Abstract
Background
The mechanism underlying cognitive impairment after hearing loss (HL) remains unclear. N6-methyladenosine (m6A) is involved in many neurodegenerative diseases; however, its role in cognitive impairment after HL has not yet been investigated. Therefore, we aimed to analyze the m6A modification profile of the mouse hippocampus after HL exposure. A mouse model of neomycin-induced HL was established. An auditory brainstem-response test was utilized for detecting hearing threshold. The passive avoidance test was served as the mean for evaluating cognitive function. The m6A-regulated enzyme expression levels were analyzed by using reverse transcription quantitative real-time polymerase chain reaction and western blot analyses. RNA sequencing (RNA-Seq) and methylated RNA immunoprecipitation sequencing (MeRIP-Seq) were performed with the aim of investigating gene expression differences and m6A modification in the mouse hippocampus.
Results
Neomycin administration induced severe HL in mice. At four months of age, the mice in the HL group showed poorer cognitive performance than the mice in the control group. METTL14, WTAP, and YTHDF2 mRNA levels were downregulated in the hippocampi of HL mice, whereas ALKBH5 and FTO mRNA levels were significantly upregulated. At the protein level, METTL3 and FTO were significantly upregulated. Methylated RNA immunoprecipitation sequencing analysis revealed 387 and 361 m6A hypermethylation and hypomethylation peaks, respectively. Moreover, combined analysis of mRNA expression levels and m6A peaks revealed eight mRNAs with significantly changed expression levels and methylation.
Conclusions
Our findings revealed the m6A transcriptome-wide profile in the hippocampus of HL mice, which may provide a basis for understanding the association between HL and cognitive impairment from the perspective of epigenetic modifications.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-023-09697-4.
Keywords: Hearing loss, Cognition impairment, Hippocampus, m6A methylation, MeRIP-Seq, RNA-Seq
Background
As the most common sensory deficit, hearing loss (HL) affects more than one billion people worldwide and has become the third leading cause of disability [1]. In children, HL can hinder daily communications, lower the quality of life, potentially delay spoken language development, and lead to cognitive impairment later in life [2]. Children with mild HL show impaired learning and memory function [3], supporting the link between HL and cognitive impairment. In rodent models of HL, pathological changes indicative of cognitive impairment (e.g., inhibited neurogenesis, increased tau protein phosphorylation, and elevated neuroinflammation) have been found in the hippocampus [4, 5]. Despite the abundance of evidence supporting a link between HL and cognitive dysfunction, however, the specific molecular mechanism still remains unsolved.
Most recently, RNA methylation-mediated RNA posttranscriptional modification has attracted much attention as an important epigenetic modification [6]. N6-methyladenosine (m6A) methylation, the most frequent modification of eukaryotic RNA, has considered to be an important epigenetic modification and plays essential roles in regulating the location, stability, splicing, translation, transport of mRNA [7, 8]. In the brains of mammals, m6A methylation can be regulated reversibly and dynamically by demethylases (erasers) and methyltransferases (writers), where m6A methylation is recognized by other proteins (readers) [9]. Erasers (α-ketoglutarate-dependent dioxygenase alkB homolog 5 [ALKBH5] and fat-mass and obesity-associated protein [FTO]) catalyze the removal of m6A, writers (methyltransferase-like 14 [METTL14], methyltransferase-like 3 [METTL3], and Wilms tumor 1-associating protein [WTAP]) catalyze the addition of m6A, and readers (YTH domain family, like YTHDF1, YTHDF2, and YTHDF3) respond or recognize m6A specifically [10–13].
An increasing number of studies highlighted the critical significance of m6A modification in the development al nervous system, and its dysregulation has been shown to be related to neurodegenerative and neurodevelopmental diseases [14, 15]. m6A participates in neurobiological processes, such as neurogenesis, neurodevelopment, synaptic plasticity, learning, and memory [16–19]. Furthermore, various m6A players have been found to be mutated or dysregulated both in neurological degeneration disease, such as AD, HD, and depression [20–22]. In the mammalian central nervous system, stimulus-dependent m6A regulation occurs in response to learning process, injury, and sensory experience [14, 23], indicating that m6A modifications may be critical for the pathogenesis of cognitive impairment after sensory deprivation, such as HL. However, the involvement of m6A methylation in cognitive impairment after HL has not yet been investigated.
In this experiment, we sought to build up a mouse model of neomycin-induced HL and perform RNA sequencing (RNA-seq) and methylated RNA immunoprecipitation sequencing (MeRIP-seq) to observe mRNA expression patterns and transcriptome-wide m6A changes in hippocampi of HL and control (Con) mice. To predict the functions of differentially expressed RNAs, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted. This study might provide insights on m6A-modified transcripts that are key for the development of cognitive impairment after HL.
Results
Establishing a mouse model of neomycin-induced HL
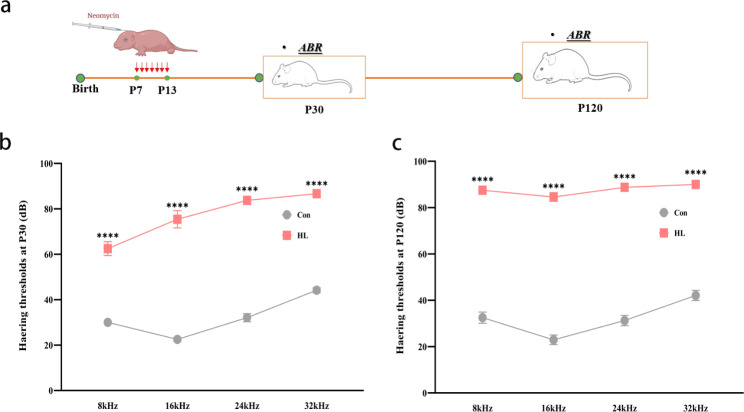
To examine whether neomycin induces HL in mice, we performed auditory brainstem response (ABR) testing at postnatal day (P) 30 and P120 (Fig. 1a). At P30, HL mice had higher ABR thresholds than the Con group at different frequencies: 8 kHz (62.50 ± 3.05 vs. 30.00 ± 0.87 dB, P < 0.0001), 16 kHz (75.42 ± 3.82 vs. 22.50 ± 2.61 dB, P < 0.0001), 24 kHz (83.75 ± 1.52 vs. 32.08 ± 1.79 dB, P < 0.0001), and 32 kHz (86.67 ± 3.89 vs. 44.17 ± 1.04 dB, P < 0.0001), as shown in Fig. 1b. At P120, the ABR thresholds were also elevated at different frequencies in the experimental group: 8 kHz (87.5 ± 1.44 vs. 32.50 ± 2.42 dB, P < 0.0001), 16 kHz (84.58 ± 1.89 vs. 22.92 ± 2.08 dB, P < 0.0001), 24 kHz (88.75 ± 0.65 vs. 31.25 ± 2.23 dB, P < 0.0001), and 32 kHz (90 ± 0 vs. 42.08 ± 2.17 dB, P < 0.0001) (Fig. 1c). These findings indicate that neomycin successfully induced severe and irreversible HL in mice.
Fig. 1.
