Abstract
The patterns of synonymous codon choices of the completely sequenced genome of the bacterium Chlamydia trachomatis were analysed. We found that the most important source of variation among the genes results from whether the sequence is located on the leading or lagging strand of replication, resulting in an over representation of G or C, respectively. This can be explained by different mutational biases associated to the different enzymes that replicate each strand. Next we found that most highly expressed sequences are located on the leading strand of replication. From this result, replicational-transcriptional selection can be invoked. Then, when the genes located on the leading strand are studied separately, the correspondence analysis detects a principal trend which discriminates between lowly and highly expressed sequences, the latter displaying a different codon usage pattern than the former, suggesting selection for translation, which is reinforced by the fact that Ks values between orthologous sequences from C.trachomatis and Chlamydia pneumoniae are much smaller in highly expressed genes. Finally, synonymous codon choices appear to be influenced by the hydropathy of each encoded protein and by the degree of amino acid conservation. Therefore, synonymous codon usage in C.trachomatis seems to be the result of a very complex balance among different factors, which rises the problem of whether the forces driving codon usage patterns among microorganisms are rather more complex than generally accepted.
INTRODUCTION
The history of the ideas concerning the factors shaping codon usage patterns and the mutations at the synonymous sites illustrates the complexity of this topic. In the late 1960s it was suggested that most, if not all, of the nucleotide changes at the third codon positions were neutral or nearly neutral with respect to natural selection (1). Hence, in principle, it could be thought that all codons coding for the same amino acid should be equally frequent if a large sample of genes is studied. Subsequent work showed that a huge interspecific variation exists, and the ‘genome hypothesis’ was proposed (2). This variation from equal usage could be interpreted as the influence of different mutational biases in different genomes. Then it was shown that biased codon usage is not only species specific but in some organisms there is a clear intragenomic variability. For microorganisms, this was explained as the effect of natural selection acting at the level of translation, which resulted in the preferential usage of a subset of ‘major’ codons (3). This hypothesis was reinforced by the finding that in Escherichia coli (4) and Saccharomyces cerevisiae (5,6) the preferred codons in highly expressed sequences are recognised by the most abundant tRNAs. For these species, it was proposed that codon choices in lowly expressed sequences are the result of the mutational biases characteristic of each genome, since these genes are less constrained by translational pressures (7,8). Among prokaryotes with extremely biased genomic compositions, synonymous codon choices appear to be determined exclusively by the biased mutational pressures. Well known examples are Mycoplasma capricolum (GC% = 25), Rickettsia prowazekii (GC% = 29) and Micrococcus luteus (GC% = 72) (9–12). Therefore, the most accepted hypothesis for the unequal usage of synonymous codons among microorganisms states that it is the result of the mutational biases and natural selection acting at the level of translation.
In the last few years several analyses on complete prokaryotic genomes allowed the detection of several new factors shaping codon usage. For instance, the physical location of each sequence determines GC3 (and hence codon usage patterns) in Mycoplasma genitalium genes, which might be related to the replication process (13,14). Second, in Borrelia burgdorferi the variation in codon usage among genes seems to be the result of selective pressures acting at the replicational and transcriptional levels (15). Similar results were recently reported by Lafay et al. in Treponema pallidum (16). Third, in Mycobacteria, the hydropathy of each encoded protein is a major factor shaping codon usage (17).
In this paper we show that the pattern of synonymous codon choices in the completed sequenced genome of the bacterium Chlamydia trachomatis could be the result of at least four different forces: (i) strand-specific mutational biases, (ii) natural selection acting at the levels of replication, transcription and translation, (iii) the hydropathy level of each protein, and (iv) the level of amino acids conservation. We discuss if this complex pattern can be reduced to the ‘mutational bias-translational efficiency’ hypothesis.
MATERIALS AND METHODS
The complete genomes and coding sequences of C.trachomatis and Chlamydia pneumoniae (18,19) were obtained from http://chlamydia-www.berkeley.edu:4231/ . Codon usage, correspondence analysis (COA) (20), GC3 (the frequency of codons ending in C or G, excluding Met, Trp and stop codons), the relative synonymous codon usage (RSCU) (21) and the frequency of optimal codons (FOP) (4) were calculated using the program CodonW 1.3 (written by John Peden and available from ftp://molbiol.ox.ac.uk/Win95.codonW.zip ). The GC skew [(G–C)/(G+C)] along the DNA sequence was calculated using a sliding window of 50 kb and a step of 10 kb. The Ks and Ka (estimated number of synonymous and non-synonymous substitutions) were calculated according to the method of Li (22) as modified by Comeron (23). For these estimations, the coding sequences from both species were translated, and then aligned using ClustalW (24). Subsequently, the alignments were back translated to the known DNA sequences. The Ks was calculated only on those pairs of sequences longer than 99 amino acids displaying a minimal value of 60% of identity at the amino acid level, increasing the probability of comparing only orthologous genes. The analyses were performed with the pairs of sequences displaying Ks values ≤2.0. The final data set comprised 210 pairs of genes.
