Abstract
The synthesis and triplex stabilizing properties of oligodeoxyribonucleotides functionalized at the 5′- and/or 3′-termini with a naphthalene diimide-based (NDI) intercalator is described. The NDI intercalator was prepared in a single step from the corresponding dianhydride and was attached to the 5′-terminus of an oligodeoxyribonucleotide following a reverse coupling procedure. The DMT protecting group was removed and the sequence phosphitylated to generate the phosphoramidite derivative on the 5′-terminus of the support-bound oligodeoxyribonucleotide. The NDI intercalator with a free hydroxyl was then added in the presence of tetrazole. Attachment of the NDI to the 3′-terminus relied upon a tethered amino group that could be functionalized first with the naphthalene dianhydride, which was subsequently converted to the diimide. Using both procedures, an oligonucleotide conjugate was prepared having the NDI intercalator at both the 5′- and 3′-termini. Thermal denaturation studies were used to determine the remarkable gain in stability for triplexes formed when the NDI-conjugated oligonucleotide was present as the third strand in the complex.
INTRODUCTION
Sequence-specific DNA triplex formation is generally the result of bidentate hydrogen bonding interactions between the third strand (either a polypurine or a polypyrimidine sequence) and the Hoogsteen face of a polypurine strand within a target DNA duplex (1–3). However the complexation of three negatively charged polyphosphate strands also results in significant destabilizing effects as a result of charge–charge repulsion. These destabilizing forces can be mediated in part by divalent salts or polyamines such as spermine that act to shield phosphates and reduce charge–charge effects (4–6). In addition to salt effects, the use of non-nucleotide ligands can also be effective in binding to nucleic acids and stabilizing selected structures (7). In some cases triplex-specific ligands have been identified that introduce additional binding interactions (8–10), usually through intercalation, to provide enhanced stabilization of the complex. Tethering such ligands to the third strands (11,12) can be more effective in overstabilizing triplexes. These ligands bind by intercalation into the triplex (12) or adjacent duplex (11–13), by minor groove binding (14,15) or by less specific interactions (14,15).
Naphthalene diimides (NDI) are planar molecules known to function as intercalators (16,17) or polyintercalators (18) into DNA duplexes and have been shown to function as effective stabilizing ligands when tethered to nucleoside residues within DNA strands (19) or when bridging two hybridizing DNA sequences (20). In the present work we describe the synthesis of an asymmetrical NDI that can be used to tether a NDI intercalator to the 5′-terminus of a DNA sequence. Tethering the intercalator to the 3′-terminus requires appropriate modification of the solid support. The effects on triplex formation by conjugates containing NDI tethered at the 5′- and/or 3′-terminus of 15- residue oligonucleotides are described.
MATERIALS AND METHODS
NMR spectra were obtained on a Varian spectrometer (400 MHz) and contained trimethylsilane as an internal standard. HRMS, LRMS (FAB) and MALDI-Voyager mass spectra were obtained from the Mass Spectrometry Laboratory, School of Chemical Sciences, University of Illinois (Urbana, IL). MALDI-TOF mass spectra were obtained from the Mass Spectrometry Laboratory, Merkert Chemistry Center, Boston College (Chestnut Hill, MA). Rotary evaporations were performed under reduced pressure utilizing Buchi systems. Thin layer chromatography (TLC) was performed on Silica Gel 60 F254 pre-coated on aluminum sheets (EM Separations Technology). Anhydrous solvents and starting materials were purchased from Aldrich Chemical Co. and used without further purification. UV measurements were obtained using a Beckman DU 640 spectrometer. Oligodeoxyribonucleotides were synthesized on an Applied Biosystems 381A DNA Synthesizer. 2′-Deoxyribonucleotide phosphoramidites, fast deprotecting N-acetyl-2′-deoxycytidine phosphoramidite (Ac-dC), 3′-terminal nucleoside controlled pore glass support (CPG) and 3′ amino modifier C7-CPG were purchased from Glen Research (Sterling, VA). High performance liquid chromatography (HPLC) was performed on a Beckman HPLC system using C-18 reversed phase columns (ODS-Hypersil, 5 µm particle size, 120 Å pore) with detection at 260 and 383 nm. Fast Flow HPLC was performed using Oligo R3 columns (Perseptive Biosytems, Framingham, MA) with detection at 260 nm. Oligodeoxyribonucleotides were desalted with Econo-Pac 10DG disposable chromatography columns (Bio-Rad, Hercules, CA). Tm measurements were performed on an AVIV Spectrometer, model 14DS UV/Vis.