Successful establishment of a mouse model of neomycin-induced hearing loss (HL). (a) Time points for administering neomycin and performing the ABR test. (b) ABR thresholds of P30 mice. At P30, mice in the HL group had lower ABR thresholds than those in the Con group at 8, 16, 24, and 32 kHz. (c) ABR thresholds of P120 mice. At P120, mice in the HL group had lower ABR thresholds than those in the Con group at 8, 16, 24, and 32 kHz. The data shown were analyzed using an unpaired t-test and represent the mean ± SEM. ****P < 0.0001. n = 9 mice per group. Con, control; HL, hearing loss. ABR, auditory brainstem response; kHz, 1000 Hz; dB, decibel
Occurrence of cognitive impairment after HL
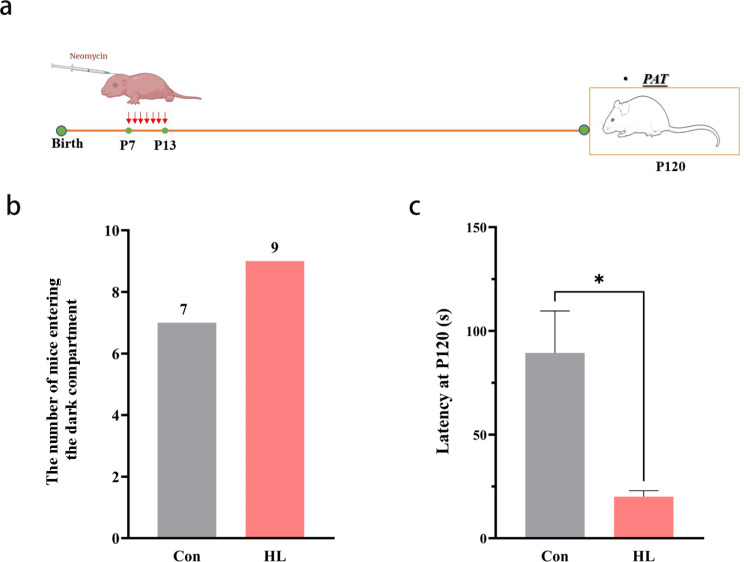
We conducted a passive avoidance test (PAT) in order to assess the cognitive function of mice at P120 (Fig. 2a). During the training period, the latencies of both groups were comparable. During the test period, seven mice (77.8%) in the Con group got into the dark compartment within the assigned time (300 s), while nine mice (100%) in the HL group (Fig. 2b). Thus, the HL mice had shorter latency (P < 0.05; Fig. 2c). These results mean that cognitive impairment occurred after HL.
Fig. 2.
Occurrence of cognitive impairment after HL. (a) Time points for administering neomycin and conducting the PAT. (b) At P120, nine and seven mice in the HL and Con groups entered the dark room, respectively. (c) Latency of P120 mice. The latency of the HL group was significantly lower than that of the Con group. The data shown were analyzed using an unpaired t-test and represent the mean ± SEM. *P < 0.05. n = 9 mice per group. Con, control; HL, hearing loss
Overview of m6A RNA methylation in mouse hippocampi after HL
MeRIP-seq was performed to investigate the transcriptome-wide m6A-seq analysis. An average of 8.47 Gb sequencing data of the input samples along with 7.86 Gb sequencing data in the MeRIP-seq were obtained (Additional file 1). After discarding low-quality reads and bases, adaptor sequences, and poly-N sequences, the input and the MeRIP-seq samples contained (on average) 6.44 and 6.97 Gb of sequencing data, respectively (Additional file 1). The majority (95.03%) of the left high-quality reads in each sample could be mapped to the reference mouse genome (GRCm39). Clean reads that had unique alignments to the mouse genome (89.41%) were reserved for further analyses, and clean reads with multiple alignments (5.89%) were eliminated (Additional file 2).
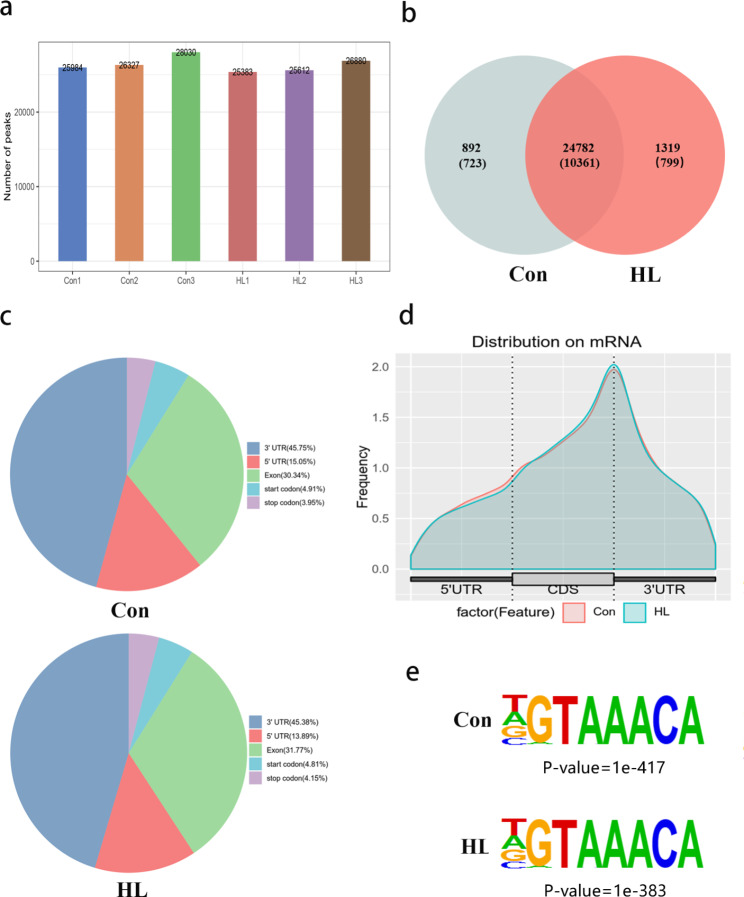
MeRIP-seq analysis revealed that the Con and HL group had an average of 26,780 and 25,958 peaks, respectively (Fig. 3a). We identified 25,674 MeRIP regions enriched in m6A peaks, and these peaks were identified within 11,084 mRNA transcripts of coding genes. In addition, 26,101 m6A peaks within 11,160 mRNAs were detected in HL group. Among the genes with m6A peaks, 10,361 mRNAs overlapped between both groups (Fig. 3b), showing that m6A methylation was abundant in the hippocampus. The distribution patterns regarding m6A methylation peaks within mRNAs were analyzed for the entire transcriptome. In both the Con and HL groups, we observed that the m6A peaks were predominantly located within the coding sequences (CDS), 5ʹ-untranslated region (UTR), and 3ʹ-UTR of the mRNA molecules (Fig. 3c, d). HOMER software was applied to observe if the m6A peaks owned the classic “RRACH” motif (R: G/A, H: U/ A/ C). In this research, the representative “AAACA” motif was enriched among the m6A peaks detected in both groups (Fig. 3e).
Fig. 3.
Overview of m6A RNA methylation in mouse hippocampi after HL. (a) The number of peaks in each sample in both groups. An average of 26,780 and 25,958 peaks were detected in the Con and HL group, respectively. (b) Overlapping mRNA m6A peaks between the Con and HL groups are displayed with a Venn diagram. (c) The distribution of m6A peaks in the transcripts is displayed with a density curve for the 5′-UTR, CDS, and 3′-UTR. (d) The ratios of m6A-peak distributions in the Con and HL samples are displayed in pie charts. (e) Detection of a common motif enriched across the m6A peaks in both groups
Altered m6A modifications in mouse hippocampi after HL
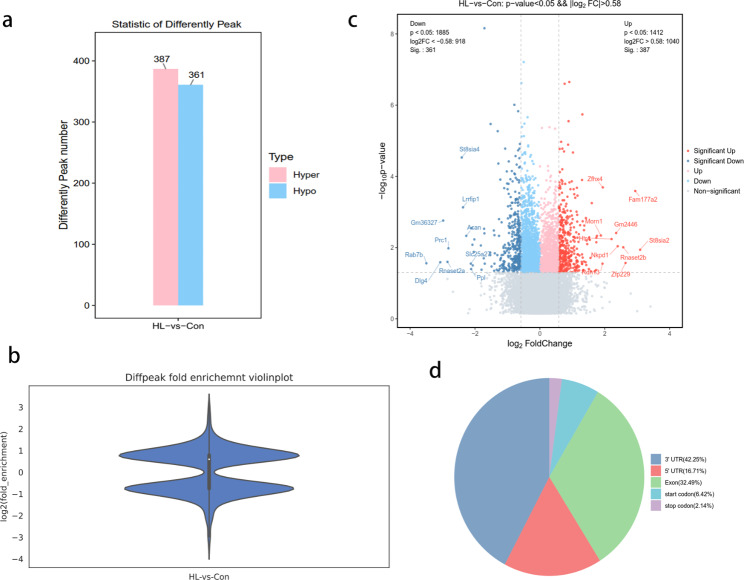
We identified 748 peaks that showed differential m6A methylation between the HL and Con groups (|log2 fold change (FC)| > 0.58 and P < 0.05; Additional file 3). Among them, 387 m6A peaks were significantly hypermethylated, while 361 were significantly hypomethylated in the HL group (Fig. 4a). Table 1 presents the top 20 genes with hypermethylated and hypomethylated m6A methylation region. Furthermore, significantly hypomethylated and hypermethylated mRNAs are depicted by the violin plot and volcano plot (Fig. 4b, c). The distribution patterns of m6A peaks were classified into five distinct types of transcript parts: the 3ʹ-UTR, CDS, stop codon segment, start codon segment, and 5ʹ-UTR. Our findings indicated that those m6A sites were typically located in the 3ʹ-UTR and CDS regions (Fig. 4d).
