Abstract
Canine infectious respiratory disease complex (CIRDC) is the primary cause of respiratory disease in the canine population and is caused by a wide array of viruses and bacterial pathogens with coinfections being common. Since its recognition in late 2019, Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) has been reported to cause respiratory disease in dogs. Therefore, the rapid detection and differentiation of SARS-CoV-2 from other common viral and bacterial agents is critical from a public health standpoint. Here, we developed and validated a panel of four one-step multiplex qPCR/RT-qPCR assays for the detection and identification of twelve pathogens associated with CIRDC (canine adenovirus-2, canine distemper virus, canine herpesvirus-1, canine influenza A virus, canine parainfluenza virus, canine pneumovirus, canine respiratory coronavirus, SARS-CoV-2, Bordetella bronchiseptica, Streptococcus equi subsp. zooepidemicus, Mycoplasma cynos, and M. canis), as well as the identification of three main CIV subtypes (i.e., H3N2, H3N8, and H1N1). All developed assays demonstrated high specificity and analytical sensitivity. This panel was used to test clinical specimens (n = 76) from CIRDC-suspected dogs. M. canis, M. cynos, and CRCoV were the most frequently identified pathogens (30.3%, 25.0%, and 19.7% of samples, respectively). The newly emerging pathogens CPnV and SARS-CoV-2 were detected in 5.3% of samples and coinfections were identified in 30.3%. This new multiplex qPCR/RT-qPCR panel is the most comprehensive panel developed thus far for identifying CIRDC pathogens, along with SARS-CoV-2.
Keywords: multiplex reverse-transcriptase qPCR (RT-qPCR), canine respiratory pathogens, canine infectious respiratory disease complex (CIRDC), SARS-CoV-2, influenza virus A
1. Introduction
Canine infectious respiratory disease complex (CIRDC) is a contagious disease syndrome, commonly referred to as “kennel cough” or “canine cough”, which is caused by a wide array of etiologic agents [1,2]. Outbreaks of CIRDC have been reported all around the globe [3,4,5,6,7,8] and most commonly occur when dogs are housed or concentrated together, such as in boarding facilities, animal shelters, dog daycare, racing facilities, and obedience training classes [3,9]. Cases of CIRDC have also been reported in veterinary hospitals [10] and individually owned dogs [3]. Common clinical signs include coughing, nasal or ocular discharge, sneezing, and respiratory distress that typically lasts one to two weeks [2]. This multifactorial disease complex is associated with several pathogens, including viruses and bacteria [2,11]. The pathogens traditionally associated with CIRDC include canine adenovirus-2 (CAdV-2), canine distemper virus (CDV), canine herpesvirus-1 (CHV-1), canine parainfluenza virus (CpiV), and Bordetella bronchiseptica [2,12,13,14,15,16]. Since the early 2000s, it has been demonstrated that other pathogens are also involved in CIRDC, including Canine Respiratory Coronavirus (CRCoV), Streptococcus equi subsp. zooepidemicus, Mycoplasma spp., and canine influenza A virus (CIV). CRCoV was first detected in 2003 in dogs with respiratory distress [17] and is now well-recognized to be associated with CIRDC [18]. Streptococcus equi subsp. zooepidemicus is highly contagious and associated with severe cases of respiratory disease [19,20]. Mycoplasma spp. are part of the bacteria flora of the dog’s upper respiratory airways; among the 15 known species of Mycoplasma reported in dogs, only M. cynos and M. canis were associated with respiratory disorders [12,21,22,23]. While the pathogenic role of M. cynos was recently supported by meta-analyses, the role of M. canis was not. [24]. Among CIV subtypes, H3N8 and H3N2 are the most prevalent in the canine population around the world [25]. CIV H3N8, derived from equine influenza A virus H3N8, was first identified in 2004 in Florida, USA [26]. CIV H3N2, suspected to be derived from an avian influenza A virus in China and Korea [27], was first detected in the USA in a 2015 CIRDC outbreak in Chicago, Illinois [28]. Moreover, following the 2009 H1N1 human pandemic, cases of H1N1 infection in dogs have also been documented [25,29,30]. Canine pneumovirus (CPnV) is an emerging pathogen first isolated in the USA in 2010 from a dog with respiratory disease [31,32]. Since then, CPnV has been reported in dogs with CIRDC in North America, Europe, and Asia [3,4,8,33,34,35,36,37]. Since the causative agents of CIRDC induce similar clinical signs, confirmatory diagnosis relies on laboratory testing. Additionally, co-infections by two or more viral and/or bacterial pathogens is commonly observed and can lead to more severe clinical signs when compared to single infections [8,12,38].
Numerous reports have emerged during the COVID-19 pandemic of SARS-CoV-2 transmission from humans to a number of farm, wild, zoo, and companion animals, including dogs and cats, during the COVID-19 pandemic [39,40,41]. Among companion animals (i.e., dogs and cats), SARS-CoV-2 infections can either be subclinical or associated with respiratory (e.g., nasal discharge, coughing, dyspnea) and/or gastrointestinal (e.g., vomiting and diarrhea) clinical signs [42,43,44,45,46]. Nevertheless, cases of SARS-CoV-2 infection in dogs typically result in none to mild clinical manifestations and are not considered significant contributors to the spread of the virus [47]. While studies conducted on dogs experimentally infected with SARS-CoV-2 have revealed that they do not display signs of illness [44,48,49], transmission from experimentally infected dogs to naive dogs (sentinel controls) has been documented [50]. This raises concerns about the possible spillover between humans and dogs, requiring continuous surveillance to monitor SARS-CoV-2 infection in companion animals. Furthermore, the clinical signs induced by SARS-CoV-2 are not unique; thus, veterinarians must rule out more common causes of respiratory in animals by seeking laboratory diagnosis. Thus, rapid detection and differentiation of SARS-CoV-2 from other common viral and bacterial agents is critical for controlling COVID-19 and implementing appropriate biosecurity measures to prevent transmission.
Most of the veterinary diagnostic laboratories offer singleplex real-time PCR (qPCR) and reverse transcriptase-qPCR (RT-qPCR) assays to detect various respiratory pathogens (viral and bacterial) in clinical specimens of animals. However, using multiplex assays for the identification of two or more targets in a single reaction has several advantages. Firstly, a multiplex qPCR and RT-qPCR reduces the amount of valuable clinical samples needed for testing. Secondly, multiplexing reduces the cost by amplifying two or more targets (up to four) in one well, saving reagents and the technical time and effort needed to set up the tests and analyze the results. Finally, the amplification of multiple genes in the same well improves precision by minimizing pipetting errors. One of the limitations of multiplexing is related to the overlapping emission and spectra that some florescent dyes have, which limits their use in multiplex scenarios. This issue has been overcome in recent years with a new generation of fluorescent dyes by several manufacturers. [51,52,53,54].
Thus, in this study, we developed and evaluated the performance of a panel of four four-plex one-step qPCR/RT-qPCR assays for the simultaneous detection and differentiation of pathogens associated with CIRDC (e.g., viruses and bacteria) and SARS-CoV-2, namely canine respiratory assay 1 (CRA_1), CRA_2, and CRA_3, as well as an RT-qPCR assay to differentiate CIV H3N2, H3N8, and H1N1subtypes (CRA_4). This new panel was then used to test clinical samples submitted to the Louisiana Animal Disease Diagnostic Laboratory (LADDL) from CIRDC-suspected dogs collected in Louisiana, USA, between 2020 and 2023. Overall, this new highly sensitive and specific panel of multiplex qPCR/RT-qPCR assays developed in this study can simultaneously detect all CIRDC pathogens and SARS-CoV-2 in respiratory specimens.
2. Materials and Methods
2.1. Viruses and Bacteria
A panel of reference (prototype) viruses and bacteria associated with CIRDC and genetically related pathogens was used to assess the specificity (inclusivity/exclusivity) of each assay in singleplex and multiplex format (Table 1). RNA of CIV H3N2 VSL-1355 and CRCoV VSL-1471 were kindly provided by Dr. Diego Diel (Department of Population Medicine and Diagnostic Sciences, Cornell University College of Veterinary Medicine, Ithaca, NY, USA). RNA of CIV H3N8 A/Ca/FL/15592/04 and A/Ca/FL/61156.2/07 were kindly provided by Dr. Edward Dubovi. All other prototype strains were obtained from the American Type Culture Collection (ATCC®; Manassas, VA, USA) or BEI Resources (Manassas, VA, USA).
Table 1.