N-[2-(2-hydroxyethoxy)ethyl]-N′-[2-(N,N-dimethylamino)-ethyl]-1,4,5,8-naphthalene tetracarboxylic diimide (1)
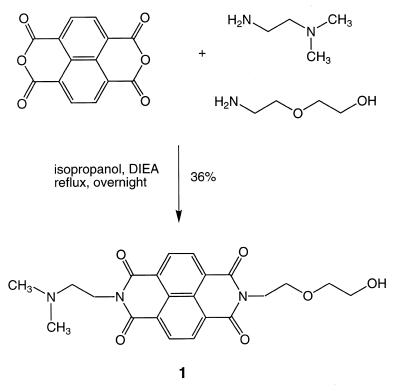
The synthesis of 1 was performed following a general procedure for the preparation of asymmetrical NDIs (18). To 1.0 g (3.73 mmol) of 1,4,5,8-naphthalenetetracarboxylic dianhydride was added 0.38 ml (3.73 mmol) of 2-(2-aminoethoxy)ethanol and 0.41 ml (3.73 mmol) of N,N-dimethylethylenediamine in 20 ml of isopropanol. The reaction mixture was refluxed overnight, the volatiles were removed by rotary evaporation and the desired product was purified by silica gel column chromatography using a gradient of methanol in chloroform (3%). The asymmetrical diimide eluted as the middle compound between the two symmetrical secondary products, obtained roughly in a 1:2:1 ratio. For 1 571 mg (1.34 mmol) of product was obtained (yield 36%) as a yellow solid.
Rf (methanol/dichloromethane, 1/9): 0.49; melting point: 178–180°C. UV (methanol): λmax = 234, 340 (sh), 357, 377 (ɛ = 3.04 × 104 cm–1 M–1) nm. 1H NMR (DMSOd6): δ = 2.20 (s, 6H, N-CH3); 2.52 (t, J = 8.0 Hz, 2H, CH2); 3.45 (m, 4H, CH2); 3.66 (t, J = 8.0 Hz, 2H, CH2); 4.15 (t, J = 6.4 Hz, 2H, CH2); 4.24 (t, J = 6.8 Hz, 2H, CH2); 4.55 (t, J = 4.0 Hz, 1H, OH); 8.64 (s, 4H, Ar-H) p.p.m. 13C NMR (DMSOd6): δ = 45.9; 56.5; 60.4; 66.9; 72.3; 118.0; 126.2; 126.3; 130.7; 162.7; 162.8 p.p.m. HRMS calculated for C22H24N3O6 (M + H+): 426.1665; found: 426.1663.
DNA synthesis
The native 25mer target sequences, 5′-d(GCGCGAAAGAAAAGAGAGAACCCGG)-3′ and 5′-d(CCGGGTTCTCTCTTTTCTTTCGCGC)-3′, the two 40mer target sequences, 5′-d(CGCCGCGCGCGCGAAAGAAAAGAGAGAACCCGGCGCGCGC)-3′ and 5′-d(GC-GCGCGCCGGGTTCTCTCTTTTCTTTCGCGCGCGCGGCG)-3′, the two 40mers containing three pyrimidines within the polypurine target, 5′-d(CGCCGCGCGCGCGAAATAAATGAGTGAACCCGGCGCGCGC)-3′ and 5′-d(GCGCGCGCCGGGTTCACTCATTTATTTCGCGCGCGCGGCG)-3′, and the control 15mer sequence, 5′-d(TTTCTTTTCTCTCTT)-3′, were prepared by solid phase DNA synthesis using conventional protocols (21). The products were characterized by MALDI-Voyager mass spectrometry. For example, 5′-d(TTTCTTTTCTCTCTT)-3′ calculated 15mer: 4442; found 15mer: 4444.
Introduction of the NDI intercalator to the 5′-terminus
The phosphoramidite derivative of 1 was difficult to store or to use effectively in coupling reactions owing to its relatively rapid decomposition. Synthesis of the 15mer functionalized at the 5′-terminus with the naphthalene intercalator was performed following the described reverse coupling procedure (22).
The oligomer was synthesized in the normal fashion and the DMT protecting group was removed after assembly of the 15mer. Subsequently 135 µl (0.425 mmol) of 2-cyanoethyl tetraisopropylphosphoramidite diluted in 300 µl of freshly distilled acetonitrile was delivered to the column to react with the free 5′-OH of the 15mer over a 1 h period of time. The column was then flushed with acetonitrile for 1 min and then removed from the synthesizer and the following step was performed manually. An aliquot of 21 mg (50 µmol) of 1, dissolved in 250 µl of anhydrous dimethylformamide (DMF), was delivered together with 300 µl of a solution of sublimated 1H-tetrazole (as the activating agent) in acetonitrile using a syringe. The coupling reaction between the naphthalene moiety and the terminal phosphoramidite was allowed to occur overnight (~18 h). The column was then reconnected to the synthesizer, rinsed with acetonitrile and oxidized in the normal manner.