Fig. 4.
The distribution of peaks reflecting differential m6A methylation in mouse hippocampi after HL. (a) The number of peaks related to differential m6A methylation after HL. We detected 387 and 361 peaks reflecting m6A hypermethylation and hypomethylation, respectively (P < 0.05, |log2 FC| > 0.58) in mouse hippocampi after HL. (b) Violin plot displaying the fold-enrichments of peaks reflecting differential m6A methylation. (c) Volcano plot displaying peaks reflecting differential m6A methylation (P < 0.05, |log2 FC| > 0.58) in mouse hippocampi after HL. Red plots, peaks reflecting m6A hypermethylation; blue plots, peaks reflecting m6A hypomethylation. (d) Pie charts displaying the ratios of peaks reflecting differential m6A methylation and m6A peak distributions are displayed by pie charts. Diffpeak, differentially methylated m6A peak; FC, fold-change
Table 1.
The representative changed m6A methylation peak. (Top 20)
| Gene Id | Chromsome | P value | Log2fold change | Start | End | Width | Regulation |
|---|---|---|---|---|---|---|---|
| St8sia2 | 7 | 0.011481536 | 3.09 | 73,592,844 | 73,592,994 | 150 | UP |
| Fam177a2 | 12 | 0.00025704 | 2.94 | 55,263,655 | 55,263,854 | 199 | UP |
| Zfp229 | 17 | 0.026915348 | 2.64 | 21,966,955 | 21,967,105 | 150 | UP |
| Rnaset2b | 17 | 0.009772372 | 2.57 | 7,259,127 | 7,265,303 | 194 | UP |
| Nkpd1 | 7 | 0.009120108 | 2.4 | 19,258,485 | 19,258,635 | 150 | UP |
| Gm2446 | 12 | 0.003890451 | 2.35 | 55,263,649 | 55,264,059 | 196 | UP |
| Htr4 | 18 | 0.005754399 | 2.21 | 62,635,349 | 62,635,499 | 150 | UP |
| Zfhx4 | 3 | 0.000204174 | 1.94 | 5,467,216 | 5,467,416 | 200 | UP |
| Rdh13 | 7 | 0.028183829 | 1.93 | 4,449,241 | 4,449,392 | 151 | UP |
| Morn1 | 4 | 0.004570882 | 1.88 | 155,184,691 | 155,184,842 | 151 | UP |
| Rab7b | 1 | 0.027542287 | -3.5 | 131,640,859 | 131,641,010 | 151 | Down |
| Dlg4 | 11 | 0.025703958 | -3.07 | 69,919,726 | 69,919,927 | 201 | Down |
| Gm36327 | 6 | 0.001737801 | -2.98 | 47,655,432 | 47,655,582 | 150 | Down |
| Rnaset2a | 17 | 0.025118864 | -2.85 | 8,347,603 | 8,353,740 | 241 | Down |
| Prc1 | 7 | 0.010471285 | -2.82 | 79,959,186 | 79,960,588 | 150 | Down |
| St8sia4 | 1 | 2.95E-05 | -2.41 | 95,555,283 | 95,555,433 | 150 | Down |
| Lrrfip1 | 1 | 0.00074131 | -2.37 | 91,043,657 | 91,043,858 | 201 | Down |
| Acan | 7 | 0.004677351 | -2.27 | 78,749,026 | 78,749,227 | 201 | Down |
| Slc25a27 | 17 | 0.028183829 | -2.13 | 43,956,575 | 43,956,725 | 150 | Down |
| Ppl | 16 | 0.039810717 | -2.12 | 4,907,292 | 4,907,442 | 150 | Down |
GO and KEGG pathway analyses of genes with different methylation
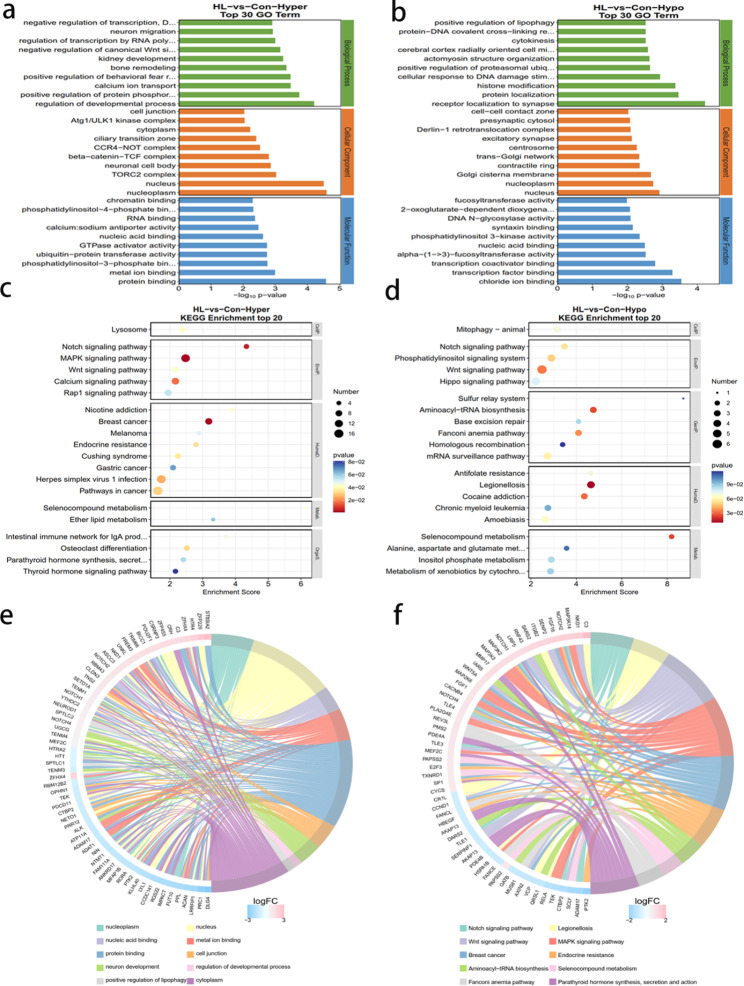
GO and KEGG pathway analyses of genes with different m6A methylation peaks were performed for determining the potential role of m6A modifications in the hippocampus after HL. The primary GO terms and KEGG pathways associated with hypermethylation are demonstrated in Fig. 5a and c, respectively. The primary GO terms and KEGG pathways associated with hypomethylation were demonstrated in Fig. 5b and d, respectively. GO analysis revealed peaks related to hypermethylation in HL mice that were significantly associated with regulating developmental processes, chromatin binding, and protein binding (Fig. 5a). Conversely, hypomethylation-associated peaks correlated significantly with receptor localization at synapses, fucosyl transferase activity, and chloride ion binding (Fig. 5b). KEGG analysis indicated that the hypermethylation-associated peaks in HL mice correlated significantly with the Notch-signaling and MAPK-signaling pathways (Fig. 5c). The hypomethylation-associated peaks correlated significantly with Legionellosis and the Wnt-signaling pathway (Fig. 5d). In summary, the GO analyses chord diagram demonstrated that the differentially methylated mRNAs between the HL and Con groups correlated with protein binding and regulating developmental progress (Fig. 5e). Additionally, KEGG analysis chord diagram demonstrated that mRNAs that exhibited altered m6A methylation were enriched in MAPK-signaling pathway (Fig. 5f).
Fig. 5.
