Abstract
Expression of heterologous proteins in Dictyostelium discoideum presents unique research opportunities, such as the functional analysis of complex human glycoproteins after random mutagenesis. In one study, human chorionic gonadotropin (hCG) and human follicle stimulating hormone were expressed in Dictyostelium. During the course of these experiments, we also investigated the role of codon usage and of the DNA sequence upstream of the ATG start codon. The Dictyostelium genome has a higher AT content than the human, resulting in a different codon preference. The hCG-β gene contains three clusters with infrequently used codons that were changed to codons that are preferred by Dictyostelium. The results reported here show that optimizing the first 5–17 codons of the hCG gene contributes to 4- to 5-fold increased expression levels, but that further optimization has no significant effect. These observations suggest that optimal codon usage contributes to ribosome stabilization, but does not play an important role during the elongation phase of translation. Furthermore, adapting the 5′-sequence of the hCG gene to the Dictyostelium ‘Kozak’-like sequence increased expression levels ~1.5-fold. Thus, using both codon optimization and ‘Kozak’ adaptation, a 6- to 8-fold increase in expression levels could be obtained for hCG.
INTRODUCTION
The soil amoeba Dictyostelium discoideum is an attractive heterologous expression host for several classes of proteins (1–3). While Dictyostelium can be manipulated with much the same ease as bacteria or yeast, it is capable of expressing functional complex heterologous proteins, which are glycosylated (3), secreted (4) or inserted into the membrane (5,6). Combined with the ability to screen randomly mutagenized proteins in Dictyostelium (7), this system provides a powerful tool for the analysis and design of new complex proteins. We have used this system to study human gonadotropins (8,9), which are secreted proteins with complex folding features, such as the formation of cysteine knots (10). In most heterologous expression studies it is important to reach the highest possible expression levels. In this paper, we have expressed variants of human chorionic gonadotropin (hCG) to study parameters that are important for expression in Dictyostelium.
One parameter that has to be considered for efficient protein expression is the phenomenon called codon bias. Codon bias is the observation that organisms do not use all codons for one amino acid at the same frequency and codon preferences vary between different organisms. Whether or not codon bias serves a regulatory function is unclear. Based on the observation that most low usage codons are found within the first 25 codons in Escherichia coli, it has been proposed that such codons play a role in expression regulation (11–14). However, another study from E.coli suggests that codon bias is unlikely to be an important mode of modulating gene expression (15). For Dictyostelium, a correlation between codon usage and expression levels has been proposed (16), indicating that optimal codon usage may contribute to high expression levels. The Dictyostelium genome has an AT content of >75% and codon usage is highly biased towards AT-rich codons (16; see also Table 1). In contrast, the human genome has an AT content of ~60% and human genes are likely to contain many codons that are sub-optimal for expression in Dictyostelium.