COA of RSCU values was carried out to determine the major source of variation among genes. RSCU is the observed frequency of a codon divided by the frequency expected if all synonyms coding for that amino acid are used equally; therefore RSCU values close to 1.0 indicate a lack of bias for that codon. Hence, each sequence is described by a vector of 59 variables, which is the number of codons for which there are synonyms. COA plots these genes in a multidimensional space of 59 axes. Then a certain number of new axes, through the cloud of points, are identified. These axes represent the most prominent factors contributing to the variation among genes. FOP is the frequency of optimal codons in each sequence, and these are defined as those codons that occur significantly more often (relative to their synonyms) in highly than in lowly expressed genes.
RESULTS
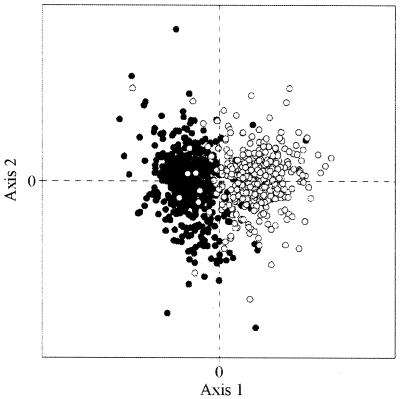
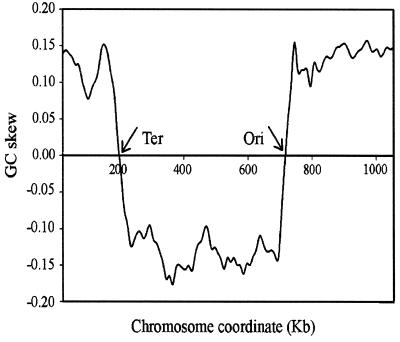
We conducted a COA of RSCU values on all genes of C.trachomatis (18), and Figure 1 shows the position of the genes on the plane defined by the first and second axes, which accounted for 9.9 and 5.7%, respectively, of the total variation. The first principal component described a rather small amount of the variation, which suggests that in C.trachomatis the major trend in codon choices is not as strong as in other species. The principal axis separated the genes into two clusters with little overlap between them. A similar scatter of points was found previously in B.burgdorferi (15), where the first axis detected genes that are transcribed either in the leading or in the lagging strand of replication. Hence, we studied this possibility in C.trachomatis. However, while in B.burgdorferi the genome is linear and the replication begins in the middle and proceeds towards the telomeres, which makes unambiguous the location of each sequence, this is not the case in C.trachomatis, where the chromosome has the usual circular permutation. To overcome this difficulty, we analysed the GC skew along the DNA of C.trachomatis, which is shown in Figure 2. For several prokaryotic genomes it has been shown that there is a change in the sign of the skew near the origin of replication (25–28). In Figure 2 it can be seen that there is an excess of C over G between positions 200 000 and 720 000 and an excess of G over C in the rest of the genome. In C.trachomatis the origin of replication is located between bases 719 988 and 720 258 (18) and the total length of the chromosome is 1 042 519 bp. If replication proceeds in opposite directions at identical rates, the replication forks should encounter each other near position 199 000, which is in agreement with the switch of the skew around base 200 000. We note that an excess of G over C in the leading strand is rather common among prokaryotes (15,28). Further evidence to support this point of encounter comes from the observation that in the majority of prokaryotic genomes there are more genes located in the leading than in the lagging strand (28). In C.trachomatis, between bases 185 673 and 194 844, there is a cluster of nine genes placed in the W strand (in that region, putatively leading strand of replication) and there are four sequences on the C strand, which are located between bases 195 091 and 196 662. Since there is no other cluster near position 200 000 where changing the strands there are as many as 13 genes located in the leading strand, we assumed that the two forks of replication probably encounter each other between bases 194 845 and 195 090. These analyses allowed us to locate each sequence either in the leading or lagging strands, and the total figures are 499 (56%) and 394 (44%) ORFs, respectively. This excess of genes in the leading strand was noted previously in several genomes (15,25,28), and probably implies that the orientation of the genes, either away from the origin of replication (on the leading strand) or towards the origin (on the lagging strand), is not selectively neutral. For instance, it has been postulated that selection acting at the levels of replication and transcription is responsible for the asymmetry in the distribution of sequences in B.burgdorferi (15).
Figure 1.
Plot of the two most prominent axes generated by the COA of the RSCU values from the C.trachomatis genome. The open circles and the filled circles correspond to the genes transcribed in the leading and lagging strand of replication, respectively.