Panel of prototype canine respiratory viruses and bacteria, SARS-CoV-2 variants of concern, and other canine pathogens used to assess the specificity of each qPCR and RT-qPCR assay.
| Pathogens | Reference Strain | Source |
|---|---|---|
| Canine Herpesvirus 1 (CHV-1) | VR-552TM | ATCC® |
| Canine Adenovirus 2 (CAdV-2) | VR-800TM | ATCC® |
| Canine Parainfluenza Virus (CPiV) | VR-399TM | ATCC® |
| Canine Respiratory Coronavirus (CRCoV) | VSL-1471 | Cornell Universitya |
| Canine Distemper Virus (CDV) Lederle Avirulent | NR-3845 | BEI Resources |
| Murine Pneumonia virus (MPV) | VR-1819 | ATCC® |
| SARS-CoV-2 USA-WA1/2020 | NR-52281 | BEI Resources |
| SARS-CoV-2 Alpha (Lineage B.1.1.7) | NR-54020 | BEI Resources |
| SARS-CoV-2 Beta (Lineage B.1.351) | NR-55282 | BEI Resources |
| SARS-CoV-2 Delta (Lineage B.1.617.2) | NR-55671 | BEI Resources |
| SARS-CoV-2 Omicron (Lineage B.1.1.529) | NR-56461 | BEI Resources |
| Canine Influenza A (CIV) H3N2 | VLS-1355 | Cornell University a |
| CIV H3N8 | A/Ca/FL/15592/04 | Cornell University b |
| CIV H3N8 | A/Ca/FL/61156.2/07 | Cornell University b |
| Influenza A virus, A/California/04/2009 (H1N1)pdm09 | NR-13658 | BEI Resources |
| Bordetella bronchiseptica, E014 | NR-44164 | BEI Resources |
| Streptococcus equi susb. zooepidemicus Farrow and Collins | 700400TM | ATCC® |
| Mycoplasma cynos Rosendal | 27544TM | ATCC® |
| Mycoplasma canis, PG 14 | NR-3865 | BEI Resources |
| Canine Adenovirus 1 (CAdV-1) | VR-293TM | ATCC® |
| Canine Enteric Coronavirus (CECoV), UCD1 | NR-868 | BEI Resources |
| Mycoplasma felis Cole et al. | 23391TM | ATCC® |
a Kindly provided by Dr. Diego Diel; b Kindly provided by Dr. Edward Dubovi; ATCC®: American Type Culture Collection; LADDL: Louisiana Animal Disease Diagnostic Laboratory.
2.2. Clinical Specimens
A total of 76 specimens including nasal swabs (n = 38), pharyngeal swabs (n = 29), and pools of tissues (n = 2) submitted for routine diagnostic testing at the Louisiana Animal Disease Diagnostic Laboratory (LADDL) between 2020 and 2023 were included in this study. These specimens were collected from a total of 50 dogs, with nasal swabs and pharyngeal swabs concurrently collected in 26/50 dogs (Table S1). Swabs were either submitted to the LADDL for CIRDC diagnosis or collected from three shelters located in and around Baton Rouge, LA, USA. Specimens were collected using sterile oropharyngeal/nasal swabs (VMRD, Pullman, WA) and all swab samples were resuspended in either 2 mL of BHI Broth (Hardy Diagnostics, Santa Maria, CA, USA) or 2 mL of PrimeStore® molecular transport medium (VMRD). Swab samples were then vortexed and centrifuged for clarification and the supernatant was stored at 4 °C until use. Tissues (i.e., kidney, liver, lung, spleen) were collected from dogs submitted for necropsy at the LADDL and were homogenized using the Bead Ruptor Elite (Omni, Inc, Dallas, TX, USA) in a 1:9 ratio with 1× phosphate-buffered saline (PBS) by performing two cycles of 30 s at 4.00 m/s. Samples were then clarified by centrifugation at 4000× g for 10 min at 4 °C.
2.3. Nucleic Acid Extraction
Nucleic acid extraction was performed using the tacoTM mini nucleic acid automatic extraction system (GeneReach, Taichung, Taiwan) following manufacturer’s recommendations. One hundred microliters of swab or 10% tissue suspensions were extracted and eluted in equal volume of elution buffer. The extracted nucleic acid samples were stored at −80 °C until used.
2.4. Primers and Probe Design
Specific forward and reverse primers and probes used for specific amplification of CPiV nucleocapsid (N), CDV phosphoprotein (P), CPnV-N, CRCoV-N, M. canis tuf gene, CIV H3N2 neuraminidase (NA), and CIV H1N1-NA assays were designed using Geneious R6 software (v.6.1.8, Auckland, New Zealand) and IDT’s PrimerQuest tool (https://www.idtdna.com/Primerquest/home/Index, accessed on 1 September 2022) from sequences available on the GenBank nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore/, accessed on 1 September 2022) and Influenza Research Database (https://www.fludb.org, accessed on 1 September 2022) (Table 2). The primers and probe sequences specificity were further validated in silico using the NCBI Basic Local Alignment Search Tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome, accessed on 1 September 2022). Self-annealing sites, hairpin loop formation, and 3’ complementarity were analyzed using IDT’s OligoAnalyzer tool (https://www.idtdna.com/calc/analyzer, accessed on 1 September 2022). Sequences of primers and probes for SARS-CoV-2 [55], CHV-1 [56], Bordetella bronchiseptica [57], M. cynos [58], Streptococcus equi subsp. Zooepidemicus [59], and CIV H3N8 [60,61] detection were used as previously published (Table 2). CIV [62] and CAdV-2 [63] primers and probes were used as previously published with addition of nucleotide degeneracy in the sequences of the CIV reverse primer 1 (CIV_M-R1 position 1: T→Y) and CAdV-2 probe (CAdV2_H-P position 10: T→Y), respectively (Table 2).
Table 2.
Primers and probe sequences used for the detection of pathogens associated with CIRDC and SARS-CoV-2.
| Target (Gene) | Oligonucleotide ID | Primers and Probe Sequences (5’–3’) | Nucleotide Position |
Product Size(bp) | GenBank Accession | Reference |
|---|---|---|---|---|---|---|
| CPiV (Nucleoprotein) |
CPiV_N-F CPiV_N-R CPiV_N-P |
ACCATCAGCCACAATGCTCA | 298–317 | EF543648.1 | This article | |
| AGCGGAATGATCCCTCCTCA | 401–382 | 104 | ||||
| FAM-AGCTGACCAGTCACCAGAAGC-QSY | 331–351 | |||||
| Canine Influenza A virus (CIV) (Matrix protein) |
CIV_M-F CIV_M-R1 a CIV_M-R2 a CIV_M-P |
AGATGAGTCTTCTAACCGAGGTCG | 24–47 | MF173222.1 | [62,64] with modification |
|
| YGCAAAGACATCTTCAAGTCTCTG TGCAAAGACACTTTCCAGTCTCTG |
124–101 124–101 |
101 | ||||
| VIC-TCAGGCCCCCTCAAAGCCGA-QSY | 74–93 | |||||
| CAdV-2 (Hexon) |
CAdV2_H-F CAdV2_H-R CAdV2_H-P |
AGTAATGGAAACCTAGGGG | 17,821–17,839 | U77082.1 | [63] with modification |
|
| TCTGTGTTTCTGTCTTGC | 17,900–17,883 | 80 | ||||
| ABY-TCAGTCATCYCAGCTCAATGCCGTG-QSY | 17,874–17,850 | |||||
| CDV (Phosphoprotein) |
CDV_P-F CDV_P-R CDV_P-P |
ACTATTGAGAGACCTCCAGCTGAAA | 1296–1320 | AB028914.1 | This article | |
| TGCGGTATCCTTCGGTTTGT | 1374–1355 | 79 | ||||
| JUN-CCGATTGCCGAGCTAGACTCTTTGTCA-QSY | 1352–1326 | |||||
| SARS-CoV-2 (Nucleocapsid) |
2019-nCoV_N1-F 2019-nCoV_N1-R 2019-nCoV_N1-P |
GACCCCAAAATCAGCGAAAT | 28,287–28,306 | MN985325.1 | [55] | |
| TCTGGTTACTGCCAGTTGAATCTG | 28,358-28,335 | 72 | ||||
| FAM-ACCCCGCATTACGTTTGGTGGACC-QSY | 28,309–28,332 | |||||
| CPnV (Nucleocapsid) |
CPnV_N-F CPnV_N-R CPnV_N-P |
CAGGACAAGTTATGCTRAGGT | 1825–1845 | NC_025344.1 | This article | |
| CTCAACCACCTGTTCCATCTC | 1925–1905 | 101 | ||||
| VIC-AGCTTGAACACTAGCATGGCCTAGC-QSY | 1904–1880 | |||||
| CRCoV (Nucleocapsid) |
CRCoV_N-F CRCoV_N-R CRCoV_N-P |
CCTCTGGAAATCGTTCTGGTAA | 8273–8294 | DQ682406.1 | This article | |
| GCTTGGGTTGAGCTCTTCTA | 8371–8352 | 99 | ||||
| ABY-ACTGATCGGCCCACTTAAGGATGC-QSY | 8320–8297 | |||||
| CHV-1 (Glycoprotein B) |
CHV_gB-F CHV_gB-R CHV_gB-P |
ACAGAGTTGATTGATAGAAGAGGTATG | 439–465 | AF361073.1 | [56] | |
| CTGGTGTATTAAACTTTGAAGGCTTTA | 574–548 | 136 | ||||
| JUN-TCTCTGGGGTCTTCATCCTTATCAAATGCG-QSY | 539–510 | |||||
|
Bordetella bronchiseptica (Intergenomic region between flaA and fliA B) |
Fla2-F Fla12-R Fla-P |
AGGCTCCCAAGAGAGAAAGGCTT | 1,140,858–1,140,880 | 118 | CP019934.1 | [57] |
| AAACCTGCCGTAATCCAGGC | 1,140,975–1,140,956 | |||||
| FAM-ACCGGGCAGCTAGGCCGC-QSY | 1,140,887–1,140,904 | |||||
|
Mycoplasma cynos (tuf) |
Mcynos_tuf-F Mcynos_tuf-R Mcynos_tuf-P |
TCTTCGTATTTAGCATCACCTTCAAGT | 8234–8260 | FJ896395.1 | [58] | |
