Abstract
We report the development of a nested-PCR-based assay for the detection of Cryptococcus neoformans in cerebrospinal fluid. The specificity and sensitivity of the test were assessed. The technique was then applied to 40 cerebrospinal fluid samples. We obtained positive reactions for all 21 clinical samples from patients who had been previously diagnosed as having cryptococcal meningitis by conventional techniques and negative reactions for all 19 negative controls. Nested PCR is here compared with other diagnostic methods currently used in patients’ follow-up exams during anticryptococcal therapy.
Cryptococcus neoformans is a yeastlike fungus that is present in the environment worldwide and responsible for one of the most common infections of the central nervous systems of AIDS patients. Infection is acquired by inhalation of C. neoformans, and in an immunocompetent host it is confined to the lungs and is usually self-limiting and asymptomatic (6). In AIDS patients, on the other hand, C. neoformans usually tends to disseminate, the most common site of extrapulmonary infection being the meninges (1). The incidence of cryptococcal meningoencephalitis among patients with AIDS has been estimated to be 6 to 10% in developed countries (9). Laboratory diagnosis of cerebrospinal fluid (CSF) is traditionally based on microscopic examination of India ink preparations and on the detection of cryptococcal capsular polysaccharide antigen by a latex agglutination test. Direct microscopic examination is a rapid but quite insensitive test and strongly depends on the operator’s skills. The latex agglutination test is a more sensitive method but may still yield false-positive and false-negative results with either serum or CSF (2–4, 11). Moreover, the simple culture of CSF samples on Sabouraud agar is time-consuming; in fact, at least 4 days is necessary to detect positive cultures of C. neoformans (7). An enzyme-linked immunosorbent assay kit for the detection of capsular antigen is also available, with a sensitivity comparable to those of agglutination tests (13). PCR techniques would greatly improve diagnosis of cryptococcal meningitis, but no amplification protocol proposed so far is sensitive and/or specific enough to be used directly with CSF. In fact, application of DNA probes and PCR techniques to the identification of previously isolated C. neoformans strains has recently been described (5, 10), but the detection of the microorganism by PCR directly in clinical samples is documented only for pulmonary cryptococcosis (12). The aim of the present work was to design a PCR for specific detection of C. neoformans directly in CSF specimens.
To optimize the amplification procedure, we used purified DNA, extracted from two previously isolated C. neoformans var. neoformans serotype A strains (SS-12 and SS-22) and from one C. neoformans var. gattii serotype B strain (CR-53-UCSC). Yeasts were cultured in YEPD medium (1% yeast extract, 2% peptone, 2% dextrose) in a shaking incubator at 30°C for 48 h, and then an aliquot of 200 μl was harvested, pelleted at 600 × g, washed twice, and resuspended in 200 μl of buffered saline. DNA extraction was carried out according to the procedure described by Sandhu et al. (10). Briefly, 500 μl of GPT reagent (6 M guanidine thiocyanate dissolved in 50 mM Tris [pH 8.3]) and 500 μl of phenol buffered in Tris (pH 8.0) were added to a washed-yeast suspension in a screw-cap tube and boiled for 15 min; 250 μl of chloroform-isoamyl alcohol was then added, and the aqueous phase was separated by centrifugation, mixed with 500 μl of 100% isopropanol, and placed at −20°C for at least 1 h. Samples were centrifuged at 14,000 × g for 15 min, and the nucleic acid pellet was washed with ice-cold 70% ethanol, dried, and resuspended in sterile double-distilled water at a concentration of 200 μg/ml.
Two nested-primer pairs specific for internal transcribed spacer regions of ribosomal DNA of C. neoformans (5) were sequentially used. In the first amplification, we used the primers ITS-1 (3′-TCCGTAGGTGAACCTGCGG-5′) and CN-4 (3′-ATCACCTTCCCACTAACACATT-5′), which resulted in an amplicon of 415 bp. One microliter of DNA was amplified in a final volume of 25 μl, containing 10 mM Tris-HCl (pH 8.80), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 200 μmol of each deoxyribonucleotide, 12.5 pmol of each primer, and 0.5 U of DynaZyme thermostable DNA polymerase (FynnZymes, Oy, Finland). Amplification conditions consisted of 20 cycles of consecutive denaturation, annealing, and DNA extension (96°C for 45 s, 55°C for 1 min, and 72°C for 1 min, respectively). One microliter of the amplification product was then used as the template for the second reaction, carried out for 30 cycles under the same conditions as the first cycle but in the presence of the primers CN-5 (3′-GAAGGGCATGCCTGTTTGAGAG-5′) and CN-6 (3′-TTTAAGGCGAGCCGACGTCCTT-5′), which produced an amplicon of 116 bp. Amplification products were electrophoresed through a 1% agarose gel and visualized with a UV transilluminator after ethidium bromide staining. Standard procedures to prevent carryover and contamination were performed (8). DNAs from both C. neoformans varieties gave positive amplification.
