campbell DE, FPIES Study Group. Food protein-induced enterocolitis syndrome in Australia: a population-based study, 2012–2014. J Allergy Clin Immunol 2017;140(5):1323–1330. [DOI] [PubMed] [Google Scholar]
- 4.Nowak-Wegrzyn A, Warren CM, Brown-Whitehorn T, Cianferoni A, Schultz-Matney F, Gupta RS. Food protein-induced enterocolitis syndrome in the US population-based study. J Allergy Clin Immunol 2019;144(4):1128–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonso SB, Ezquiaga JG, Berzal PT, et al. Food protein-induced enterocolitis syndrome: increased prevalence of this great unknown-results of the PREVALE study. J Allergy Clin Immunol 2019;143(1):430–433. [DOI] [PubMed] [Google Scholar]
- 6.Blackman AC, Anvari S, Davis CM, Anagnostou A. Emerging triggers of food protein-induced enterocolitis syndrome: lessons from a pediatric cohort of 74 children in the United States. Ann Allergy Asthma Immunol 2019;122(4):407–411. [DOI] [PubMed] [Google Scholar]
- 7.Su KW, Patil SU, Stockbridge JL, et al. Food aversion and poor weight gain in food protein-induced enterocolitis syndrome: a retrospective study. J Allergy Clin Immunol 2020;145(5):1430–1437. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruffner MA, Ruymann K, Barni S, Cianferoni A, Brown-Whitehorn T, Spergel JM. Food protein-induced enterocolitis syndrome: insights from review of a large referral population. J Allergy Clin Immunol Pract 2013;1(4):343–349. [DOI] [PubMed] [Google Scholar]
- 9.Dupont C, Heyman M. Food protein-induced enterocolitis syndrome: laboratory perspectives. J Pediatr Gastroenterol Nutr 2000;30 Suppl:S50-S57. [DOI] [PubMed] [Google Scholar]
- 10.Du YJ, Nowak-Wegrzyn A, Vadas P. FPIES in adults. Ann Allergy Asthma Immunol 2018;121(6):736–738. [DOI] [PubMed] [Google Scholar]
- 11.Goswami R, Blazquez AB, Kosoy R, Rahman A, Nowak-Wegrzyn A, Berin MC. Systemic innate immune activation in food protein-induced enterocolitis syndrome. J Allergy Clin Immunol 2017;139(6):1885–1896.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehr S, Lee E, Hsu P, et al. Innate immune activation occurs in acute food protein-induced enterocolitis syndrome reactions. J Allergy Clin Immunol 2019;144(2):600–602.e2. [DOI] [PubMed] [Google Scholar]
- 13.Berin MC, Lozano-Ojalvo D, Agashe C, Baker MG, Bird JA, Nowak-Wegrzyn A. Acute FPIES reactions are associated with an IL-17 inflammatory signature. J Allergy Clin Immunol 2021;148(3):895–901.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su KW, Shreffler WG, Yuan Q. Gastrointestinal immunopathology of food protein-induced enterocolitis syndrome and other non-immunoglobulin E-mediated food allergic diseases. Ann Allergy Asthma Immunol 2021;126(5):516–523. [DOI] [PubMed] [Google Scholar]
- 15.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med 2016;375(24):2369–2379. [DOI] [PubMed] [Google Scholar]
- 16.Khan S, Luck H, Winer S, Winer DA. Emerging concepts in intestinal immune control of obesity-related metabolic disease. Nat Commun 2021;12(1):2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmermann P, Messina N, Mohn WW, Finlay BB, Curtis N. Association between the intestinal microbiota and allergic sensitization, eczema, and asthma: a systematic review. J Allergy Clin Immunol 2019;143(2):467–485. [DOI] [PubMed] [Google Scholar]
- 18.Fazlollahi M, Chun Y, Grishin A, et al. Early-life gut microbiome and egg allergy. Allergy 2018;73(7):1515–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bunyavanich S, Shen N, Grishin A, et al. Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol 2016;138(4):1122–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumagai H, Maisawa S, Tanaka M, et al. Intestinal microbiota and secretory immunoglobulin A in feces of exclusively breastfed infants with blood-streaked stools. Microbiol Immunol 2012;56(10):657–663. [DOI] [PubMed] [Google Scholar]
- 21.Harris JK, Fang R, Wagner BD, et al. Esophageal microbiome in eosinophilic esophagitis. PLoS One 2015;10(5):e0128346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin VM, Virkud YV, Dahan E, et al. Longitudinal disease-associated gut microbiome differences in infants with food protein-induced allergic proctocolitis. Microbiome 2022;10(1):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin VM, Virkud YV, Seay H, et al. Prospective assessment of pediatrician-diagnosed food protein-induced allergic proctocolitis by gross or occult blood. J Allergy Clin Immunol Pract 2020;8(5):1692–1699.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho D, Imai K, King G. Matching as nonparametric preprocessing for reducing model dependence in parametric causal inference. Polit Anal 2007;15(3):199–236. [Google Scholar]
- 25.Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37(8):852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallick H, McIver LJ, Rahnavard A, et al. Multivariable Association in Population-scale Meta-omics Studies. PLoS Comput Biol 2021;17(11):e1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12(6):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandal S, Van Treuren W, White RA, Eggesbo M, Knight R, Peddada SD. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis 2015;26:27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16 S rRNA marker gene sequences. Nat Biotechnol 2013;31(9):814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn M Building predictive models in R using the caret package. J Stat Softw 2008;28(5):1–26.27774042 [Google Scholar]
- 31.Wolin MJ, Zhang Y, Bank S, Yerry S, Miller TL. NMR detection of 13CH313COOH from 3–13C-glucose: a signature for Bifidobacterium fermentation in the intestinal tract. J Nutr 1998;128(1):91–96. [DOI] [PubMed] [Google Scholar]
- 32.Caubet JC, Ford LS, Sickles L, et al. Clinical features and resolution of food protein-induced enterocolitis syndrome: 10-year experience. J Allergy Clin Immunol 2014;134(2):382–389. [DOI] [PubMed] [Google Scholar]
- 33.Arrieta MC, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 2015;7(307):307ra152. [DOI] [PubMed] [Google Scholar]
- 34.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013;504(7480):446–450. [DOI] [PubMed] [Google Scholar]
- 35.Stefka AT, Feehley T, Tripathi P, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A 2014;111(36):13145–13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henrick BM, Rodriguez L, Lakshmikanth T, et al. Bifidobacteria-mediated immune system imprinting early in life. Cell 2021;184(15):3884–3898.e11. [DOI] [PubMed] [Google Scholar]
- 37.Caparrós E, Cenit MC, Muriel J, et al. Intestinal microbiota is modified in pediatric food protein–induced enterocolitis syndrome. J Allergy Clin Immunol 2022;1:217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyer J, Scuderi V. Comparison of the gut microbiome between food protein-induced enterocolitis sydrome (FPIES) infants and allergy-free infants. Ann Allergy Asthma Immunol 2017;119:e3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