| TGATGGAGATAATGCGCCAAT | 8305–8285 | 72 | ||||
| VIC-CTTTTAAAGCTGAACCACG-QSY | 8262–8280 | |||||
|
Streptococcus equi subsp. zooepidemicus (sodA) |
SodA-F SodA-R SodA-Bd 4116/06-R SodA-P |
AGAGCAATTCACAGCAGCA | 246–264 | JN631988.1 | [59] | |
| ACCAGCCTTATTCACAACCA ACCGGCTTGGTTAACCACTA |
318–299 318–299 |
73 | ||||
| ABY-CAGGCCCAACCTGAGCCAAA-QSY | 296–277 | |||||
|
Mycoplasma canis (tuf) |
Mcanis_tuf-F Mcanis_tuf-R Mcanis_tuf-P |
CAACAGCATCCATTAATTCCAT | 305–326 | FJ896394.1 | This article | |
| ACGGATTTGACGGAGATAAC | 412–393 | 108 | ||||
| JUN-TGAAGCTGATCCACGGATAATTGGAGC-QSY | 366–392 | |||||
| Canine H3N2 (Neuraminidase) |
H3N2_NA-F H3N2_NA-R H3N2_NA-P |
CCGTTGAAGGCAAAAGCTGT | 1251–1270 | MF173401.1 | This article | |
| TCTCTTGTGGCCCTCCTCTT | 1319–1300 | 69 | ||||
| FAM-AATAGGTGTTTTTATGTGGAGTTGAT-QSY | 1274–1299 | |||||
| Canine H3N8 (Hemagglutinin) |
H3N8_HA3-F H3N8_HA3-R H3N8_HA3-P |
TCACATGGACAGGTGTCACTCA | 448–469 | MF173285.1 | [60,61] | |
| GGCTGATCCCCTTTTGCA | 506–489 | 59 | ||||
| JUN-AACGGAAGAAGTGGAGC-QSY | 471–487 | |||||
| Canine H1N1 (Neuraminidase) |
H1N1_NA-FH1N1_NA-R H1N1_NA-P |
GCGGGCAATTCCTCTCTC | 256–276 | MG254090.1 | This article | |
| CTTGGAACCGATTCKTACACTRT | 333–311 | 78 | ||||
| ABY-TGYCCTGTTAGTGGATGGGCTATATACAGT-QSY | 274–303 |
ABY, ABYTM dye; F: forward primer; FAM, 6-carboxyfluorescein dye; JUN, JUN™ dye; P: probe; QSY, QSY™ quencher; R: reverse primer; VIC, VIC™ dye; a CIV_M-R1 and CIV_M-R2 were used at equimolar amount (200 nM).
2.5. Specific Multiplex TaqMan® Quantitative PCR (qPCR) and Reverse Transcription PCR (RT-qPCR) Assays for Canine Respiratory Pathogens
Four four-plex RT-qPCR assays were developed and designated as follows: CRA_1 (detection of CIV, CDV, CpiV, and CAdV-2), CRA_2 (detection of CRCoV, CPnV, CHV-1, and SARS-CoV-2), CRA_3 (detection of B. bronchiseptica, S. equi subsp. zooepidemicus, M. cynos, and M. canis) and CIV_4 for identification of CIV and its most prevalent subtypes in dogs (i.e., CIV-H3N2, CIV-H3N8, CIV-H1N1). RT-qPCR assays were performed in a total volume of 25 μL containing 12.5 μL of 2× QuantiTectTM Multiplex RT-PCR Master Mix (QIAGEN, Hilden, Germany), 0.25 μL of QuantiTectTM RT Mix, 1.25 μL of primers and fluorogenic probes mix (200 nM each), 6 μL of RNase free water, and 5 μL of template DNA/RNA. A 7500 Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) was used with the following thermal profile: a reverse transcription step (20 min at 50 °C) followed by an initial activation step (15 min of at 95 °C) and 40 cycles of denaturation and annealing/extension (45 s at 94 °C and 75 s at 60 °C). The complete step-by-step protocols have been deposited on protocol.io platform (DOIs: dx.doi.org/10.17504/protocols.io.kxygx9x7zg8j/v1; dx.doi.org/10.17504/protocols.io.14egn2o9pg5d/v1).
2.6. Synthesis of In Vitro Transcribed RNA and DNA
Specific in vitro transcribed (IVT) RNA and plasmid DNA were synthesized in order to determine the analytical sensitivity of each multiplex RT-qPCR assay as previously described, with minor modifications [53]. Four inserts containing the target regions of each assay flanked by PstI and HindIII restriction enzymes were chemically synthesized and cloned into the pGEM®-3Z vector (Promega, Madison, WI, USA) downstream of the T7 promoter by GeneArt Gene Synthesis (Thermo Fisher Scientific, Waltham, MA, USA). Transformed Escherichia coli DH10β cells were cultured overnight at 37 °C with agitation at 270 rpm. Plasmid DNA was extracted using QIAprep Spin Miniprep kit (QIAGEN). Plasmid DNA was linearized using HindIII restriction enzyme and concentration was measured using Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific). When IVT RNA was needed, linearized plasmid DNA were subjected to in vitro transcription using the Megascript® T7 Transcription Kit (Thermo Fisher Scientific) following manufacturer’s recommendations. Subsequently, DNase treatment was performed with TURBOTM DNase (Thermo Fisher Scientific) for 15 min at 37 °C. The IVT RNA products were purified using MEGAclearTM Transcription Clean-Up Kit (Thermo Fisher Scientific) and quantified using Qubit RNA BR Assay Kit (Thermo Fisher Scientific). The number of plasmid DNA and IVT RNA (copies/μL) were calculated according to the following formula:
IVT RNA and DNA plasmid molecular weight was calculated using Molbiotools website (https://molbiotools.com/dnacalculator.php, accessed on 1 October 2022). Each IVT RNA and plasmid DNA concentration was adjusted to 107 copies/μL in nuclease-free water and stored at −80 °C until used. Then, 40 ng/μL of yeast tRNA (Thermo Fisher Scientific) was added in IVT RNA preparations. Ten-fold serial dilutions of IVT RNA and plasmid DNA were directly used for determining the analytical sensitivity of the qPCR assays.
2.7. Analytical Parameter Determination and Statistical Analysis
Analytical parameters were determined as previously described [53], with minor modifications. Standard curves were generated using ten-fold dilutions of plasmid DNA and IVT RNA (107 to 101 copies/μL) in triplicate. Coefficients of determination (R2) were used to assess curve fitness. Amplification efficiency [E (%)] was calculated after regression analysis using the following formula: E = [10−1/slope − 1] × 100. Limit of detection with 95% confidence (LOD95%) of each assay was determined by statistical probit analysis (non-linear regression model) using SPSS 14.0 software (SPSS Inc., Chicago, IL, USA) from twelve replicates per dilution ranging from 103 to 100 copies/μL. Cycle threshold (Ct) cut-off values were determined using the following formula: Ct cut-off = Average Ct values of 12 replicates of the endpoint dilution + (3 × standard deviation [SD]) [53,54,65]. Intra-run imprecision was determined by performing 12 replicates of plasmid DNA/IVT RNA containing 105 to 103 copies/μL on the same run and inter-run imprecision was determined by using three replicates of plasmid DNA/IVT RNA containing 105 to and 103 copies/μl on two independent runs. The coefficient of variation (%CV) was calculated using the following formula: %CV = 100 × (standard deviation of replicates [log10 copies/µL] ÷ average of replicates [log10 copies/µL]). Data were graphically represented using GraphPad Prism v9.3.1 statistical analysis software (GraphPad, San Diego, CA, USA) and UpSetR package [66].
3. Results
3.1. Analytical Specificity of Singleplex and Multiplex Assays for the Detection of Canine Respiratory Pathogens
The analytical specificity (inclusivity/exclusivity) of all singleplex and four-plex qPCR/RT-qPCR assays was first evaluated using a panel of reference viruses and bacteria associated with respiratory disease in dogs, unrelated pathogens (i.e., CAdV-1, Canine enteric coronavirus [CECoV], and M. felis), and different SARS-CoV-2 variants of concern (VOCs). The previously published assays [55,56,57,58,59,60,61,62,63] and the news assays developed in this study showed exclusive specificity for the respective targets and did not cross-react between each other when used under multiplex conditions (Figure S1). As no prototype CPnV strain was available, the closely related murine pneumonia virus strain 15 (ATCC®, VR-1819TM) was used [67] and detected by the newly developed CPnV (N) assay. The CAdV-2 (H) assay did not amplify the CAdV-1 reference strain, the CRCoV (N) assay did not amplify the CECoV reference strain, the M. cynos (tuf) assay did not amplify the M. canis and M. felis reference strains, and the M. canis (tuf) assay did not amplify the M. cynos and M. felis reference strains.