Figure 2.
GC skew along the C.trachomatis genome. It was calculated using a sliding window of 50 kb and a step of 10 kb.
In Figure 1, the points marked with an open circle correspond to the genes transcribed in the leading strand while those indicated with a filled circle are transcribed in the lagging strand. A careful inspection showed that the majority of open circles (462/499 = 92.6%) are placed in the left quadrants while in the right quadrants are placed the greatest proportion (376/394 = 95.4%) of the sequences transcribed in the lagging strand. Therefore, the two groups of genes separated by the first axis are defined by their direction of transcription. On the other hand, the second axis was significantly correlated (r = 0.36, P < 0.0001) with GC3, and in Figure 1 in the lower quadrants are placed the GC3-poorest sequences.
The cumulative codon usage corresponding to the genes located in each strand is shown in Table 1. In total, 168 152 and 144 703 codons were analysed in the leading and lagging strands, respectively. A χ2 test was applied to evaluate the differences in codon usage between the two categories of genes. The differences were found to be significant (P < 0.01) for 49/59 of the synonymous codons, and are marked with an * or a - in Table 1. This analysis shows that 27 codons were used at the highest frequency in the leading strand, while 22 triplets were used most frequently in the lagging strand. Of the preferred codons on the leading strand 12 are either T- or G- ending, while three display an A in their third positions; finally, no preferred codon is C- ending. On the lagging strand, 16 of the preferred codons are C- ending, five are A- ending while one and none are T- or G- ending, respectively. These results confirm that there is a bias towards G and T in the leading strand, and towards C in the lagging strand. Therefore, it can be concluded that in C.trachomatis, as is the case in several prokaryotes (25–29) the leading and lagging strands of replication display an asymmetry in the mutational biases, and, as shown in B.burgdorferi (15), this difference is the most important source of variation in codon usage.
Table 1. Codon usage for genes located in the leading and lagging strands on the C.trachomatis genome, and preferred codons on highly expressed sequences.
| AA | Codon | Leading | Lagging | ||
|---|---|---|---|---|---|
| N | RSCU | N | RSCU | ||
| Phe | TTT* | 5450 | 1.37 | 4149 | 1.16 |
| TTC- | 2491 | 0.63 | 3008 | 0.84 | |
| Leu | TTA* | 5922 | 1.96 | 4215 | 1.49 |
| TTG* | 4254 | 1.41 | 1930 | 0.68 | |
| CTT- | 3311 | 1.10 | 3835 | 1.36 | |
| CTC- | 1241 | 0.41 | 2568 | 0.91 | |
| CTA- | 1846 | 0.61 | 2817 | 1.00 | |
| CTG | 1553 | 0.51 | 1563 | 0.55 | |
| Ile | ATT* | 5793 | 1.69 | 5105 | 1.48 |
| ATC- | 2555 | 0.74 | 3652 | 1.06 | |
| ATA* | 1954 | 0.57 | 1567 | 0.46 | |
| Met | ATG | 3747 | 1.00 | 2623 | 1.00 |
| Val | GTT* | 4843 | 1.56 | 2702 | 1.41 |
| GTC- | 1616 | 0.52 | 1618 | 0.85 | |
| GTA | 3228 | 1.04 | 2046 | 1.07 | |
| GTG* | 2709 | 0.87 | 1291 | 0.67 | |
| Tyr | TAT* | 3742 | 1.46 | 2724 | 1.23 |
| TAC- | 1398 | 0.54 | 1705 | 0.77 | |
| TER | TAA | 250 | 1.50 | 243 | 1.85 |
| TAG | 182 | 1.09 | 87 | 0.66 | |
| His | CAT* | 2654 | 1.53 | 2436 | 1.31 |
| CAC- | 804 | 0.47 | 1296 | 0.69 | |
| Gln | CAA- | 3868 | 1.14 | 4612 | 1.47 |
| CAG* | 2935 | 0.86 | 1651 | 0.53 | |
| Asn | AAT* | 3981 | 1.48 | 3588 | 1.29 |
| AAC- | 1391 | 0.52 | 1966 | 0.71 | |
| Lys | AAA- | 6034 | 1.24 | 6503 | 1.58 |
| AAG* | 3707 | 0.76 | 1743 | 0.42 | |
| Asp | GAT* | 6422 | 1.62 | 4417 | 1.43 |
| GAC- | 1516 | 0.38 | 1741 | 0.57 | |
| Glu | GAA- | 6931 | 1.15 | 6250 | 1.46 |
| GAG* | 5101 | 0.85 | 2317 | 0.54 | |
| Ser | TCT | 5270 | 2.49 | 5237 | 2.49 |
| TCC- | 1418 | 0.67 | 2403 | 1.14 | |
| TCA | 1499 | 0.71 | 1474 | 0.70 | |
| TCG* | 1238 | 0.58 | 883 | 0.42 | |
| Pro | CCT | 3584 | 2.24 | 3950 | 2.19 |
| CCC- | 639 | 0.40 | 1153 | 0.64 | |
| CCA* | 1476 | 0.92 | 1531 | 0.85 | |
| CCG* | 701 | 0.44 | 589 | 0.33 | |
| Thr | ACT | 2453 | 1.32 | 2842 | 1.33 |
| ACC- | 1016 | 0.55 | 1818 | 0.85 | |
| ACA | 2450 | 1.32 | 2813 | 1.32 | |
| ACG* | 1495 | 0.81 | 1079 | 0.50 | |
| Ala | GCT* | 6299 | 1.96 | 4926 | 1.86 |
| GCC- | 1388 | 0.43 | 1818 | 0.69 | |
| GCA | 3335 | 1.04 | 2766 | 1.04 | |
