Abstract
The molecular mechanisms connecting environmental exposures to adverse endpoints are often unknown, reflecting knowledge gaps. At the Comparative Toxicogenomics Database (CTD), we developed a bioinformatics approach that integrates manually curated, literature-based interactions from CTD to generate a “CGPD-tetramer”: a 4-unit block of information organized as a step-wise molecular mechanism linking an initiating Chemical, an interacting Gene, a Phenotype, and a Disease outcome. Here, we describe a novel, user-friendly tool called CTD Tetramers that generates these evidence-based CGPD-tetramers for any curated chemical, gene, phenotype, or disease of interest. Tetramers offer potential solutions for the unknown underlying mechanisms and intermediary phenotypes connecting a chemical exposure to a disease. Additionally, multiple tetramers can be assembled to construct detailed modes-of-action for chemical-induced disease pathways. As well, tetramers can help inform environmental influences on adverse outcome pathways (AOPs). We demonstrate the tool’s utility with relevant use cases for a variety of environmental chemicals (eg, perfluoroalkyl substances, bisphenol A), phenotypes (eg, apoptosis, spermatogenesis, inflammatory response), and diseases (eg, asthma, obesity, male infertility). Finally, we map AOP adverse outcome terms to corresponding CTD terms, allowing users to query for tetramers that can help augment AOP pathways with additional stressors, genes, and phenotypes, as well as formulate potential AOP disease networks (eg, liver cirrhosis and prostate cancer). This novel tool, as part of the complete suite of tools offered at CTD, provides users with computational datasets and their supporting evidence to potentially fill exposure knowledge gaps and develop testable hypotheses about environmental health.
Keywords: database tool, chemical-disease pathway, molecular mechanisms, environmental health, knowledge gaps, adverse outcome pathway
Environmental health-related research seeks to understand how chemical exposures influence human health (Grondin et al., 2016). While associations between exposures and adverse outcomes (AOs) can be documented, there typically remain knowledge gaps with respect to the related molecular mechanisms connecting the 2. One approach to ascertaining these mechanisms is to use bioinformatics to integrate content from public databases and computationally identify potential connections (Davis et al., 2019, 2020). Toward that end, we developed the Comparative Toxicogenomics Database (CTD; http://ctdbase.org/) as a resource to explore how environmental chemicals interact with genes and influence biological processes to affect human health (Davis et al., 2023). Since 2005, CTD biocurators have manually curated scientific articles using controlled vocabularies and ontologies to help harmonize environmental health data and capture literature-based interactions relating chemicals, gene products, phenotypes, anatomical terms, and diseases from comparative organisms (Davis et al., 2015). Currently, CTD includes over 3.3 million manually curated interactions describing relationships between 17 200 chemicals, 54 000 gene products, 6300 phenotypes, 960 anatomical terms, and 4100 diseases, curated from over 142 000 research papers (http://ctdbase.org/about/dataStatus.go). In turn, CTD integrates these curated interactions with one another and with select external data sources to compute “inferences,” ie, predictive associations based upon shared intermediates (Davis et al., 2008, 2016). For example, if chemical C1 has a directly curated interaction with gene G1, and, independently, gene G1 has a directly curated association with disease D1, then chemical C1 can be inferred to have a relationship with disease D1 via the shared gene G1 (King et al., 2012). CTD inferences enable the postulation of novel connections that might not otherwise be apparent, as well as provide the potential molecular mechanisms that make the connection: chemical C1 might play a role in disease D1 via interaction with the intermediate gene G1.
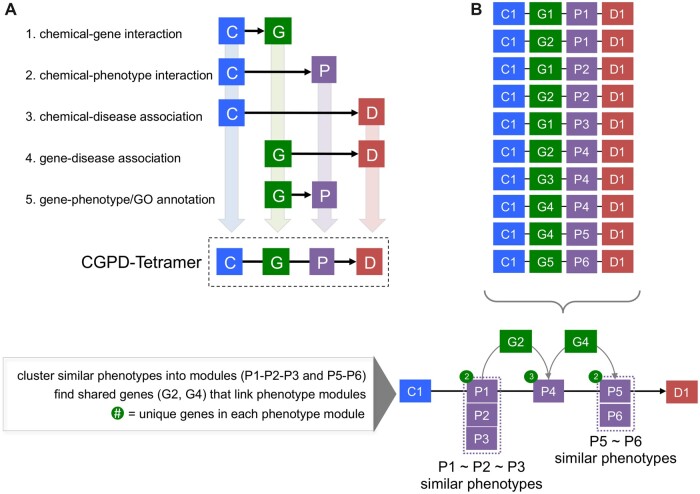
Expanding upon this approach, we developed a novel method that leverages curated interactions in CTD to generate a “CGPD-tetramer,” which is defined as a 4-unit computational block of information that relates an initiating Chemical with an interacting Gene and a modulated Phenotype to a Disease outcome (Davis et al., 2020). In this approach, 5 independently curated relationships in CTD are combined: a chemical-gene interaction, a chemical-phenotype interaction, a chemical-disease interaction, a gene-disease interaction, and a gene-phenotype/Gene Ontology (GO) annotation (Figure 1A). A CGPD-tetramer is generated only if all 5 curated statements exist in CTD as supporting evidence. If any one of the statements do not exist in CTD, the tetramer cannot be generated. In turn, the evidence-backed CGPD-tetramers that do emerge can be aligned by shared intermediate genes and similar phenotypes, allowing them to be assembled into potential mechanistic chemical-disease pathways that fill exposure knowledge gaps (Figure 1B).
Figure 1.
CGPD-tetramers. A, CGPD-tetramers are computationally generated information units that interrelate 4 data types from CTD: a chemical (C), gene product (G), phenotype (P), and disease (D). To generate a CGPD-tetramer by data integration, 5 individual lines of supporting evidence are required as directly curated interactions between the 4 data types: chemical-gene, chemical-phenotype, chemical-disease, gene-disease, and gene-phenotype/GO. If any 1 line of supporting evidence is lacking, the tetramer is not generated. B, Maximizing the shared gene and phenotype overlaps between individual tetramers allows them to be constructed into longer, more detailed molecular mechanistic pathways (see Davis et al. [2020] for details). Here, 10 tetramers involving 1 chemical (C1), 5 unique genes (G1–G5), 6 phenotypes (P1–P6), and 1 disease outcome (D1) can be combined. Similar phenotypes (eg, P1–P2–P3 and P5–P6) can be clustered into separate modules. Shared genes are used to bridge the modules through shared molecular mechanisms: gene G2 connects phenotype module P1–P2–P3 to phenotype P4, and then gene G4 bridges phenotype P4 with phenotype module P5–P6. Some examples of similar phenotypes might include all those involving cell death, such as “apoptotic process” (GO:0006915), “apoptotic signaling pathway” (GO:0097190), “programmed cell death” (GO:0012501), “positive regulation of apoptotic process” (GO:0043065), “cell death” (GO:0008219), etc.
Applying CTD content to fill knowledge gaps between exposure and adverse endpoints has been used to propose potential mechanisms and pathways for an array of environmental health events, including pesticide-induced prostate cancer (Grondin et al., 2016), 4-nonylphenol-associated Parkinson disease (Kosnik et al., 2019), ozone-induced myocardial infarction (Davis et al., 2020), sulforaphane-triggered AOs (Bozic et al., 2023), heavy metal-associated depression (Nguyen and Kim, 2023), air pollution/metal-induced Alzheimer disease (Davis et al., 2023), seizure risks associated with pesticides in medical cannabis (Pinkhasova et al., 2021), bisphenol A and benzo(a)pyrene-induced cell signaling modes-of-action for lung cancer (Stanic et al., 2021), arsenic-induced reproductive defects (Chai et al., 2021), and respiratory diseases resulting from e-cigarette chemicals (Grondin et al., 2021), among many others.
A major advantage to this computational approach is that no a priori knowledge is needed regarding relationships between a chemical, gene, phenotype, or disease to collect the datasets. Users simply input any chemical, gene, phenotype, and/or disease of interest, and CTD automatically constructs the relevant tetramers containing the data of interest. Previously, this process required complex manual integration of data content freely available from CTD (http://ctdbase.org/downloads/). Here, we introduce a new, user-friendly, online tool called CTD Tetramers that performs the necessary integration to generate and display computed CGPD-tetramers with their supporting lines of evidence. We demonstrate this novel tool’s utility with numerous relevant use cases.