3.2. Analytical Sensitivity of Singleplex and Multiplex Assays for the Detection of Canine Respiratory Pathogens
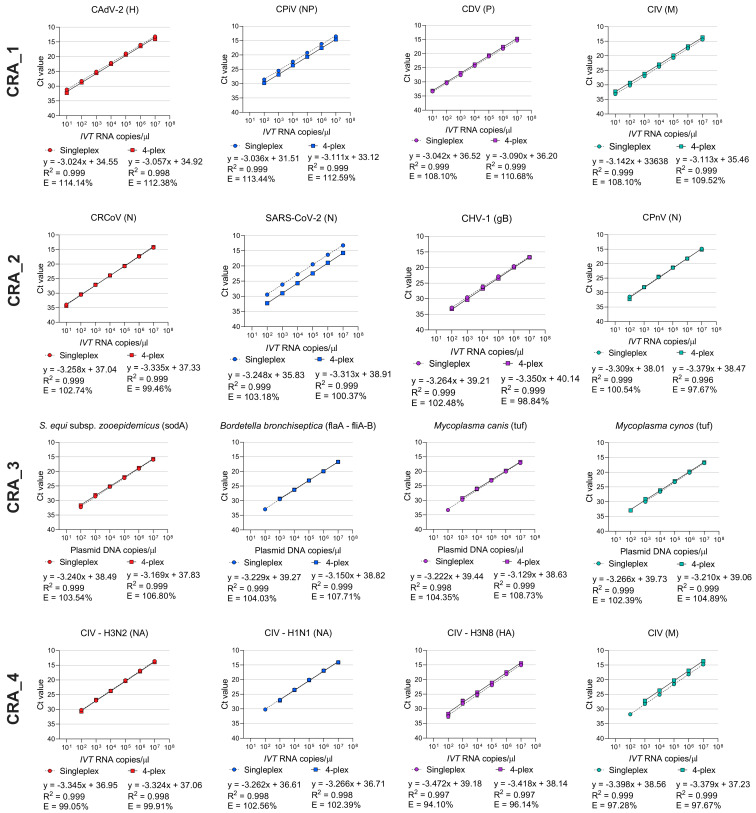
The analytical sensitivity of all assays in singleplex and in multiplex were determined using ten-fold dilutions (107 copies/µL to 102 copies/µL) of the specific plasmid DNA/IVT RNA containing the target sequences. Linear standards curves were generated for each assay in singleplex and four-plex with a coefficient of linear regression (R2) ≥ 0.997 (Figure 1 and Table 3). The amplification efficiency for each singleplex and four-plex assay was similar and ranged between 94.10% and 114.35% (Table 3).
Figure 1.
Comparison of analytical sensitivity of each singleplex and multiplex qPCR and RT-qPCR assays for the detection of pathogens associated with CIRDC and SARS-CoV-2. Ct: cycle threshold; IVT RNA: in vitro transcribed RNA; R2: linearity; E: efficiency.
Table 3.
Analytical performance of singleplex and four-plex qPCR/RT-qPCR assays for the detection of pathogens associated with CIRDC and SARS-CoV-2.
| Assay | Target | Parameter | Slope | Linearity (R2) | Efficiency (%) | LOD95% (Copies/µL) | Detection Rate Limit (Copies/µL) | Ct Cut-Off |
|---|---|---|---|---|---|---|---|---|
| CRA_1 | CAdV-2 (H) | Singleplex | −3.024 | 0.999 | 114.14 | 4 | 10 | 34 |
| Four-plex | −3.057 | 0.998 | 112.38 | 5 | 10 | 35 | ||
| CPiV (NP) | Singleplex | −3.037 | 0.999 | 113.44 | 5 | 10 | 35 | |
| Four-plex | −3.053 | 0.999 | 112.59 | 5 | 10 | 34 | ||
| CDV (P) | Singleplex | −3.042 | 0.999 | 113.17 | 4 | 10 | 36 | |
| Four-plex | −3.090 | 0.999 | 110.68 | 4 | 10 | 35 | ||
| CIA (M) | Singleplex | −3.142 | 0.999 | 108.10 | 4 | 10 | 37 | |
| Four-plex | −3.113 | 0.999 | 109.52 | 4 | 10 | 34 | ||
| CRA_2 | CRCoV (N) | Singleplex | −3.258 | 0.999 | 102.74 | 10 | 100 | 35 |
| Four-plex | −3.335 | 0.999 | 99.46 | 8 | 10 | 34 | ||
| SARS-CoV-2 (N1) | Singleplex | −3.248 | 0.999 | 103.18 | 5 | 10 | 39 | |
| four-plex | −3.313 | 0.999 | 100.37 | 5 | 10 | 37 | ||
| CHV-1 (gB) | Singleplex | −3.264 | 0.999 | 102.48 | 12 | 100 | 35 | |
| Four-plex | −3.350 | 0.999 | 98.84 | 14 | 100 | 38 | ||
| CPnV (N) | Singleplex | −3.309 | 0.999 | 100.54 | 6 | 10 | 40 | |
| Four-plex | −3.379 | 0.996 | 97.67 | 6 | 10 | 36 | ||
| CRA_3 | S. equi subsp. zooepidemicus (sodA) | Singleplex | −3.240 | 0.999 | 103.54 | 6 | 10 | 34 |
| Four-plex | −3.169 | 0.998 | 106.80 | 15 | 100 | 35 | ||
| B. bronchiseptica (flaA-fliA-B) | Singleplex | −3.229 | 0.999 | 104.03 | 6 | 10 | 35 | |
| Four-plex | −3.150 | 0.999 | 107.71 | 43 | 100 | 40 | ||
| M. canis (tuf) | Singleplex | −3.222 | 0.999 | 104.35 | 28 | 100 | 35 | |
| Four-plex | −3.129 | 0.999 | 108.73 | 60 | 100 | 31 | ||
| M. cynos (tuf) | Singleplex | −3.266 | 0.999 | 102.39 | 43 | 100 | 38 | |
| Four-plex | −3.210 | 0.998 | 104.89 | 53 | 100 | 35 | ||
| CRA_4 | CIA - H3N2 (NA) | Singleplex | −3.345 | 0.999 | 99.05 | 6 | 10 | 37 |
| Four-plex | −3.324 | 0.998 | 99.91 | 8 | 10 | 34 | ||
| CIA - H1N1 (NA) | Singleplex | −3.262 | 0.998 | 102.56 | 5 | 10 | 35 | |
| Four-plex | −3.266 | 0.998 | 102.39 | 12 | 100 | 32 | ||
| CIA - H3N8 (HA) | Singleplex | −3.472 | 0.997 | 94.10 | 8 | 10 | 39 | |
| Four-plex | −3.418 | 0.997 | 96.14 | 8 | 10 | 35 | ||
| CIA (M) | Singleplex | −3.398 | 0.999 | 97.28 | 6 | 10 | 39 | |
| Four-plex | −3.379 | 0.999 | 97.67 | 8 | 10 | 38 |
CRA: canine respiratory assay; R2: linearity; LOD95% limit of detection 95%; Ct: cycle threshold.
The detection rate limit (100%) was equal to 10–100 copies/µL for all assays when ran in singleplex and in four-plex conditions. The limit of detection (LOD95%) calculated using a probit analysis was ≤15 copies/µL for all singleplex assays, except for M. canis (tuf) and M. cynos (tuf) where the LOD95% was slightly higher with values of 28 and 43 copies/µL, respectively (Table 3 and Figure S2). A similar LOD95% was observed for most of these assays when performed in a multiplex format. An increase in the LOD95% was noted for B. bronchiseptica (flaA-fliA-B) but was less than 1 Log10.
3.3. Repeatability and Reproducibility of Multiplex qPCR/RT-qPCR Assays for Detection of Canine Respiratory Pathogens
The repeatability and reproducibility of each four-plex assay was determined by measuring the intra-run and inter-run variability, respectively. Three concentrations of plasmid DNA/IVT RNA were used: 105 copies/µL (high concentration), 104 copies/µL (medium concentration), and 103 copies/µL (low concentration). The coefficients of variability (CV) are presented in Table 4. For all assays, the intra-run variability was <2% at high concentration, <4% at medium concentration, and <6% at low concentration. Similarly, the inter-run variability was <2% at high concentration, <3% at medium concentration, and < 5% at low concentration.
Table 4.