Biological information related to mRNA m6A methylation identified via GO and KEGG analyses. (a) GO analyses of genes with peaks related to m6A hypermethylation. (b) GO analyses of genes with peaks indicating m6A hypomethylation. (c) KEGG analyses of peaks related to m6A hypermethylation. (d) KEGG analyses of peaks reflecting m6A hypomethylation. (e) GO analyses chord diagram of genes with differentially methylated m6A peaks. (f) KEGG analyses chord diagram of genes with differentially methylated m6A peaks. BP, biological process; MF, molecular function; CC, cellular component; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes
Altered mRNAs in mouse hippocampi after HL
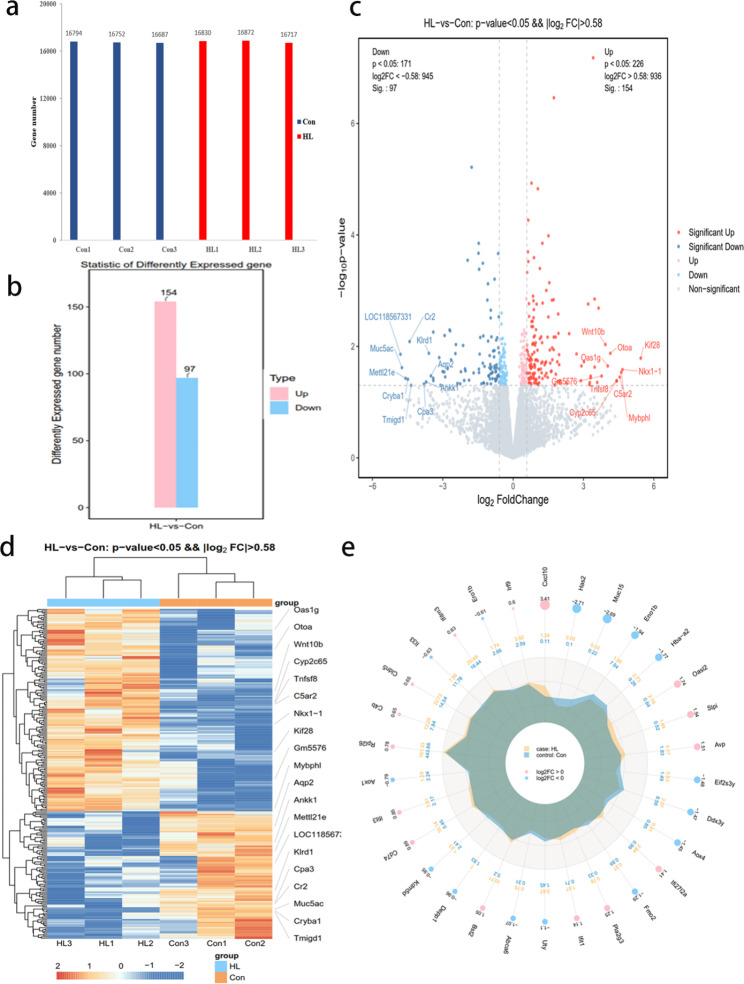
The RNA-seq data from the input experiments were used to acquire transcriptome profiles (Additional file 4). On average, 16,806 and 16,744 mRNA transcripts were detected in the HL and Con mice, respectively (Fig. 6a). We identified 251 differentially expressed mRNAs in the HL group with 154 and 97 genes being upregulated and downregulated, respectively (P < 0.05, |log2 FC| > 0.58; Fig. 6b). Table 2 lists the top 20 most differentially expressed mRNAs. The volcano chart displays all significantly downregulated and upregulated mRNAs in the HL mice (Fig. 6c). We identified the top 10 most significantly upregulated (Kif28, Nkx1-1, Mybphl, C5ar2, Cyp2c65, Tnfsf8, Otoa, Oas1g, Wnt10b, and Gm5576) and downregulated genes (LOC118567331, Muc5ac, Mettl21e, Cryba1, Cr2, Tmigd1, Cpa3, Ankk1, Klrd1, and Aqp2). The heatmap shows the relative mRNA expression levels were comparable among samples in either group (Fig. 6d). The radar map displays the top 30 most differentially expressed genes between both groups (P < 0.05; Fig. 6e).
Fig. 6.
mRNA expression levels in mouse hippocampi after HL. (a) The number of genes with detectable mRNA levels in each sample in both groups. We detected the expression of an average of 16,744 and 16,806 genes in the Con and HL group, respectively. (b) The number of significantly different mRNA expression levels after HL, including 154 upregulated and 97 downregulated mRNAs (P < 0.05, |log2 FC| > 0.58) in mouse hippocampi after HL. (c) A volcano plot displaying significantly upregulated and downregulated mRNAs after HL. Red, upregulated genes; green, downregulated genes. (d) A heatmap displaying relative mRNA expression levels in both groups. Similar mRNA expression levels were found among the samples in both groups. (e) A radar map showing the 30 genes with the most significantly different expression levels between groups (P < 0.05)
Table 2.
The representative changed gene. (Top 20)
| Gene_id | Chromsome | FoldChange | Log2FoldChange | P value | Regulation |
|---|---|---|---|---|---|
| Kif28 | 1 | 43.45455655 | 5.441435558 | 0.0162293 | Up |
| Nkx1-1 | 5 | 25.33432945 | 4.663021739 | 0.0260227 | Up |
| Mybphl | 3 | 24.4256914 | 4.610327495 | 0.0291509 | Up |
| C5ar2 | 7 | 23.35309053 | 4.545541583 | 0.0353948 | Up |
| Cyp2c65 | 19 | 21.39526886 | 4.419219904 | 0.0416854 | Up |
| Tnfsf8 | 4 | 21.29017576 | 4.412115955 | 0.0411565 | Up |
| Otoa | 7 | 17.6677598 | 4.143047218 | 0.0132915 | Up |
| Oas1g | 5 | 16.38921182 | 4.03467457 | 0.0224271 | Up |
| Wnt10b | 15 | 15.29475475 | 3.934965068 | 0.0092452 | Up |
| Gm5576 | 6 | 13.67752784 | 3.773735587 | 0.0339823 | Up |
| LOC118567331 | 1 | 0.035875729 | -4.800848029 | 0.0138397 | Down |
| Muc5ac | 7 | 0.037414001 | -4.740277948 | 0.0240738 | Down |
| Mettl21e | 1 | 0.041680633 | -4.584479016 | 0.0373792 | Down |
| Cryba1 | 11 | 0.044582308 | -4.487384889 | 0.0387728 | Down |
| Cr2 | 1 | 0.046931171 | -4.413309729 | 0.0081753 | Down |
| Tmigd1 | 11 | 0.049098081 | -4.348189557 | 0.0498687 | Down |
| Cpa3 | 3 | 0.07175006 | -3.800876158 | 0.0472127 | Down |
| Ankk1 | 9 | 0.076489594 | -3.708592703 | 0.0433636 | Down |
| Klrd1 | 6 | 0.082967284 | -3.591313635 | 0.0132689 | Down |
| Aqp2 | 15 | 0.087996741 | -3.506406101 | 0.0337882 | Down |
Combined analysis of the RNA-seq and MeRIP-seq
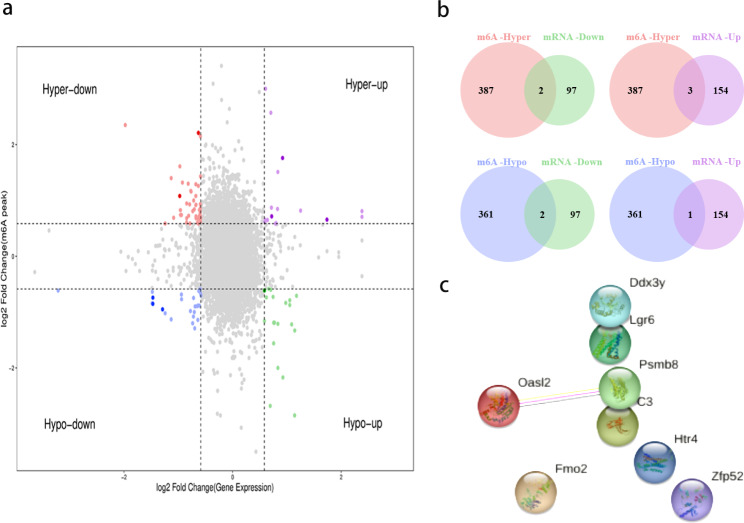
RNA-seq (P < 0.05, |log2 FC| > 0.58) and MeRIP-seq data (P < 0.05, |log2 FC| > 0.58) were analyzed together. We identified eight genes with significantly altered m6A peaks and mRNA expression. Among them, three genes showed upregulated mRNA expression and m6A peaks, two showed downregulated mRNA expression and m6A peaks, another two showed upregulated mRNA expression with downregulated m6A peaks, and one showed downregulated mRNA expression with upregulated m6A peaks (Additional file 5). The quadrant graph and Venn diagram demonstrated relative changed genes (Fig. 7a, b). Based on these results, we speculate that these eight genes may be relative to the development of cognitive impairment after HL. A network of protein–protein interactions (PPI) was employed for the purpose of illustrating the relationship between the proteins that these eight genes encode (Fig. 7c).