| GCG* | 1861 | 0.58 | 1085 | 0.41 | |
| Cys | TGT* | 1969 | 1.41 | 1219 | 1.07 |
| TGC- | 832 | 0.59 | 1059 | 0.93 | |
| TER | TGA | 67 | 0.40 | 64 | 0.49 |
| Trp | TGG | 1728 | 1.00 | 1256 | 1.00 |
| Arg | CGT* | 2626 | 1.75 | 1445 | 1.43 |
| CGC- | 944 | 0.63 | 1459 | 1.44 | |
| CGA | 1734 | 1.15 | 1235 | 1.22 | |
| CGG* | 943 | 0.63 | 382 | 0.38 | |
| Ser | AGT* | 2085 | 0.98 | 1176 | 0.56 |
| AGC- | 1207 | 0.57 | 1431 | 0.68 | |
| Arg | AGA | 2164 | 1.44 | 1342 | 1.33 |
| AGG* | 604 | 0.40 | 203 | 0.20 | |
| Gly | GGT* | 2408 | 0.82 | 1446 | 0.71 |
| GGC- | 1284 | 0.44 | 1405 | 0.69 | |
| GGA- | 4826 | 1.65 | 3772 | 1.86 | |
| GGG* | 3180 | 1.09 | 1474 | 0.73 | |
AA, amino acid; N, count of codons. Codons marked with an * or - are statistically more frequent in the leading or lagging strand of replication, respectively (P < 0.01). Underlined codons are those more frequent (P < 0.05) in highly expressed sequences.
In order to understand if codon usage patterns are further determined by other factors, and, in particular, to investigate whether highly expressed genes do prefer a subset of codons (i.e., if there is selection for codon usage at the level of translation), we conducted a COA of RSCU values on the genes located on the leading strand of replication, since >75% of the highly expressed sequences are located in that strand. The position of the genes along the first axis generated by the analysis was associated with expressivity, since at one extreme were clustered sequences coding for ribosomal proteins, elongation factors, outer membrane proteins, heat-shock proteins, histone-like proteins, single-stranded DNA binding proteins; while genes presumably expressed at lowest levels were scattered all through the distribution. This allowed us to compare codon usage patterns in the sequences displaying the most extreme values at both ends of the first axis of the COA (49 genes each), and the result of this analysis is shown in Table 1. In order to test the differences in codon usage between the two groups of sequences a χ2 test was applied. There are 17 codons whose usage is significantly higher among the highly expressed genes, which code for 14 different amino acids.
As mentioned above, this analysis was performed only for the genes located in the leading strand of replication, which is characterised by a mutational bias towards G and T. For several amino acids this bias seems to determine the preferred codon, specially among quartets, but interestingly the base composition of third codon positions among several of these preferred codons is against the bias (Table 1). This is the case among duets, where C- and A- ending triplets are preferred. Although in C.trachomatis the concentrations of tRNAs are not known, it is important to notice that in the case of duets and Ile the most frequent codon among highly expressed sequences matches without wobbling with the only tRNA for the corresponding amino acid (18). Furthermore, in the case of His and Cys there is an increment (although not significant) of the C- ending codons, which are again the only triplets that pair without wobbling with the only tRNA for those amino acids. These findings suggest that in this bacterium, superimposed to the mutational biases characteristic of each strand of replication, selection is acting at the level of translation.
To test this possibility, we used two different approaches. First, we calculated the FOP for each sequence, and the genes displaying the highest values were sequences with putative high or very high expression levels, including ribosomal proteins, a histone-like protein, HSP 60, ompA, tsf, dnaK, fusA, ssb and subunits of the RNA pol. Although the majority of these genes are located on the leading strand of replication, some of them are placed in the lagging strand. Hence, although the FOP was calculated considering genes located in the leading strand only, the ‘optimal codons’ detected by our analysis are more frequent in highly expressed sequences independently of their orientation in the genome. Remarkably, there is a strong correlation (r = –0.65, P < 0.0001) between the FOP in each gene and the respective position on the second axis generated by the COA carried out on all the genes. This firmly suggests that the second axis discriminates expression levels.
