Abstract
The synthesis of a 5′-O-BzH–2′-O-ACE-protected pseudouridine phosphoramidite is reported [BzH, benzhydryloxy-bis(trimethylsilyloxy)silyl; ACE, bis(2-acetoxyethoxy)methyl]. The availability of the phosphoramidite allows for reliable and efficient syntheses of hairpin RNAs containing single or multiple pseudouridine modifications in the stem or loop regions. Five 19-nt hairpin RNAs representing the 1920-loop region (G1906–C1924) of Escherichia coli 23S rRNA were synthesized with pseudouridine residues located at positions 1911, 1915 and 1917. Thermodynamic parameters, circular dichroism spectra and NMR data are presented for all five RNAs. Overall, three different structural contexts for the pseudouridine residues were examined and compared with the unmodified RNA. Our main findings are that pseudouridine modifications exhibit a range of effects on RNA stability and structure, depending on their locations. More specifically, pseudouridines in the single-stranded loop regions of the model RNAs are slightly destabilizing, whereas a pseudouridine at the stem–loop junction is stabilizing. Furthermore, the observed effects on stability are approximately additive when multiple pseudouridine residues are present. The possible relationship of these results to RNA function is discussed.
INTRODUCTION
Ribonucleic acids perform a wide range of cellular functions, including carrying genetic information and catalyzing protein synthesis (1). Furthermore, RNAs must function with exceptional fidelity and precision, and sometimes under extreme conditions such as high temperatures or high salt. This vast array of biologically significant functions under a wide range of conditions will require structural variance among the different types of RNAs. One way that RNA can be diversified is through the use of nucleoside modifications. Each of the 95 known natural modifications has the potential for unique and important roles in the maintenance of global or local RNA structure and in the regulation of RNA function (2).
Pseudouridine (Ψ) (Fig. 1) is the most common modified nucleotide found in RNA to date (3). It comprises up to 8% of all uridines found in large subunit (LSU) ribosomal RNAs (4), and its locations have been associated with areas of functional importance, most notably the peptidyl transferase center (PTC) (5–8). The clustering of Ψ residues at the PTC appears to be a conserved feature over a broad range of organisms (4,7). In Escherichia coli there are nine Ψ residues in the LSU RNA (6,7). Recently, the synthase RluD was identified which is responsible for the in vivo synthesis of three Ψ residues in domain IV of 23S rRNA, namely Ψ1911, Ψ1915 and Ψ1917 (9). Interestingly, pseudouridines at positions 1915 and 1917 are universally conserved among all organisms examined to date (4) and the absence of RluD is detrimental to E.coli (9). The implications of these results are that the Ψ1911, Ψ1915 and Ψ1917 residues are essential for protein synthesis in all organisms. Recent crystallographic work has indicated that the 1920 loop protrudes from the surface of the 50S subunit and likely plays an important role in subunit association (10).
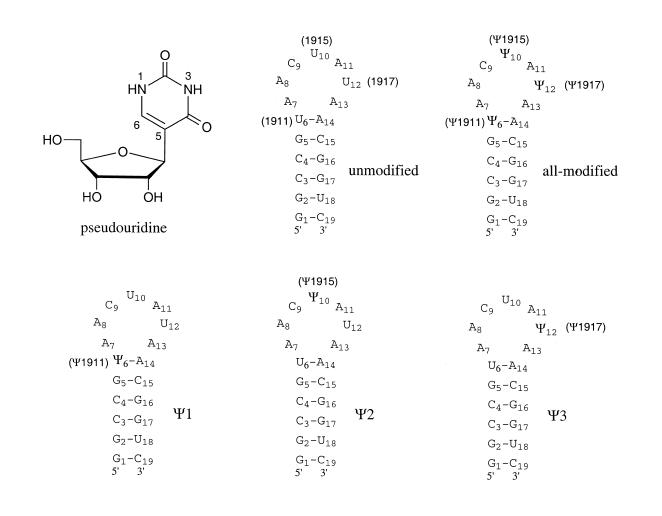
Figure 1.
The structure of pseudouridine is shown. Secondary structure representations of the synthetic RNA hairpins based on the 1920 stem–loop region of E.coli 23S rRNA, in which positions 1911, 1915 or 1917 are modified with pseudouridine (Ψ), are shown. In the native rRNA, position 1915 contains a methylated pseudouridine (m3Ψ) (25). The positions have been renumbered consecutively from G1 to C19 for NMR analysis.
Many questions still remain regarding the structural and functional roles of pseudouridine in the LSU rRNA and in other RNAs. The goals of the research described here were to develop a reliable and efficient method for incorporation of pseudouridine residues into RNA model systems (e.g., hairpin structures) and to examine the influence of pseudouridines on structure and stability of RNA in a systematic manner. A new approach towards RNA synthesis was recently introduced which involves a 5′-O-silyl-2′-O-orthoester protection system (11). For this study, a modified version of this protection system was used to generate a 5′-O-BzH–2′-O-ACE-pseudouridine-3′-phosphoramidite [BzH, benzhydryloxy-bis(trimethylsilyloxy)silyl; ACE, bis(2-acetoxyethoxy)methyl] and incorporate pseudouridine site-specifically into RNA. Five hairpin RNAs were synthesized which represent the 1920 region of E.coli 23S rRNA and contain one to three pseudouridine residues at positions 1911, 1915 and 1917, or remain unmodified. The stabilities and structures of the five RNAs were examined by using thermal melting, circular dichroism (CD) and NMR spectroscopy.
MATERIALS AND METHODS
Preparation of the 5′-O-BzH–2′-O-ACE-pseudouridine-3′-phosphoramidite
Tris(2-acetoxyethoxy)orthoformate and BzH-Cl were prepared as reported elsewhere (11) (S.A.Scaringe, N.Westwood and M.H.Caruthers, manuscript in preparation).