Materials and methods
CTD data version and analysis
Analysis was performed using CTD public data available in March 2023 (revision 17024). CTD is updated with new content on a monthly basis (http://ctdbase.org/about/dataStatus.go); consequently, query results described in this text may vary over time. For analysis, CTD data pages and tetramer query results were downloaded (available formats: CSV, Excel, XML, or TSV) into spreadsheets and the sorting, advanced filtering, and subtotaling functions provided in Excel were used to survey and count the unique data types. Venn diagrams were constructed using CTD’s MyVenn tool (https://ctdbase.org/tools/myVenn.go) and the publicly available “Venny” tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html). For use cases, official CTD terms were input as query terms in the CTD Tetramers tool for chemicals, genes, phenotypes, and diseases, with the filtering parameter set to Direct Relationships (“return data for exact input query term only”), unless otherwise indicated in the text. The official CTD terms (and their CTD accession identifiers) include: case 1: “Fluorocarbons” (MESH:D005466) and “Asthma” (MESH:D001249); case 2: “bisphenol A” (MESH:C006780), “Obesity” (MESH:D009765), “Diabetes Mellitus, Type 2” (MESH:D003924), and “Hypertension” (MESH: D006973); case 3: “apoptotic process” (GO:0006915), “spermatogenesis” (GO:0007283), and “Infertility, Male” (MESH:D007248); and case 4: “Carbon Tetrachloride” (MESH:D002251), “Liver Cirrhosis” (MESH: D008103), “cell death” (GO:0008219), “cell activation” (GO:0001775), “inflammatory response” (GO:0006954), and “collagen biosynthetic process” (GO:0032964).
Phenotype versus disease
CTD operationally distinguishes between the concept of phenotype and disease based upon the source vocabulary used by a CTD biocurator to annotate the chemical-induced endpoint reported in the literature (Davis et al., 2018). If the chemical-induced outcome is a term in the MEDIC disease vocabulary (http://ctdbase.org/voc.go?type=disease), then the information is curated and annotated as a disease data type in CTD using that MEDIC disease term (Davis et al., 2012). Consequently, any chemical-induced endpoint that does not exist in MEDIC is considered, de facto, a phenotype and instead is curated and annotated using a term from the GO (http://ctdbase.org/voc.go?type=go), which CTD uses as a controlled vocabulary for chemical-induced molecular and biological phenotypes (Davis et al., 2018). For example, “breast neoplasms” exists in MEDIC (MESH: D001943) and is therefore curated and treated as a disease data type in CTD, while “blood vessel endothelial cell migration” does not exist in MEDIC, and is consequently treated as a phenotype using the GO (GO:0043534). In this sense, phenotypes are observable biological events that could potentially be intermediary processes informing and influencing the normal and predisease state prior to clinical manifestation.
CTD Tetramer tool
CTD’s tetramer-generating functionality was initially written as standalone processes for internal (Davis et al., 2020; Grondin et al., 2021) and third-party analyses (Pinkhasova et al., 2021); these processes are now converted and fully integrated into ctdbase.org for public use (http://ctdbase.org/query.go?type=tetramer). CTD tetramers are generated by backend processes on a real-time basis, as are summaries of the supporting lines of evidence. The CTD architecture of front-facing and backend processes has been described elsewhere (Davis et al., 2011; Grondin et al., 2018). Briefly, ctdbase.org is primarily a Jakarta EE-based, Model-View-Controller architecture with PostgreSQL database management systems, all within the context of Linux and Apache/Tomcat.
AO term mapping
The “Key Event Relationships” file (https://aopwiki.org/downloads/aop_ke_ker.tsv) from the public AOP-Wiki database (https://aopwiki.org/) was downloaded (April 15, 2022), and 179 terms defined as “AOs” by AOP-Wiki were extracted. A CTD biocurator reviewed the individual AO terms and manually mapped them to corresponding terms and accession identifiers in either CTD Disease (http://ctdbase.org/voc.go?type=disease) or CTD GO/Phenotype (http://ctdbase.org/voc.go?type=go). Of the official 179 AOs, 153 (85%) mapped to 1 or more terms in CTD, including: 98 AOs resolved to a single corresponding CTD disease term, 29 AOs mapped to a single corresponding CTD phenotype, and 26 AOs mapped to 1 or more phenotype and/or disease terms. These mapped AOs are used in 297 AOPs. A few AO terms mapped to multiple terms in CTD; for example, the AO term “Hyperinflammation” (AO:1868) was mapped to both the CTD disease term “Inflammation” (MESH:D007249) and to the CTD phenotype term “positive regulation of inflammatory response” (GO: 0050729). Of the 179 AO terms, 26 (15%) could not be confidently mapped to a corresponding CTD term because they described concepts not found in CTD, eg, “Increased, Population” (AO:1164), “Inflammatory events in light-exposed tissues” (AO:1599), and “impaired, Hive thermoregulation” (AO:568).
Results and discussion
CTD Tetramers tool
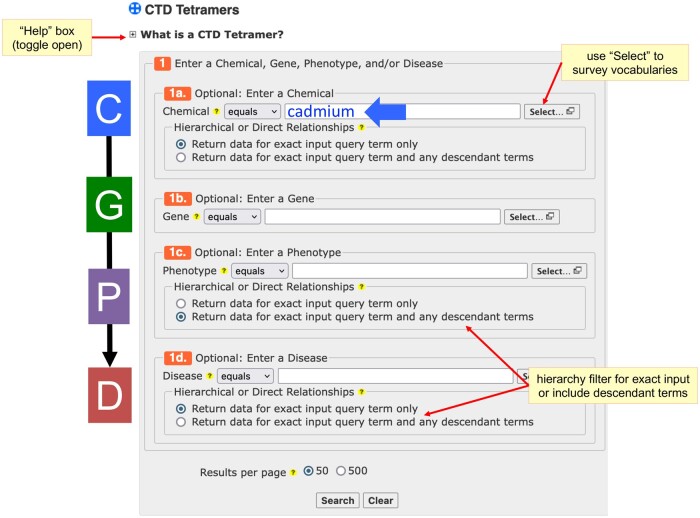
The new CTD Tetramers tool is available under the “Analyze” menu bar at CTD (http://ctdbase.org/query.go?type=tetramer). The query form simply requires a user to input any data types of interest, ie, a chemical, gene, phenotype, or disease (Figure 2). Since the CTD Chemical, CTD Phenotype, and CTD Disease vocabularies are hierarchical, users have the option to return tetramers for the exact input term only (eg, diabetes mellitus) or include the descendant to the input term (eg, diabetes mellitus, as well as the descendant terms gestational diabetes, type 1 diabetes, type 2 diabetes (T2D), diabetic neuropathies, diabetic angiopathies, diabetic ketoacidosis, etc.). Users can input their search terms directly into the fields or first explore the available terms from the CTD vocabularies by using the “Select” pop-up boxes for each data type.
Figure 2.
CTD Tetramers query page. CTD’s new tool CTD Tetramers (http://ctdbase.org/query.go?type=tetramer). Users simply enter at least 1 data type of interest in either the chemical, gene, phenotype, and/or disease query boxes. The “Select” pop-up windows allow users to first survey terms from the available vocabularies for each data type. As well, users can filter to retrieve either direct or hierarchical relationships for chemicals, phenotypes, and diseases. A toggle box at the top (“What is a CTD Tetramer?”) provides additional information about how tetramers are computed and how to use the tool.