The second approach was to estimate the Ks between 210 orthologous sequences from C.trachomatis and C.pneumoniae, which is a related bacterium whose genome is completely sequenced (19). Although the Ks values are relatively high, two results concerning this analysis support the hypothesis that selection acting at the level of translation contributes to codon usage in Chlamydia. First, when the sequences are sorted according to the Ks, the genes displaying the lowest values are presumably highly or very highly expressed sequences: ribosomal proteins, tufA, abundant membrane proteins like groES and ompA, HSP 60, nusA, pfrA, rpoD and B and elongation factors. This indicates that highly expressed genes have diverged less at the synonymous sites than lowly expressed ones since the split of this two bacterial species from their last common ancestor. In turn, this suggests that selection (i) is acting at the synonymous sites, and (ii) is more effective on the sequences with highest expression levels, as is the case, among other examples, in Enterobacteria and Mycobacteria (17,30). Second, there is a negative and significant correlation (r = –0.47, P < 0.0001) between Ks and FOP, which indicates that the genes which diverged less are the sequences which display the highest frequencies of optimal codons. Therefore, we conclude that selection acting at the level of translation is indeed contributing to codon choices in C.trachomatis.
A striking result from the analysis of the COA was the significant correlation found between the second component and the hydropathy of each protein, using the Kyte-Doolittle (KD) scale (r = 0.20, P < 0.0001). To understand how this feature is related to codon usage, we compared the codon usage patterns of the genes encoding the most hydrophilic (KD < –0.5) with that of the most hydrophobic (KD > 0.5) proteins, comprising 78 and 86 ORFs, respectively. We found that there is a significant (as evaluated through a χ2 test) increment of five codons within the ‘hydrophilic group’: GTA (Val), CCA (Pro), AAG (Lys), CGT (Arg) and GGT (Gly). Of these amino acids, all but Val are hydrophilic. Interestingly, three of these codons (CCA, CGT and GGT) are among the triplets preferred by the highly expressed sequences (Table 1). On the other hand, nine codons corresponding to seven amino acids were found to be significantly more frequent among genes coding for hydrophobic proteins: TCC (Ser), CCC (Pro), ACC (Thr), GCC (Ala), AAA (Lys), GGG (Gly) and CGC, CGG and AGA (Arg). Among these, only AGA was detected as a more frequent codon in highly expressed sequences. Of these amino acids all but Ala are hydrophilic. To understand if these results are influenced by the strand-specific mutational biases, we compared the codon usage figures between the genes coding for hydrophilic and hydrophobic proteins only in the sequences located in the leading strand; and then the same was done for the genes placed in the lagging strand. The differences were evaluated with a χ2 test. We found that among the ‘hydrophilic group of codons’, GTA, CCA, CGT and GGT are independent of the mutational biases; and the same happens with TCC, CCC, ACC, GCC, GGG, CGC and CGG within the ‘hydrophobic group’. Therefore, we conclude that these groups of four and seven codons are indeed more frequent in the ORFs coding for hydrophilic and hydrophobic proteins, respectively.
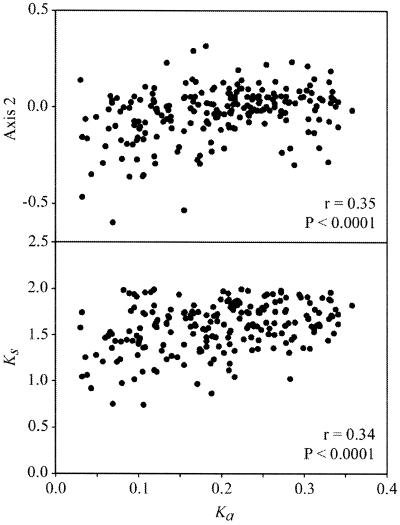
Other results that are important for understanding the diversity of factors shaping codon usage in C.trachomatis are the significant correlations found (i) between the Ka and the second axis of the COA, and (ii) between Ks and Ka (Fig. 3A and B). From these correlations two conclusions can be reached. First, in this species the second axis generated by the COA is influenced by features as diverse as expression levels, hydropathy and synonymous and non-synonymous substitution rates, which suggests that there may be a common factor underlying these features. Second, the correlation between Ks and Ka, which has been detected in both prokaryotes (30,31) and mammals (32–34), suggests a relationship between amino acid conservation with codon bias. The negative correlation that exists between Ka and FOP (r = –0.38, P < 0.0001), indicates that the genes which diverged less at the non-synonymous sites are the sequences which display the highest frequencies of optimal codons.