3′,5′-O-(1,1,3,3-tetraisopropyl-1,3-disiloxanediyl)pseudouridine [1]. Pseudouridine (Sigma, St Louis, MO) (8.19 mmol, 2.00 g) was dried by co-evaporation with 40 ml pyridine and then dissolved in 40 ml pyridine. 1,3-Dichloro-1,1,3,3-tetraisopropyldisiloxane (TIPDSCl2, Aldrich, Milwaukee, WI) (8.19 mmol, 2.58 ml) was added dropwise over 1 h at 0°C. The reaction was then stirred for 1 h at ambient temperature and stopped by addition of 2 ml of water. The pyridine was removed by evaporation followed by co-evaporation with 50 ml toluene. The crude residue was dissolved in 100 ml dichloromethane and washed with 5% sodium bicarbonate followed by saturated sodium chloride. The product was purified via silica gel chromatography using a hexane and ethyl acetate gradient to give a white solid (3.45 g, 86%). Exact mass calculated for C21H38N2O7Si2: 486.22; found by LC-MS (ES+): [M+H]+ 487.02, [M+Na]+ 508.90.
2′-O-[bis(2-acetoxyethoxy)methyl]pseudouridine (2′-O-ACE-pseudouridine) [2]. Compound 1 (7.10 mmol, 3.45 g) was suspended in 40 ml dioxane. Tris(2-acetoxyethoxy)orthoformate (16.3 mmol, 5.25 g), pyridinium p-toluenesulfonate (1.42 mmol, 0.356 g) and 4-(tert-butyldimethylsilyloxy)-3-penten-2-one (12.8 mmol, 3.04 ml) were added and the reaction stirred at 55°C for 14 h. The reaction was cooled to room temperature and neutralized with 1 ml N,N,N′,N′-tetramethylethylenediamine (TEMED). The crude reaction was diluted with 100 ml dichloromethane and loaded onto 200 g of silica gel and eluted with 35% hexanes and 65% ethyl acetate. After removal of the solvent, the slightly purified product was treated with a freshly prepared solution of TEMED (42.0 mmol, 6.30 ml) and 48% hydrofluoric acid (27.8 mmol, 1.00 ml) in acetonitrile (50 ml) for 6 h. The product was isolated in 81% yield (2.68 g) as a white solid via silica gel chromatography using an ethyl acetate and methanol gradient with 0.1% TEMED. Exact mass calculated for C18H26N2O12: 462.15; found by LC-MS (ES+): [M+Na]+ 485.08.
5′-O-[benzhydryloxy-bis(trimethylsilyloxy)silyl]–2′-O-[bis(2-acetoxyethoxy)methyl]-pseudouridine (5′-O-BzH–2′-O-ACE-pseudouridine) [3]. Compound 2 (5.79 mmol, 2.68 g) was suspended in 25 ml dichloromethane and diisopropylamine (5.79 mmol, 0.811 ml) and stirred at 0°C. In a separate flask, BzH-Cl (14.4 mmol, 6.15 g) was diluted with 12 ml of dichloromethane. Diisopropylamine (5.79 mmole, 0.811 ml) was added slowly to the BzH-Cl solution over 1 min. The silylating solution was then added slowly to the nucleoside solution in 3 ml aliquots until the starting material was completely consumed. The reaction was washed first with 5% sodium bicarbonate and then with saturated sodium chloride. The product was purified as a clear oil in 82% yield (4.06 g) via silica gel chromatography using a hexane and ethyl acetate gradient with 20% acetone. Exact mass calculated for C37H54N2O15Si3: 850.28; found by LC-MS (ES+): [M+Na]+ 872.84.
5′-O-[Benzhydryloxy-bis(trimethylsilyloxy)silyl]–2′-O-[bis(2-acetoxyethoxy)methyl]-pseudouridine-3′-(methyl-N,N-diisopropylphosphoramidite) (5′-O-BzH–2′-O-ACE-pseudouridine-3′-phosphoramidite) [4]. Compound 3 (4.77 mmol, 4.06 g) was dissolved in 20 ml of dichloromethane. Methyl tetraisopropylphosphorodiamidite (13.3 mmol, 3.50 g) was added followed immediately by tetrazole (3.81 mmol, 0.267 g) and the reaction stirred for 14 h. Thin-layer chromatography (TLC) analysis showed formation of the product and two additional spots moving faster than the product, and no starting material was observed. The reaction was washed first with 5% sodium bicarbonate and then with saturated sodium chloride. The organic layer was dried over sodium sulfate and filtered. After standing for 24 h, the faster moving spots disappeared. The product was then purified via silica gel chromatography using a solvent mixture of 65% hexanes, 25% acetone, 10% triethylamine to give a clear oil (3.78 g, 77%). Exact mass calculated for C44H70N3O16PSi3: 1011.38; found by FAB+: [M+Na]+ 1034.40; 31P NMR (CDCl3) δ (p.p.m.) (mixture of diastereomers): 150.71, 152.05; 1H NMR (CDCl3) δ (p.p.m.) (mixture of diastereomers): 0.04 [18H, m, Si(CH3)3], 1.17 [12H, m, two CH(CH3)3], 2.06 (6H, m, two COCH3), 3.40 (3H, m, OCH3), 3.54–3.63 [2H, m, two CH(CH3)3], 3.80–3.93 [6H, m, OCH(Ph)2, H2′, 3′, 4′, 5′, 5′′], 4.04–4.33 (8H, m, two OCH2CH2), 4.95 (1H, s, H1′), 5.98 (1H, s, OCH), 7.25–7.38 (10H, m, Ph), 7.43 (1H, d, H6, J = 1.6 Hz).