Fig. 7.
Combined MeRIP-seq and RNA-seq analyses of hippocampal samples after HL. a, b) The relationships between m6A-methylated mRNAs and their expression levels are displayed in a four-quadrant graph (a) and in a Venn diagram (b). c) Connections between differentially m6A-methylated mRNAs and the encoded proteins were analyzed by constructing a PPI network
Expression of m6A methylation regulators altered in mouse hippocampi after HL
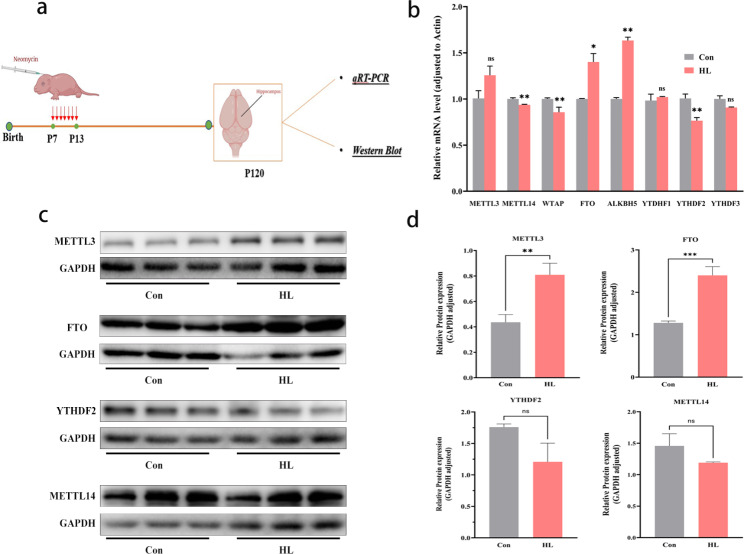
We examined the levels of key m6A regulators (METTL14, METTLL3, WTAP, FTO, ALKBH5, YTDHF1, YTDHF2, and YTDHF3) in mouse hippocampi (Fig. 8a). Reverse transcription quantitative real-time polymerase chain reaction (qRT-PCR) analysis showed that METTL14, WTAP, and YTHDF2 levels were significantly lower (P < 0.05); FTO and ALKBH5 levels were significantly higher (P < 0.05); and METTL3, YTDHF1, and YTHDF3 levels were not significantly different in HL mice than in control mice (P<0.05; Fig. 8b). Furthermore, the protein expression levels of METTL3, METTL14, FTO, and YTHDF2 in the hippocampus was measured via western blot. In HL group, METTL3 and FTO protein levels were significantly upregulated (P < 0.05), whereas METTL14 and YTHDF2 levels were downregulated (P<0.05), but not significantly (Fig. 8c, d). These results indicate that abnormal m6A methylation may be due to changes in METTL3 and FTO expression.
Fig. 8.
Regulated expression levels of m6A-methylated mRNAs in mouse hippocampi after HL. (a) Time points for qRT-PCR and western blot analyses after inducing HL. (b) qRT-PCR analysis showed that the hippocampal mRNA expression levels of METTL14, WTAP, and YTHDF2 were significantly lower in the HL group than in the Con group and those of FTO and ALKBH5 were significantly higher. c, d) Western blotting results showed that the hippocampal protein expression levels of FTO and METTL3 were higher in the HL group than in the Con group, whereas those of METTL14 and YTHDF2 were comparable between both groups. The data shown were analyzed using an unpaired t-test and are presented as the mean ± SEM. Not significant (ns), P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001. n = 6 mice per group. Con, control; HL, hearing loss
Discussion
As far as we know, this research is the first to reveal the m6A epitranscriptome profiles of the mouse hippocampus following HL using MeRIP-seq analysis. We observed 748 altered m6A peaks including 387 peaks showing hypermethylation and 361 peaks showing hypomethylation. The differentially methylated mRNA molecules were functionally predicted using GO and KEGG pathway analyses.
Neomycin-induced HL in mice is a commonly used model of sensorineural HL [24]. In this experiment, the mice that received neomycin from P7 to P13 exhibited severe HL at P30 and P120 (Fig. 1). In a previous study, neonatal mice that experienced noise exposure could suffered from severe HL with decreased neurogenesis, chronic spatial learning, and memory deficits in adulthood [25]. We used the PAT, a widely used behavioral test to detect learning and memory function in animals [26] to examine the cognitive function of mice at P120. Mice in the HL group showed a shorter latency period in the PAT test than the mice in the Con group (Fig. 2), suggesting that learning and memory deficits occurred after HL. Theories involving sensory deprivation, information degradation, common pathologies, and social isolation [27].
m6A methylation has been considered as novel epigenetic modifications related with various diseases, like cancers, inflammation, and cerebral ischemic disease [28–30]. With significant progress in sequencing technology, it has been identified that m6A is highly abundant in the mammalian brain [31]. The m6A is an abundant RNA modification in the brain, participating in neurodegenerative diseases. In mice with Huntington’s disease (HD), the alterations of m6A RNA methylation in hippocampal affected the expression levels of specific genes involved in synaptic plasticity and memory consolidation. Changes in expression and function of m6A RNA methyltransferase METTL3 and the m6A reader protein YTHDF1 involved were associated with memory deficits in HD mice [21]. In AD patients, the expression level of m6A RNA demethylase FTO decreased. For m6A RNA binding protein, the expression levels of YTHDC1, YTHDF1, FMR1, IGF2BP2, and HNRNPA2B1 rose, while YTHDF2 and EIF3H expression dropped [32]. In 3xTg AD mice, FTO has been shown to promote insulin-deficiency-associated AD by decreasing TSC complex subunit 1 (TSC1) mRNA levels, activating the mammalian target of the rapamycin (mTOR) signaling pathway, and promoting tau protein phosphorylation [15].
In this research, we examined m6A methylation profiles that may be involved in the mechanism of cognitive impairment after HL. According to the MeRIP-seq analysis, we revealed epitranscriptomic m6A dysregulation in the hippocampus of HL mice. By analyzing the m6A peak distribution profiles of mRNAs, we found that they were typically distributed in the CDS, 5′-UTR, and 3′-UTR region. Furthermore, the main variations in the m6A locations between the control and HL group were concentrated in the 3′-UTR (42.25%) (Fig. 4), which implied the potential target of m6A modifications in hippocampus after HL. Several cis elements responsible for posttranscriptional gene regulation are located in 3′ UTR [33]. As a critical region for RNA regulation, the 3’ UTR, plays a significant role in influencing translation efficiency, RNA stability, subcellular localization, and translation regulation [34]. Additionally, the 3’ UTR contains target sites for many RNA-binding proteins, which are key regulators of these important processes, which can bind to specific structural motifs or consensus sequences within the 3’ UTR [31]. Our results indicate m6A RNA methylation in the hippocampi may account for cognitive impairment in HL mice.
According to the result of GO analysis, those differentially methylated genes were primarily associated with synaptic and protein binding (Fig. 5). Synapse loss can induce cognitive impairment in many pathological processes such as AD [35], and impaired hippocampal synapses have been observed in patients with dementia [36]. According to the result of KEGG pathway analyses, those key differentially methylated genes were predominantly related to MAPK, Notch, and calcium-signaling pathways (Fig. 5). As a prominent intracellular signaling pathway, the MAPK pathway is of great importance in several critical physiological processes, including learning, memory, development, and cell differentiation [37, 38]. The MAPK cascade is required for spatial and contextual learning in mice and is associated with hippocampal neurodegeneration [39, 40]. It is noteworthy that Notch signaling pathways exhibited a correlation with both hyper- and hypomethylation of distinct genes (Fig. 5c, d), indicating a diverse role of m6A in the regulation of specific molecular processes. The Notch-signaling pathway participates in many biological processes, and deficiency in the pathway is detrimental to hippocampal neural-stem cell maintenance [41]. In addition, the key AD gene Notch 2, a genetic biomarker, was closely related to m6A regulators and might be the important targets in m6A methylation during the progression of AD [32]. In this study, the MeRIP-Seq analysis showed that there was hypermethylated peak of Notch2 in the region of the exons on chromosome 3 (Table S3). This genetic dynamic may impair cognitive function of the hearing loss patient and Notch2 may serve as a genetic marker for diagnosis.