Figure 3.
Plots of the coordinate of each gene on the second axis generated by the COA of the RSCU values of each gene (A) and Ks (B) against Ka. The correlation coefficients (r) and level of significance (P) are shown.
We stress that a correlation between two quantities does not necessarily indicate a cause–effect relationship, it could be an indirect consequence of independent correlations with the same variable. The feasibility of both possibilities is discussed below.
DISCUSSION
The most accepted hypothesis for the unequal usage of synonymous codons among microorganisms states that it is the result of mutational biases and natural selection acting at the level of translation. The first factor is assumed to be (i) selectively neutral, and (ii) the main cause of the dominant bias, that can be either towards GC- or AT- ending triplets. Natural selection should act mainly on highly expressed sequences, and be the cause of the preferential usage of some translationally optimal codons. Since these two forces, although with different intensity and directions were detected in several systems, this hypothesis was accepted as a paradigm (35,36). However, several recent reports, analysing complete prokaryotic genomes, show that codon usage is a rather more complex trait. In this paper we present evidence suggesting that the pattern of synonymous codon choices in the bacterium C.trachomatis appears to be the result of a complex equilibrium between different forces, namely strand-specific mutational biases, natural selection probably acting at the three levels of the informational process (replication, transcription and translation), the hydropathy level of each protein and the level of amino acids conservation. Therefore, it seems important to understand if these results tackle the ‘paradigm’ or can be reduced to it. In other words, are the factors that we detected in C.trachomatis just pleiotropic effects of the mutational bias characteristic of this genome and of translational selection, or do they constitute truly new independent factors?
The discovery of the ‘GC skew’ (25) and its influence on codon usage (15) was not possible until the availability of complete genomes or very long contigs containing the origin of replication. Hence, a GC (or AT) bias was logically assumed to be the same for the two strands. Therefore, it seems important to discuss if this strand asymmetry is nothing but a more complex (but still selectively neutral) pattern of mutational biases. If it is neutral, then it becomes necessary to explain why almost all bacterial genomes, including species that diverged more than 2 billion years ago (this is probably the case for Gram+ and Gram– bacteria) still share the same pattern, namely towards G and T in the leading strands and towards A and C in the lagging strands (28). There are not trivial reasons why a given mutational bias, always in the same direction and independent of the genomic GC%, should be intrinsic to the enzymatic apparatus that replicates each strand. Although it may appear reasonable to argue that since in bacterial genomes there are usually more genes (specially highly expressed) located in the leading strand of replication, the mutations induced by transcription will be more frequent in that strand; but the issue is not with the different rates of mutations of each strand but with their conserved direction through the evolution of bacteria. Is it possible to speculate that natural selection may be the cause of this asymmetry? Although there are no obvious selective advantages for this almost universal feature, it is worth noting that theoretical approaches have suggested that a disparity in mutational biases can be advantageous at the population level (37). However, even if this is the case, the conserved direction of mutations remains unsolved. Related to this point is the asymmetrical distribution of sequences. It was suggested that this feature is the consequence of selection acting at the transcription–replication levels, since it should be convenient for an organism to maintain most of its genes on the leading strand for two reasons: (i) it reduces the probability of head-on collisions between the enzymes involved in the replicational and transcriptional processes, and (ii) transcription might not be aborted by the replication complex (38). However, the idea that selection at this level is the cause of a given codon usage pattern (15) is doubtful. Indeed, the consequence of selection acting at these levels is the asymmetry in the distribution of genes, and therefore codon usage may only be the ‘passive’ result of each strand-specific mutational pattern, whatever being their causes.
To understand why hydropathy affects codon usage is not simple. Two recent results deal with this problem. (i) Among prokaryotes it has been shown that there is a positive correlation between mean GC3 values and hydropathy, i.e., as long as prokaryotic genomes become GC- richest, its encoded amino acids (mean values) are more hydrophobic (39). (ii) In Mycobacterium species de Miranda et al. (17) have reported that in nearly all quartets there is a decrease in C- ending codons and an increase in G- ending triplets as long as hydrophobicity increases. The first result, although demonstrative of a whole genome relationship, since only deals with mean values is not indicative of an intragenomic variability in codon usage associated with hydropathy. The second paper describes an intragenomic variation in codon choices, and, as our own results, indicates that hydropathy does influence the final pattern. However, the changes in codon usage associated with the variation in hydropathy are not the same in the two species. The implications of these differences are not clear, although one explanation may be that there are not ‘universal codons’ associated with certain levels of hydropathy, as do exist for translational selection (among duets of the type NNY, the NNC codons seem to be optimal in many different organisms). Another possibility comes from the inspection of which are the preferred triplets in genes coding for hydrophilic and hydrophobic proteins in C.trachomatis. Among the former there are four significantly increased codons. Of these, three are at the same time ‘translationally optimal’, and the other (GTA) is incremented in highly expressed sequences, although not significantly. On the other hand, among genes coding for hydrophobic proteins there are seven significantly incremented triplets, and all of them are less frequent among highly expressed sequences. This might imply that the genes coding for hydrophobic proteins tend to prefer translationally non-optimal codons. This could make sense if the process of folding hydrophobic proteins (or hydrophilic regions within hydrophobic proteins) must be slower than in hydrophilic proteins. This may explain why the majority of significantly incremented triplets in genes coding for hydrophobic proteins code for hydrophilic amino acids. If this is the case, since translational ‘optimal’ codons may change in different organisms, the ‘non-optimal’ preferred triplets may change too. A similar analysis in several bacterial species should easily verify this hypothesis.