Table 1. Codon usage table for Dictyostelium (November 1999).
| Amino acid | Codon | Usage (%) | Amino acid | Codon | Usage (%) |
|---|---|---|---|---|---|
| Phe (F) | TTT | 58.6 | Ala (A) | GCT | 40.5 |
| TTC | 41.4 | GCC | 18.8 | ||
| Leu (L) | TTA | 55.5 | GCA | 39.4 | |
| TTG | 14.9 | GCG | 1.3 | ||
| CTT | 14.5 | His (H) | CAT | 78.7 | |
| CTC | 9.7 | CAC | 21.3 | ||
| CTA | 4.7 | Gln (Q) | CAA | 97.0 | |
| CTG | 0.7 | CAG | 3.0 | ||
| Ile (I) | ATT | 60.4 | Asn (N) | AAT | 81.7 |
| ATC | 22.7 | AAC | 18.3 | ||
| ATA | 16.9 | Lys (K) | AAA | 79.4 | |
| Met (M) | ATG | 100 | AAG | 20.6 | |
| Val (V) | GTT | 56.0 | Asp (D) | GAT | 88.6 |
| GTC | 14.1 | GAC | 11.4 | ||
| GTA | 25.6 | Glu (E) | GAA | 83.9 | |
| GTG | 4.3 | GAG | 16.1 | ||
| Ser (S) | TCT | 19.9 | Cys (C) | TGT | 85.2 |
| TCC | 6.5 | TGC | 14.8 | ||
| TCA | 47.9 | Trp (W) | TGG | 100 | |
| TCG | 2.6 | Arg (R) | CGT | 33.9 | |
| AGT | 19.9 | CGC | 0.6 | ||
| AGC | 3.2 | CGA | 1.9 | ||
| Pro (P) | CCT | 11.9 | CGG | 0.3 | |
| CCC | 2.5 | AGA | 59.9 | ||
| CCA | 84.3 | AGG | 3.4 | ||
| CCG | 1.3 | Gly (G) | GGT | 80.0 | |
| Thr (T) | ACT | 39.6 | GGC | 4.6 | |
| ACC | 19.7 | GGA | 14.1 | ||
| ACA | 39.2 | GGG | 1.3 | ||
| ACG | 1.5 | Ter (*) | TAA | 89.5 | |
| Tyr (Y) | TAT | 75.1 | TAG | 5.3 | |
| TAC | 24.9 | TGA | 5.2 |
Codon usage frequencies (%) for each amino acid in Dictyostelium, calculated from 483 cDNAs (261 653 codons) on November 12, 1999. For current codon tables see http://www.kazusa.or.jp/codon/ . The bold codons indicate codons that at the start of the project (January 1998) appeared to be used <1% and were optimized in the three regions of hCG (see Fig. 1).
The general consensus from the literature on heterologous gene expression and codon use is that optimizing codon sequences will increase expression levels in organisms as diverse as E.coli (reviewed in 17–19), yeast (20,21) and mammalian cells (22–26). Studies of expression regulation in E.coli indicate that rare codons are most effective in reducing expression levels when they are positioned in the 5′-region of a gene (12,13,27), but it is not clear whether optimizing the 5′-region alone is sufficient to reach maximal expression levels. In this paper we have addressed this question in Dictyostelium by optimizing the codons of various regions in hCG.
A second parameter for efficient expression is the regulation of ribosome assembly at the start codon (28). Initiation sites are most efficient if they conform to a specific sequence motif. In vertebrates this sequence is GCCRCCATGG, which is commonly referred to as the Kozak sequence (28). The ATG start codon in Dictyostelium genes is generally preceded by several A residues (3,29) and it has been suggested (3) that this sequence serves the same function as the mammalian Kozak sequence. Thus, non-A sequences may not provide an optimal environment for translation initiation and in this paper we have adapted the translation initiation sequence of the hCG gene to AAAAA, the Dictyostelium ‘Kozak’-like sequence, to study its influence on expression levels.
MATERIALS AND METHODS
Plasmids and DNA cloning
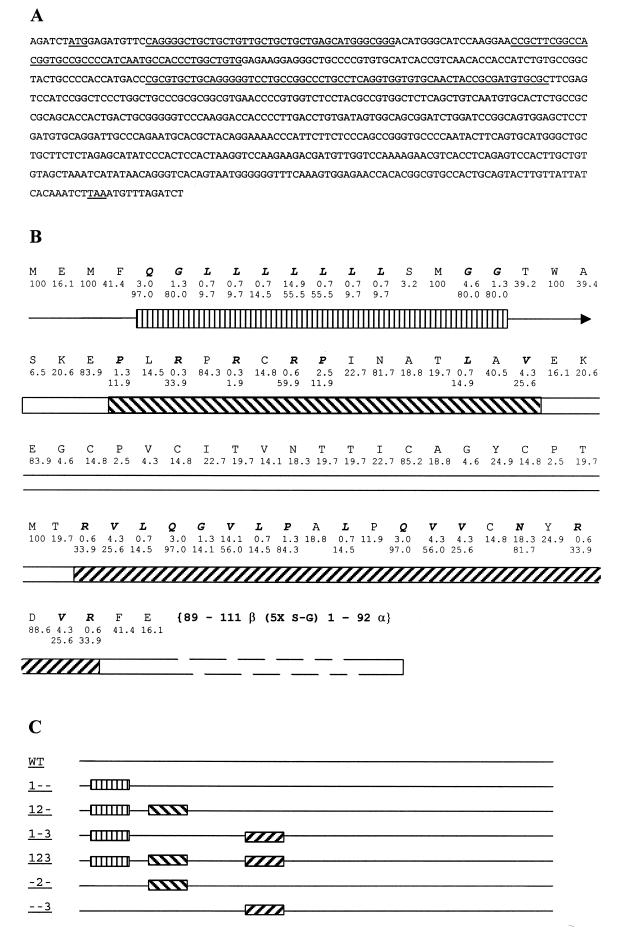