The instability of mRNA can be determined by m6A modifications. To obtain a more comprehensive understanding of m6A methylation in the hippocampus after HL, conjoint analyses of MeRIP-seq and RNA-seq were utilized to screen all differentially m6A-methylated with differentially expressed mRNA. We had identified eight mRNAs that had undergone significant alterations in distribution of m6A peak and mRNA expression levels. However, we did not find significant correlations between mRNA expression and m6A methylation in the PPI network (Fig. 7c). The protein expression levels of those differentially m6A-methylated mRNAs should be determined by further researches.
m6A modification is a dynamic and reversible process driven via “writers,” removed via “erasers,” and recognized via “readers” [42]. Thus, we screened for several m6A regulators in Con and HL groups via performing qRT-PCR and western blot analyses. Among those m6A regulators, FTO is highly abundant in brain tissues and is related to neurotransmitter delivery and nervous system development [18, 43]. FTO dysfunction has been reported for participating in several mRNA processes including splicing, stability, translocation, and in protein translation. Several genome-wide association studies showed the importance of FTO in memory processing and its overexpression can significantly upregulate neuronal protein phosphorylation levels in mice with AD [15]. Exercise can decrease m6A levels by upregulating FTO in the mouse prefrontal cortex, and previous data showed that FTO knockdown in the prefrontal cortex led to enhanced consolidation of cued fear memory [14]. In this research, Western blot and qRT-PCR showed that the expression of FTO in the HL group increased compared to that in the control group. It is speculated that abnormal m6A modification in the hippocampus is due to the altered FTO expression after HL.
The current study has some limitations. First, relatively few mice hippocampal samples were used for MeRIP-seq analysis (three HL and Con samples each). Hence, the mRNAs showing altered m6A modifications should be further validated by performing MeRIP-PCR analysis with more samples. Second, we focused on some of the common key regulators of m6A and tested them by performing western blotting and qRT-PCR analyses. However, further in vitro and in vivo research are necessary to examine the specific roles behind m6A modification regulators (including “writers,” “erasers,” and “readers”) in greater detail to explore the mechanism underlying cognitive impairment after HL.
Conclusions
In summary, this is the first study to present the m6A transcriptome-wide map about the mouse hippocampus following HL. The results provide novel insights into the dynamics of m6A modifications after HL, with indication that the FTO might play a critical role. Those findings expand our understanding of the role about m6A modifications and the underlying mechanisms contributing to cognitive impairment following HL from an epigenetic perspective. Further studies on the involvement of abnormal m6A mRNA methylation during the pathological process of hippocampal neurodegeneration following HL are warranted.
Methods
Animals
Male, P7 Kunming mice were acquired from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). Those mice were housed at the Animal Experimental Center of Fudan University in a temperature-controlled room (22–23 °C) with a 12-hour light/dark cycle. Water and food were both provided ad libitum. The Animal Care and Use Committee of Fudan University approved all procedures. The corresponding protocols were conducted to comply with the National Institutes of Health Guide for the Care and Use of Laboratory Animals to minimize potential animal suffering. The mice were randomly sorted into Con and HL groups for ABR testing (9 mice per group), PAT analysis (9 mice per group), MeRIP-seq and RNA-seq (9 mice per group), qRT-PCR analysis (3 mice per group), and western blot analysis (6 mice per group).
Mouse model of neomycin-induced severe HL
The HL mouse model was established as previously described [24]. On P7, the experimental mice began receiving one daily dose of neomycin (Sangon Biotech, Shanghai, China) for seven consecutive days (200 mg/kg, subcutaneous, P7–P13). A corresponding dose of sterile saline was administered to the control group mice. ABR tests were performed as previously described [44] on P30 and P120 after anesthetizing the mice with xylazine (25 mg/kg) and esketamine (50 mg/kg). The average response to 1,000 repetitive stimuli at each frequency were magnified (100,000 times), filtered (0.3–3.0 kHz), and then digitized with an analog-to-digital converter (Tucker-Davis Technologies, Alachua, FL, USA). Specific evoked brainstem responses were calculated at frequencies of 8, 16, 24, and 32 kHz. At each frequency, the stimulus intensity was initiated from 90 dB and increased at 5 dB intervals until 20 dB.
Cognitive function assessment with the PAT
Cognitive function was determined using the PAT [45, 46] at P120. During the training period, mice were arranged in the left light box. The door opened automatically after an adaptation period (30 s), and the system recorded the latency before each mouse crossed into the dark zone. Once the mice entered the right dark zone, automatic door would be closed immediately, and the electrical shock (0.7 mA, 2 s) was released. Electrical shock stimuli were not applied during the test period. The maximum latency did not exceed 300 s.
Western blot analysis
The mice were sacrificed at P120 under isoflurane anesthesia, and the bilateral hippocampi were quickly obtained. Western blotting was conducted as previously described [44]. Radioimmunoprecipitation Assay lysis buffer (Beyotime, Shanghai, China) containing phenylmethylsulfonyl fluoride was used to extract the total protein. Primary antibodies against METTL14 (1:1000 dilution; ABclonal, Wuhan, China; A8530), METTL3 (1:1000 dilution; ABclonal; A19079), FTO (1:500 dilution; Proteintech, Wuhan, China; 27226-1-AP), YTHDF2 (1:4000 dilution; Proteintech; 24744-1-AP), and GAPDH (1:5000 dilution; Proteintech; HRP-60,004) were used. The horseradish peroxidase-conjugated goat anti-rabbit IgG (1:10000 dilution; Jackson, Pennsylvania, USA; 111-035-003) were used as the secondary antibody. The expression levels of YTHDF2, FTO, METTL14, and METTL3 were normalized to those of GAPDH to control for loading the difference amount of total protein in the gels.
qRT-PCR analysis
The mouse hippocampi were harvested at P120. Total RNA of hippocampi was obtained via using the TRIzol method (Invitrogen, Waltham, MA, USA) and detected by NanoDrop 2000 instrument (Thermo Fisher Scientific, Waltham, MA, USA). In reverse transcription procession, a PrimeScript™ RT Reagent Kit With gDNA Eraser (Perfect Real Time, Takara Bio, Shiga, Japan; RR047A) was applied to synthesize the complementary DNA (cDNA). TB Green Master Mix (Tli RNase H Plus, Takara Bio, Shiga, Japan; RR420A) in an ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) was used for qRT-PCR analysis. As a normalization control, β-actin was chosen and the relative genes expression were calculated by the 2−ΔΔCT method. The primer sequences are listed in Additional file 6.
MeRIP-seq and RNA-Seq with data processing
MeRIP-seq and RNA-Seq
The hippocampal samples were harvested at P120. Each sample was collected from the hippocampi of three mice each in the Con and HL group. The number of samples in each group was as follows: Con, n = 3; HL, n = 3”. Total RNA was extracted and isolated using magnetic beads with poly-T oligonucleotides and then chemically fragmented into 150-nucleotide oligonucleotides. RNA quantity and quality were analysed by an Agilent 2100 Bioanalyzer (Agilent Technology, Santa Clara, California, USA) and a NanoDrop 2000 instrument (Thermo Fisher Scientific, Waltham, MA, USA). The MeRIP-seq and RNA-seq analyses were conducted by OE Biotech Co., Ltd. (Shanghai, China).
For MeRIP-seq, the cleaved RNA fragments were incubated for 2 h at 4 °C with an m6A-specific antibody (Synaptic Systems, Germany; 202,003) in the immunoprecipitation (IP) buffer. The mixture was then incubated with protein A beads and eluted with elution buffer (1× IP buffer containing 6.7 mM m6A). Eluted m6A-containing fragments (IP fraction) and untreated input control fragments were converted to final strand-specific cDNA libraries in the presence of dUTP. The average insert size of the paired-end libraries was approximately 150 base pairs. We then performed paired-end 2 × 150 bp sequencing using an Illumina NovaSeq 6000 platform in accordance with the manufacturer’s recommended protocol.