Preparation of RNAs
Five hairpin RNAs, based on the 1920 loop of E.coli 23S rRNA, were synthesized chemically on a 1.0 µmol scale and deprotected by using methods described previously (11). A modified 380B (PE-ABI) instrument was used (12). Each cycle consisted of the following reactions and appropriate washes: 30 s 5′ deprotection with 1.1 M HF, 1.6 M triethylamine in DMF; 60 s coupling with 15 eq. amidite (0.05 M) and 30 eq. S-ethyl tetrazole (0.25 M); 30 s 10% acetic anhydride/10% N-methylimidazole; 40 s oxidation with 1.5 M tert-butylhydroperoxide in toluene. The synthesis cycle required 9.1 min for three parallel syntheses. Syntheses were performed on polystyrene supports (Pharmacia, Piscataway, NJ) loaded with the appropriate nucleoside through a succinate linker (5–18 µmol/g). The sequences of the five RNAs are as follows: 5′-G1906GCCGU1911AACU1915AU1917AACGGUC1924-3′ (unmodified), 5′-G1906GCCGΨ1911AACΨ1915AΨ1917AACGGUC1924-3′ (all-modified; Ψ, pseudouridine), 5′-G1906GCCGΨ1911AACUAUAACGGUC1924-3′ (Ψ1), 5′-G1906GCCGUAACΨ1915AUAACGGUC1924-3′ (Ψ2) and 5′-G1906GCCGUAACUAΨ1917AACGGUC1924-3′ (Ψ3) with numbering according to the full-length E.coli 23S rRNA. All of the RNAs were obtained in high yields (typically 130–150 A260 units per 1.0 µmol synthesis) and the crude material was >95% pure as determined by 32P-labeling followed by analysis on high-resolution, 20% denaturing polyacrylamide gels and phosphorimaging. High-performance liquid chromatography (HPLC) analysis revealed some additional impurities (~10% of the product) that were not apparent on the gels. The deprotected oligoribonucleotides were purified by HPLC on a Waters BondapakTM C-18 Prep column (7.8 × 300 mm) in 10 mM ammonium acetate, pH 7.0, with a gradient of 0–40% CH3CN over 35 min. The purified RNAs were desalted by extensive dialysis for 2 days against RNase-free water.
Melting studies
The melting curves (absorbance versus temperature profiles) were obtained using an Aviv 14DS UV-vis spectrophotometer with a five-cuvette thermoelectric controller as described previously (13,14). Microcuvettes with 0.1 or 0.2 cm path lengths (60 and 120 µl volumes, respectively) were employed. Each measurement was taken in duplicate or triplicate. The buffer employed contained 15 mM NaCl, 20 mM sodium cacodylate and 0.5 mM Na2EDTA, pH 7.0. When the temperature was at 95°C, the absorbances of each sample were measured at 260 nm and used to determine the RNA concentrations. Single-strand extinction coefficients were calculated as described by Richards (15) except that the extinction coefficient for uridine (1.0 × 104 cm–1 M–1 at pH 7.0) was used instead of pseudouridine (8.1 × 103 cm–1 M–1 at pH 7.0) (16) for all RNAs because the nearest-neighbor extinction coefficients for pseudouridine are unknown, therefore some error will exist for the pseudouridine-containing RNAs. The absorbances were measured at 260 nm from 0 to 95°C with a heating rate of 0.8°C/min. The thermodynamic parameters were obtained from the absorbance versus temperature profiles using a van’t Hoff analysis, assuming a two-state model (17).
Circular dichroism (CD) spectroscopy
CD spectra were measured on a Jasco J600 spectropolarimeter from 220 to 330 nm at ambient temperature. The buffer used was 15 mM NaCl, 20 mM sodium cacodylate and 0.5 mM Na2EDTA at pH 7.0. Taking the RNA strand concentration into consideration, the measured CD spectra were converted to molar ellipticity (Δɛ) (18), except that values are expressed in moles of RNA molecules rather than moles of individual residues.
NMR methods
The crude RNAs were desalted by extensive dialysis for 3 days against RNase-free water and used without further purification for all NMR studies. All five RNAs were dissolved in 30 mM NaCl, 10 mM sodium phosphate and 0.5 mM Na2EDTA, pH 6.5, to final concentrations of 0.8–1 mM. For the imino proton studies, a solvent of 90% H2O and 10% D2O was employed. Spectra were obtained at 3°C on a Varian UNITY 500 MHz spectrometer using previously described methods (19).
RESULTS AND DISCUSSION
Synthesis of a novel pseudouridine phosphoramidite
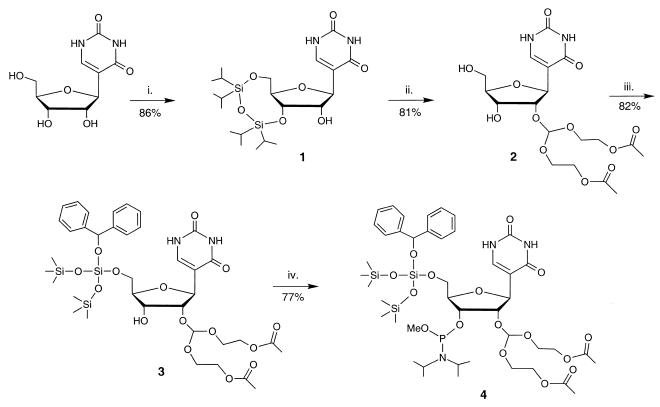
The 5′-O-BzH–2′-O-ACE-pseudouridine-3′-phosphoramidite 4 was synthesized in five steps with an overall yield of 44% (Scheme 1). This RNA building block contains 5′-O-BzH ether and 2′-O-ACE orthoester protecting groups. The 3′ hydroxyl was functionalized as the methyl-N,N-diisopropylphosphoramidite. The final amidite product 4 was fully characterized by 1H NMR, 31P NMR and FAB mass spectroscopy. The remaining unmodified bases (G, C, A and U) were converted to their corresponding 5′-O-SIL–2′-O-ACE-phosphoramidites as described previously (11).