Three sections in the β part of single chain (sc) hCG were identified which contain clusters of non-optimal codons for expression in Dictyostelium. These clusters comprise codons 5–17 (cluster 1), 24–38 (cluster 2) and 63–83 (cluster 3), respectively. We created optimized versions of these three clusters, which were used as modules in designing sc-hCG variants (see Results, Table 1 and Fig. 1 for the optimizing strategy). The template used for all these constructs was sc-hCG variant β-(1–111)-(Ser-Gly)5-α-(1–92) (30), which contains the original human codon usage and which we therefore call wild-type (wt) in this study. In addition to sc-hCG (wt), six sc-hCG constructs were designed: 1--, adapted cluster 1; -2-, adapted cluster 2; 12-, adapted clusters 1 and 2; --3, adapted cluster 3; 1-3, adapted clusters 1 and 3; 123, adapted clusters 1–3 (see Fig. 1 for details). Using PCR with mutant oligonucleotides, combined with site-specific mutagenesis and synthetic DNA approaches (31), each module was synthesized and combined with other modules or wild-type sequences. The resulting variant sc-hCG genes were subcloned either in the unique BglII site of the PCR2.1 TOPO vector (Invitrogen, Leek, The Netherlands) or the pSP72 vector (Invitrogen) in which the PvuII restriction site was removed (pSP72ΔPvuII). The sequences of the final constructs were verified using an automated DNA sequencer (ABI). After digestion with BglII, all variants were cloned into the unique BglII site in the expression vector MB12n (8). The sequence of the wild-type gene is shown in Figure 1A, as well as the amino acid positions of the optimized codons for each region (Fig. 1B).
Figure 1.
(A) DNA sequence of the sc-hCG (30). The start codon ATG, the three optimized regions (see B and C) as well as the stop codon are underlined. (B) Amino acid sequence of the optimized region, in which the original (top line) and optimized percent usage (bottom line) are indicated (see also Table 1). The shaded boxes indicate regions 1 (straight hashes), 2 (slash forward) and 3 (slash backward) described in the text. (C) Schematic view of the different constructs used. Boxes are as in (B).
Three constructs were used for investigation of the effect of the Dictyostelium ‘Kozak’-like sequence on expression levels. The sequence AAAAA was inserted in front of the start codon of the variant sc-hCG genes, changing GGAGATCTATGG to GGAGATCTAAAAAATGG (start codon underlined), using PCR amplification with a mutant oligonucleotide. These constructs were named Awt, A1- and A12, following the nomenclature in the preceding paragraph. During assembly of these constructs, one sequence was fortuitously generated which contained an AAAAA sequence in front of the wild-type gene but which had codon 5 optimized (Q = CAG, usage 3.0%, substituted with CAA, usage 97.0%). This construct was labeled A*-.
Expression of recombinant hormones
DNA was prepared for all constructs and Dictyostelium was transformed by electroporation as described (8). Selection with blasticidin (10 µg/ml) was introduced 5 h after electroporation. Medium was replaced every 3–4 days, while maintaining selective conditions. Cells for each electroporation were aliquoted over four dishes, which were grown to confluence. These cells were continually passaged and an equal amount of fresh medium was added 4 days before harvest. The time from addition of the medium to harvest needs to be consistent throughout the experiment, since hCG is continually being expressed by the cells and the concentration increases with time (8). Medium was harvested at days 14, 25 and 36 after electroporation. Each medium sample was assayed twice for hCG concentration by a sandwich ELISA.