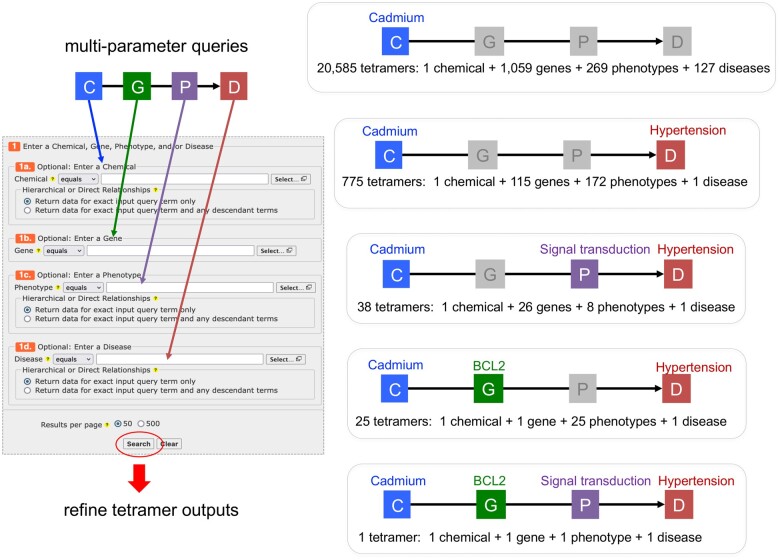
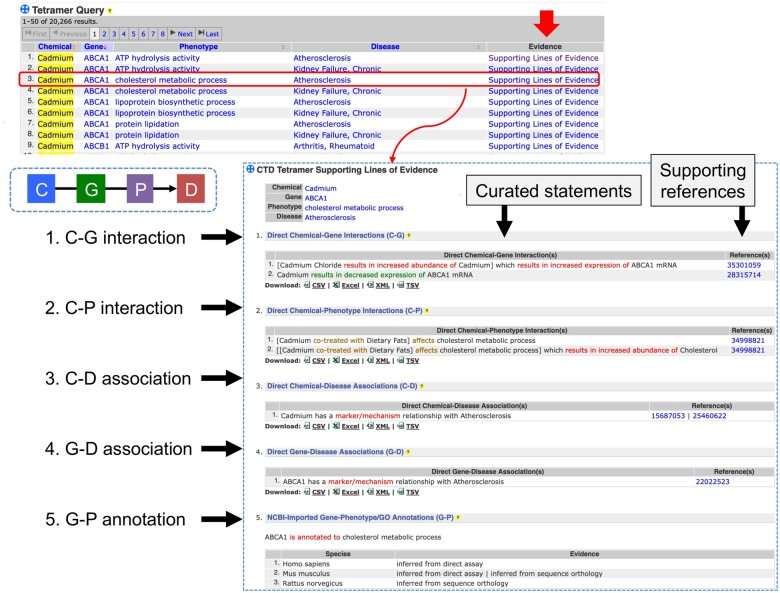
With 4 distinct types of data input, a user can perform simple searches or construct multiparameter queries (Figure 3). Searching with a single environmental chemical, such as “cadmium,” returns 20 585 tetramers linking this heavy metal with 1059 genes, 269 phenotypes, and 127 unique diseases. By clicking the “Revise query” button at the top of the output page, a user can modify the query by adding another data type, such as “hypertension” as the disease outcome, which reduces the output to 775 tetramers. A further query revision adding a phenotype such as “signal transduction” and its descendants refines the output to 38 tetramers for cadmium, 26 genes, 8 different signal transducing phenotypes, and hypertension. This iterative process allows users to survey the resulting data landscape for each query and make changes to refine or expand the results. Importantly, a user can initiate the query for any data type of interest; it does not have to be a chemical. For example, a user might be interested in a specific gene (eg, heme oxygenase HMOX1 yields over 19 700 tetramers composed of more than 790 chemicals, 45 phenotypes, and 60 diseases) or a particular disease or cellular phenotype (eg, “neuron projection development” generates over 4000 tetramers composed of more than 45 chemicals, 80 genes, and 300 diseases) to see how they can be connected to environmental health. Importantly, all returned tetramers include an “Evidence” column which documents the 5 supporting lines of evidence necessary to computationally construct each unique tetramer, providing both comprehensive traceability and transparency for the user, and allowing them the ability to assess the plausibility and strength of the relationships (Figure 4).
Figure 3.
Multiparameter queries. Users can enter up to 4 data types as input values simultaneously to perform multiparameter queries. A single input term (eg, cadmium as a chemical) generates over 20 500 tetramers composed of 1059 genes, 269 phenotypes, and 127 diseases. Including an additional data type (eg, hypertension as a disease outcome) refines the output to 775 tetramers composed of 115 genes and 172 phenotypes. Additional input terms further restrict the tetramer outputs, enabling users to navigate the data landscape by expanding or focusing the number of tetramers and data components.
Figure 4.
CTD tetramer transparency and traceability. Every generated CGPD-tetramer includes an “Evidence” column with a hyperlinked “Supporting Lines of Evidence,” providing both transparency and traceability as to how the tetramer was generated. The first 4 supporting lines of evidence (C-G, C-P, C-D, and G-D) are direct interactions annotated by CTD biocurators from the literature, and all the supporting interaction statements as well as the PubMed article identifiers (from whence the interactions were curated) are listed. The fifth supporting line of evidence (G-P) is electronically imported from gene-GO annotations provided by NCBI.
The CTD Tetramers tool generates data for constructing potential mechanistic modes-of-action for environmental health. In the use cases below, we demonstrate how the tool can be leveraged to retrieve datasets that enable users to survey environmental health events and help design testable hypotheses. The results below are not meant to be taken as finalized, resolved knowledge pathways, but are merely illustrative examples of how such hypotheses can be rapidly designed using results from this new online tool.
Use case 1: 1 chemical-1 disease: exploring perfluoroalkyl substances and asthma
Per- and polyfluoroalkyl substances (PFAS) are widespread fluorocarbon contaminants from industrial and consumer products. They are often dubbed “forever chemicals” due to their degradation-resistant molecular configuration, leading to environmental persistence and bioaccumulation (Berhanu et al., 2023; Langenbach and Wilson, 2021). Recently, PFAS exposure in children has been associated with immune system defects, including asthma, but the evidence is equivocal and a mode-of-action remains unknown (Beck et al., 2019; Jackson-Browne et al., 2020; Rappazzo et al., 2017; von Holst et al., 2021).
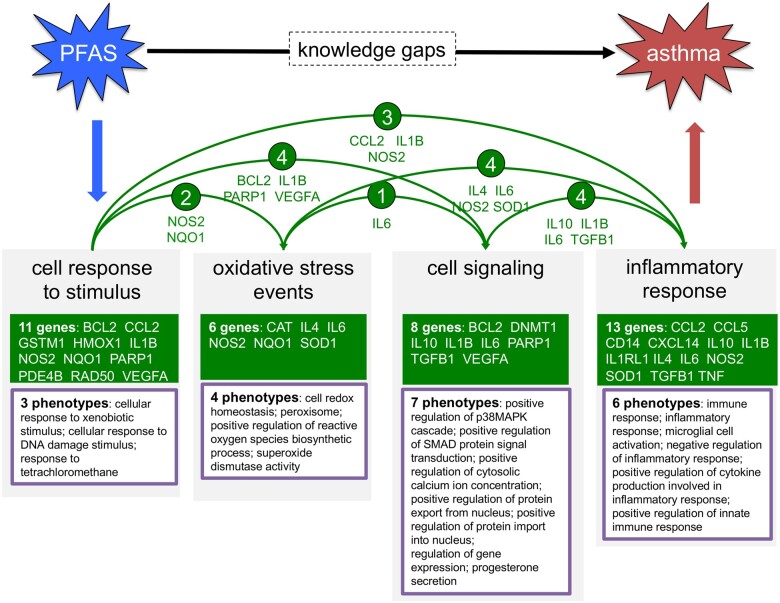
To see if we could fill such knowledge gaps with potential molecular intermediates and provide a testable model for verifying a relationship between PFAS exposure and asthma, we queried CTD Tetramers using “fluorocarbons” as the chemical parent input (and selected the hierarchy filter to include descendant chemicals) and “asthma” as the disease outcome. The query generated 203 tetramers that included only 2 PFAS chemicals (perfluorooctane sulfonic acid [PFOS] and perfluoro-n-nonanoic acid [PFNA]), 28 genes, and 70 phenotypes connected to asthma (Supplementary Table 1). This initial analysis provides a starting point for identifying potential molecular intermediates (Table 1), many of which have recognized mechanistic signaling roles in immune and inflammatory responses (eg, IL1B, TNF, IL4, IL6, IL10, IL1RL1, CCL2, CCL5, CXCL14). Next, following our previously described protocol for manually clustering similar phenotypes into groups (Davis et al., 2020), we assigned the 70 phenotypes to 14 broad modules, including the modules “Cell Response to Stimulus,” “Oxidative Stress Events,” “Cell Signaling,” and “Inflammatory Response” (Table 2). Starting with only those 4 modules, the data can be assembled into an initial framework using overlapping genes to establish mechanistic connections between the 4 modules, and many of these genes have a role in immune and inflammatory responses, making them candidates as molecular intermediates for PFAS-driven asthma in children (Figure 5). Importantly, additional phenotype modules can be incorporated into the framework. For example, a recent study observed that elevated serum triglycerides are associated with and might represent an unrecognized trait contributing to asthma (van Zelst et al. 2021). Interestingly, several of the computed phenotypes generated by CTD Tetramers are about lipid, cholesterol, and triglyceride metabolism and they involve shared genes/proteins (eg, CAT, IL4, and IL6) that enable the “Metabolism” module to be connected with the “Oxidative Stress Events,” “Cell Signaling,” and “Inflammatory Response” modules. Similarly, the role of apoptosis in asthma (Hough et al., 2020) enables the “Cell Death” module to be integrated into the same framework, with many overlapping genes/proteins (eg, BCL2, IL1B, IL6, NOS2, PARP1, SOD1, TGFB1, TNF, and VEGFA) to provide the molecular connections. In this approach, CTD Tetramers provide the initial datasets to help users construct and tailor chemical-disease pathways that include modules of interest into testable frameworks for environmental health.
Table 1.