For RNA-seq, the input sample without IP were used for sequencing library generation with the NEBNext® Ultra II Directional RNA Library Prep Kit (New England Biolabs, Inc., USA).
Bioinformatic analysis
Raw reads obtained from RNA sequences were used to statistical and quality control analyses. Adaptor, low quality bases and poly-N sequences were trimmed using FastP [47] to obtain clean data. After further removing rRNA reads, HISAT2 was applied to make clean reads map to reference genome [48], and only high-mapping unique reads were retained. To access the quality of MeRIP-seq, the deepTools [49] and Guitar R package [50] were used. In each m6A-IP sample, the enriched m6A peaks were identified via MeTDiff peak-calling software [51]. The threshold criteria for calling peaks showing differential m6A methylation were an absolute log2 FC value of > 0.58 and P value of < 0.05. Peaks showing differential methylation between groups were annotated using ChIPseeker software.
Differential gene expression was analyzed by using DESeq2. P < 0.05 and |log2 FC| > 0.58 was considered as the threshold for significant differentially expressed genes (DEGs). Hierarchical cluster analysis of DEGs was performed using R (v 3.2.0) to demonstrate the gene expression pattern in different groups and samples. The radar map of top 30 genes was drawn to show the expression of upregulated or downregulated DEGs using R packet ggradar.
Functional enrichment analysis
The identified and differential peaks with the hypergeometric distribution were analyzed by R software by performing GO and KEGG pathway analyses [52–54]. Both MEME and DREME software were applied to observe sequence motifs which were annotated via Tomtom software.
Combined analysis of MeRIP-seq and RNA-seq data
Those mRNAs with different expression levels were categorized as upregulated or downregulated based on the RNA-seq data. Differential regulation of gene methylation was identified based on changes found in m6A peak abundances following MeRIP-seq. The Python Script (v.2.7.12) software was used to perform correlation analysis on data obtained from both sequencing approaches to compare transcription and methylation levels simultaneously. To construct a PPI network, we utilized the Search Tool for the Retrieval of Interacting Genes database (https://www.string-db.org).
Statistical analysis
The data generated in this experiment were analyzed with GraphPad Prism® 9 (San Diego, CA, USA) and were displayed as the mean ± standard error of the mean (SEM). The unpaired Student’s t test was employed to assess differences between the two groups. A P value < 0.05 was used to representative a statistically significant difference.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
List of abbreviations
- ABR
auditory brainstem response
- ALKBH5
α-ketoglutarate-dependent dioxygenase alkB homolog 5
- BP
biological process
- CC
cellular component
- cDNA
complementary DNA
- CDS
coding sequence
- Con
control
- Diffpeak
peak reflecting differential m6A methylation
- FC
fold-change
- FTO
fat-mass and obesity-associated protein
- GO
Gene Ontology
- HL
hearing loss
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MeRIP-seq
methylated RNA immunoprecipitation sequencing
- METTL14
methyltransferase-like 14
- METTL3r
methyltransferase-like 3
- MF
molecular function
- ns
not significant
- P
postnatal day
- PAT
passive-avoidance test
- RNA-seq
RNA sequencing
- PPI
protein–protein interaction
- qRT-PCR
reverse transcription quantitative real-time polymerase chain reaction
- SEM
standard error of the mean
- SPL
sound pressure level
- UTR
untranslated region
- WTAP
Wilms tumor 1-associating protein
- YTHDF
YTH domain family
Authors’ contributions
SX, ZX, and JL designed the experiments. ZX, JL, and LY performed the experiments. ZX, LW, and WY analyzed the data and drew the figures. SX and ZX drafted the manuscript. SX revised the manuscript accordingly. All authors read and approved the final manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (grant numbers 81671045 and 82171184).
Data Availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center (https://ngdc.cncb.ac.cn/gsa/s/2Mz4btx4) under the accession number: CRA011232.
Declarations
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
All procedures were approved by the Animal Care and Use Committee of Fudan University, and the relevant protocols were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All animals were handled in accordance with the Animal Research: Reporting of in vivo Experiments (ARRIVE) guidelines.
Consent for publication
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xuehua Zhou and Lin Jin contributed equally to this work.
References
- 1.Collaborators GBDHL. Hearing loss prevalence and years lived with disability, 1990–2019: findings from the global burden of Disease Study 2019. Lancet. 2021;397:996–1009. doi: 10.1016/S0140-6736(21)00516-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin FR. Hearing loss and cognition among older adults in the United States. J Gerontol A Biol Sci Med Sci. 2011;66:1131–6. doi: 10.1093/gerona/glr115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronnberg J, Hygge S, Keidser G, Rudner M. The effect of functional hearing loss and age on long- and short-term visuospatial memory: evidence from the UK biobank resource. Front Aging Neurosci. 2014;6:326. doi: 10.3389/fnagi.2014.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen Y, Hu H, Fan C, Wang Q, Zou T, Ye B, Xiang M. Sensorineural hearing loss may lead to dementia-related pathological changes in hippocampal neurons. Neurobiol Dis. 2021;156:105408. doi: 10.1016/j.nbd.2021.105408. [DOI] [PubMed] [Google Scholar]
- 5.Park SY, Kim MJ, Kim HL, Kim DK, Yeo SW, Park SN. Cognitive decline and increased hippocampal p-tau expression in mice with hearing loss. Behav Brain Res. 2018;342:19–26. doi: 10.1016/j.bbr.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Song P, Tayier S, Cai Z, Jia G. RNA methylation in mammalian development and cancer. Cell Biol Toxicol. 2021;37:811–31. doi: 10.1007/s10565-021-09627-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garbo S, Zwergel C, Battistelli C. m6A RNA methylation and beyond - the epigenetic machinery and potential treatment options. Drug Discov Today. 2021;26:2559–74. doi: 10.1016/j.drudis.2021.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteom Bioinf. 2013;11:8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)a mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–4. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bi Z, Liu Y, Zhao Y, Yao Y, Wu R, Liu Q, Wang Y, Wang X. A dynamic reversible RNA N(6) -methyladenosine modification: current status and perspectives. J Cell Physiol. 2019;234:7948–56. doi: 10.1002/jcp.28014. [DOI] [PubMed] [Google Scholar]
- 12.Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–76. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Zhao JC. Update: mechanisms underlying N(6)-Methyladenosine modification of eukaryotic mRNA. Trends Genet. 2016;32:763–73. doi: 10.1016/j.tig.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Widagdo J, Zhao QY, Kempen MJ, Tan MC, Ratnu VS, Wei W, Leighton L, Spadaro PA, Edson J, Anggono V, et al. Experience-dependent Accumulation of N6-Methyladenosine in the Prefrontal cortex is Associated with memory processes in mice. J Neurosci. 2016;36:6771–7. doi: 10.1523/JNEUROSCI.4053-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Ren Y, Mao K, Hua F, Yang Y, Wei N, Yue C, Li D, Zhang H. FTO is involved in Alzheimer’s disease by targeting TSC1-mTOR-Tau signaling. Biochem Biophys Res Commun. 2018;498:234–9. doi: 10.1016/j.bbrc.2018.02.201. [DOI] [PubMed] [Google Scholar]
- 16.Chang M, Lv H, Zhang W, Ma C, He X, Zhao S, Zhang ZW, Zeng YX, Song S, Niu Y et al. Region-specific RNA m(6)a methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 2017;7. [DOI] [PMC free article] [PubMed]