Deprotection of the NDI conjugate used concentrated ammonium hydroxide at room temperature for 4 h. The product proved to be labile to longer reaction conditions or to heat, conditions commonly used for DNA deprotection. Purification of the conjugated oligonucleotide from failed sequences and unconjugated 15mer was accomplished by fast flow HPLC (4.6 × 100.0 mm column), starting with 0% B for 1 min, then using a linear gradient of 0–40% B over 4.5 min (A, 20 mM KH2PO4, pH 5.5; B, 20 mM KH2PO4, pH 5.5, in 70% methanol). The desired sequence was collected as the latest eluting peak. This product was then desalted (Sephadex G-10) and stored at –20°C. MALDI-Voyager MS calculated for 5′-NDI-TTTCTTTTCTCTCTT-3′ (15mer+): 4930; found: 4933.
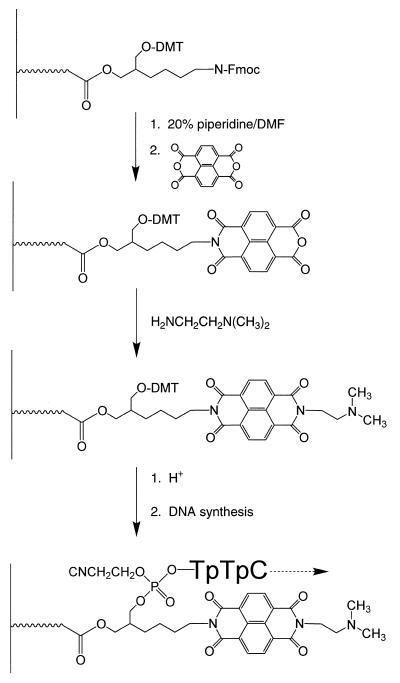
Introduction of the NDI intercalator to the 3′-terminus
Conjugation of the NDI to the 3′-terminus of an oligonucleotide was accomplished using a special ‘amino modifier’ DNA synthesis support. Removal of the Fmoc protecting group from 40 mg (1.92 µmol) of the 3′ amino modifier C7-CPG was performed by treating the CPG beads with 20% piperdine in DMF for 30 min. The beads were then dried in vacuo and transferred to a small glass vial into which 300 µl of isopropanol was added. Following that, 3.6 mg (13.44 µmol) of 1,4,5,8-naphthalenetetracarboxylic dianhydride was added and the vial was shaken vigorously. The vial was then sealed with parafilm, wrapped entirely in aluminum foil and allowed to incubate at 80°C for 18 h. The contents of the vial were then filtered through the DNA synthesizer column to collect the beads and remove the isopropanol solution. A small quantity of the beads was treated with a solution of 0.2% ninhydrin in ethanol and heated. The beads remained off-white in color, indicating that no free primary amino groups were present at this point. For comparison the same test was conducted on 3′ amino modifier CPG after removal of the Fmoc protection and the beads assumed the typical bright purple color. The filtered beads were washed with acetonitrile, dried in vacuo and again transferred to a small glass vial to which 300 µl of isopropanol and 3.2 µl (28.8 µmol) of N,N-dimethylethylenediamine were added. The vial was then shaken vigorously, wrapped as described above and incubated at 80°C for an additional 18 h. The product was then filtered as described and flushed with acetonitrile for 1 min. Automated DNA synthesis was then carried out using the fast-deprotecting Ac-dC and dT phosphoramidites and allowing extra time (~1 min) for coupling of the first nucleoside to the conjugated solid support. The remaining part of the DNA synthesis was carried out using conventional procedures. Upon completion of the DNA synthesis, the beads were dried in vacuo and deprotected in 5 ml of 0.05 M K2CO3 in methanol for 2 h at ambient temperature. The pH was then adjusted to ~6.5 with 2% acetic acid. The solution was filtered to remove the CPG beads and concentrated in vacuo to remove methanol.
Isolation of the 3′-conjugated oligomer was carried out by HPLC (4.6 × 250 mm column) with a linear gradient of 0–100% B over 60 min (A, 20 mM KH2PO4, pH 5.5; B, 20 mM KH2PO4, pH 5.5, in 70% methanol). The desired product eluted at ~30 min. The product was desalted utilizing disposable desalting columns and stored at –20°C. MALDI-TOF MS calculated for 5′-TTTCTTTTCTCTCTT-NDI-3′ (15mer–): 4969; found: 4977.