Genes from 203 CGPD-tetramers connecting PFAS exposure with asthma
| Gene | No. associated tetramers | No. associated phenotypes |
|---|---|---|
| BCL2 | 18 | 15 |
| IL1B | 18 | 15 |
| SOD1 | 14 | 13 |
| TGFB1 | 14 | 14 |
| TNF | 14 | 11 |
| IL6 | 13 | 10 |
| VEGFA | 13 | 11 |
| IL4 | 9 | 7 |
| NOS2 | 9 | 7 |
| PARP1 | 9 | 9 |
| CAT | 7 | 5 |
| CCL2 | 6 | 6 |
| CCL5 | 6 | 6 |
| HMOX1 | 6 | 5 |
| ICAM1 | 6 | 6 |
| DNMT1 | 5 | 5 |
| IL10 | 5 | 5 |
| NQO1 | 5 | 5 |
| CYP2E1 | 4 | 2 |
| GSTM1 | 4 | 4 |
| MMP9 | 4 | 4 |
| CD14 | 3 | 2 |
| CXCL14 | 3 | 3 |
| GSTP1 | 3 | 3 |
| BGLAP | 2 | 2 |
| IL1RL1 | 1 | 1 |
| PDE4B | 1 | 1 |
| RAD50 | 1 | 1 |
Table 2.
Phenotypes from 203 CGPD-tetramers connecting PFAS exposure with asthma, manually grouped into phenotype modules
| Phenotype | No. associated tetramers | No. associated genes | Phenotype module |
|---|---|---|---|
| Cellular response to xenobiotic stimulus | 8 | 8 | Cell response to stimulus |
| Cellular response to DNA damage stimulus | 3 | 3 | Cell response to stimulus |
| Response to tetrachloromethane | 1 | 1 | Cell response to stimulus |
| Cell redox homeostasis | 3 | 3 | Oxidative stress events |
| Peroxisome | 3 | 3 | Oxidative stress events |
| Superoxide dismutase activity | 2 | 2 | Oxidative stress events |
| Positive regulation of reactive oxygen species biosynthetic process | 1 | 1 | Oxidative stress events |
| Regulation of gene expression | 4 | 4 | Cell signaling |
| Positive regulation of p38MAPK cascade | 2 | 2 | Cell signaling |
| Positive regulation of protein import into nucleus | 2 | 2 | Cell signaling |
| Positive regulation of SMAD protein signal transduction | 2 | 2 | Cell signaling |
| Positive regulation of cytosolic calcium ion concentration | 1 | 1 | Cell signaling |
| Positive regulation of protein export from nucleus | 1 | 1 | Cell signaling |
| Progesterone secretion | 1 | 1 | Cell signaling |
| Inflammatory response | 14 | 9 | Immune/inflammatory response |
| Immune response | 8 | 8 | Immune/inflammatory response |
| Negative regulation of inflammatory response | 3 | 3 | Immune/inflammatory response |
| Microglial cell activation | 2 | 2 | Immune/inflammatory response |
| Positive regulation of cytokine production involved in inflammatory response | 2 | 2 | Immune/inflammatory response |
| Positive regulation of innate immune response | 1 | 1 | Immune/inflammatory response |
| Apoptotic process | 9 | 9 | Cell death |
| Positive regulation of apoptotic process | 9 | 9 | Cell death |
| Positive regulation of cell death | 2 | 1 | Cell death |
| Activation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 1 | Cell death |
| Positive regulation of neuron death | 1 | 1 | Cell death/nervous system |
| Positive regulation of cell migration | 6 | 6 | Cell migration |
| Cell chemotaxis | 4 | 4 | Cell migration |
| Cell migration | 4 | 4 | Cell migration |
| Chemotaxis | 3 | 3 | Cell migration |
| Cell adhesion | 3 | 3 | Cell process |
| Cell-cell adhesion | 3 | 3 | Cell process |
| Positive regulation of epithelial to mesenchymal transition | 3 | 3 | Cell process |
| Establishment of endothelial barrier | 1 | 1 | Cell process |
| Establishment of Sertoli cell barrier | 1 | 1 | Cell process |
| Positive regulation of cell population proliferation | 13 | 7 | Cell proliferation |
| Negative regulation of cell population proliferation | 7 | 7 | Cell proliferation |
| Cell population proliferation | 5 | 3 | Cell proliferation |
| Negative regulation of mitotic cell cycle | 3 | 3 | Cell proliferation |
| Cell growth | 2 | 1 | Cell proliferation |
| Stem cell proliferation | 2 | 2 | Cell proliferation |
| Mitotic cell cycle | 1 | 1 | Cell proliferation |
| DNA methylation | 1 | 1 | DNA methylation |
| DNA methylation involved in embryo development | 1 | 1 | DNA methylation |
| DNA methylation on cytosine within a CG sequence | 1 | 1 | DNA methylation |
| Cholesterol metabolic process | 4 | 2 | Metabolism |
| Lipid metabolic process | 4 | 3 | Metabolism |
| Triglyceride metabolic process | 4 | 2 | Metabolism |
| Glutathione metabolic process | 3 | 3 | Metabolism |
| Glucose homeostasis | 2 | 1 | Metabolism |
| Glutathione derivative biosynthetic process | 2 | 2 | Metabolism |
| Metabolic process | 2 | 2 | Metabolism |
| Negative regulation of protein catabolic process | 2 | 2 | Metabolism |
| Aerobic respiration | 1 | 1 | Metabolism |
| ATP biosynthetic process | 1 | 1 | Metabolism |
| Negative regulation of ATP biosynthetic process | 1 | 1 | Metabolism |
| Regulation of mitochondrial membrane potential | 2 | 2 | Mitochondrial |
| Mitochondrion organization | 1 | 1 | Mitochondrial |
| Cognition | 2 | 1 | Nervous system |
| Sensory perception of sound | 2 | 2 | Nervous system |
| Locomotory behavior | 1 | 1 | Nervous system |
| Memory | 1 | 1 | Nervous system |
| Regulation of blood pressure | 6 | 3 | Physiology/development |
| Determination of adult lifespan | 2 | 2 | Physiology/development |
| Bone mineralization | 1 | 1 | Physiology/development |
| Developmental growth | 1 | 1 | Physiology/development |
| Negative regulation of adipose tissue development | 1 | 1 | Physiology/development |
| Ovarian follicle development | 4 | 4 | Reproduction |
| Decidualization | 1 | 1 | Reproduction |
| Ovulation | 1 | 1 | Reproduction |
| Spermatogenesis | 1 | 1 | Reproduction |
Figure 5.
Use case 1: computing mechanistic framework to fill knowledge gaps for PFAS and asthma. The CTD Tetramers tool generated 203 CGPD-tetramers connecting 2 PFAS chemicals to asthma (Supplementary Table 1). Similar phenotypes were manually grouped and binned into modules (Table 2), such as “oxidative stress events” made up of 4 distinct phenotypes. Here, 4 modules are shown (“cell response to stimulus,” “oxidative stress events,” “cell signaling,” and “immune/inflammatory response”), composed of 23 total genes and 20 unique phenotypes. These modules are connected by shared genes (arrows). Importantly, additional modules (eg, “cell death” and “triglyceride metabolism” from Table 2) can be incorporated to further expand the chemical-disease pathway. The computed molecular mechanisms offer potential solutions that fill knowledge gaps between PFAS and asthma.
Use case 2: 1 chemical-multiple diseases: discovering shared mechanisms for bisphenol A-associated metabolic outcomes
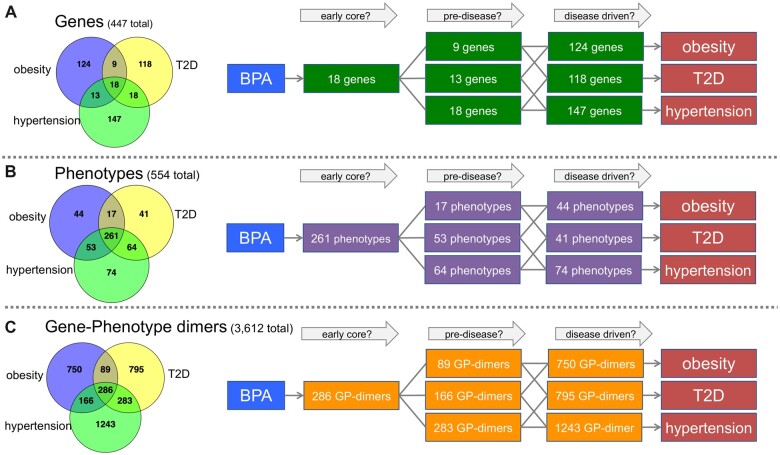
Bisphenol A is a commercial chemical that can function as an endocrine disrupter (Kahn et al., 2020), with the potential to influence numerous health outcomes, including several metabolic diseases (Boudalia et al., 2021; Provvisiero et al., 2016; Wehbe et al., 2020). We used CTD Tetramers to quickly generate tetramers connecting bisphenol A to 3 metabolic endpoints: obesity, T2D, and hypertension. These queries independently resulted in 1291 tetramers (164 genes and 375 phenotypes) for obesity, 1453 tetramers (163 genes and 383 phenotypes) for T2D, and 1978 tetramers (196 genes and 452 phenotypes) for hypertension. To identify potential shared mechanisms, we compared by Venn analysis the sets of genes (447 total), phenotypes (554 total), and gene-phenotype dimers (3612 total) across all 3 disease outcomes. The intersections of associated gene and phenotype datasets allows a dissection of potentially shared and unique mechanistic steps connecting bisphenol A to the 3 AOs (Figure 6). The genes/phenotypes shared by all 3 diseases could potentially represent common molecular mechanisms, perhaps reflecting core biological processes in either the normal or predisease state. Other mechanisms shared by only 1 or 2 diseases might potentially contribute to more specific disease-defining events. While these hypothetical pathways are highly speculative, the point is that CTD Tetramers provides users with the ability to rapidly collect complex data relationships and the associated lines of evidence to start building mechanistic frameworks for further analysis and improved understanding of how the same chemical might contribute to different environmental diseases.