Precision assessment of each four-plex qPCR and RT-qPCR assay.
| Assay | Target | Intra-Run Variability CV (%) # |
Inter-Run Variability CV (%) # |
||||
|---|---|---|---|---|---|---|---|
| 105 Copies/µL |
104 Copies/µL |
103 Copies/µL |
105 Copies/µL |
104 Copies/µL |
103 Copies/µL |
||
| CRA_1 | CAdV-2 (H) | 0.55 | 2.58 | 2.00 | 1.19 | 0.83 | 1.92 |
| CPiV (NP) | 0.56 | 2.33 | 0.95 | 1.44 | 0.65 | 0.78 | |
| CDV (P) | 1.01 | 2.50 | 3.05 | 1.60 | 1.65 | 2.56 | |
| CIA (M) | 0.65 | 2.27 | 0.69 | 1.69 | 0.57 | 0.35 | |
| CRA_2 | CRCoV (N) | 1.09 | 1.19 | 2.66 | 0.88 | 1.05 | 1.72 |
| SARS-CoV-2 (N1) | 0.84 | 1.14 | 1.79 | 0.85 | 0.73 | 1.13 | |
| CHV-1 (gB) | 1.66 | 3.96 | 2.95 | 1.58 | 1.61 | 3.80 | |
| CPnV (N) | 0.66 | 0.89 | 2.49 | 1.77 | 1.57 | 1.79 | |
| CRA_3 | S. equi subsp. zooepidemicus (sodA) | 0.82 | 2.29 | 3.66 | 1.00 | 2.67 | 3.07 |
| B. bronchiseptica (flaA–fliA-B) | 0.43 | 1.86 | 2.96 | 0.57 | 1.14 | 4.83 | |
| M. canis (tuf) | 1.29 | 3.23 | 5.70 | 1.66 | 2.68 | 4.23 | |
| M. cynos (tuf) | 0.57 | 1.53 | 3.54 | 0.96 | 2.06 | 2.85 | |
| CRA_4 | CIA-H3N2 (NA) | 0.82 | 0.78 | 1.09 | 0.50 | 0.60 | 3.07 |
| CIA-H1N1 (NA) | 0.67 | 0.58 | 1.29 | 0.90 | 1.48 | 1.49 | |
| CIA-H3N8 (HA) | 1.42 | 1.80 | 3.93 | 1.20 | 1.42 | 4.19 | |
| CIA (M) | 0.28 | 0.43 | 0.86 | 1.23 | 1.45 | 2.67 | |
# CV (%): Coefficient of variation = (standard deviation of replicates [log10 copies/µL] ÷ Average of replicates [log10 copies/µL]) × 100.
3.4. Use of Multiplex Assays on Biological Specimens from CIRDC-Suspected Dogs
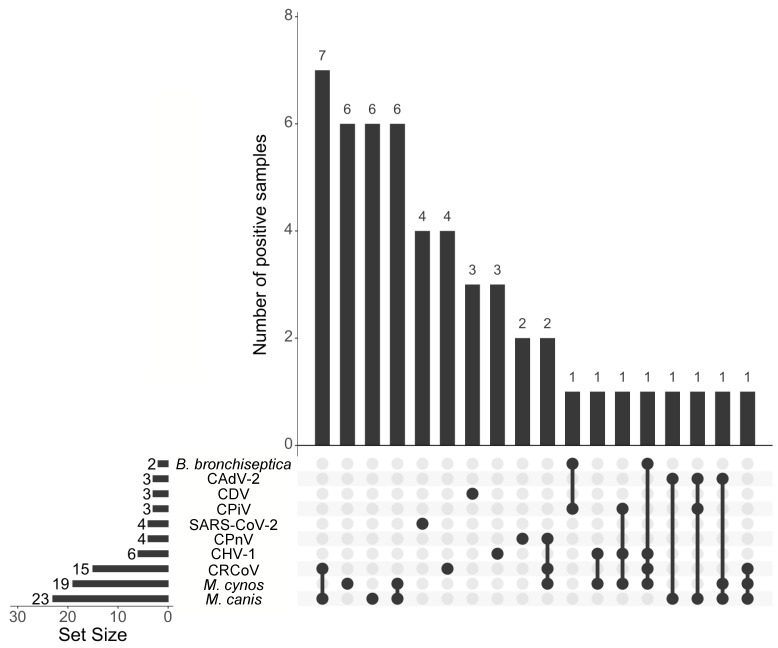
The panel of multiplex assays was used to test 76 clinical samples collected from dogs that displayed respiratory disease between 2020 and 2023 in Louisiana, USA. Among the 76 samples, 51 (67.1%) were positive for at least one of the 12 pathogens tested, M. canis (n = 23; 30.3%), M. cynos (n = 19; 25.0%), and CRCoV (n = 15; 19.7%) were the most commonly identified pathogens (Figure 2; Table S2). No samples were positive for S. equi subsp. zooepidemicus or CIV. SARS-CoV-2 was detected in four samples (5.3%) collected in August 2021. The first SARS-CoV-2-positive sample was a nasal swab collected from a 4-year-old female Goldendoodle presenting with leukocytosis, fever, and lethargy. The second SARS-CoV-2 positive sample was a pharyngeal swab collected from a six-year-old male Cocker Spaniel with a cough. The respective owners of these dogs have previously tested positive for COVID-19. No record of the dogs’ clinical signs and owner status was available for the two other samples collected from a 7-year-old female Schnauzer and a one-year-old female Goldendoodle. Additionally, four samples were positive for CPnV RNA (5.3%); these samples were collected from two dogs located in the same shelter (nasal and pharyngeal swabs).
Figure 2.
UpSet plot summarizing the number of CIRDC pathogens and SARS-CoV-2 detected in dogs using the newly developed panel. The number samples with single infection or co-infection are shown as vertical bars. The bottom left horizontal bar graph labeled Set Size shows the total number of positive samples for each specific CIRDC pathogens and SARS-CoV-2.
Co-infections were identified in 23 samples (30.3%) and ranged from two (16 samples, 21.1%) to up to four agents (one sample, 1.3%) (Figure 2; Table S3). Mycoplasma species were typically the most common co-infecting agent.
4. Discussion
CIRDC is a complex infectious disease in dogs caused by one or a combination of several viruses and bacteria [2]. A rapid detection of the implicated pathogen(s) is important in order to provide the most appropriate treatment and implement proper biosecurity measures to prevent the spread of disease. Moreover, the detection of emerging pathogens, such as CPnV and SARS-CoV-2, is essential to understand their epidemiology and, in the case of SARS-CoV-2 and influenza A virus A, to inform the owners and public health officials. Multiplex one-step qPCR or RT-qPCR assays allow the rapid identification of up to four targets in a single reaction. Therefore, this technique is now widely adopted in veterinary diagnostic laboratories [51,52,53,54]. Panels of qPCR/RT-qPCR and multiplex qPCR/RT-qPCR were previously developed for the detection of CIRDC-associated pathogens but they neither include all of the CIRDC-associated pathogens nor do they incorporate SARS-CoV-2 for its simultaneous detection [4,8,21]. In this study, the new qPCR/RT-qPCR panel developed for the detection of pathogens associated with CIRDC (including eight viruses and four bacteria), influenza A virus (H1N1, H3N2, and H3N8), and SARS-CoV-2 shows high analytical specificity and sensitivity for all targets tested.
With the expansion of next-generation sequencing in the past decade, pathogen sequence data have dramatically increased, leading to the discovery of new variants and strains. Additionally, pathogens, especially RNA viruses, are subject to point mutations leading to rapid evolution, as illustrated by the recent SARS-CoV-2 pandemic [68]. It is therefore important to validate and modify existing assays for the optimal detection of circulating strains. All assays in this study were developed to target most of the strains published on the GenBank nucleotide database and Influenza Research Database. In addition, degeneracy has been added to previously published assays to target circulating strains. The addition of one nucleotide degeneracy in the previously published CAdV-2_H probe (position 10: T→Y) [63] allows our assay to match in silico at 100% with all the CAdV-2 sequences available without affecting the specificity of the assay. Using online Influenza virus A databases, a similar approach was applied on the CIV_M reverse primer (position 1: T→Y) [62].
Different species within the Mycoplasma genus were isolated from dogs but only two of them, M. cynos and M. canis, were reported to induce respiratory disease [12,21,22,23]. In order to differentiate these two strains, the qPCR assays specific to M. cynos and M. canis developed by Tallmadge et al. [58] were evaluated. Although the M. cynos assay was specific for the M. cynos reference strain DNA, the amplification of both the M. cynos (27544 Rosendal, ATCC®) and M. canis (NR-3865, ATCC®) reference strain DNA was observed with the M. canis assay. For this reason, we developed a different M. canis qPCR assay targeting the elongation factor tuf gene (M.canis_tuf). This assay was specific for M. canis without the amplification of the M. cynos reference strain DNA. An in silico analysis was also performed for the development of new assays for the specific detection of CDV, CPiV, CPnV, CRCoV, CIV H3N2, and CIV H1N1 subtypes; all of those assays have shown an excellent specificity.
The analytical sensitivity of each multiplex qPCR and RT-qPCR assay was evaluated in this study and compared to singleplex assay formats. The nearly perfect linearity (R2 ≥ 0.997) and high amplification efficiency (>94%) demonstrate the high sensitivity of our panel of multiplex RT-qPCR assays for the detection of canine respiratory pathogens, without a loss of analytical sensitivity when used in multiplex conditions. Additionally, the detection of low genome copy numbers in clinical specimens is critical to the assay’s sensitivity. Here, a high analytical sensitivity was observed for most of our assays when used in multiplex with a LOD95 ≤ 15 copies/μL. While a lower sensitivity was observed for B. bronchiseptica, M. canis, and M. cynos (LOD95 ≤ 60 copies/μL), these results remained robust. Additionally, the excellent intra-run repeatability and inter-run reproducibility at high to low concentrations of target demonstrated that our panel of four multiplex qPCR/RT-qPCR assays ensure a high quality diagnostic and reproducibility of the results. In view of our data, we have demonstrated that using our assays in a multiplex format did not affect the analytical parameters. Additionally, these parameters were similar to those from other assays recently developed in our laboratory for the detection of canine enteric viruses and feline respiratory pathogens [69,70]. In this study, two to three log10 improvement of the LOD95 was determined when compared to the previously published multiplex PCR assays for the detection of CAdV-2, CDV, CIV, and CPiV [38]. However, the LOD95 determined in this study was in the same range of the recent three-panel triplex qPCR assays for the detection of nine pathogens associated with CIRDC (CAdV-2, CHV-1, CPiV, CDV, CIA, CRCoV, M. cynos, M. canis, and B. bronchiseptica) [21]. To our knowledge, no previous panel of multiplex qPCR/RT-qPCR assays was developed for the simultaneous detection of twelve canine respiratory pathogens and associated with the detection of SARS-CoV-2.