Introduction of the NDI intercalator to both the 5′- and 3′-termini
The 5′,3′-bisconjugated oligonucleotide was synthesized by performing the procedures described above first to introduce the naphthalene diimide to the 3′-terminus, followed by those procedures designed to introduce the conjugate to the 5′-terminus. The bisconjugate was deprotected and isolated following the milder conditions and procedures used for the 3′-terminal conjugate; the desired product eluted at ~32 min. The yields for the bisconjugate were low owing to the number of chemical steps performed. In a typical reaction we obtained 30 A260 units of crude material from which ~5 A260 units of pure material was isolated. MALDI-TOF MS calculated for 5′-NDI-TTTCTTTTCTCTCTT-NDI-3′ (15mer–): 5459; found: 5459.
Nucleoside analysis
The 25mers, 40mers and 15mer control sequences were digested with a combination of snake venom phosphodiesterase and alkaline phosphatase and the deoxynucleosides analyzed by HPLC. A 20 µl reaction mixture containing 0.5 A260 units of oligomer, 50 mM Tris–HCl, pH 8.0, 100 mM MgCl2, 1 U snake venom phosphodiesterase and 1 U alkaline phosphatase was incubated for 2 h at 37°C. An aliquot of this mixture was analyzed by HPLC (4.6 × 250 mm column) using a linear gradient of 0–100% B over 60 min (A, 20 mM KH2PO4, pH 5.5; B, 20 mM KH2PO4, pH 5.5, in 70% methanol).
The conjugated 15mers were digested with a combination of S1 nuclease and alkaline phosphatase. A 10 µl reaction mixture containing 0.5 A260 units of oligomer, 200 mM NaCl, 5 mM MgCl2, 0.1 mM ZnSO4, 25 mM sodium acetate, pH 5.9, 1 U S1 nuclease was incubated overnight at 37°C. To this mixture was added 7 µl of 0.1 M Tris–HCl, pH 8.0, and 1 U calf intestinal alkaline phosphatase and the reaction was incubated for an additional 60 min at 37°C. An aliquot of this mixture was then analyzed by HPLC using the conditions described above.
The analysis described above produced an unknown peak for conjugates containing a 5′-terminal diimide thought to reflect a linkage that was refractory to enzymatic cleavage. To confirm the nature of this peak, the simple standard was prepared by reverse coupling of the NDI to a support-bound T residue. The product was isolated by HPLC. MALDI-Voyager MS calculated for 5′-NDIpT-3′ (M+): 731.2; found: 731.0.
Thermal melting analyses
Tm values were obtained for complexes containing a 1:1:1 mixture of oligonucleotides at a concentration of 1 µM in 50 mM NaCl, 10 mM MgCl2 and 10 mM PIPES (pH 6.4 or 7.0). Solutions were heated in 1°C steps and absorbances were recorded after temperature stabilization using an AVIV 14DS spectrophotometer. Absorbance and temperature readings were plotted using Igor Pro software. Tm values were determined from first order derivatives as well as graphically from absorbance versus temperature plots. Differences greater than ±1°C were not observed when using the two procedures.
RESULTS AND DISCUSSION
We chose NDI as a simple, readily accessible and effective intercalator to tether to the termini of DNA sequences involved in triplex formation. The design required the preparation of an asymmetrical diimide with one moiety terminating in a hydroxyl that would permit tethering of the intercalator to the DNA sequence through a phosphodiester linkage. The second moiety terminates in a tertiary amine that is likely to be protonated under conditions of neutral pH and such sites of protonation are a common feature of effective intercalators. Presumably, the positively charged site can interact effectively with the negatively charged phosphate(s) at or near the site of intercalation.
Synthesis
Preparation of an asymmetrical NDI was simple and the procedure employed has been described previously (18). With a mixture of the two amine starting materials, the two symmetrical diimides as well as the asymmetrical derivative (see Scheme 1) were obtained in an approximate ratio of 1:1:2. Conversion of 1 (Scheme 1) to the corresponding phosphoramidite derivative could be accomplished, but it was relatively unstable and could not be stored. For incorporation of the linker into the 5′-terminus of the oligonucleotide, it was more effective to use a reverse coupling protocol (22) in which the DMT protecting group was removed from the support-bound oligonucleotide; the unmasked 5′-hydroxyl was converted to the phosphoramidite derivative and then 1 in the presence of tetrazole was added. This procedure does not result in the high yield couplings common for conventional phosphoramidite procedures. In a conventional coupling procedure, the phosphoramidite species is the reagent in solution and is added in excess. Trace quantities of water reduce the excess of the incoming phosphoramidite but do not necessarily result in a failed coupling. In the reversed coupling procedure the active phosphoramidite species is the limiting reagent and is associated with the support-bound oligonucleotide. Trace quantities of water can drastically reduce the coupling yield by reacting with the phosphoramidite. On the basis of HPLC analyses of the products, this final coupling yield with the NDI (1) was estimated to be ~50%. This procedure produced sufficient material to conduct analyses for characterization of the conjugate and to determine Tm values for the corresponding triple helices. Owing to the relative lability of the diimide conjugates, we employed a fast-deprotecting dC building block for all DNA syntheses described.