Figure 6.
Use case 2: surveying 3 diseases for bisphenol A exposure. Three tetramer datasets were independently generated using the CTD Tetramers tool for bisphenol A (BPA) and the metabolic diseases obesity, T2D, and hypertension. The relationships between the resulting (A) 447 genes, (B) 554 phenotypes, and (C) 3612 gene-phenotype dimers were compared. Venn analysis can help develop schematic and hypothetical pathways as a starting point to explore and understand the commonality and uniqueness of potential mechanisms for each outcome. For example, the 18 genes, 261 phenotypes, and 286 gene-phenotype dimers shared by all 3 diseases might be hypothesized as representing common processes, while elements shared by 2 diseases might be involved in predisease events, and other unique mechanisms might potentially be more disease-driven processes.
Use case 3: multiple chemicals-1 disease: surveying how different exposures converge on the same molecular mechanisms
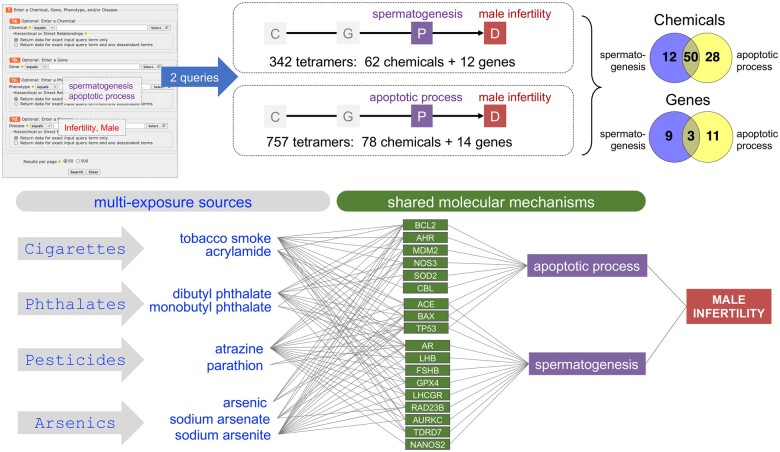
Various environmental factors have been associated with male infertility (Rodprasert et al., 2023; Szabó et al., 2023), consistent with the idea that diseases can be the result of multifactorial components, both genetic and environmental, often from an assortment of stressors over a lifetime. Understanding the interplay between multiple low-dose chemical exposures and an array of genetic factors is necessary to formulate testable, realistic models for environmental health (McHale et al., 2018; Virolainen et al., 2023). To survey how different types of chemical exposures could converge to potentially influence male infertility, we utilized CTD Tetramers to independently generate 342 tetramers (62 chemicals and 12 genes) and 757 tetramers (78 chemicals and 14 genes), respectively, connecting the phenotypes of “spermatogenesis” and “apoptotic process” (and descendants) to the disease outcome of male infertility (Figure 7). There are 50 shared chemicals between the 2 sets of tetramers, and several of them can be grouped into different exposure sources, including cigarettes, phthalates, pesticides, and arsenics, that have interactions with shared genes modulating apoptosis and spermatogenesis, and in the case of 3 genes (ACE, BAX, and TP53), both phenotypes (Figure 7). This type of analysis enables users to build testable pathways that incorporate various multiexposure sources that converge upon shared molecular mechanisms to derive more complex environmental and genetic contributions toward male infertility.
Figure 7.
Use case 3: discovering shared molecular mechanisms for multiple chemical exposures. CTD Tetramers is used to independently generate tetramers that connect the phenotypes “spermatogenesis” and “apoptotic process” to male infertility. Comparing the data types from these 2 sets of tetramers discovers 50 shared chemicals and 3 shared genes. Many of the chemicals can be grouped into different sources of exposure, including cigarettes, phthalates, pesticides, and arsenics; yet these diverse chemicals have interactions with shared genes that modulate the 2 phenotypes apoptosis and spermatogenesis, suggesting that different exposures from different sources can potentially converge upon the same molecular mechanisms toward the same potential disease outcome. Note: for simplicity, not all genes or shared chemicals are shown.
Use case 4: applying tetramers to inform AO pathways
CGPD-tetramers are computed as a 4-unit block, initiated by a chemical interacting with a gene product to trigger an intermediate biological phenotype that ultimately can be linked to a disease outcome (Davis et al., 2018). This framework is complementary to the AO pathway (AOP), a conceptual module that connects a molecular initiating event (MIE) with a linked series of ordered key events (KEs) that ultimately lead to an AO. The KEs often reflect broad molecular, cellular, or tissue/organ level concepts, and are assumed to have underlying causal subnetworks involving gene products and biological reactions that define them. AOPs have become an increasingly useful method to formalize a series of mechanistic steps leading to any adverse endpoint (Ankley and Edwards, 2018), and the AOP-Wiki (https://aopwiki.org/) is an official resource for researchers to construct, store, and publish their designed AOPs for the scientific community. Currently, AOP-Wiki contains 423 AOP records; however, 224 of them (53%) have the status of “Do Not Cite” (as they are either under development or not open for comments), and only 161 (38%) are open to citation or are still under development but open to comments, suggesting that many of these stored AOPs still need valuable input, validation, and refinement from the scientific community. While AOPs are defined as being chemical-agnostic, some include prototypical chemical stressors, and integrating extraneous chemical exposure data to defined AOPs is a widely used practice to help inform and provide potential insight into environmental diseases, chemical safety assessment, and human health risk management (Bajard et al., 2023; Edwards et al., 2016; Kan et al., 2022; Lambert, 2022; Li et al., 2022; Mortensen et al., 2018; Paini et al., 2022; Perkins et al., 2022; Pogrmic-Majkic et al., 2022; Saarimäki et al., 2023). CTD Tetramers is a valuable tool to identify and provide literature-based evidence for potential gene and phenotype intermediates that can help inform, refine, and provide evidence for mechanistic causal subnetworks for AOPs, especially with respect to environmental influences on human health.
Integrating environmental exposure information often first requires harmonization across a variety of data type platforms (Davis et al., 2019; Holmgren et al., 2021; Thessen et al., 2020). To facilitate using AO terms as input queries in the CTD Tetramer tool, we manually mapped the 179 official AO terms from the AOP-Wiki to corresponding CTD phenotype and disease terms used for curation and construction of CGPD-tetramers (Supplementary Table 2).
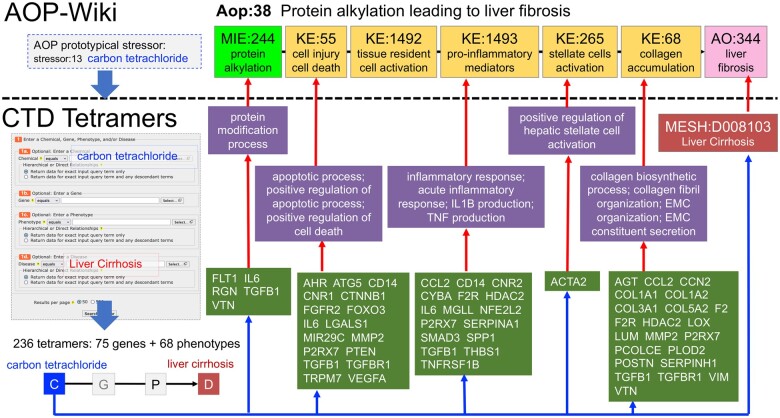
To demonstrate how CTD tetramers can be used to help inform AOPs in various ways, we used Aop:38 (“Protein alkylation leading to liver fibrosis”; https://aopwiki.org/aops/38) as a test case. This established AOP is composed of 1 MIE, 5 KEs, and an AO, with several prototypical stressors proposed, including carbon tetrachloride (Figure 8). Since the associated AO “Liver fibrosis” (AO:344) maps to the CTD disease term “Liver Cirrhosis” (MESH:D008103), we queried CTD Tetramers with “carbon tetrachloride” and “liver cirrhosis” to return 236 tetramers composed of 75 genes and 68 phenotypes. Many of the computed phenotypes from these CGPD-tetramers map to similar concepts denoted by the KEs in the AOP (Figure 8), providing users with a plethora of specific gene-phenotype molecular mechanisms that provide literature-based evidence to support the causal subnetworks of KEs (Perkins et al., 2022). This approach can aid the development of new or unpublished AOPs by providing literature-based evidence and support for creating new KE relationships (Jeong and Choi, 2022; Jeong et al., 2023). This is important because the quality of an AOP depends upon the strength of the underlying evidence used to support the KE relationships, and, since AOPs are chemical-agnostic, the evidence to build an AOP can come from any type of chemical stressor (Chauhan et al., 2021). Additionally, this approach furnishes a list of potential genes for subsequent analysis regarding environmentally induced AOs (Virolainen et al., 2023).