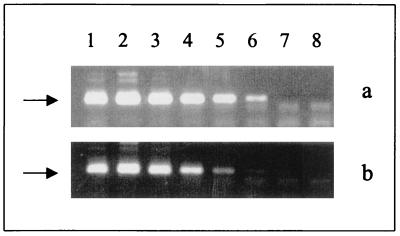
Since amplification techniques can present both sensitivity and specificity problems (8), we have addressed such problems in our experiments. Serial dilutions of DNA extracted from controlled amounts of C. neoformans cells (numbers of microorganisms, 106, 105, 104, 103, 102, 10, and 1) were subjected to nested PCR. The sensitivity of the technique was extremely high: it easily detected purified DNA corresponding to as few as 10 cryptococcal cells (Fig. 1a). Since amplification of specific DNAs extracted directly from clinical samples is known to be limited by the presence of several inhibitors (8), we also tested the sensitivity of the nested PCR using as the template DNA from a CSF sample known to be negative for C. neoformans that had been previously seeded with controlled amounts of cryptococcal cells (10-fold dilutions ranging from 106 cells to 1 cell per reaction). A weak-positive amplification was obtained in the presence of 10 yeast cells, as demonstrated by the appearance of a faint band in lane 7 of Fig. 1b. The specificity of the reaction was verified with, as the templates, DNAs extracted from different microorganisms that can be found in CSF, namely, Candida albicans, Candida glabrata, Candida guilliermondii, Candida famata, Candida krusei, Candida tropicalis, Trichosporon beigelii, Blastoschizomyces capitatus, Staphylococcus aureus, Escherichia coli, and Mycobacterium tuberculosis, and DNA from human cells. In all cases the nested PCR gave negative results.
FIG. 1.
Sensitivity of nested PCR with, as the templates, serial dilutions of DNAs extracted from cultured C. neoformans (a) and from CSF samples previously seeded with decreasing numbers of yeast cells (b). Lanes 1 to 7 show amplicons with cryptococcal cells numbering 106, 105, 104, 103, 102, 10, and 1, respectively. Lane 8 represents the negative control.
The technique was then validated with clinical samples; 21 CSF samples from AIDS patients with acute cryptococcal meningitis and 19 CSF samples from patients diagnosed with bacterial or viral meningitis were tested. All specimens were obtained at the Infectious Diseases Institute of the University of Sassari; an aliquot of 200 μl was immediately stored at −80°C for DNA extraction. Diagnosis of cryptococcal meningitis was made by microscopic observation of at least 1 yeast cell/field in India ink preparations and confirmed by isolation of C. neoformans on Sabouraud agar. All CSF samples from patients with cryptococcal meningitis showed an antigenic titer of at least 1:1,024 when they were tested by the CALAS polyclonal latex agglutination kit (Meridian Diagnostics, Cincinnati, Ohio). The stored aliquots of each CSF sample were subjected to DNA extraction, according to the protocol described above. For each sample, extracted DNA was resuspended in 25 μl of double-distilled water and 1 μl was subjected to nested PCR. Both DNA extraction and amplification were done in triplicate. All 21 samples obtained from patients with acute cryptococcal meningitis were positive when they were subjected to nested PCR, while all 19 DNA samples from the control group were negative.
Nested PCR was then compared with the other currently used diagnostic techniques in the follow-up of a patient with cryptococcal meningitis, starting from the first diagnosis and continuing through the course of anticryptococcal therapy. Table 1 shows the results obtained. Antigenic titers decreased very slowly both in serum and in CSF, despite the fact that no viable cryptococcal cell could be detected by cultivation on Sabouraud agar. The one yeast cell microscopically observed in CSF sample 3 could be explained (i) by the persistence of inviable yeast cells, which are morphologically indistinguishable from living cells, or (ii) simply as an artifact. Nested PCR is able to monitor the presence of living forms of C. neoformans better than microscopic examination or the latex agglutination test. In fact, the cryptococcal antigen titer was still high even 5 months after adequate therapy. The positive results obtained by the latex agglutination test are probably due to antigen persistence rather than to the presence of living forms of the yeast.
TABLE 1.