Scheme 1.
Incorporation of the NDI into the 3′-terminus of a support-bound oligonucleotide should have been relatively simple. Modified supports such as the amino support provide both an Fmoc-protected primary amine and a DMT-protected hydroxyl. By preparing the asymmetrical diimide with a terminal carboxylic acid in place of the alcohol, simple activation of the acid for amide formation permitted attachment of the diimide to the unmasked amino group. A ninhydrin test confirmed that after the amide coupling reaction, no significant amount of the primary amine remained. Unfortunately, after completion of the DNA synthesis, and under a variety of mild deprotection conditions, significant amounts of the 3′-conjugate could not be obtained. We cannot account for this extreme lability of the amide-coupled diimide. We then examined a second procedure in which the naphthalene tetracarboxylic dianhydride was coupled directly to the unmasked amino group of the modified support (Scheme 2). Again, a ninhydrin test was used to confirm the absence of any primary amino groups after this reaction. To the putative imide/anhydride intermediate product (see Scheme 2) was added N,N-dimethylethylenediamine and the reaction was permitted to proceed overnight. The diimide-modified support was then treated with acid and, after appropriate washings, the first nucleoside phosphoramidite was added to the unmasked hydroxyl (Scheme 2). It is likely that during reaction with the anhydride trace amounts of acid resulted in some loss of the support-bound DMT protecting group. Although we did not confirm this possibility, we observed that if the support was reacted with an acetic anhydride capping reagent prior to initiating DNA synthesis, the yield of oligonucleotide was significantly reduced, presumably as a result of acetylating some of the free hydroxyls generated on the support. During these support modification procedures, we relied upon the ninhydrin test to confirm that essentially all of the amino groups had reacted and then we capped the support only after addition of the first nucleoside residue.
Scheme 2.
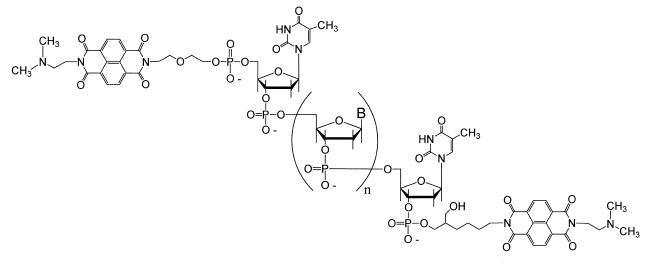
Preparation of the 3′,5′-bisconjugated oligodeoxyribonucleotides, those containing an NDI at both the 3′- and 5′-termini (Fig. 1), was achieved simply by a combination of the two individual procedures. The bisconjugate was obtained in the lowest yield because of the number of reactions, in addition to the conventional DNA couplings, that needed to take place at both termini of the conjugate. In spite of the number of steps, enough purified material could be isolated for the necessary Tm measurements.
Figure 1.
Structure of the 3′,5′-NDI bisconjugate with NDI intercalators conjugated to both the 3′- and 5′-termini of an oligonucleotide.
Two different procedures were employed for deprotection of the various conjugates. For those conjugates containing the NDI tethered only to the 5′-terminus, the support-bound oligonucleotide was treated with concentrated aqueous ammonia at ambient temperature for 4 h. This time was sufficient to remove phosphate and base protecting groups and did not result in significant hydrolysis of the diimide. However, for those conjugates containing a NDI at the 3′-terminus these conditions largely decomposed the tethered NDI. In these cases, the support-bound oligonucleotide was treated with 5 ml of 0.05 M K2CO3 in methanol for 2 h at ambient temperature. These conditions were shown previously (19) to effectively deprotect DNA containing fast-deprotecting building blocks.