Scheme 1. Synthesis of the 5′-O-BzH–2′-O-ACE-pseudouridine phosphoramidite 4: (i) TIPDSCl2, pyridine, 0°C to room temperature; (ii) a) tris(2-acetoxyethyl)orthoformate, pyridinium p-toluenesulfonate, 4-(tert-butyldimethylsilyloxy)-3-penten-2-one, dioxane, 55°C; b) TEMED-HF, CH3CN; (iii) BzH-Cl, diisopropylamine, CH2Cl2, 0°C; (iv) methyl tetraisopropylphosphorodiamidite, tetrazole, CH2Cl2, room temperature.
Phosphoramidite 4 offers several advantages over pseudouridine phosphoramidites reported previously (20–23). The removal of the BzH-protective group is facile and occurs under neutral conditions (S.A.Scaringe, N.Westwood and M.H.Caruthers, manuscript in preparation). The ACE group is removed under extremely mild conditions in aqueous buffers (11) and its by-products are easily removed from the RNA product by dialysis. In addition, the required coupling times of these amidites are relatively short (~60 s) with high coupling efficiencies (>99%). Furthermore, the synthesis can be carried out on a multigram scale to produce enough of amidite 4 for numerous couplings. We also reported recently on a method that allows for the multigram synthesis of pseudouridine which can be converted to 4 (24).
Synthesis and analysis of RNAs from the 1920 region of E.coli 23S rRNA
Five RNAs were synthesized using the 5′-O-BzH–2′-O-ACE-pseudouridine-3′-phosphoramidite. Each RNA was synthesized and deprotected in high yield (~4 mg per 1 µmol synthesis). The presence of pseudouridine residues was confirmed by P1 nuclease digestion and alkaline phosphatase treatment, followed by reverse-phase HPLC analysis (see Supplementary Material), and their positions were confirmed by using chemical sequencing methods (data not shown) (6). Overall, the availability of the 5′-O-BzH–2′-O-ACE-pseudouridine-3′-phosphoramidite allows for efficient synthesis of short pseudouridine-modified RNAs (~20 nt in length) in >70% overall yields.
Figure 1 shows the RNA sequences used in this study which represent the 1920 region of E.coli 23S rRNA (with the numbering system based on full-length rRNA). For NMR analysis, the residues are numbered consecutively from G1 to C19. This region of rRNA was selected because of its functional significance; one or more of the pseudouridines at positions 1911, 1915 and 1917 are essential for survival of E.coli (4,9), although the natural RNA sequence has a methylated pseudouridine at position 1915 (25). Uridine positions 1911, 1915 or 1917 in the unmodified RNA were substituted with pseudouridine such that the all-modified RNA has three pseudouridine residues and Ψ1, Ψ2 and Ψ3 RNAs each contain one pseudouridine.
The effects of pseudouridine on stability of the 1920-region hairpin RNAs
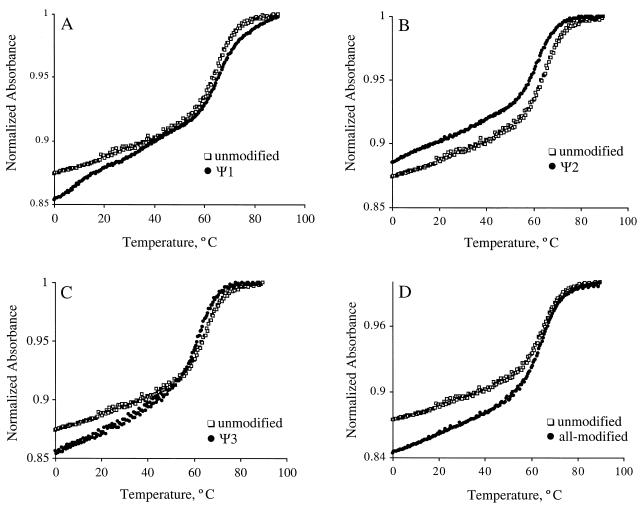
For the five RNAs (unmodified, all-modified, Ψ1, Ψ2 and Ψ3), absorbance versus temperature profiles were obtained at pH 7.0 in low salt conditions (35 mM Na+) and analyzed in terms of the melting temperature (Tm), ΔH°, ΔS°, ΔG°37 and ΔG°60 (14,17,26). The normalized absorbance plots at single RNA concentrations are shown in Figure 2. The melting transitions are monophasic for all RNAs, except Ψ1 (Fig. 2A), with Tms in the range of 62–68°C. Despite the large loop size of these RNAs, they all form hairpin structures. The helix-to-coil transitions were all independent of RNA concentration over a range of ~5–200 µM. The melting curve for Ψ1 is biphasic with transitions at 0–30 and 50–90°C. The higher melting transition was concentration independent, consistent with unimolecular unfolding of a hairpin structure. In contrast, the lower temperature transition was concentration dependent, suggesting the formation of a bimolecular complex such as a duplex, or a loop–loop interaction. Thermodynamic parameters for the five RNAs are listed in Table 1. The all-modified and unmodified variants were each synthesized twice and independent melting curves showed similar results. The thermodynamic parameters obtained from separate experiments were within the assumed error limits (3% for ΔG°60, 5% for ΔG°37, 7% for ΔH° and 8% for ΔS°; values not shown) (27,28). The data in Table 1 indicate that the Ψ1 variant is more stable than the unmodified RNA. In contrast, the Ψ2 and Ψ3 variants are slightly less stable than the unmodified RNA. The all-modified RNA exhibits almost equal stability to the unmodified RNA.
Figure 2.