Comparison of results obtained by nested PCR and standard techniques with samples from a patient with cryptococcal meningitis
| Samplea | Date of sample (day/mo/yr) | Test resultb with India ink | CSF antigen titer | Serum antigen titer | Test resultc by:
|
|
|---|---|---|---|---|---|---|
| Cultivation | Nested PCR | |||||
| 1 | 28/5/97 | +++ | 1:2,048 | 1:2,048 | + | + |
| 2 | 10/6/97 | − | 1:2,048 | 1:2,048 | − | − |
| 3 | 26/6/97 | + | 1:128 | 1:512 | − | − |
| 4 | 11/7/97 | − | 1:128 | 1:128 | − | − |
| 5 | 6/9/97 | − | 1:32 | 1:128 | − | − |
| 6 | 11/11/97 | − | 1:8 | 1:128 | − | − |
Sample 1 is the sample obtained before the patient started therapy. Sample 2 was obtained during pharmacological therapy, and samples 3 to 6 were obtained after therapy.
+++, more than four C. neoformans cells per field; +, only one yeast cell observed in 10 fields; −, no yeast cells observed.
+, positive; −, negative.
These results demonstrate that the developed nested PCR is a sensitive, specific, and reproducible technique and that it represents a promising method to be used in the analysis of CSF samples from patients suspected of having cryptococcal meningitis. Moreover, since in the course of treatment of cryptococcosis the duration of therapy is still controversial, usually depending on the time needed for clearance of cryptococcal antigen from the CSF as demonstrated by the latex agglutination test, the nested PCR may be a useful tool not only for the rapid diagnosis of acute cryptococcosis but also for monitoring during therapy the clearance of the parasite from patients in follow-up exams.
Acknowledgments
This work was supported in part by grants from the Regione Autonoma della Sardegna (Progetto Biotecnologie) and MURST 60%.
We thank Giulia Morace for kindly providing Cryptococcus and Trichosporon strains.
REFERENCES
- 1.Chuck S L, Sande M A. Infections with Cryptococcus neoformans in the acquired immunodeficiency syndrome. N Engl J Med. 1989;321:794–799. doi: 10.1056/NEJM198909213211205. [DOI] [PubMed] [Google Scholar]
- 2.Engler H D, Shea Y R. Effect of potential interference factors on performance of enzyme immunoassay and latex agglutination assay for cryptococcal antigen. J Clin Microbiol. 1994;32:2307–2308. doi: 10.1128/jcm.32.9.2307-2308.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamilton J R, Noble A, Denning D W, Stevens D A. Performance of Cryptococcus antigen latex agglutination kits on serum and cerebrospinal fluid specimens of AIDS patients before and after pronase treatment. J Clin Microbiol. 1991;29:333–339. doi: 10.1128/jcm.29.2.333-339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hay R J, Mackenzie D W R. False positive latex tests for cryptococcal antigen in cerebrospinal fluid. J Clin Pathol. 1982;35:244–245. doi: 10.1136/jcp.35.2.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell T G, Freedman E Z, White T J, Taylor J W. Unique oligonucleotide primers in PCR for identification of Cryptococcus neoformans. J Clin Microbiol. 1994;32:253–255. doi: 10.1128/jcm.32.1.253-255.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell T G, Perfect J R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris A J, Byrne T C, Madden J F, Reller L B. Duration of incubation of fungal cultures. J Clin Microbiol. 1996;34:1583–1585. doi: 10.1128/jcm.34.6.1583-1585.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Persing D H, Smith T F, Tenover F C, White T J. Diagnostic molecular microbiology. Principles and applications. Washington, D.C: American Society for Microbiology; 1993. [Google Scholar]
- 9.Powderly W G. Cryptococcal meningitis and AIDS. Clin Infect Dis. 1993;17:837–842. doi: 10.1093/clinids/17.5.837. [DOI] [PubMed] [Google Scholar]
- 10.Sandhu G S, Kline B C, Stockman L, Roberts G D. Molecular probes for diagnosis of fungal infections. J Clin Microbiol. 1995;33:2913–2919. doi: 10.1128/jcm.33.11.2913-2919.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamm A M, Polt S S. False-negative cryptococcal antigen test. JAMA. 1980;244:1359. [PubMed] [Google Scholar]
- 12.Tanaka K, Miyazaki T, Maesaki S, Mitsutake K, Kakeya H, Yamamoto Y, Yanagihara K, Hossain M A, Tashiro T, Kohno S. Detection of Cryptococcus neoformans gene with pulmonary cryptococcosis. J Clin Microbiol. 1996;34:2826–2828. doi: 10.1128/jcm.34.11.2826-2828.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanner D C, Weinstein M P, Fedorciw B, Joho K L, Thorpe J J, Reller L B. Comparison of commercial kits for detection of cryptococcal antigen. J Clin Microbiol. 1994;32:1680–1684. doi: 10.1128/jcm.32.7.1680-1684.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]