Analyses
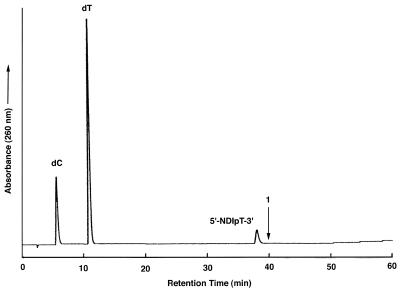
The conjugates were analyzed by both mass spectrometry and HPLC both before and after digestion with phosphodiesterase and phosphatase. All three conjugates were purified to homogeneity as judged by their elution as single peaks from a reversed phase C-18 column. After enzymatic digestion, those conjugates containing a 3′-terminal NDI resulted in two peaks corresponding to dC and dT, respectively, in the expected ratio of 4:11. We did not observe a peak for the NDI itself, largely due to the low extinction coefficient of this molecule at 260 nm. But its presence could be confirmed by MALDI-TOF analyses and UV absorbance (see below). Conjugates containing a 5′-NDI intercalator exhibited peaks for dC and dT (at a ratio of 4:10) and a third peak eluting slightly earlier than would have been expected for compound 1 (Fig. 2). The absence of a dT residue suggested that the third peak was undigested 5′-NDIpT-3′. Preparation of the monomer conjugate by coupling NDI to a single dT residue (5′-NDIpT-3′) confirmed that the material observed in the digest contained the naphthalene residue and the 5′-terminal dT nucleoside and indicated that this phosphodiester linkage was refractory to cleavage by snake venom phosphodiesterase and S1 nuclease.
Figure 2.
HPLC analysis of the S1 nuclease- and alkaline phosphatase-treated 5′-conjugated NDI oligonucleotide. The retention time for compound 1 (see Scheme 1) is marked.
UV analysis also confirmed the presence of the NDI moiety in all cases, with characteristic peaks at 361 and 383 nm. MALDI-TOF analyses confirmed the expected molecular ions for all conjugates. With those samples containing a 3′-terminal conjugate, a small mass peak was present with a slightly smaller mass that corresponded to the intermediate imide/anhydride product (see Scheme 2). This analysis suggested that trace amounts of the monoimide conjugate may have been present in some samples, but we could not confirm this observation on the basis of HPLC analyses of the fully digested material.
Tm analyses of the DNA triplexes
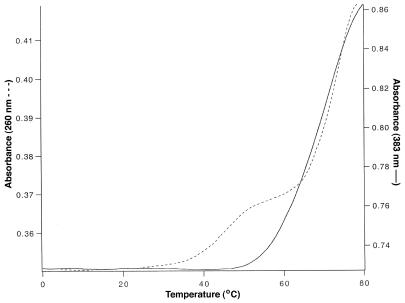
We examined the Tm values for a 15mer third strand conjugated to one or two NDI intercalators. The 15mer polypurine target sequence was embedded within a 25mer duplex. The target sequence contained 11 dA residues and four dC residues. The Tm value for the native sequence (260 nm) lacking an intercalator was 30°C at pH 6.4 and 22°C at pH 7.0 (Table 1). A subsequent transition at 71°C was also present and this latter transition was also observed when the target was analyzed in the absence of a third strand and reflects the duplex→coil transition. With the introduction of the NDI intercalator at the 3′-terminus the Tm values for the first transition increased by 18°C at pH 6.4 and by 15°C at pH 7.0 (Table 1). Similar increases of 17 and 13°C were observed for the sequence having the NDI tethered at the 5′-terminus for pH values of 6.4 and 7.0, respectively (Fig. 3). In all cases the initial transition was followed by a second transition at 71 or 72°C for the target duplex.
Table 1. Tm values for NDI-conjugated triplexesa.
| Sequence | Triplex Tm (°C)b measured at 260 nm |
NDI unstacking Tm (°C)c measured at 383 nm | |
|---|---|---|---|
| pH 6.4 | pH 7.0 | pH 6.4 | |
| Native | 30 | 22 | –d |
| 3′-NDI | 48 | 37 | 56 |
| 5′-NDI | 47 | 35 | 59 |
| 3′,5′-NDI | –e | –e | 71 |
5′-NDI-TTTCTTTTCTCTCTT-NDI-3′
5′-GCGCGAAAGAAAAGAGAGAACCCGG-3′
3′-CGCGCTTTCTTTTCTCTCTTGGGCC-5′
aTm values obtained at strand concentrations of 1 µM.
bOnly the temperature for the early transition is noted. In all cases a transition was observed at 70 or 71°C for the duplex→random coil transition.
cFor each sample monitored at 383 nm only a single transition was present.
dNo transition detected without NDI intercalator.
eNo transition detected other than that at 71°C for the duplex→random coil transition.
Figure 3.
Absorbance versus temperature plots for the 5′-NDI conjugate measured at 260 nm (dotted line) and the 3′,5′-NDI bisconjugate measured at 383 nm (solid line).
These values are comparable with those obtained previously for third strands tethering a single acridine where a 13°C increase for a 5′-conjugated 16mer at pH 7.0 was reported (23). The corresponding 3′-conjugate to an α-oligodeoxynucleotide exhibited a Tm value reduced by ~8°C relative to the 5′-derivative in this study. These differences likely reflect the variation in the manner in which the intercalator was tethered to either the 5′- or 3′-terminus and differences in α- versus β-oligodeoxynucleotides.