Figure 8.
Use case 4: providing mechanistic insights for AOPs. Aop:38 relates how protein alkylation can lead to liver fibrosis (AO:344), which maps to liver cirrhosis (MESH:D008102) in CTD, with several prototypical stressors, including carbon tetrachloride (AOP stressor:13). Querying CTD Tetramers with carbon tetrachloride and liver cirrhosis generates 236 tetramers composed of 75 genes and 68 phenotypes, with many of the returned phenotypes reflecting the individual KE modules, providing potential mechanistic insights and literature-supported evidence for the causal subnetworks for each KE.
Next, the molecular mechanisms identified can be used subsequently to query CTD Tetramers for other environmental chemicals that might act as additional stressors for the same AOP. For example, a multiparameter query of CTD Tetramers using the phenotype “collagen biosynthetic process” (part of a computed causal subnetwork for KE:68, “collagen accumulation”; Figure 8) and the disease term “liver cirrhosis” returns over 190 tetramers (https://bit.ly/ctdcollagenLC). These tetramers are composed of more than 40 additional environmental chemicals, including acetaminophen, particulate matter, dimethylnitrosamine, and dietary fats that can be explored for influencing this same AOP for liver cirrhosis.
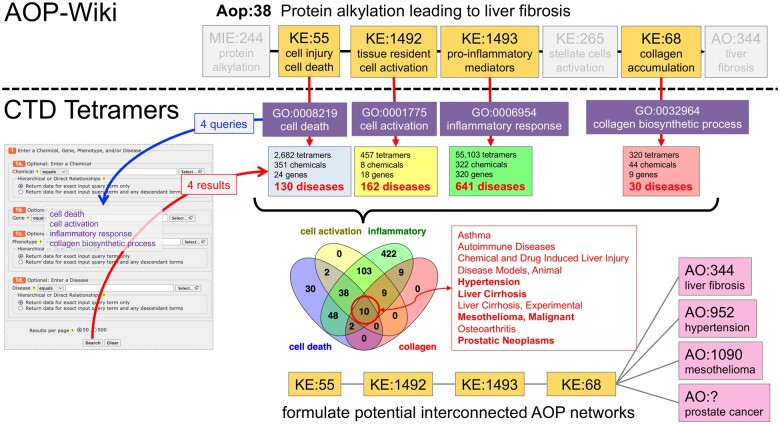
Finally, the KE terms can be used to discover other CTD diseases, enabling interconnected AOP networks to be proposed. We selected 4 CTD phenotypes that reflect 4 of the KEs for this AOP: CTD phenotype “cell death” (GO:0008219) for KE “cell injury/cell death” (KE:55), CTD phenotype “cell activation” (GO:0001775) for KE “tissue resident cell activation” (KE:1492), CTD phenotype “inflammatory response” (GO:00006954) for KE “pro-inflammatory mediators” (KE:1493), and CTD phenotype “collagen biosynthetic process” (GO:0032964) for KE “collagen accumulation” (KE:68). These 4 CTD phenotypes were independently used to query CTD Tetramers to generate tetramers with 130, 162, 641, and 30 diseases, respectively (Figure 9). We identified 10 diseases shared by all 4 phenotypes/KEs, including hypertension, malignant mesothelioma, and prostate cancer, some of which can be mapped to official AOP events (Supplementary Table 2). This method allows users to formulate sets of interconnected AOP networks that use shared KEs but result in different disease outcomes (Figure 9), and could be used to help compute and model possible comorbidities (Hidalgo et al., 2009).
Figure 9.
Use case 4: proposing AOP networks. Four KEs (highlighted) used in Aop:38 are matched to corresponding CTD phenotypes, and these phenotypes are used to query CTD Tetramers independently to return 4 sets of tetramers for each phenotype. The diseases associated with each phenotype from the 4 sets of returned tetramers are compared in Venn analysis to discover 10 overlapping diseases shared by all 4 queries. Among the 10, hypertension corresponds to AO:952 and mesothelioma corresponds to AO:1090 in the AOP-Wiki; currently, there is no AO term for prostate cancer. These 4 KEs may be used to create new or augment existing independent AOPs leading to 4 different diseases, allowing an interconnected AOP network to be postulated. (Note: the additional MIEs and KEs necessary for each proposed independent AOP are not known/shown).
Caveats, limitations, and potential future considerations
An important caveat to our methodology lies in the requirement that CGPD-tetramers generated by the tool CTD Tetramers must have 5 statements of supporting evidence (Figure 1A). Because of this requirement, the computed tetramers represent only a subset of all CTD data relationships found in the database. For example, currently in CTD, there are 288 unique chemicals that affect the phenotype “spermatogenesis” (http://ctdbase.org/detail.go?type=go&acc=GO%3A0007283&view=phenotype); however, in use case 3 (Figure 7), only 62 chemicals were retrieved as part of the tetramer analysis that connects “spermatogenesis” to “male infertility” because only those 62 of the 288 chemicals fulfill the requirement of having all 5 evidence statements curated in CTD. That is not to say the remaining 226 chemicals might not play a role in male infertility, but only that, currently, all 5 of the required evidence statements do not yet exist in CTD for those additional 226 chemicals to become part of a tetramer. Nonetheless, tetramers provide a quick and easy starting point to initiate the construction of chemical-disease pathways, and this new tool now helps to bring these datasets with the most evidence to the forefront for users. If necessary, pathways derived from tetramer analysis can be enhanced manually by the user with additional genes, chemicals, and phenotypes that have other relationships in CTD but do not show up in tetramers because one or more of the required evidential statements is currently lacking. Importantly, every month CTD biocurators manually curate over 250 new peer-reviewed articles, adding over 18 000 interactions into the database that can be leveraged as new evidence statements by the CTD Tetramers tool to continually expand the number of tetramer computations.
Another limitation, as demonstrated in many of the use cases, is that the follow-up study of the computed tetramers often requires manual analysis by the user. While querying for and retrieving the computed CGPD-tetramers themselves does not require any a priori knowledge about the data types, evaluating the tetramer outputs can involve ancillary manual tasks. For example, in use case 1 (Figure 5), tetramer-derived phenotypes are first manually evaluated and binned into modules to help construct a potential chemical-disease pathway. In use case 2 (Figure 6), Venn analysis is necessary to derive the shared and unique molecular operations for different disease outcomes, and in use case 3 (Figure 7), chemicals are manually surveyed and grouped into categories to evaluate the shared molecular mechanisms from different exposure sources. Future considerations to facilitate these manual processes could include using network models developed by GO-CAM (Thomas et al., 2019) that might enable easier and automatic mapping and binning of CTD phenotypes by their GO accession identifiers into structured GO pathways as well as help reduce potential bias introduced by manual clustering. Constructing a chemical classifier, similar to the MEDIC-Slim classifier developed for diseases (Davis et al., 2013), would help group chemicals into broader categories for easier meta-analysis of exposure sources.
We are appraising other potential functionalities to add to CTD Tetramers. For example, since CTD curation of chemical-phenotype interactions also include anatomy terms (Davis et al., 2021), we are testing ways to add an optional filter to enable users to explore tetramers from an anatomical perspective. As well, we are investigating ways to incorporate visualization tools that transform the extensive tetramer query outputs into more user-friendly illustrations for users to export as a network graph. For instance, the 203 tetramers depicting the complex PFAS-asthma dataset from use case 1 can be visualized to help highlight the salient relationships between certain key chemicals, genes, phenotypes, and diseases (Supplementary Figure 1). Finally, we are considering ways to weight the 5 evidentiary statements necessary for each generated tetramer, such as basing it upon the number of curated scientific articles used to support each statement; this type of weighted scoring could help rank, sort, and prioritize the tetramers in the future.
Summary
Here, we describe a novel, user-friendly tool called CTD Tetramers that enables scientists to quickly retrieve CGPD-tetramers, which are 4-unit data modules computationally generated by integrating 5 lines of curated evidence from CTD. Importantly, this method does not require a priori knowledge for any of the data types: users simply enter any chemical, gene, phenotype, or disease of interest, and the tool automatically generates all possible CGPD-tetramers and provides the supporting lines of evidence from the literature. These tetramers, in turn, can be used in a variety of ways to fill exposure knowledge gaps with potential molecular mechanisms for chemical-disease pathways and form testable hypotheses about environmental health.