hCG ELISA assay
The immunoassay for hCG was a sandwich-type ELISA, employing an α-subunit-specific mAb as capture antibody and a peroxidase-labeled β-specific mAb as detector antibody. Wells of microtiter plates (Organon Teknika, Boxtel, The Netherlands) were coated overnight with 1 µg/ml anti-α (48A2) mAb. After washing and blocking with PBS containing Tween, the wells were incubated with serial dilutions of the hCG-containing samples, washed again, incubated with the β-specific mAb 119A-HRP (1 µg/ml) in PBS/Tween, incubated for 1 h and washed again. End point determination was carried out using UPO-TMB substrate as originally described by Bos et al. (32). Using a standard curve for a dilution series of hCG protein (Ebo Bos, Organon NV), sc-hCG concentrations in the samples were calculated by the parallel line assay principle. For each construct the electroporated cell population was plated over four dishes, giving four independent pools of transformed cells. Each plate was assayed in duplicate and at three different time points after electroporation (14, 25 and 36 days post-electroporation). The amount of wild-type expression was taken as 100% in each series.
RESULTS
Codon optimization
At the onset of the project we compiled a codon bias table for Dictyostelium from the available data (16; information available on the Internet). We identified 11 codons that were used <1%. These 11 codons were replaced in our optimization strategy. Recently, we updated our codon bias table from all the available information in the database (Table 1) and found that some of these codons are used slightly more than 1%.
hCG consists of two subunits, encoded by α and β genes. In the α gene, 17% of the codons are used <1% in Dictyostelium, while the β gene contains 40% codons that are infrequently used in Dictyostelium. The gene for a single chain variant of hCG (sc-hCG) starts encoding the β chain, followed by a Ser-Gly linker and the α chain (30). Thus sc-hCG is well suited for codon optimization studies in Dictyostelium, since 40 out of the first 100 codons are used <1%.
For this study we started with the original, non-optimized sc-hCG sequence (30; Fig. 1A), which is referred to as wt (wild-type). Based on the sequence information of sc-hCG and Table 1, genes were designed which contain different optimized regions. Region one (codon 5–17), in which 11 of 13 codons are optimized, covers the signal peptide. In region 2 (codon 24–38), seven of 15 codons are optimized. It should be noted that although codon 28 is not the most optimal (1.9%), it is still used six times more frequently than the naturally occurring codon. Region three (codon 63–83), in which 16 of 21 codons are optimized, is more downstream (Fig. 1B). Each of these regions was optimized singly or in combination (Fig. 1C) so that the contribution of each area could be determined.
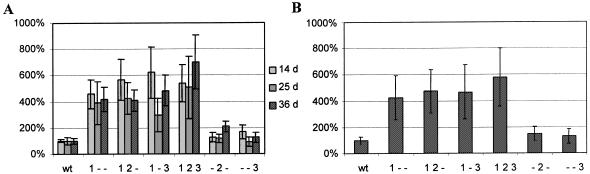
All genes were electroporated into Dictyostelium and, after selection of transformed cells, the production of hCG was measured using a sandwich ELISA assay (see Materials and Methods). Figure 2 presents the data of these experiments for each time point (Fig. 2A), as well as the average of the time points (Fig. 2B).
Figure 2.
Expression levels of sc-hCG genes. (A) Activity at three different harvest times after electroporation: light shading, 14 days; gray, 25 days; dark shading, 36 days. The data show some variation per experiment as discussed in the text. Each experiment by itself supports the general conclusion that optimizing the first region appears to be most influential. Full 100% activity in this experiment is equal to 4.2 ng/ml. (B) Average of the three time points for each construct, n = 24. The bar graphs give the mean and the error bars indicate the standard deviation.
Although some variation in the data is observed, which may reflect small differences in growth conditions or variation in the ELISA, it is clear that only the optimization of cluster 1 contributes significantly to a higher expression level. Compared to wild-type expression, an increase of 4- to 5-fold is seen by optimizing the first cluster. Further optimizing of clusters 2 and 3 does not appear to increase expression levels significantly. More revealing is the fact that optimization of cluster 2 or 3 by themselves did not raise expression levels, even though cluster 3 contains 16 of 21 unfavorable codons. These observations hint at the importance of optimal codon usage at the 5′-end of the gene for high expression levels.