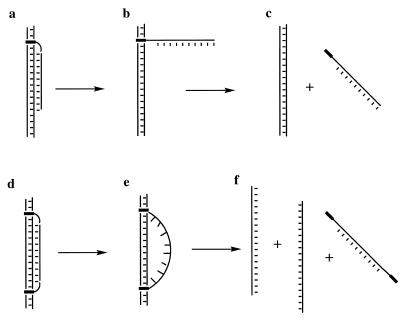
When these same transitions were monitored at 383 nm (the absorbance maximum for the NDI intercalator) a single cooperative transition was observed with a hyperchromic effect, suggesting that the intercalators undergo a thermally induced unstacking process in much the same manner as do the DNA bases. However, the midpoints of these transitions differed significantly from those determined at 260 nm. For example, at pH 6.4 the conjugate with the 3′-tethered NDI exhibited a Tm of 56°C, some 8°C higher than the value obtained with detection at 260 nm. Similarly, the 5′-conjugate resulted in a Tm at 383 nm that was 12°C higher than that measured at 260 nm. These results suggest two processes, one in which the DNA bases undergo denaturation from the target duplex and then relatively late in the transition the NDI intercalator is unstacked from the duplex. The first process can be envisaged to occur as illustrated in Figure 4a and b; the bases have largely undergone denaturation with the NDI still bound to the target. In the second process the NDI unstacks from the duplex to complete the denaturation process (Fig. 4b and c).
Figure 4.
Illustration of potential melting pathways for a 3′- or 5′-NDI conjugate (a–c) or the 3′,5′-NDI bisconjugate (d–f).
The melting results for the sequence containing two terminal NDI intercalators appeared initially to be different from those for the monoconjugates. No significant Tm value was observed at 260 nm except that resulting from denaturation of the target duplex (~71°C). With detection at 383 nm a single transition was also observed at 71°C (Table 1). Both of these assays suggest that the triplex remains largely complexed to the target duplex until the duplex target itself undergoes denaturation. Denaturation may occur as a single process or continue to occur, in part, through two processes, beginning with some disruption of the base triplets (Fig. 4d and e) before duplex denaturation and ending with the unstacking of the NDI intercalators (Fig. 4e and f).
To ensure that dissociation of the third strand from the duplex does not occur at higher temperatures, we redesigned the target duplex to be longer and form a more thermally stable complex. We chose a 40mer sequence that exhibited a duplex Tm of 82°C at 260 nm. Thermal denaturation studies were performed employing this 40mer as the target duplex and the 3′,5′-bisconjugate. Tm values obtained at 383 nm confirmed that dissociation of the third strand from the duplex occurs at 71°C, exactly the same result as that obtained from studies utilizing the 25mer target sequences. Thermally induced transitions of the triplex with the 40mer target at 260 nm yielded a very broad transition, consistent with some loss of base stacking interactions before complete denaturation of the bisconjugate. This result is consistent with a complex that undergoes a partial structural transition (Fig. 4d and e) before complete dissociation of the third strand.
Since the stabilization afforded by either the mono- or bisconjugated NDI sequences is quite significant, it is possible, particularly in the latter case, that the sequence functions simply as a tether between two NDI intercalators. To address this issue, we prepared an additional 40mer target in which we introduced three pyrimidines (mismatches) into the polypurine target. At 260 nm we observed only a sharp single transition corresponding to the duplex→random coil transition for both the 5′-NDI conjugate and the 5′,3′-bisconjugate, suggesting the absence of any DNA triplex in both cases. At 383 nm both conjugates exhibited a transition near 60°C, essentially the same as that observed for the 5′-conjugate with the polypurine target, but lower than that observed for the 5′,3′-bisconjugate (see Table 1). This result is consistent with a mode of binding in which non-specific intercalation by the NDI of the monoconjugates (see Fig. 4b) or by one of the NDI residues of the bisconjugate occurs without any triplex formation. Had the bisconjugate functioned as simply a bisintercalator (see Fig. 4e) then a transition near 70°C should have been apparent when monitored at 383 nm. The higher Tm values observed with the cognate sequence indicates that the NDI conjugates are capable of discriminating cognate from non-cognate sequences.
CONCLUSIONS
The results for the described conjugates indicate that NDI intercalators are very effective as ligands to enhance binding of the third strand to the target duplex in DNA triplexes. The presence of a single conjugated NDI results in two transitions, the first with a ΔTm of 17°C represents denaturation of the third strand while the second with a ΔTm of at least 26°C represents unstacking of the NDI conjugate from the target. Complexes with two terminal NDI conjugates exhibit Tm enhancements as great as 41°C. NDI conjugates should be effective in stabilizing triplexes in the presence of competing macromolecules.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by a grant from the NIH (GM 53201).