Supplementary Material
Contributor Information
Allan Peter Davis, Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
Thomas C Wiegers, Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
Jolene Wiegers, Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
Brent Wyatt, Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
Robin J Johnson, Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
Daniela Sciaky, Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
Fern Barkalow, Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
Melissa Strong, Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
Antonio Planchart, Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA; Center for Human Health and the Environment, North Carolina State University, Raleigh, North Carolina 27695, USA.
Carolyn J Mattingly, Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA; Center for Human Health and the Environment, North Carolina State University, Raleigh, North Carolina 27695, USA.
Data availability
CTD is publicly available at http://ctdbase.org/ and the CTD Tetramers tool is available at http://ctdbase.org/query.go?type=tetramer.
Declaration of conflicting interests
The authors declared no conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary data
Supplementary data are available at Toxicological Sciences online.
Funding
National Institute of Environmental Health Sciences (U24 ES033155). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Ankley G. T., Edwards S. W. (2018). The adverse outcome pathway: a multifaceted framework supporting 21st century toxicology. Curr. Opin. Toxicol. 9, 1–7. 10.1016/j.cotox.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajard L., Adamovsky O., Audouze K., Baken K., Barouki R., Beltman J. B., Beronius A., Bonefeld-Jørgensen E. C., Cano-Sancho G., de Baat M. L., et al. (2023). Application of AOPs to assist regulatory assessment of chemical risks—case studies, needs and recommendations. Environ. Res. 217, 114650. 10.1016/j.envres.2022.114650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck I. H., Timmermann C. A. G., Nielsen F., Schoeters G., Jøhnk C., Kyhl H. B., Høst A., Jensen T. K. (2019). Association between prenatal exposure to perfluoroalkyl substances and asthma in 5-year-old children in the Odense child cohort. Environ. Health 18, 97. 10.1186/s12940-019-0541-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berhanu A., Mutanda I., Taolin J., Qaria M. A., Yang B., Zhu D. (2023). A review of microbial degradation of per- and polyfluoroalkyl substances (PFAS): Biotransformation routes and enzymes. Sci. Total Environ. 859, 160010. 10.1016/j.scitotenv.2022.160010 [DOI] [PubMed] [Google Scholar]
- Boudalia S., Bousbia A., Boumaaza B., Oudir M., Lavier M. C. C. (2021). Relationship between endocrine disruptors and obesity with a focus on bisphenol A: A narrative review. Bioimpacts 11, 289–300. 10.34172/bi.2021.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic D., Živančević K., Baralić K., Miljaković E. A., Djordjević A. B., Ćurčić M., Bulat Z., Antonijević B., Đukić-Ćosić D. (2023). Conducting bioinformatics analysis to predict sulforaphane-triggered adverse outcome pathways in healthy human cells. Biomed. Pharmacother. 160, 114316. 10.1016/j.biopha.2023.114316 [DOI] [PubMed] [Google Scholar]
- Chai Z., Zhao C., Jin Y., Wang Y., Zou P., Ling X., Yang H., Zhou N., Chen Q., Sun L., et al. (2021). Generating adverse outcome pathway (AOP) of inorganic arsenic-induced adult male reproductive impairment via integration of phenotypic analysis in comparative toxicogenomics database (CTD) and AOP wiki. Toxicol. Appl. Pharmacol. 411, 115370. 10.1016/j.taap.2020.115370 [DOI] [PubMed] [Google Scholar]
- Chauhan V., Sherman S., Said Z., Yauk C. L., Stainforth R. (2021). A case example of a radiation-relevant adverse outcome pathway to lung cancer. Int. J. Radiat. Biol. 97, 68–84. 10.1080/09553002.2019.1704913 [DOI] [PubMed] [Google Scholar]
- Davis A. P., Murphy C. G., Rosenstein M. C., Wiegers T. C., Mattingly C. J. (2008). The comparative toxicogenomics database facilitates identification and understanding of chemical-gene-disease associations: Arsenic as a case study. BMC Med. Genomics. 1, 48. 10.1186/1755-8794-1-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Wiegers T. C., Rosenstein M. C., Murphy C. G., Mattingly C. J. (2011). The curation paradigm and application tool used for manual curation of the scientific literature at the comparative toxicogenomics database. Database (Oxford) 2011, bar034. 10.1093/database/bar034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Wiegers T. C., Rosenstein M. C., Mattingly C. J. (2012). MEDIC: A practical disease vocabulary used at the comparative toxicogenomics database. Database (Oxford) 2012, bar065. 10.1093/database/bar065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Murphy C. G., Johnson R., Lay J. M., Lennon-Hopkins K., Saraceni-Richards C., Sciaky D., King B. L., Rosenstein M. C., Wiegers T. C., et al. (2013). The comparative toxicogenomics database: update 2013. Nucleic Acids Res. 41, D1104–D1114. 10.1093/nar/gks994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Grondin C. J., Lennon-Hopkins K., Saraceni-Richards C., Sciaky D., King B. L., Wiegers T. C., Mattingly C. J. (2015). The comparative toxicogenomics database’s 10th year anniversary: update 2015. Nucleic Acids Res. 43, D914–D920. 10.1093/nar/gku935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Wiegers T. C., King B. L., Wiegers J., Grondin C. J., Sciaky D., Johnson R. J., Mattingly C. J. (2016). Generating gene ontology-disease inferences to explore mechanisms of human disease at the comparative toxicogenomics database. PLoS One 11, e0155530. 10.1371/journal.pone.0155530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Wiegers T. C., Wiegers J., Johnson R. J., Sciaky D., Grondin C. J., Mattingly C. J. (2018). Chemical-induced phenotypes at CTD help inform the predisease state and construct adverse outcome pathways. Toxicol. Sci. 165, 145–156. 10.1093/toxsci/kfy131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Wiegers J., Wiegers T. C., Mattingly C. J. (2019). Public data sources to support systems toxicology applications. Curr. Opin. Toxicol. 16, 17–24. 10.1016/j.cotox.2019.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Wiegers T. C., Grondin C. J., Johnson R. J., Sciaky D., Wiegers J., Mattingly C. J. (2020). Leveraging the comparative toxicogenomics database to fill in knowledge gaps for environmental health: a test case for air pollution-induced cardiovascular disease. Toxicol. Sci. 177, 392–404. 10.1093/toxsci/kfaa113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Wiegers T. C., Wiegers J., Grondin C. J., Johnson R. J., Sciaky D., Mattingly C. J. (2021). CTD anatomy: Analyzing chemical-induced phenotypes and exposures from an anatomical perspective, with implications for environmental health studies. Curr. Res. Toxicol. 2, 128–139. 10.1016/j.crtox.2021.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Wiegers T. C., Johnson R. J., Sciaky D., Wiegers J., Mattingly C. J. (2023). Comparative toxicogenomics database: update 2023. Nucleic Acids Res. 51, D1257–D1262. 10.1093/nar/gkac833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. W., Tan Y. M., Villeneuve D. L., Meek M. E., McQueen C. A. (2016). Adverse outcome pathways—organizing toxicological information to improve decision making. J. Pharmacol. Exp. Ther. 356, 170–181. 10.1124/jpet.115.228239 [DOI] [PubMed] [Google Scholar]
- Grondin C. J., Davis A. P., Wiegers T. C., King B. L., Wiegers J. A., Reif D. M., Hoppin J. A., Mattingly C. J. (2016). Advancing exposure science through chemical data curation and integration in the comparative toxicogenomics database. Environ. Health Perspect. 124, 1592–1599. 10.1289/ehp174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin C. J., Davis A. P., Wiegers T. C., Wiegers J. A., Mattingly C. J. (2018). Accessing an expanded exposure science module at the comparative toxicogenomics database. Environ. Health Perspect. 126, 014501. 10.1289/ehp2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin C. J., Davis A. P., Wiegers J. A., Wiegers T. C., Sciaky D., Johnson R. J., Mattingly C. J. (2021). Predicting molecular mechanisms, pathways, and health outcomes induced by juul e-cigarette aerosol chemicals using the comparative toxicogenomics database. Curr. Res. Toxicol. 2, 272–281. 10.1016/j.crtox.2021.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo C. A., Blumm N., Barabasi A.-L., Christakis N. A. (2009). A dynamic network approach for the study of human phenotypes. PLoS Comput. Biol. 5, e1000353. 10.1371/journal.pcbi.1000353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren S. D., Boyles R. R., Cronk R. D., Duncan C. G., Kwok R. K., Lunn R. M., Osborn K. C., Thessen A. E., Schmitt C. P. (2021). Catalyzing knowledge-driven discovery in environmental health sciences through a community-driven harmonized language. Int. J. Enviorn. Res. Public Health 18, 8985. 10.3390/ijerph18178985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough K. P., Curtiss M. L., Blain T. J., Liu R.-M., Trevor J., Deshane J. S., Thannickal V. J. (2020). Airway remodeling in asthma. Front. Med. (Lausanne) 7, 191. 10.3389/fmed.2020.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Browne M. S., Eliot M., Patti M., Spanier A. J., Braun J. M. (2020). PFAS (per- and polyfluoroalkyl substances) and asthma in young children: NHANES 2013-2014. Int. J. Hyg. Environ. Health. 229, 113565. 10.1016/j.ijheh.2020.113565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Choi J. (2022). Advancing the adverse outcome pathway for PPARgamma inactivation leading to pulmonary fibrosis using Bradford–Hill consideration and the comparative toxicogenomics database. Chem. Res. Toxicol. 35, 233–243. 10.1021/acs.chemrestox.1c00257 [DOI] [PubMed] [Google Scholar]