Representative normalized UV melting curves for the unmodified and modified RNAs taken in 15 mM NaCl, 20 mM sodium cacodylate, 0.5 mM EDTA, pH 7.0 are shown. The curve for unmodified RNA is shown in (A–D) (open squares), and the curves for modified RNAs (closed circles) Ψ1, Ψ2, Ψ3 and all-modified are shown in (A), (B), (C) and (D), respectively. The melting curves were normalized at 95°C.
Table 1. Thermodynamics of 1920-loop RNAs, 5′-GGCCGXAACYAZAACGGUC-3′, with Ψ modifications.
| X, Y, Z | ΔG°60 (kcal/mol)a | ΔG°37 (kcal/mol)a | ΔH° (kcal/mol)a | ΔS° (e.u.)a | Tm (°C) |
|---|---|---|---|---|---|
| U, U, U (unmodified) | –0.7 | –4.9 | –62.2 | –184.8 | 63.7 |
| Ψ, U, U (Ψ1) | –1.5 | –5.9 | –66.1 | –193.9 | 67.2 |
| U, Ψ, U (Ψ2) | –0.5 | –4.2 | –55.3 | –164.7 | 62.5 |
| U, U, Ψ (Ψ3) | –0.6 | –4.6 | –59.0 | –175.3 | 63.2 |
| Ψ, Ψ, Ψ (all-modified) | –1.0 | –4.8 | –57.0 | –168.3 | 65.6 |
aConservative estimates of standard errors for ΔG°60, ΔG°37, ΔH° and ΔS° are 3, 5, 7 and 8%, respectively (27,28). Best fits were obtained by assuming hairpin formation (the melting profiles were concentration independent). The buffer conditions were 15 mM NaCl, 20 mM sodium cacodylate and 0.5 mM Na2EDTA, pH 7.0.
Overall, the observed order of stability of the RNAs is Ψ1 > all-modified ≥ unmodified ≥ Ψ3 ≥ Ψ2 (in order of increasing ΔG°37 and ΔG°60). The ΔG°37 values for the unmodified and all-modified variants differ by ~0.1 kcal/mol, indicating that the overall RNA stability is not influenced by the presence of the pseudouridine residues. In contrast, a single pseudouridine residue in the stem–loop junction is stabilizing by ~1 kcal/mol, whereas single Ψs in the loop regions are slightly destabilizing (ΔΔG°37 = 0.3–0.7 kcal/mol). Thus, it is apparent from the melting curve analysis that single modifications have different effects on RNA stability depending on their structural and sequence contexts. Furthermore, the thermodynamic effects appear to be additive such that the all-modified RNA has almost identical stability to the unmodified RNA.
The stabilizing effects of the pseudouridine in Ψ1 RNA were not surprising. Previous studies have already shown that pseudouridines stabilize RNA structure (typically with free energy differences of ~0.3–2 kcal/mol) (29–32). In addition, the ΨN1 proton is exchanged slowly in several different sequence and structural contexts (30,32–34). It has been suggested from NMR studies (30,35), and shown in X-ray crystal structures of tRNAGln and tRNAAsp (36,37) and molecular dynamics simulations (38) that a water molecule can coordinate between the pseudouridine base (ΨN1H) and the RNA backbone (Ψn and n – 1 phosphates). In contrast, the destabilizing effects of Ψ1915 and Ψ1917 observed in this study (in the Ψ2 and Ψ3 RNAs) are unusual and suggest that novel interactions or conformations are occurring in the loop regions of RNAs containing those sites of modification. For this reason, further structural characterization of these RNAs was desirable.
The effects of pseudouridine on structure of the 1920-region hairpin RNAs
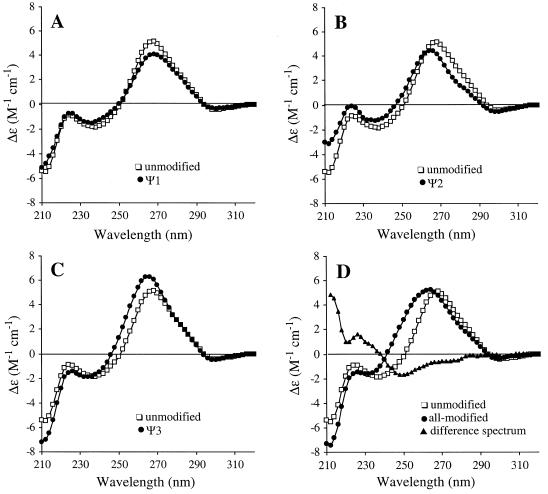
The CD spectra of the 1920-region hairpin RNAs were measured in order to compare the effects of single or multiple pseudouridines on the folded structures. The spectra of the unmodified and modified RNAs all have maxima centered around 265 nm and minima near 240 nm, similar to other A-form RNAs. The CD spectra in Figure 3 show that the pseudouridine-containing RNAs exhibit subtle differences in the maximum and minimum wavelengths, peak heights (molar ellipticity) and crossover points, suggesting that the pseudouridine residues are influencing each of the folded RNA structures in a unique manner. These differences are most apparent between the all-modified and unmodified variants (Fig. 3D). The peak maximum for the all-modified RNA is 262 nm compared to 268 nm for the unmodified RNA, and the crossover points differ by 8 nm (242 versus 250 nm for all-modified and unmodified RNAs, respectively). The CD spectrum of the variant with three pseudouridine residues (all-modified) is also different from each of the singly modified RNA spectra. A difference spectrum was obtained using equation 1 in order to determine if the structural changes induced by pseudouridine are additive. In part A of equation 1, the difference spectrum for the singly modified RNAs and unmodified RNA is set equal to the difference spectrum of triply modified and unmodified RNA. If the effects of pseudouridine on structure are additive, then the total difference spectrum should be equal to zero (equation 1, part B).
Figure 3.