REFERENCES
- 1.Cooney M., Czernuszewicz,G., Postel,E.H., Flint,S.J. and Hogan,M.E. (1988) Science, 241, 456–459. [DOI] [PubMed] [Google Scholar]
- 2.Moser H.E. and Dervan,P.B. (1987) Science, 238, 645–650. [DOI] [PubMed] [Google Scholar]
- 3.LeDoan T., Perrouault,L., Praseuth,D., Habhoub,N., Decout,J.L., Thoung,N.T., Lhomme,J. and Helene,C. (1987) Nucleic Acids Res., 15, 7749–7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hampel K.J., Crosson,P. and Lee,J.S. (1991) Biochemistry, 30, 4455–4459. [DOI] [PubMed] [Google Scholar]
- 5.Thomas T. and Thomas,T.J. (1993) Biochemistry, 32, 14069–14074. [Google Scholar]
- 6.Tung C.H., Breslauer,K.J. and Stein,S. (1993) Nucleic Acids Res., 21, 5489–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soyfer V.N. and Potaman,V.N. (1995) Triple Helical Nucleic Acids. Springer Verlag, New York, NY.
- 8.Escude C., Sun,J.S., Nguyen,C.H., Bisagni,E., Garestier,T. and Helene,C. (1996) Biochemistry, 35, 5735–5740. [DOI] [PubMed] [Google Scholar]
- 9.Escude C., Nguyen,C.H., Kukreti,S., Janin,Y., Sun,J.S., Bisagni,E., Garestier,T. and Helene,C. (1998) Proc. Natl Acad. Sci. USA, 95, 3591–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigoriev M., Praseuth,D., Robin,P., Hemar,A., Saisonbehmoaras,T., Dautryvarsat,A., Thuong,N.T., Helene,C. and Harelbellan,A. (1992) J. Biol. Chem., 267, 3389–3395. [PubMed] [Google Scholar]
- 11.Helene C. and Thuong,N.T. (1989) Genome, 31, 413–421. [DOI] [PubMed] [Google Scholar]
- 12.Silver G.C., Nguyen,C.H., Boutorine,A.S., Bisagni,E., Garestier,T. and Helen,C. (1997) Bioconjugate Chem., 8, 15–22. [DOI] [PubMed] [Google Scholar]
- 13.Zhou B.W., Puga,E., Sun,J.S., Garestier,T. and Helene,C. (1995) J. Am. Chem. Soc., 117, 10425–10428. [Google Scholar]
- 14.Robles J., Rajur,S.B. and McLaughlin,L.W. (1996) J. Am. Chem. Soc., 118, 5820–5821. [Google Scholar]
- 15.Robles J. and McLaughlin,L.W. (1997) J. Am. Chem. Soc., 119, 6014–6021. [Google Scholar]
- 16.Matsugo S., Kawanishi,S., Sugiyama,H., Matsuura,T. and Saito,I. (1991) Angew. Chem. Int. Edn Engl., 30, 1351–1353. [Google Scholar]
- 17.Liu Z.R., Hecker,K.H. and Rill,R.L. (1996) J. Biomol. Struct. Dyn., 14, 331–339. [DOI] [PubMed] [Google Scholar]
- 18.Lokey R.S., Kwok,Y., Guelev,V., Pursell,C.J., Hurley,L.H. and Iverson,B.L. (1997) J. Am. Chem. Soc., 119, 7202–7210. [Google Scholar]
- 19.Gianolio D.A. and McLaughlin,L.W. (1999) J. Am. Chem. Soc., 121, 6334–6335. [Google Scholar]
- 20.Bevers S., O’Dea,T.P. and McLaughlin,L.W. (1998) J. Am. Chem. Soc., 120, 11004–11005. [Google Scholar]
- 21.Matteucci M.D. and Caruthers,M.H. (1981) J. Am. Chem. Soc., 103, 3185–3191. [Google Scholar]
- 22.Gianolio D.A. and McLaughlin,L.W. (1999) Nucl. Nucl., 18, 1751–1769. [DOI] [PubMed] [Google Scholar]
- 23.Sun J.S., Giovannangeli,C. Francois,J.C., Kurfurst,R., Montenay-Garestier,T., Asseline,U., Saison-Behmoaras,T., Thuong,N.T. and Helene,C. (1991) Proc. Natl Acad. Sci. USA, 88, 6023–6027. [DOI] [PMC free article] [PubMed] [Google Scholar]