Sequences for efficient translation initiation
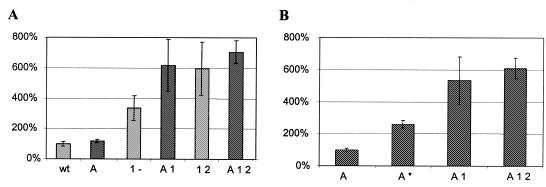
Since the sequence 5′ of the start codon of sc-hCG was based on the BglII restriction recognition site, GATCT, we investigated whether further optimizing could be achieved by inserting an optimal sequence, AAAAA. Several constructs used in the codon optimization experiments were altered by inserting five A residues preceding the start ATG. The expression of sc-hCG in Dictyostelium from these constructs was compared to the level of expression directed by the corresponding construct without the ‘Kozak’ adaptation. After electroporation, Dictyostelium cells were grown as above and sc-hCG expression levels were measured. The results (Fig. 3A) show that replacement of the 5′-GATCT sequence with 5′-AAAAA did not dramatically change the expression levels. For the wt gene no significant increase was observed, while for the codon-optimized genes only a 1.7-fold (A1- versus 1-) or a 1.3-fold (A12 versus 12) increase was observed.
Figure 3.
(A) Expression levels of sc-hCG genes after ‘Kozak’ adaptation. Light shaded bars are control non-‘Kozak’ adapted constructs, dark shaded bars are adapted counterparts; n = 5. Original wt is taken as 100% activity, equal to 6.3 ng/ml. (B) Effect of single codon optimization (A*, see text for details) in a ‘Kozak’ adapted background; n = 5. ‘Kozak’ adapted wt is taken as 100% activity, equal to 7.1 ng/ml. Note that in this experiment (n = 5) an increase can be seen between optimizing region 1 alone (3.4-fold increase) and optimizing both regions 1 and 2 (6.0-fold increase), whereas the data in Figure 2 show little difference between 1 and 12. However, the expression levels (based on five data points, n = 5) still fall within the range indicated by the standard deviation of the experiment in Figure 2 (n = 24). Thus, these data are still consistent with the notion that optimal 5′ codon usage is most important for high expression levels.
During the experiments investigating the role of the AAAAA sequence, we also fortuitously generated a plasmid in which the first non-optimal codon, CAG (codon 5, usage 3%), was replaced by the optimal codon, CAA (usage 97%). By comparing the expression level of this construct with the expression level of other AAAAA optimized constructs, we found that merely optimizing codon 5 increased expression levels 2.6-fold (Fig. 3B). This observation again suggests that the 5′ region of the gene is most sensitive to codon usage in relation to expression levels.
A comparison of the results with the ‘non-Kozak’ constructs versus the ‘Kozak’ adapted constructs illustrate that in the case of sc-hCG some increase may be gained from adapting the 5′ region of the gene, but that this increase is modest in comparison to the effects of codon optimization. The increase in expression gained by adapting a 5′ sequence to the Dictyostelium ‘Kozak’-like sequence will ultimately depend on the nucleotide sequence that is replaced. The increase may be larger for GC-rich sequences than for AT-rich sequences. It is also possible that in the case of sc-hCG an increased translation efficiency is obscured due to the fact that the third codon in sc-hCG is also an ATG, which may be used as an alternative start site through leaky scanning of the ribosome (28).
DISCUSSION
In this paper we have investigated to what extent optimizing codon usage and Kozak sequence in hCG increases expression levels in Dictyostelium. The data from Figure 2 clearly show that optimizing the first 17 codons causes a 4-fold increase in expression levels and that optimizing more downstream codons has a very limited additional effect. Even more telling is the fact that if the latter are optimized without optimizing the first 17 codons, no increase in expression level is seen. This suggests that once translation is firmly established, being determined by the first 10 or 20 codons, no significant increase in expression levels can be achieved by improving downstream codons.
The data from Figure 3B show that when only the first non-optimal codon in sc-hCG (codon 5) is optimized, a 2.6-fold increase in expression level is seen. It is not possible to investigate the role of the first four codons in hCG, since codons 1 and 3 encode methionine (100%), while the least used codon for glutamic acid (codon 2) is GAG (16.1%) and for phenylalanine (codon 4) it is TTC (41.4%). Combined with the data from Figure 2 we suggest that, in general, optimizing the first 10 or 15 codons may be sufficient to yield as high an expression level as is attainable through codon optimization.