CD spectra of the unmodified and modified RNAs. The molar ellipticities are normalized to RNA concentrations (3.7, 3.8, 4.3, 4.5 and 5.3 × 10–6 M in molecules of RNA for unmodified, Ψ1, Ψ2, Ψ3 and all-modified, respectively). Each spectra is an average of four scans. The CD spectrum of the unmodified variant (open squares) is shown in (A–D) with overlays (closed circles) of the Ψ1, Ψ2, Ψ3 and all-modified spectra in (A), (B), (C) and (D), respectively. The total difference spectrum (Ψ1 + Ψ2 + Ψ3 – 2 × unmodified – all-modified) is shown in (D) (closed triangles).
(A) Ψ1 + Ψ2 + Ψ3 – 3 × unmodified = all-modified – unmodified 1
(B) Ψ1 + Ψ2 + Ψ3 – 2 × unmodified – all-modified = 0
As shown in Figure 3D, the total difference spectrum is not equal to zero and distinct structural features can be observed, particularly near the crossover points, although errors in the strand concentration could partially explain some of the residual ellipticity. Thus, the structural effects are unique for the individual Ψ residues in Ψ1, Ψ2 and Ψ3 RNAs and for the combination of three Ψs in the all-modified RNA. Together, the CD results demonstrate that the single Ψ modifications have unique local influences on each of the folded RNA structures and in combination they induce further structural variations.
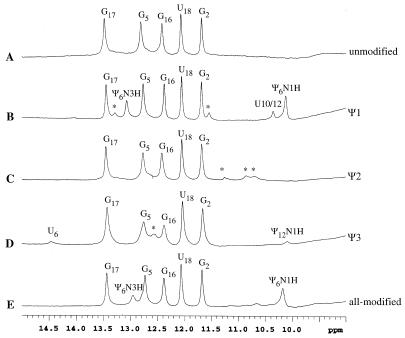
NMR spectroscopy was employed to examine hydrogen-bonding interactions in the five RNAs. The 1H NMR spectra of the RNAs are shown in Figure 4 (imino proton region). The spectrum of unmodified RNA (A) shows five imino proton resonances. Assignments were determined by using 1D NOE difference spectroscopy (see Supplementary Material) and indicate the formation of three G–C base pairs and one G·U mismatch. The imino protons for the terminal G1–C19 pair and the loop closing U6–A14 pair were not observed, presumably due to solvent exchange.
Figure 4.
The 1D imino proton (uridine H3/guanine H1/pseudouridine N1H and N3H) NMR spectra at 3°C of unmodified, Ψ1, Ψ2, Ψ3 and all-modified RNA sequences [from top to bottom (A–E)] dissolved in 90% H2O and 10% D2O (0.8–1 mM). The buffer employed for all samples contained 30 mM NaCl, 10 mM sodium phosphate and 0.5 mM Na2EDTA, at pH 6.5. Nucleotide assignments based on 1D-NOE difference spectroscopy (Supplementary Material) are indicated above each resonance (the residues are numbered consecutively from G1 to C19).
The Ψ1 RNA (Fig. 4B) spectrum exhibits 10 imino resonances. The additional resonance at 13.1 p.p.m. is assigned to the Ψ6–A14 pair. Assignments of the Ψ6N3 and Ψ6N1 imino proton peaks at 13.1 and 10.1 p.p.m., respectively, were confirmed by NOESY spectroscopy using the strong NOE from Ψ6N1H to Ψ6H6 (data not shown). The presence and chemical shift locations of these resonances are also consistent with previous studies on Ψ-containing RNAs (29,30,32–34). The peak at 10.4 p.p.m. is assigned as a loop residue (U10 or U12) based on a lack of NOEs to other residues. The imino proton resonances marked by asterisks at 13.3 and 11.5 p.p.m. are probably from G1 and either U10 or U12. These additional resonances are not apparent at temperatures >3°C, indicating solvent accessibility of these protons (see Supplementary Material). Interestingly, these resonances are too broad to be observed in the unmodified RNA (Fig. 4A). This observation suggests that the stabilization conferred by the Ψ6–A14 pair in Ψ1 RNA results in a structural change in the loop region as evident by the reduction in chemical exchange at the loop imino protons (from U10 or U12).
The 1H NMR spectrum for Ψ2 RNA (Fig. 4C) is similar to the unmodified RNA spectrum, except that three additional resonances at 10.7, 10.9 and 11.2 p.p.m. are observed. These resonances could not be assigned based on the 1D NOE difference spectra, but nonetheless indicate that the Ψ2 RNA structure differs from the unmodified and Ψ1 RNA structures.
The spectrum for Ψ3 RNA (Fig. 4D) shows eight imino proton resonances. Six of those resonances indicate the presence of four Watson–Crick base pairs (including a U6–A14 pair) and a G·U mismatch. The assignment for U6 at 14.5 p.p.m. is based on comparison to other RNAs with closing A–U pairs (31). The resonance at 10.1 p.p.m. shows no NOEs to other residues and is therefore assigned as Ψ12N1H because of its chemical shift location and a strong NOE to Ψ12H6 (data not shown). Further experiments are necessary to assign the peak at 12.6 p.p.m. (marked by * in Fig. 4D). As observed with Ψ1 and Ψ2 RNAs, the Ψ3 RNA must have unique structural features in the loop region which lead to additional resonances in the imino proton region. In addition, the G5 and G16 imino proton resonances are broadened in the Ψ2 and Ψ3 RNAs suggesting conformational exchange in the stem propagated by destabilization of the loop region. This effect is not observed in the unmodified RNA or Ψ1 RNA, thus indicating further differences between the singly modified and unmodified RNA structures.
The all-modified RNA (Fig. 4E) 1H NMR spectrum exhibits seven imino resonances that are associated with five base pairs, including a Ψ6–A14 pair and a G·U mismatch. The Ψ6N3H assignment at 12.9 p.p.m. was confirmed by using 1D NOE difference spectroscopy (see Supplementary Material) and the Ψ6N1H assignment was made by comparison with the Ψ1 RNA spectrum. Overall, the 1D NMR spectra for the all-modified, unmodified, Ψ1, Ψ2 and Ψ3 RNAs reveal observable structural differences between the RNAs.