- 17.Walters BJ, Mercaldo V, Gillon CJ, Yip M, Neve RL, Boyce FM, Frankland PW, Josselyn SA. The role of the RNA demethylase FTO (Fat Mass and Obesity-Associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology. 2017;42:1502–10. doi: 10.1038/npp.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Zang L, Zhang F, Chen J, Shen H, Shu L, Liang F, Feng C, Chen D, Tao H, et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum Mol Genet. 2017;26:2398–411. doi: 10.1093/hmg/ddx128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merkurjev D, Hong WT, Iida K, Oomoto I, Goldie BJ, Yamaguti H, Ohara T, Kawaguchi SY, Hirano T, Martin KC, et al. Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat Neurosci. 2018;21:1004–14. doi: 10.1038/s41593-018-0173-6. [DOI] [PubMed] [Google Scholar]
- 20.Zhao F, Xu Y, Gao S, Qin L, Austria Q, Siedlak SL, Pajdzik K, Dai Q, He C, Wang W, et al. METTL3-dependent RNA m(6)a dysregulation contributes to neurodegeneration in Alzheimer’s disease through aberrant cell cycle events. Mol Neurodegener. 2021;16:70. doi: 10.1186/s13024-021-00484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pupak A, Singh A, Sancho-Balsells A, Alcala-Vida R, Espina M, Giralt A, Marti E, Orom UAV, Gines S, Brito V. Altered m6A RNA methylation contributes to hippocampal memory deficits in Huntington’s disease mice. Cell Mol Life Sci. 2022;79:416. doi: 10.1007/s00018-022-04444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Xiu J, Zhu C, Meng K, Li C, Han R, Du T, Li L, Xu L, Liu R, et al. Fat mass and obesity-associated protein regulates RNA methylation associated with depression-like behavior in mice. Nat Commun. 2021;12:6937. doi: 10.1038/s41467-021-27044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng YL, Wang X, An R, Cassin J, Vissers C, Liu Y, Liu Y, Xu T, Wang X, Wong SZH et al. Epitranscriptomic m(6)a regulation of Axon Regeneration in the adult mammalian nervous system. Neuron. 2018;97:313 – 25 e6. [DOI] [PMC free article] [PubMed]
- 24.Yu H, Lin Q, Wang Y, He Y, Fu S, Jiang H, Yu Y, Sun S, Chen Y, Shou J, et al. Inhibition of H3K9 methyltransferases G9a/GLP prevents ototoxicity and ongoing hair cell death. Cell Death Dis. 2013;4:e506. doi: 10.1038/cddis.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao S, Liu L, Shi L, Li X, Shen P, Xun Q, Guo X, Yu Z, Wang J. Spatial learning and memory deficits in young adult mice exposed to a brief intense noise at postnatal age. J Otol. 2015;10:21–8. doi: 10.1016/j.joto.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGaugh JL, Roozendaal B. Drug enhancement of memory consolidation: historical perspective and neurobiological implications. Psychopharmacology. 2009;202:3–14. doi: 10.1007/s00213-008-1285-6. [DOI] [PubMed] [Google Scholar]
- 27.Lin FR, Albert M. Hearing loss and dementia - who is listening? Aging Ment Health. 2014;18:671–3. doi: 10.1080/13607863.2014.915924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klungland A, Dahl JA, Greggains G, Fedorcsak P, Filipczyk A. Reversible RNA modifications in meiosis and pluripotency. Nat Methods. 2016;14:18–22. doi: 10.1038/nmeth.4111. [DOI] [PubMed] [Google Scholar]
- 29.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, et al. M(6)a RNA methylation regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem cells. Cell Rep. 2017;18:2622–34. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han M, Liu Z, Xu Y, Liu X, Wang D, Li F, Wang Y, Bi J. Abnormality of m6A mRNA methylation is involved in Alzheimer’s Disease. Front Neurosci. 2020;14:98. doi: 10.3389/fnins.2020.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Cheng W, Zhang L, Zhou C, Peng X, Yu S, Zhang W. Novel Roles of RNA m6A Methylation Regulators in the Occurrence of Alzheimer’s Disease and the Subtype Classification. Int J Mol Sci. 2022;23. [DOI] [PMC free article] [PubMed]
- 33.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3’ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–33. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, Schultz RM, Wang PJ. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14:e1007412. doi: 10.1371/journal.pgen.1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mecca AP, O’Dell RS, Sharp ES, Banks ER, Bartlett HH, Zhao W, Lipior S, Diepenbrock NG, Chen MK, Naganawa M, et al. Synaptic density and cognitive performance in Alzheimer’s disease: a PET imaging study with [(11) C]UCB-J. Alzheimers Dement. 2022;18:2527–36. doi: 10.1002/alz.12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng L, Bestard-Lorigados I, Song W. The synapse as a treatment avenue for Alzheimer’s Disease. Mol Psychiatry. 2022;27:2940–9. doi: 10.1038/s41380-022-01565-z. [DOI] [PubMed] [Google Scholar]
- 37.Gyurko MD, Stetak A, Soti C, Csermely P. Multitarget network strategies to influence memory and forgetting: the Ras/MAPK pathway as a novel option. Mini Rev Med Chem. 2015;15:696–704. doi: 10.2174/1389557515666150219144336. [DOI] [PubMed] [Google Scholar]
- 38.Albert-Gasco H, Ros-Bernal F, Castillo-Gomez E, Olucha-Bordonau FE. MAP/ERK signaling in developing cognitive and emotional function and its effect on pathological and neurodegenerative processes. Int J Mol Sci. 2020;21. [DOI] [PMC free article] [PubMed]
- 39.Wang C, Li Y, Yi Y, Liu G, Guo R, Wang L, Lan T, Wang W, Chen X, Chen S, et al. Hippocampal microRNA-26a-3p deficit contributes to neuroinflammation and behavioral disorders via p38 MAPK signaling pathway in rats. J Neuroinflammation. 2022;19:283. doi: 10.1186/s12974-022-02645-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selcher JC, Atkins CM, Trzaskos JM, Paylor R, Sweatt JD. A necessity for MAP kinase activation in mammalian spatial learning. Learn Mem. 1999;6:478–90. doi: 10.1101/lm.6.5.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwarz TJ, Ebert B, Lie DC. Stem cell maintenance in the adult mammalian hippocampus: a matter of signal integration? Dev Neurobiol. 2012;72:1006–15. doi: 10.1002/dneu.22026. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Xiao J, Bai J, Tian Y, Qu Y, Chen X, Wang Q, Li X, Zhang Y, Xu J. Molecular characterization and clinical relevance of m(6)a regulators across 33 cancer types. Mol Cancer. 2019;18:137. doi: 10.1186/s12943-019-1066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma C, Chang M, Lv H, Zhang ZW, Zhang W, He X, Wu G, Zhao S, Zhang Y, Wang D, et al. RNA m(6)a methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018;19:68. doi: 10.1186/s13059-018-1435-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan X, Liu H, Li Y, Li W, Yu H, Shen X. Ribbon synapses and hearing impairment in mice after in utero sevoflurane exposure. Drug Des Devel Ther. 2020;14:2685–93. doi: 10.2147/DDDT.S253031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JS, Lee HJ, Lee S, Lee HS, Jeong YJ, Son Y, Kim JM, Lee YJ, Park MH. Conductive hearing loss aggravates memory decline in Alzheimer Model mice. Front Neurosci. 2020;14:843. doi: 10.3389/fnins.2020.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwon W, Kim HS, Jeong J, Sung Y, Choi M, Park S, Lee J, Jang S, Kim SH, Lee S, et al. Tet1 overexpression leads to anxiety-like behavior and enhanced fear memories via the activation of calcium-dependent cascade through Egr1 expression in mice. FASEB J. 2018;32:390–403. doi: 10.1096/fj.201601340RR. [DOI] [PubMed] [Google Scholar]
- 47.Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i90. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui X, Wei Z, Zhang L, Liu H, Sun L, Zhang SW, Huang Y, Meng J. Guitar: an R/Bioconductor Package for Gene Annotation guided transcriptomic analysis of RNA-Related genomic features. Biomed Res Int. 2016;2016:8367534. doi: 10.1155/2016/8367534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–91. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui X, Zhang L, Meng J, Rao MK, Chen Y, Huang Y, MeTDiff A Novel Differential RNA methylation analysis for MeRIP-Seq Data. IEEE/ACM Trans Comput Biol Bioinform. 2018;15:526–34. doi: 10.1109/TCBB.2015.2403355. [DOI] [PubMed] [Google Scholar]
- 52.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–51. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51:D587–D92. doi: 10.1093/nar/gkac963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center (https://ngdc.cncb.ac.cn/gsa/s/2Mz4btx4) under the accession number: CRA011232.