It has been found in E.coli that consecutive low usage codons inserted at the 5′-end of a gene can slow or block translation (11–14). Whether this decreased translation efficiency is due to the stalling of ribosomes (13,33) or due to an increased drop-off rate of the ribosomes (34) is not yet clear, but the effect is 5′ specific (13,34). It has been suggested that this positional effect is due to an effect on the stability of translation complexes near the beginning of a message (12). Low usage codons may cause slow translation and increase the drop-off rate during this stage. Indeed, it has been found that translation initiation factors play a role in ribosome instability during the early stages of translation and that instability correlates with peptide length (35). Thus, although speculative, it may be argued that ribosomes are not stable until a peptide is synthesized of 5–10 residues and that low usage codons in this synthesis may decrease the rate of stable ribosome assembly. Further analysis of the architecture of the ribosome itself may provide more clues as to what processes are involved (36).
Dictyostelium genes do not share the same translation initiation recognition sequence with vertebrates, the so-called ‘Kozak’ sequence (3,28). We changed the sequence in sc-hCG to the Dictyostelium ‘Kozak’-like sequence and observed a modest increase in expression levels. As already discussed, the increase in expression may be dependent on the sequence that was replaced and we, like others (3), adapt this sequence whenever possible.
Optimizing heterologous gene expression is in many cases an important step that precedes the production and study of the protein of interest. The results we present here indicate that for Dictyostelium optimizing the first 10–15 codons is most effective in increasing expression levels. Adding a ‘Kozak’-like sequence will add a modest further increase, to obtain a combined 6- to 8-fold increase in expression. Further codon optimization in more 3′-parts of the gene has no significant effect and appears not to be required for heterologously expressed genes in Dictyostelium. Our work, together with previous studies in E.coli (13,34,35), suggest that this may be a universal concept which may apply to other heterologous expression systems as well.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Nathascha Hanzen and Petra Werler for experimental assistance, Dr Ebo Bos for providing expertise and reagents for the hCG ELISA, Dr Barend Kraal for helpful discussions and Drs Karen Prowse and Jaco Knol for critically reading the manuscript.
REFERENCES
- 1.Dittrich W., Williams,K.L. and Slade,M.B. (1994) Biotechnology, 12, 614–618. [DOI] [PubMed] [Google Scholar]
- 2.Jung E. and Williams,K.L. (1997) Biotechnol. Appl. Biochem., 25, 3–8. [DOI] [PubMed] [Google Scholar]
- 3.Slade M.B., Emslie,K.R. and Williams,K.L. (1997) Biotechnol. Genet. Eng. Rev., 14, 1–35. [DOI] [PubMed] [Google Scholar]
- 4.Dingermann T., Troidl,E.M., Broker,M. and Nerke,K. (1991) Appl. Microbiol. Biotechnol., 35, 496–503. [DOI] [PubMed] [Google Scholar]
- 5.Cohen N.R., Knecht,D.A. and Lodish,H.F. (1996) Biochem. J., 315, 971–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voith G. and Dingermann,T. (1995) Pharmazie, 50, 758–762. [PubMed] [Google Scholar]
- 7.Kim J.Y., Caterina,M.J., Milne,J.L., Lin,K.C., Borleis,J.A. and Devreotes,P.N. (1997) J. Biol. Chem., 272, 2060–2068. [DOI] [PubMed] [Google Scholar]
- 8.Heikoop J.C., Grootenhuis,P.D., Blaauw,M., Veldema,J.S., Van Haastert,P.J. and Linskens,M.H. (1998) Eur. J. Biochem., 256, 359–363. [DOI] [PubMed] [Google Scholar]