Based on the NMR data, decreased stability of Ψ2 is likely due to the lack of a U6–A14 pair at the stem–loop junction. Formation of a U6/Ψ6–A14 base pair is observed in the Ψ3, Ψ1 and all-modified RNAs. The results for Ψ1 are consistent with previous studies that showed stabilization of closing base pairs involving Ψ residues (29,31,32). In addition, Davis showed that the ΨN1 imino proton in the sequence context AAΨA exchanged slowly with solvent. A role for the ΨN1 proton in stabilizing the RNA conformation through hydrogen bonding or increased base-stacking interactions was suggested (30). Interestingly, Davis’ study revealed that the pseudouridine effects on structure and stability could be propagated through the single-stranded helix to stabilize the stacking of neighboring residues. In the case of Ψ3 RNA, the formation of a U6–A14 pair appears to be favorable even in the absence of a Ψ6 modification. Thus, the structure could be stabilized instead by the neighboring Ψ12 residue (in the context of 5′-A11Ψ12A13A14-3′). The presence of additional imino proton peaks with different chemical shifts for all of the Ψ-containing RNAs suggests the formation of alternate conformations. Furthermore, comparison of singly and triply modified RNAs with unmodified RNA reveals distinct and unique structural changes induced by the pseudouridine residues which are not additive.
Conclusions
Several conclusions can be made from these studies regarding the relative stabilities and structures of RNAs with single and multiple pseudouridine residues. First, the influence of pseudouridines were examined in three different structural contexts, two in single-stranded loop regions and one at the closing pair of a stem–loop junction. Small differences in free energy values for the singly modified variants suggest their importance in determining stability of the overall RNA structure. Furthermore, the effects of multiple pseudouridines on stability are additive such that little or no overall effect may be observed. We have also considered the possible differences in RNA structure for the 1920-region RNA in the presence and absence of pseudouridines. CD data indicate that the unmodified and singly or triply modified RNAs all exist in solution as A-form helices, but they display different local conformations. The presence of pseudouridines in specific locations appears to influence the ability to form a base pair at the stem–loop junction (positions 6–14) and affects the loop structure.
Previously Raychaudhuri and co-workers demonstrated the functional importance of the Ψ residues in the 1920 region of E.coli 23S rRNA (9). The exact functional roles of Ψ1911, Ψ1915 and Ψ1917 in the LSU RNA of E.coli are still unclear, even though they are known to occur in a highly conserved region of the 23S rRNA. The stabilizing effects of Ψ1911 are consistent with previous studies (29–32). In addition, a large percentage (~30%) of naturally occurring pseudouridines are located at similar closing base-pair positions (39). In contrast, the slight destabilizing effects of Ψ1915 and Ψ1917 are unprecedented and surprising given that they are more highly conserved than Ψ1911. Thus, it is tempting to speculate that Ψ1915 and Ψ1917 may have specific functional roles, whereas Ψ1911 serves to compensate for their destabilizing effects.
The presence of Ψ1915 and Ψ1917 may be important for generating a specific loop structure and mediating interactions with other regions of the ribosome. A number of studies have implicated the 1920 region in subunit association. This region of 23S rRNA has been placed near the decoding region and the 790 loop of the small ribosomal subunit by cross-linking, directed probing and nucleotide protection studies (40–43), as well as in the X-ray crystal structure of a 70S complex (10). Thus, contacts between those regions of RNA may be responsible for communication between the ribosomal subunits and ultimately for protein synthesis. The possible roles of pseudouridine in mediating these interactions remains to be determined, but may involve specific hydrogen-bonding interactions with the 16S rRNA through its unique N1 proton.
Detailed NMR studies with these modified RNAs are currently in progress and will be necessary in order to gain a complete understanding of the relative contributions of pseudouridine to local conformations and RNA folding. In addition, further structure studies in the presence of magnesium chloride will be necessary in order to understand the functional relationships under physiological conditions. We have shown here that considerations of the RNA sequences and structure will be important in such studies with RNAs containing single or multiple modifications. Furthermore, the improvement of synthetic methods for pseudouridine incorporation will allow for the synthesis of sufficiently large quantities of modified RNAs which can be used for complete structure determinations.
SUPPLEMENTARY MATERIAL
See Supplementary Material available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to H. M.-P. Chui, S. Varma and M. Ksebati for technical assistance and helpful discussions, L. Hryhorczuk and R. Hood for mass analysis, and D. Kitchen and G. Haas for RNA synthesis. This work was supported by the National Institutes of Health (GM54632 to C.S.C, GM55745 to J.S.L. and GM56044 to S.A.S.) and the National Science Foundation (DMI-9801382 to S.A.S).
REFERENCES
- 1.Noller H.F. (1999) In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 197–220.