Our panel of qPCR/RT-qPCR was used to evaluate clinical samples collected from CIRDC-suspected dogs. While 36.8% of the samples were positive for only a single pathogen, 30.3% of the samples yielded positive results for at least two pathogens, demonstrating the high rate of co-infections, as previously reported [4,8,24,71]. This observation supports the need for a simultaneous detection of all the pathogens involved in CIRDC in order to inform treatment strategies. Among the positive samples, 82.4% were positive for M. canis or M. cynos, showing the common occurrence of these bacteria in CIRDC-suffering dogs, as recently highlighted in different studies [12,21]. As M. canis was detected in 73.9% of multi-infected dogs in this study, it is hypothesized that either this pathogen provides favorable conditions for secondary infections in the upper airways, or is a normal commensal of the upper respiratory tract in both health and disease, as previously suggested [23]. B. bronchiseptica, which is one of the most common pathogens found in CIRDC-suffering dogs [4,8,21], was surprisingly detected in only 2.6% dogs in our study. The low detection rate of B. bronchiseptica in the samples tested here can be explained by the lowest analytical sensitivity of this test (LOD95 = 43 copies/μL) when compared to the other targets.
The performance of the CRA_4 assay was not evaluated using field samples as no CIV-positive samples were identified during the study period. The emerging virus, CPnV, was also detected in two CIRD-suffering dogs located in the same shelter; co-infection with CRCoV and M. cynos was observed in one of the CPnV-infected dogs. SARS-CoV-2 was detected in four dogs in the absence of co-infection with other CIRDC pathogens. This result confirms that SARS-CoV-2 may induce respiratory disorders in dogs, highlighting the importance of including it in molecular diagnostic assays for CIRDC. Additionally, as the owners of two of these dogs were known to be positive for SARS-CoV-2 before their dogs, human-to-dog transmission was highly suspected. These results underline the need for the continued surveillance of SARS-CoV-2 infections in canine populations. In addition, the versatility of this panel allows the replacement or adjustment of SARS-CoV-2 detection for the identification of other emerging pathogens in the future.
5. Conclusions
To conclude, this study highlights the robustness of our new qPCR/RT-qPCR panel for the detection of SARS-CoV-2 and the eleven most important CIRDC-associated pathogens in clinical specimens. Therefore, this panel is suitable for routine diagnostics and the rapid identification of pathogens associated with CIRDC.
Acknowledgments
The following reagents were obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/USAWA1/2020, NR-52281, Centers for Disease Control and Prevention; SARS-Related Coronavirus 2, Isolate hCoV-19/USA/CA_UCSD_5574/2020, NR-54020, contributed by Aaron Carlin; SARS-Related Coronavirus 2, Isolate hCoV-19/USA/MDHP01542/2021 (Lineage B.1.351), in Homo sapiens Lung Adenocarcinoma (Calu-3) Cells, NR-55282 (contributed by Andrew S. Pekosz); SARS-Related Coronavirus 2, Isolate hCoV-19/USA/MDHP05285/2021 (Lineage B.1.617.2; Delta Variant), NR-55671, contributed by Andrew S. Pekosz; SARS-Related Coronavirus 2, Isolate hCoV-19/USA/MDHP20874/2021 (Lineage B.1.1.529; Omicron Variant), NR-56461, contributed by Andrew S. Pekosz; Canine Distemper Virus, Lederle Avirulent, NR-3845; Influenza A Virus, A/California/04/2009 (H1N1)pdm09, Cell Isolate (Produced in Cells), NR-13658; Bordetella bronchiseptica, Strain E014, NR-44164; Mycoplasma canis, Strain PG 14, NR-3865. The following reagent was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: Canine Coronavirus, UCD1, NR-868. The authors would like to kindly acknowledge Diego Diel, Department of Population Medicine and Diagnostic Sciences, Cornell University College of Veterinary Medicine, Ithaca, NY and Edward Dubovi for providing RNA derived from CIV and CRCoV strains.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v15091881/s1, Figure S1: Assessment of the specificity of all qPCR and RT-qPCR assays using reference strain DNA/RNA; Figure S2: analytical sensitivity determination of singleplex and multiplex qPCR and RT-qPCR assays using plasmid DNA and IVT RNA; Table S1: origin of the samples used in this study; Table S2: detection rate of CIRDC-associated pathogens and SARS-CoV-2 in clinical specimens using the newly developed panel; Table S3: detection rate of single agent infections and co-infections associated with CIRDC in the clinical samples tested using the newly developed panel.
Author Contributions
Conceptualization, U.B.R.B. and L.P.; Specimen submission, W.W.; Data curation, C.J.T.; Formal analysis, C.J.T.; Funding acquisition, U.B.R.B., M.C. and L.P.; Investigation, C.J.T. and K.S.; Methodology, C.J.T., M.C. and U.B.R.B.; Software, C.J.T.; Supervision, M.C. and U.B.R.B.; Visualization, C.J.T.; Roles/Writing—original draft, C.J.T.; Writing—review and editing, C.J.T., M.C., L.P., K.S., W.W. and U.B.R.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
All samples utilized in this study were clinical samples submitted to the Louisiana Animal Disease Diagnostic Laboratory, LA, USA. No specimens from experimental animals were utilized in this study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The study is part of a project entitled “Development of multiplex real-time PCR assays for differentiating SARS-CoV-2 from other respiratory and enteric pathogens and enteric pathogens and an IgM/IgG ELISA for the serologic diagnosis of COVID-19 in dogs and cats”, and was funded by the Vet-LIRN COVID-19 Capacity, grant number 1U18FD007514, and supported by the Food and Drug Administration (FDA) of the U.S. Department of Health and Human Services (HHS) as part of a financial assistance award totaling $100,000 with 100 percent funding by FDA/HHS. The contents are those of the author(s) and do not necessarily represent the official views of, nor are an endorsement by the FDA/HHS or the U.S. Government. Reference to any commercial materials, equipment, or process does not in any way constitute approval, endorsement, or recommendation by the Food and Drug Administration. Come J. Thieulent is partially funded by the NIH-USDA NIFA R01 Research Grant Program Dual Purpose with Dual Benefit: Research in Biomedicine and Agriculture Using Agriculturally Important Domestic Animal Species, grant number 2019-67016-29102 (award number AWD-47990-1) from the USDA National Institute of Food and Agriculture.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Buonavoglia C., Martella V. Canine Respiratory Viruses. Vet. Res. 2007;38:355–373. doi: 10.1051/vetres:2006058. [DOI] [PubMed] [Google Scholar]
- 2.Reagan K.L., Sykes J.E. Canine Infectious Respiratory Disease. Vet. Clin. N. Am. Small Anim. Pract. 2020;50:405–418. doi: 10.1016/j.cvsm.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell J.A., Cardwell J.M., Leach H., Walker C.A., Le Poder S., Decaro N., Rusvai M., Egberink H., Rottier P., Fernandez M., et al. European Surveillance of Emerging Pathogens Associated with Canine Infectious Respiratory Disease. Vet. Microbiol. 2017;212:31–38. doi: 10.1016/j.vetmic.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuu A., Yabuki M., Aoki E., Iwahana M. Molecular Detection of Canine Respiratory Pathogens between 2017 and 2018 in Japan. J. Vet. Med. Sci. 2020;82:690–694. doi: 10.1292/jvms.20-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiebl A., Auer A., Bagrinovschi G., Stejskal M., Hirt R., Rümenapf H.T., Tichy A., Künzel F. Detection of Selected Viral Pathogens in Dogs with Canine Infectious Respiratory Disease in Austria. J. Small Anim. Pract. 2019;60:594–600. doi: 10.1111/jsap.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavan R., Knesl O. Prevalence of Canine Infectious Respiratory Pathogens in Asymptomatic Dogs Presented at US Animal Shelters. J. Small Anim. Pract. 2015;56:572–576. doi: 10.1111/jsap.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sowman H.R., Cave N.J., Dunowska M. A Survey of Canine Respiratory Pathogens in New Zealand Dogs. N. Z. Vet. J. 2018;66:236–242. doi: 10.1080/00480169.2018.1490214. [DOI] [PubMed] [Google Scholar]