- 9.Linskens M.H., Grootenhuis,P.D., Blaauw,M., Huisman-de Winkel,B., Van Ravestein,A., Van Haastert,P.J. and Heikoop,J.C. (1999) FASEB J., 13, 639–645. [DOI] [PubMed] [Google Scholar]
- 10.Lapthorn A.J., Harris,D.C., Littlejohn,A., Lustbader,J.W., Canfield,R.E., Machin,K.J., Morgan,F.J. and Isaacs,N.W. (1994) Nature, 369, 455–461. [DOI] [PubMed] [Google Scholar]
- 11.Rosenberg A.H., Goldman,E., Dunn,J.J., Studier,F.W. and Zubay,G. (1993) J. Bacteriol., 175, 716–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldman E., Rosenberg,A.H., Zubay,G. and Studier,F.W. (1995) J. Mol. Biol., 245, 467–473. [DOI] [PubMed] [Google Scholar]
- 13.Chen G.T. and Inouye,M. (1994) Genes Dev., 8, 2641–2652. [DOI] [PubMed] [Google Scholar]
- 14.Chen G.F. and Inouye,M. (1990) Nucleic Acids Res., 18, 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurland C.G. (1991) FEBS Lett., 285, 165–169. [DOI] [PubMed] [Google Scholar]
- 16.Sharp P.M. and Devine,K.M. (1989) Nucleic Acids Res., 17, 5029–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannig G. and Makrides,S.C. (1998) Trends Biotechnol., 16, 54–60. [DOI] [PubMed] [Google Scholar]
- 18.Zhang M.Y., Schillberg,S., Prins,M. and Fischer,R. (1999) Anal. Biochem., 271, 202–204. [DOI] [PubMed] [Google Scholar]
- 19.Hale R.S. and Thompson,G. (1998) Protein Expr. Purif., 12, 185–188. [DOI] [PubMed] [Google Scholar]
- 20.Cormack B.P., Bertram,G., Egerton,M., Gow,N.A., Falkow,S. and Brown,A.J. (1997) Microbiology, 143, 303–311. [DOI] [PubMed] [Google Scholar]
- 21.Brocca S., Schmidt-Dannert,C., Lotti,M., Alberghina,L. and Schmid,R.D. (1998) Protein Sci., 7, 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang G., Gurtu,V. and Kain,S.R. (1996) Biochem. Biophys. Res. Commun., 227, 707–711. [DOI] [PubMed] [Google Scholar]
- 23.Kim C.H., Oh,Y. and Lee,T.H. (1997) Gene, 199, 293–301. [DOI] [PubMed] [Google Scholar]
- 24.Mirzabekov T., Bannert,N., Farzan,M., Hofmann,W., Kolchinsky,P., Wu,L., Wyatt,R. and Sodroski,J. (1999) J. Biol. Chem., 274, 28745–28750. [DOI] [PubMed] [Google Scholar]
- 25.Uchijima M., Yoshida,A., Nagata,T. and Koide,Y. (1998) J. Immunol., 161, 5594–5599. [PubMed] [Google Scholar]
- 26.Nagata T., Uchijima,M., Yoshida,A., Kawashima,M. and Koide,Y. (1999) Biochem. Biophys. Res. Commun., 261, 445–451. [DOI] [PubMed] [Google Scholar]
- 27.Johansson A.S., Bolton-Grob,R. and Mannervik,B. (1999) Protein. Expr. Purif., 17, 105–112. [DOI] [PubMed] [Google Scholar]
- 28.Kozak M. (1999) Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 29.Yamauchi K. (1991) Nucleic Acids Res., 19, 2715–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heikoop J.C., van Beuningen-de Vaan,M.M., van den Boogaart,P. and Grootenhuis,P.D. (1997) Eur. J. Biochem., 245, 656–662. [DOI] [PubMed] [Google Scholar]
- 31.Higuchi R. (1989) In Erlich,H.A. (ed.), PCR Technology: Principles and Applications for DNA Amplification. Stockton Press, New York, NY, pp. 61–70.
- 32.Bos E.S., van der Doelen,A.A., van Rooy,N. and Schuurs,A.H. (1981) J. Immunoassay, 2, 187–204. [DOI] [PubMed] [Google Scholar]
- 33.Zahn K. and Landy,A. (1996) Mol. Microbiol., 21, 69–76. [DOI] [PubMed] [Google Scholar]
- 34.Gao W., Tyagi,S., Kramer,F.R. and Goldman,E. (1997) Mol. Microbiol., 25, 707–716. [DOI] [PubMed] [Google Scholar]
- 35.Karimi R., Pavlov,M.Y., Heurgue-Hamard,V., Buckingham,R.H. and Ehrenberg,M. (1998) J. Mol. Biol., 281, 241–252. [DOI] [PubMed] [Google Scholar]
- 36.Garrett R. (1999) Nature, 400, 811–812. [DOI] [PubMed] [Google Scholar]