Another factor that we detected as shaping codon usage is the level of amino acids conservation of each protein. The significant correlations found between Ka, Ks and FOP suggest that there is a common factor underlying codon choices, synonymous and non-synonymous substitutions, amino acids conservation and frequency of optimal codons. Considering that Ka and Ks are positively correlated and that Ks and FOP are negatively correlated, we can infer that the negative correlation of Ka with FOP could be just a passive consequence of its correlation with Ks. But going further in unravelling this problem, is it possible to identify a common factor behind these correlations? The most obvious choice is the absolute concentration of tRNAs and of isoacceptors, as was suggested to be the case in Drosophila species (40). If this happens to be true, in Chlamydia, selection might be operating at two different levels during translation, namely speed and accuracy.
Up to this point we have discussed the several biological features that we detected as shaping codon usage in C.trachomatis; now we shall consider if they are independent among them. The second axis of the COA correlates with several features that, at first sight, may appear not necessarily linked as Ka, Ks, codon bias, expressivity, hydropathy and GC3. However, many of these factors can probably be unified, since it is well known that as long as expressivity increases there are higher constraints on the sequences, they diverge less and display stronger codon biases. The link with hydropathy may be caused by the fact that many of the highly expressed sequences are hydrophilic just because they accomplish their function in the aqueous media of the cell. The link with GC3, finally, is the most difficult to understand, since it is the result of at least three different factors: the overall mutational bias, which determines the global genomic GC%, the strand-specific mutational biases, and the influence of translationally optimal codons. The complexity of this issue is increased if we consider that the first factor might be selectively neutral: there are doubts about the neutrality of the second while the third is clearly the result of natural selection acting probably at different levels as speed, accuracy and hydropathy, and favouring, in each case, different codons.
Summarising, as long as more completed prokaryotic genomes are studied, different factors appear to shape the pattern of codon usage. This pattern is the result of biological processes (i.e. protein structure and folding, physiological constraints, translation, replication, transcription, mutation, etc.), and hence it becomes imperative to analyse codon usage under the light of this complexity. However, it is not still possible to say that the ‘mutational bias-translational selection’ paradigm is not enough to explain codon usage in bacteria, since as discussed above, all ‘new factors’, by the moment, can be explained in terms of this paradigm, although it is certainly becoming more complex.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Fernando Alvarez and two anonymous referees for their helpful comments and criticism. We also thank Ariel Chaparro for developing the software used in part of this work.
REFERENCES
- 1.King J. and Jukes,T. (1969) Science, 164, 788–798. [DOI] [PubMed] [Google Scholar]
- 2.Grantham R., Gautier,C., Gouy,M., Mercier,R. and Pavé,A. (1980) Nucleic Acids Res., 8, r49–r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gouy M. and Gautier,C. (1982) Nucleic Acids Res., 10, 7055–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikemura T. (1981) J. Mol. Biol., 151, 389–409. [DOI] [PubMed] [Google Scholar]
- 5.Ikemura T. (1982) J. Mol. Biol., 158, 573–597. [DOI] [PubMed] [Google Scholar]
- 6.Bennetzen J. and Hall,B. (1982) J. Biol. Chem., 257, 3026–3031. [PubMed] [Google Scholar]