- 8.Decaro N., Mari V., Larocca V., Losurdo M., Lanave G., Lucente M.S., Corrente M., Catella C., Bo S., Elia G., et al. Molecular Surveillance of Traditional and Emerging Pathogens Associated with Canine Infectious Respiratory Disease. Vet. Microbiol. 2016;192:21–25. doi: 10.1016/j.vetmic.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erles K., Dubovi E.J., Brooks H.W., Brownlie J. Longitudinal Study of Viruses Associated with Canine Infectious Respiratory Disease. J. Clin. Microbiol. 2004;42:4524–4529. doi: 10.1128/JCM.42.10.4524-4529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weese J.S., Stull J. Respiratory Disease Outbreak in a Veterinary Hospital Associated with Canine Parainfluenza Virus Infection. Can. Vet. J. 2013;54:79–82. [PMC free article] [PubMed] [Google Scholar]
- 11.Priestnall S.L., Mitchell J.A., Walker C.A., Erles K., Brownlie J. New and Emerging Pathogens in Canine Infectious Respiratory Disease. Vet. Pathol. 2014;51:492–504. doi: 10.1177/0300985813511130. [DOI] [PubMed] [Google Scholar]
- 12.Maboni G., Seguel M., Lorton A., Berghaus R., Sanchez S. Canine Infectious Respiratory Disease: New Insights into the Etiology and Epidemiology of Associated Pathogens. PLoS ONE. 2019;14:e0215817. doi: 10.1371/journal.pone.0215817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day M.J., Carey S., Clercx C., Kohn B., MarsilIo F., Thiry E., Freyburger L., Schulz B., Walker D.J. Aetiology of Canine Infectious Respiratory Disease Complex and Prevalence of Its Pathogens in Europe. J. Comp. Pathol. 2020;176:86–108. doi: 10.1016/j.jcpa.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ditchfield J., Macpherson L.W., Zbitnew A. Association of Canine Adenovirus (Toronto A 26/61) with an Outbreak of Laryngotracheitis (“Kennel Cough”) Can. Vet. J. 1962;3:238–246. [PMC free article] [PubMed] [Google Scholar]
- 15.Karpas A., King N.W., Garcia F.G., Calvo F., Cross R.E. Canine Tracheobronchitis: Isolation and Characterization of the Agent with Experimental Reproduction of the Disease. Proc. Soc. Exp. Biol. Med. 1968;127:45–52. doi: 10.3181/00379727-127-32618. [DOI] [PubMed] [Google Scholar]
- 16.Appel M.J., Percy D.H. SV-5-like Parainfluenza Virus in Dogs. J. Am. Vet. Med. Assoc. 1970;156:1778–1781. [PubMed] [Google Scholar]
- 17.Erles K., Toomey C., Brooks H.W., Brownlie J. Detection of a Group 2 Coronavirus in Dogs with Canine Infectious Respiratory Disease. Virology. 2003;310:216–223. doi: 10.1016/S0042-6822(03)00160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erles K., Brownlie J. Canine Respiratory Coronavirus: An Emerging Pathogen in the Canine Infectious Respiratory Disease Complex. Vet. Clin. North Am. Small Anim. Pract. 2008;38:815–825. doi: 10.1016/j.cvsm.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chalker V.J., Brooks H.W., Brownlie J. The Association of Streptococcus Equi Subsp. Zooepidemicus with Canine Infectious Respiratory Disease. Vet. Microbiol. 2003;95:149–156. doi: 10.1016/S0378-1135(03)00155-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priestnall S., Erles K. Streptococcus Zooepidemicus: An Emerging Canine Pathogen. Vet. J. 2011;188:142–148. doi: 10.1016/j.tvjl.2010.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong J., Tsui W.N.T., Leng X., Fu J., Lohman M., Anderson J., Hamill V., Lu N., Porter E.P., Gray M., et al. Development of a Three-Panel Multiplex Real-Time PCR Assay for Simultaneous Detection of Nine Canine Respiratory Pathogens. J. Microbiol. Methods. 2022;199:106528. doi: 10.1016/j.mimet.2022.106528. [DOI] [PubMed] [Google Scholar]
- 22.Hong S., Kim O. Molecular Identification of Mycoplasma Cynos from Laboratory Beagle Dogs with Respiratory Disease. Lab. Anim. Res. 2012;28:61–66. doi: 10.5625/lar.2012.28.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chalker V.J., Owen W.M.A., Paterson C., Barker E., Brooks H., Rycroft A.N., Brownlie J. Mycoplasmas Associated with Canine Infectious Respiratory Disease. Microbiology. 2004;150:3491–3497. doi: 10.1099/mic.0.26848-0. [DOI] [PubMed] [Google Scholar]
- 24.Jambhekar A., Robin E., Le Boedec K. A Systematic Review and Meta-Analyses of the Association between 4 Mycoplasma Species and Lower Respiratory Tract Disease in Dogs. J. Vet. Intern. Med. 2019;33:1880–1891. doi: 10.1111/jvim.15568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wasik B.R., Voorhees I.E.H., Parrish C.R. Canine and Feline Influenza. Cold Spring Harb. Perspect. Med. 2021;11:a038562. doi: 10.1101/cshperspect.a038562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crawford P.C., Dubovi E.J., Castleman W.L., Stephenson I., Gibbs E.P.J., Chen L., Smith C., Hill R.C., Ferro P., Pompey J., et al. Transmission of Equine Influenza Virus to Dogs. Science. 2005;310:482–485. doi: 10.1126/science.1117950. [DOI] [PubMed] [Google Scholar]
- 27.Zhu H., Hughes J., Murcia P.R. Origins and Evolutionary Dynamics of H3N2 Canine Influenza Virus. J. Virol. 2015;89:5406–5418. doi: 10.1128/JVI.03395-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voorhees I.E.H., Glaser A.L., Toohey-Kurth K., Newbury S., Dalziel B.D., Dubovi E.J., Poulsen K., Leutenegger C., Willgert K.J.E., Brisbane-Cohen L., et al. Spread of Canine Influenza A(H3N2) Virus, United States. Emerg. Infect. Dis. 2017;23:1950–1957. doi: 10.3201/eid2312.170246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Damiani A.M., Kalthoff D., Beer M., Müller E., Osterrieder N. Serological Survey in Dogs and Cats for Influenza A(H1N1)Pdm09 in Germany. Zoonoses Public Health. 2012;59:549–552. doi: 10.1111/j.1863-2378.2012.01541.x. [DOI] [PubMed] [Google Scholar]
- 30.Dundon W.G., De Benedictis P., Viale E., Capua I. Serologic Evidence of Pandemic (H1N1) 2009 Infection in Dogs, Italy. Emerg. Infect. Dis. 2010;16:2019–2021. doi: 10.3201/eid1612.100514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renshaw R.W., Zylich N.C., Laverack M.A., Glaser A.L., Dubovi E.J. Pneumovirus in Dogs with Acute Respiratory Disease. Emerg. Infect. Dis. 2010;16:993–995. doi: 10.3201/eid1606.091778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renshaw R., Laverack M., Zylich N., Glaser A., Dubovi E. Genomic Analysis of a Pneumovirus Isolated from Dogs with Acute Respiratory Disease. Vet. Microbiol. 2011;150:88–95. doi: 10.1016/j.vetmic.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Decaro N., Pinto P., Mari V., Elia G., Larocca V., Camero M., Terio V., Losurdo M., Martella V., Buonavoglia C. Full-Genome Analysis of a Canine Pneumovirus Causing Acute Respiratory Disease in Dogs, Italy. PLoS ONE. 2014;9:e85220. doi: 10.1371/journal.pone.0085220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell J.A., Cardwell J.M., Renshaw R.W., Dubovi E.J., Brownlie J. Detection of Canine Pneumovirus in Dogs with Canine Infectious Respiratory Disease. J. Clin. Microbiol. 2013;51:4112–4119. doi: 10.1128/JCM.02312-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piewbang C., Techangamsuwan S. Phylogenetic Evidence of a Novel Lineage of Canine Pneumovirus and a Naturally Recombinant Strain Isolated from Dogs with Respiratory Illness in Thailand. BMC Vet. Res. 2019;15:300. doi: 10.1186/s12917-019-2035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.More G.D., Cave N.J., Biggs P.J., Acke E., Dunowska M. A Molecular Survey of Canine Respiratory Viruses in New Zealand. N. Z. Vet. J. 2021;69:224–233. doi: 10.1080/00480169.2021.1915211. [DOI] [PubMed] [Google Scholar]
- 37.Song X., Li Y., Huang J., Cao H., Zhou Q., Sha X., Zhang B. An Emerging Orthopneumovirus Detected from Dogs with Canine Infectious Respiratory Disease in China. Transbound. Emerg. Dis. 2021;68:3217–3221. doi: 10.1111/tbed.14291. [DOI] [PubMed] [Google Scholar]