- 2.Agris P.F. (1996) Prog. Nucleic Acids Res. Mol. Biol., 53, 79–129. [DOI] [PubMed] [Google Scholar]
- 3.Maden B.E.H. (1990) Prog. Nucleic Acids Res. Mol. Biol., 39, 241–303. [DOI] [PubMed] [Google Scholar]
- 4.Ofengand J. and Bakin,A. (1997) J. Mol. Biol., 266, 246–268. [DOI] [PubMed] [Google Scholar]
- 5.Brimacombe R., Mitchell,P., Osswald,M., Stade,K. and Bochkariov,D. (1993) FASEB J., 7, 161–167. [DOI] [PubMed] [Google Scholar]
- 6.Bakin A. and Ofengand,J. (1993) Biochemistry, 32, 9754–9762. [DOI] [PubMed] [Google Scholar]
- 7.Bakin A., Lane,B.G. and Ofengand,J. (1994) Biochemistry, 33, 13475–13483. [DOI] [PubMed] [Google Scholar]
- 8.Lane B.G., Ofengand,J. and Gray,M.W. (1995) Biochimie, 77, 7–15. [DOI] [PubMed] [Google Scholar]
- 9.Raychaudhuri S., Conrad,J., Hall,B.G. and Ofengand,J. (1998) RNA, 4, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cate J.H., Yusupov,M.M., Yusupova,G.Zh., Earnest,T.N. and Noller,H.F. (1999) Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- 11.Scaringe S.A., Wincott,F.E. and Caruthers,M.H. (1998) J. Am. Chem. Soc., 120, 11820–11821. [Google Scholar]
- 12.Scaringe S.A. (2000) Methods Enzymol., in press. [DOI] [PubMed] [Google Scholar]
- 13.SantaLucia J. Jr, Allawi,H.T. and Seneviratne,P.A. (1996) Biochemistry, 35, 3555–3562. [DOI] [PubMed] [Google Scholar]
- 14.Meroueh M. and Chow,C.S. (1999) Nucleic Acids Res., 27, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards E.G. (1975) In Fasman,G.D. (ed.), Handbook of Biochemistry and Molecular Biology: Nucleic Acids. CRC Press, Cleveland, OH, pp. 596–599.
- 16.Hall R.H. (1971) The Modified Nucleosides in Nucleic Acids. Columbia University Press, New York, NY, p. 170.
- 17.McDowell J.A. and Turner,D.H. (1996) Biochemistry, 35, 14077–14089. [DOI] [PubMed] [Google Scholar]
- 18.Cantor C.R. and Schimmel,P.R. (1980) Biophysical Chemistry. Part II: Techniques for the Study of Biological Structure and Function. W.H. Freeman and Co., San Francisco, CA, pp. 412–413.
- 19.Lee K., Varma,S., SantaLucia,J.,Jr and Cunningham,P.R. (1997) J. Mol. Biol., 269, 732–743. [DOI] [PubMed] [Google Scholar]
- 20.Hall K.B. and McLaughlin,L.W. (1992) Nucleic Acids Res., 20, 1883–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasparutto D., Livache,T., Bazin,H., Duplaa,A.-M., Guy,A., Khorlin,A., Molko,D., Roget,A. and Téoule,R. (1992) Nucleic Acids Res., 20, 5159–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pieles U., Beijer,B., Bohmann,K., Weston,S., O’Loughlin,S., Adam,V. and Sproat,B.S. (1994) J. Chem. Soc. Perkin Trans., 1, 3423–3429. [Google Scholar]
- 23.Bergmann F. and Pfleiderer,W. (1994) Helv. Chim. Acta, 77, 481–501. [Google Scholar]
- 24.Grohar P.J. and Chow,C.S. (1999) Tetrahedron Lett., 40, 2049–2052. [Google Scholar]
- 25.Kowalak J.A., Bruenger,E., Hashizume,T., Peltier,J.M., Ofengand,J. and McCloskey,J.A. (1996) Nucleic Acids Res., 24, 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.SantaLucia J. Jr, Kierzek,R. and Turner,D.H. (1992) Science, 256, 217–219. [DOI] [PubMed] [Google Scholar]
- 27.SantaLucia J. Jr and Turner,D.H. (1997) Biopolymers, 44, 309–319. [DOI] [PubMed] [Google Scholar]
- 28.Allawi H.T. and SantaLucia,J.,Jr (1997) Biochemistry, 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- 29.Davis D.R. and Poulter,C.D. (1991) Biochemistry, 30, 4223–4231. [DOI] [PubMed] [Google Scholar]
- 30.Davis D.R. (1995) Nucleic Acids Res., 23, 5020–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durant P.C. and Davis,D.R. (1999) J. Mol. Biol., 285, 115–131. [DOI] [PubMed] [Google Scholar]
- 32.Yarian C.S., Basti,M.M., Cain,R.J., Ansari,G., Guenther,R.H., Sochacka,E., Czerwinska,G., Malkiewicz,A. and Agris,P.F. (1999) Nucleic Acids Res., 27, 3543–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall K.B. and McLaughlin,L.W. (1991) Biochemistry, 30, 1795–1801. [DOI] [PubMed] [Google Scholar]
- 34.Davis D.R., Veltri,C.A. and Nielsen,L. (1998) J. Biomol. Struct. Dyn., 15, 1121–1132. [DOI] [PubMed] [Google Scholar]
- 35.Griffey R.H., Davis,D., Yamaizumi,Z., Nishimura,S., Bax,A., Hawkins,B. and Poulter,C.D. (1985) J. Biol. Chem., 260, 9734–9741. [PubMed] [Google Scholar]
- 36.Arnez J.G. and Steitz,T.A. (1994) Biochemistry, 33, 7560–7567. [DOI] [PubMed] [Google Scholar]
- 37.Westhof E. (1988) Ann. Rev. Biophys. Chem., 17, 125–144. [DOI] [PubMed] [Google Scholar]
- 38.Auffinger P. and Westhof,E. (1997) J. Mol. Biol., 269, 326–341. [DOI] [PubMed] [Google Scholar]
- 39.Ofengand J. and Fournier,M.J. (1998) In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, p. 238.
- 40.Merryman C., Moazed,D., Daubresse,G. and Noller,H.F. (1999) J. Mol. Biol., 285, 107–113. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell P., Osswald,M. and Brimacombe,R. (1992) Biochemistry, 31, 3004–3011. [DOI] [PubMed] [Google Scholar]
- 42.Joseph S., Weiser,B. and Noller,H.F. (1997) Science, 278, 1093–1098. [DOI] [PubMed] [Google Scholar]
- 43.Wilson K.S. and Noller,H.F. (1998) Cell, 92, 131–139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.