- 7.Sharp P. and Li,W.-H. (1986) Nucleic Acids Res., 14, 7737–7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shields D. and Sharp,P. (1987) Nucleic Acids Res., 15, 8023–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohkubo S., Muto,A., Kawauchi,Y., Yamao,F. and Osawa,S. (1987) Mol. Gen. Genet., 210, 314–322. [DOI] [PubMed] [Google Scholar]
- 10.Andersson S. and Sharp,P. (1996) J. Mol. Evol., 42, 525–536. [DOI] [PubMed] [Google Scholar]
- 11.Andersson S., Zomorodipour,A., Andersson,J., Sicheritz-Ponten,T., Alsmark,U., Podowski,R., Naslund,A., Eriksson,A., Winkler,H. and Kurland,C. (1998) Nature, 396, 133–140. [DOI] [PubMed] [Google Scholar]
- 12.Ohama T., Muto,A. and Osawa,S. (1990) Nucleic Acids Res., 18, 1565–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McInerney J. (1997) Microb. Compar. Genomics, 2, 1–10. [Google Scholar]
- 14.Kerr A., Peden,J. and Sharp,P. (1997) Mol. Microbiol., 25, 1177–1179. [DOI] [PubMed] [Google Scholar]
- 15.McInerney J. (1998) Proc. Natl Acad. Sci. USA, 95, 10698–10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lafay B., Lloyd,A.T., McLean,M.J., Devine,K.M., Sharp,P.M. and Wolfe,K.H. (1999) Nucleic Acids Res., 27, 1642–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Miranda A., Alvarez-Valin,F., Jabbari,K., Degrave,W. and Bernardi,G. (1999) J. Mol. Evol., 50, 45–55. [DOI] [PubMed] [Google Scholar]
- 18.Stephens R., Kalman,S., Lammel,C., Fan,J., Marathe,R., Aravind,L., Mitchell,W., Olinger,L., Tatusov,R., Zhao,Q. et al. (1998) Science, 282, 754–759. [DOI] [PubMed] [Google Scholar]
- 19.Kalman S., Mitchell,W., Marathe,R., Lammel,C., Fan,J., Hyman,R.W., Olinger,L., Grimwood,J., Davis,R.W. and Stephens,R.S. (1999) Nature Genet., 21, 385–389. [DOI] [PubMed] [Google Scholar]
- 20.Greenacre M. (1984) Theory and Applications of Correspondence Analysis. Academic, London, UK.
- 21.Sharp P., Tuohy,T. and Mosurski,K. (1986) Nucleic Acids Res., 14, 5125–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W.-H. (1993) J. Mol. Evol., 36, 96–99. [DOI] [PubMed] [Google Scholar]
- 23.Comeron J. (1995) J. Mol. Evol., 41, 1152–1159. [DOI] [PubMed] [Google Scholar]
- 24.Thompson J., Higgins,D. and Gibson,J. (1994) Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobry J. (1996) Science, 272, 745–746. [DOI] [PubMed] [Google Scholar]
- 26.Kunst F., Ogasawara,N., Moszer,I., Albertini,A., Alloni,G., Azevedo,V., Bertero,M., Bessieres,P., Bolotin,A., Borchert,S. et al. (1997) Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 27.Blattner F., Plunkett,G., Bloch,C., Perna,N., Burland,V., Riley,M., Collado-Vides,J., Glasner,J., Rode,C., Mayhew,G. et al. (1997) Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 28.McLean M., Wolfe,K. and Devine,K. (1998) J. Mol. Evol., 47, 691–696. [DOI] [PubMed] [Google Scholar]
- 29.Francino M. and Ochman,H. (1997) Trends Genet., 13, 240–245. [DOI] [PubMed] [Google Scholar]
- 30.Sharp P. and Li,W.-H. (1987) Mol. Biol. Evol., 4, 222–230. [DOI] [PubMed] [Google Scholar]
- 31.Sharp P. and Li,W.-H. (1986) J. Mol. Evol., 24, 28–38. [DOI] [PubMed] [Google Scholar]
- 32.Graur D. (1985) J. Mol. Evol., 22, 53–62. [DOI] [PubMed] [Google Scholar]
- 33.Li W.-H., Wu,C.-I. and Luo,C.-C. (1985) Mol. Biol. Evol., 2, 150–174. [DOI] [PubMed] [Google Scholar]
- 34.Wolfe K.H. and Sharp,P. (1993) J. Mol. Evol., 37, 441–456. [DOI] [PubMed] [Google Scholar]
- 35.Sharp P. and Matassi,G. (1994) Curr. Opin. Genet. Dev., 4, 851–860. [DOI] [PubMed] [Google Scholar]
- 36.Sharp P., Averof,M., Lloyd,A., Matassi,G. and Peden,J. (1995) Phil. Trans. R. Soc. Lond. B., 349, 241–247. [DOI] [PubMed] [Google Scholar]
- 37.Wada K.-N., Doi,H., Tanaka,S.-I., Wada,Y. and Furusawa,M. (1993) Proc. Natl Acad. Sci. USA, 90, 11934–11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.French S. (1992) Science, 258, 1362–1365. [DOI] [PubMed] [Google Scholar]
- 39.D’Onofrio G., Jabbari,K., Musto,H., Alvarez-Valin,F., Cruveiller,S. and Bernardi,G. (1999) Ann. N. Y. Acad. Sci., 870, 81–94. [DOI] [PubMed] [Google Scholar]
- 40.Akashi H. (1994) Genetics, 136, 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]