- 38.Hao X., Liu R., He Y., Xiao X., Xiao W., Zheng Q., Lin X., Tao P., Zhou P., Li S. Multiplex PCR Methods for Detection of Several Viruses Associated with Canine Respiratory and Enteric Diseases. PLoS ONE. 2019;14:e0213295. doi: 10.1371/journal.pone.0213295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The Proximal Origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Padilla-Blanco M., Vega S., Enjuanes L., Morey A., Lorenzo T., Marín C., Ivorra C., Maiques E., Rubio V., Rubio-Guerri C. Detection of SARS-CoV-2 in a Dog with Hemorrhagic Diarrhea. BMC Vet. Res. 2022;18:370. doi: 10.1186/s12917-022-03453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sit T.H.C., Brackman C.J., Ip S.M., Tam K.W.S., Law P.Y.T., To E.M.W., Yu V.Y.T., Sims L.D., Tsang D.N.C., Chu D.K.W., et al. Infection of Dogs with SARS-CoV-2. Nature. 2020;586:776–778. doi: 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., et al. Susceptibility of Ferrets, Cats, Dogs, and Other Domesticated Animals to SARS-Coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernández-Bastit L., Rodon J., Pradenas E., Marfil S., Trinité B., Parera M., Roca N., Pou A., Cantero G., Lorca-Oró C., et al. First Detection of SARS-CoV-2 Delta (B.1.617.2) Variant of Concern in a Dog with Clinical Signs in Spain. Viruses. 2021;13:2526. doi: 10.3390/v13122526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garigliany M., Van Laere A.-S., Clercx C., Giet D., Escriou N., Huon C., van der Werf S., Eloit M., Desmecht D. SARS-CoV-2 Natural Transmission from Human to Cat, Belgium, March 2020. Emerg. Infect. Dis. 2020;26:3069–3071. doi: 10.3201/eid2612.202223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Decaro N., Balboni A., Bertolotti L., Martino P.A., Mazzei M., Mira F., Pagnini U. SARS-CoV-2 Infection in Dogs and Cats: Facts and Speculations. Front. Vet. Sci. 2021;8:619207. doi: 10.3389/fvets.2021.619207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bosco-Lauth A.M., Hartwig A.E., Porter S.M., Gordy P.W., Nehring M., Byas A.D., VandeWoude S., Ragan I.K., Maison R.M., Bowen R.A. Experimental Infection of Domestic Dogs and Cats with SARS-CoV-2: Pathogenesis, Transmission, and Response to Reexposure in Cats. Proc. Natl. Acad. Sci. USA. 2020;117:26382–26388. doi: 10.1073/pnas.2013102117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyoo K.-S., Yeo Y.-H., Lee S.-G., Yeom M., Lee J.-Y., Kim K.-C., Song D. Susceptibility to SARS-CoV-2 and MERS-CoV in Beagle Dogs. Animals. 2023;13:624. doi: 10.3390/ani13040624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyoo K.-S., Lee H., Lee S.-G., Yeom M., Lee J.-Y., Kim K.-C., Yang J.-S., Song D. Experimental Infection and Transmission of SARS-CoV-2 Delta and Omicron Variants among Beagle Dogs. Emerg. Infect. Dis. 2023;29:782–785. doi: 10.3201/eid2904.221727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan Z., Lu J., Wang N., He W.-T., Zhang L., Zhao W., Su S. Development of a TaqMan-Probe-Based Multiplex Real-Time PCR for the Simultaneous Detection of Emerging and Reemerging Swine Coronaviruses. Virulence. 2020;11:707–718. doi: 10.1080/21505594.2020.1771980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang R., Zhang W., Ye R., Pan Z., Li G., Su S. One-Step Multiplex TaqMan Probe-Based Method for Real-Time PCR Detection of Four Canine Diarrhea Viruses. Mol. Cell. Probes. 2020;53:101618. doi: 10.1016/j.mcp.2020.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carossino M., Barrandeguy M.E., Erol E., Li Y., Balasuriya U.B.R. Development and Evaluation of a One-Step Multiplex Real-Time TaqMan® RT-QPCR Assay for the Detection and Genotyping of Equine G3 and G14 Rotaviruses in Fecal Samples. Virol. J. 2019;16:49. doi: 10.1186/s12985-019-1149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carossino M., Balasuriya U.B.R., Thieulent C.J., Barrandeguy M.E., Vissani M.A., Parreño V. Quadruplex Real-Time TaqMan® RT-QPCR Assay for Differentiation of Equine Group A and B Rotaviruses and Identification of Group A G3 and G14 Genotypes. Viruses. 2023;15:1626. doi: 10.3390/v15081626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., et al. US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020;26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Decaro N., Amorisco F., Desario C., Lorusso E., Camero M., Bellacicco A.L., Sciarretta R., Lucente M.S., Martella V., Buonavoglia C. Development and Validation of a Real-Time PCR Assay for Specific and Sensitive Detection of Canid Herpesvirus 1. J. Virol. Methods. 2010;169:176–180. doi: 10.1016/j.jviromet.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tizolova A., Brun D., Guiso N., Guillot S. Development of Real-Time PCR Assay for Differential Detection of Bordetella Bronchiseptica and Bordetella Parapertussis. Diagn. Microbiol. Infect. Dis. 2014;78:347–351. doi: 10.1016/j.diagmicrobio.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 58.Tallmadge R.L., Anderson R., Mitchell P.K., Forbes Z.C., Werner B., Gioia G., Moroni P., Glaser A., Thachil A.J., Goodman L.B. Characterization of a Novel Mycoplasma Cynos Real-Time PCR Assay. J. VET Diagn. Investig. 2020;32:793–801. doi: 10.1177/1040638719890858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Båverud V., Johansson S.K., Aspan A. Real-Time PCR for Detection and Differentiation of Streptococcus Equi Subsp. Equi and Streptococcus Equi Subsp. Zooepidemicus. Vet. Microbiol. 2007;124:219–229. doi: 10.1016/j.vetmic.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 60.Lu Z., Dubovi E.J., Zylich N.C., Crawford P.C., Sells S., Go Y.Y., Loynachan A.T., Timoney P.J., Chambers T.M., Balasuriya U.B.R. Diagnostic Application of H3N8-Specific Equine Influenza Real-Time Reverse Transcription Polymerase Chain Reaction Assays for the Detection of Canine Influenza Virus in Clinical Specimens. J. VET Diagn. Investig. 2010;22:942–945. doi: 10.1177/104063871002200614. [DOI] [PubMed] [Google Scholar]
- 61.Lu Z., Chambers T.M., Boliar S., Branscum A.J., Sturgill T.L., Timoney P.J., Reedy S.E., Tudor L.R., Dubovi E.J., Vickers M.L., et al. Development and Evaluation of One-Step TaqMan Real-Time Reverse Transcription-PCR Assays Targeting Nucleoprotein, Matrix, and Hemagglutinin Genes of Equine Influenza Virus. J. Clin. Microbiol. 2009;47:3907–3913. doi: 10.1128/JCM.00598-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spackman E., Senne D.A., Myers T.J., Bulaga L.L., Garber L.P., Perdue M.L., Lohman K., Daum L.T., Suarez D.L. Development of a Real-Time Reverse Transcriptase PCR Assay for Type A Influenza Virus and the Avian H5 and H7 Hemagglutinin Subtypes. J. Clin. Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dowgier G., Mari V., Losurdo M., Larocca V., Colaianni M.L., Cirone F., Lucente M.S., Martella V., Buonavoglia C., Decaro N. A Duplex Real-Time PCR Assay Based on TaqMan Technology for Simultaneous Detection and Differentiation of Canine Adenovirus Types 1 and 2. J. Virol. Methods. 2016;234:1–6. doi: 10.1016/j.jviromet.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 64.Slomka M.J., Densham A.L.E., Coward V.J., Essen S., Brookes S.M., Irvine R.M., Spackman E., Ridgeon J., Gardner R., Hanna A., et al. Original Article: Real Time Reverse Transcription (RRT)-Polymerase Chain Reaction (PCR) Methods for Detection of Pandemic (H1N1) 2009 Influenza Virus and European Swine Influenza A Virus Infections in Pigs: SIV and Pandemic (H1N1) 2009 Detection by RRT PCRs. Influenza Other Respir. Viruses. 2010;4:277–293. doi: 10.1111/j.1750-2659.2010.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burd E.M. Validation of Laboratory-Developed Molecular Assays for Infectious Diseases. Clin. Microbiol. Rev. 2010;23:550–576. doi: 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conway J.R., Lex A., Gehlenborg N. UpSetR: An R Package for the Visualization of Intersecting Sets and Their Properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Percopo C.M., Dubovi E.J., Renshaw R.W., Dyer K.D., Domachowske J.B., Rosenberg H.F. Canine Pneumovirus Replicates in Mouse Lung Tissue and Elicits Inflammatory Pathology. Virology. 2011;416:26–31. doi: 10.1016/j.virol.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thieulent C.J., Carossino M., Peak L., Wolfson W., Balasuriya U.B.R. Development and Validation of Multiplex One-Step QPCR/RT-QPCR Assays for Simultaneous Detection of SARS-CoV-2 and Pathogens Associated with Feline Respiratory Disease Complex. PLoS ONE. 2023 doi: 10.1371/journal.pone.0297796. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thieulent C.J., Carossino M., Peak L., Wolfson W., Balasuriya U.B.R. Multiplex One-Step RT-QPCR Assays for Simultaneous Detection of SARS-CoV-2 and Other Enteric Viruses of Dogs and Cats. Viruses. 2023 doi: 10.3390/v15091890. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Viitanen S.J., Lappalainen A., Rajamäki M.M. Co-Infections with Respiratory Viruses in Dogs with Bacterial Pneumonia. J. Vet. Intern. Med. 2015;29:544–551. doi: 10.1111/jvim.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.