- Jeong J., Kim D., Choi J. (2023). Integrative data mining approach: Case study with adverse outcome pathway network leading to pulmonary fibrosis. Chem. Res. Toxicol. 36, 838–847. 10.1021/acs.chemrestox.2c00325 [DOI] [PubMed] [Google Scholar]
- Kahn L. G., Philippat C., Nakayama S. F., Slama R., Trasande L. (2020). Endocrine-disrupting chemicals: implications for human health. Lancet Diab. Endocrinol. 8, 703–718. 10.1016/s2213-8587(20)30129-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan H.-L., Tung C.-W., Chang S.-E., Lin Y.-C. (2022). In silico prediction of parkinsonian motor deficits-related neurotoxicants based on the adverse outcome pathway concept. Arch. Toxicol. 96, 3305–3314. 10.1007/s00204-022-03376-1 [DOI] [PubMed] [Google Scholar]
- King B. L., Davis A. P., Rosenstein M. C., Wiegers T. C., Mattingly C. J. (2012). Ranking transitive chemical-disease inferences using local network topology in the comparative toxicogenomics database. PLoS One 7, e46524. 10.1371/journal.pone.0046524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosnik M. B., Planchart A., Marvel S. W., Reif D. M., Mattingly C. J. (2019). Integration of curated and high-throughput screening data to elucidate environmental influences on disease pathways. Comput.Toxicol. 12, 100094. 10.1016/j.comtox.2019.100094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J. C. (2022). Adverse outcome pathway ‘footprinting’: A novel approach to the integration of 21st century toxicology information into chemical mixtures risk assessment. Toxics 11, 37. 10.3390/toxics11010037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenbach B., Wilson M. (2021). Per- and polyfluoroalkyl substances (PFAS): Significance and considerations within the regulatory framework of the USA. Int. J. Environ. Res. Public Health 18, 11142. 10.3390/ijerph182111142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Yu Y., Sun Z., Duan J. (2022). A comprehensive understanding of ambient particulate matter and its components on the adverse health effects based from epidemiological and laboratory evidence. Part. Fibre Toxicol. 19, 67. 10.1186/s12989-022-00507-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale C. M., Osborne G., Morello-Frosch R., Salmon A. G., Sandy M. S., Solomon G., Zhang L., Smith M. T., Zeise L. (2018). Assessing health risks from multiple environmental stressors: Moving from GxE to IxE. Mutat. Res. Rev. Mutat. Res. 775, 11–20. 10.1016/j.mrrev.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen H. M., Chamberlin J., Joubert B., Angrish M., Sipes N., Lee J. S., Euling S. Y. (2018). Leveraging human genetic and adverse outcome pathway (AOP) data to inform susceptibility in human health risk assessment. Mamm. Genome 29, 190–204. 10.1007/s00335-018-9738-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. D., Kim M.-S. (2023). Interactions between cadmium, lead, mercury, and arsenic and depression: A molecular mechanism involved. J. Affect. Disord. 327, 315–329. 10.1016/j.jad.2023.02.013 [DOI] [PubMed] [Google Scholar]
- Paini A., Campia I., Cronin M. T. D., Asturiol D., Ceriani L., Exner T. E., Gao W., Gomes C., Kruisselbrink J., Martens M., et al. (2022). Towards a qAOP framework for predictive toxicology—linking data to decisions. Comput. Toxicol. 21, 100195. 10.1016/j.comtox.2021.100195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins E. J., Woolard E. A., Garcia-Reyero N. (2022). Integration of adverse outcome pathways, causal networks and 'omics to support chemical hazard assessment. Front. Toxicol. 4, 786057. 10.3389/ftox.2022.786057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkhasova D. V., Jameson L. E., Conrow K. D., Simeone M. P., Davis A. P., Wiegers T. C., Mattingly C. J., Leung M. C. K. (2021). Regulatory status of pesticide residues in cannabis: Implications to medical use in neurological diseases. Curr. Res. Toxicol. 2, 140–148. 10.1016/j.crtox.2021.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogrmic-Majkic K., Samardzija Nenadov D., Tesic B., Fa Nedeljkovic S., Kokai D., Stanic B., Andric N. (2022). Mapping DEHP to the adverse outcome pathway network for human female reproductive toxicity. Arch. Toxicol. 96, 2799–2813. 10.1007/s00204-022-03333-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provvisiero D., Pivonello C., Muscogiuri G., Negri M., de Angelis C., Simeoli C., Pivonello R., Colao A. (2016). Influence of bisphenol a on type 2 diabetes mellitus. Int. J. Environ. Res. Public Health 13, 989. 10.3390/ijerph13100989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappazzo K. M., Coffman E., Hines E. P. (2017). Exposure to perfluorinated alkyl substances and health outcomes in children: A systematic review of the epidemiologic literature. Int. J. Environ. Res. Public Health 14, 691. 10.3390/ijerph14070691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodprasert W., Toppari J., Virtanen H. (2023). Environmental toxicants and male fertility. Best Pract. Res. Clin. Obstet. Gynaecol. 86, 102298. 10.1016/j.bpobgyn.2022.102298 [DOI] [PubMed] [Google Scholar]
- Saarimäki L. A., Morikka J., Pavel A., Korpilähde S., Del Giudice G., Federico A., Fratello M., Serra A., Greco D. (2023). Toxicogenomics data for chemical safety assessment and development of new approach methodologies: An adverse outcome pathway-based approach. Adv. Sci. (Weinh.) 10, e2203984. 10.1002/advs.202203984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanic B., Nenadov D. S., Fa S., Pogrmic-Majkic K., Andric N. (2021). Integration of data from the cell-based ERK1/2 ELISA and the comparative toxicogenomics database deciphers the potential mode of action of bisphenol A and benzo[a]pyrene in lung neoplasm. Chemosphere 285, 131527. 10.1016/j.chemosphere.2021.131527 [DOI] [PubMed] [Google Scholar]
- Szabó A., Váncsa S., Hegyi P., Váradi A., Forintos A., Filipov T., Ács J., Ács N., Szarvas T., Nyirády P., et al. (2023). Lifestyle-, environmental-, and additional health factors associated with an increased sperm DNA fragmentation: A systematic review and meta-analysis. Reprod. Biol. Endocrinol. 21, 5. 10.1186/s12958-023-01054-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thessen A. E., Grondin C. J., Kulkarni R. D., Brander S., Truong L., Vasilevsky N. A., Callahan T. J., Chan L. E., Westra B., Willis M., et al. (2020). Community approaches for integrating environmental exposures into human models of disease. Environ. Health Perspect. 128, 125002. 10.1289/ehp7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. D., Hill D. P., Mi H., Osumi-Sutherland D., Van Auken K., Carbon S., Balhoff J. P., Albou L.-P., Good B., Gaudet P., et al. (2019). Gene ontology causal activity modeling (GO-CAM) moves beyond GO annotations to structured descriptions of biological functions and systems. Nat. Genet. 51, 1429–1433. 10.1038/s41588-019-0500-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zelst C. M., de Boer G. M., Türk Y., van Huisstede A., In’t Veen J. C. C. M., Birnie E., Boxma-de Klerk B. M., Tramper-Stranders G. A., Braunstahl G.-J. (2021). Association between elevated serum triglycerides and asthma in patients with obesity: An explorative study. Allergy Asthma Proc. 42, e71–e76. 10.2500/aap.2021.42.210020 [DOI] [PubMed] [Google Scholar]
- Virolainen S. J., VonHandorf A., Viel K. C. M. F., Weirauch M. T., Kottyan L. C. (2023). Gene–environment interactions and their impact on human health. Genes Immun. 24, 1–11. 10.1038/s41435-022-00192-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Holst H., Nayak P., Dembek Z., Buehler S., Echeverria D., Fallacara D., John L. (2021). Perfluoroalkyl substances exposure and immunity, allergic response, infection, and asthma in children: Review of epidemiologic studies. Heliyon 7, e08160. 10.1016/j.heliyon.2021.e08160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehbe Z., Nasser S. A., El-Yazbi A., Nasreddine S., Eid A. H. (2020). Estrogen and bisphenol a in hypertension. Curr. Hypertens. Rep. 22, 23. 10.1007/s11906-020-1022-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CTD is publicly available at http://ctdbase.org/ and the CTD Tetramers tool is available at http://ctdbase.org/query.go?type=tetramer.