ABSTRACT
The human immunodeficiency virus (HIV-1) envelope glycoprotein (Env) trimer mediates entry into host cells by binding receptors, CD4 and CCR5/CXCR4, and fusing the viral and cell membranes. In infected cells, cleavage of the gp160 Env precursor yields the mature Env trimer, with gp120 exterior and gp41 transmembrane Env subunits. Env cleavage stabilizes the State-1 conformation, which is the major target for broadly neutralizing antibodies, and decreases the spontaneous sampling of more open Env conformations that expose epitopes for poorly neutralizing antibodies. During HIV-1 entry into cells, CD4 binding drives the metastable Env from a pretriggered (State-1) conformation into more “open,” lower-energy states. Here, we report that changes in two dissimilar elements of the HIV-1 Env trimer, namely particular gp120 glycans and the gp41 fusion peptide-proximal region (FPPR), can independently modulate the stability of State 1. Individual deletion of several gp120 glycans destabilized State 1, whereas removal of a V1 glycan resulted in phenotypes indicative of a more stable pretriggered Env conformation. Likewise, some alterations of the gp41 FPPR decreased the level of spontaneous shedding of gp120 from the Env trimer and stabilized the pretriggered State-1 Env conformation. State-1-stabilizing changes were additive and could suppress the phenotypes associated with State-1-destabilizing alterations in Env. Our results support a model in which multiple protein and carbohydrate elements of the HIV-1 Env trimer additively contribute to the stability of the pretriggered (State-1) conformation. The Env modifications identified in this study will assist efforts to characterize the structure and immunogenicity of the metastable State-1 conformation.
IMPORTANCE
The elicitation of antibodies that neutralize multiple strains of HIV-1 is an elusive goal that has frustrated the development of an effective vaccine. The pretriggered shape of the HIV-1 envelope glycoprotein (Env) spike on the virus surface is the major target for such broadly neutralizing antibodies. The “closed” pretriggered Env shape resists the binding of most antibodies but is unstable and often assumes “open” shapes that elicit ineffective antibodies. We identified particular changes in both the protein and the sugar components of the Env trimer that stabilize the pretriggered shape. Combinations of these changes were even more effective at stabilizing the pretriggered Env than the individual changes. Stabilizing changes in Env could counteract the effect of Env changes that destabilize the pretriggered shape. Locking Env in its pretriggered shape will assist efforts to understand the Env spike on the virus and to incorporate this shape into vaccines.
KEYWORDS: virus entry, State-1 conformation, trimer, CD4-mimetic compound, transmembrane glycoprotein, cold sensitivity, glycosylation, glycan, mutant, stabilizing mutation
INTRODUCTION
Despite progress, an effective vaccine for human immunodeficiency virus (HIV-1) remains an unmet goal (1 – 5). A critical vaccine immunogen, the HIV-1 envelope glycoprotein (Env) trimer, mediates virus entry into cells and is the sole target for neutralizing antibodies (3 – 8). The HIV-1 Env trimer is a Class I viral fusion protein, composed of three gp120 and three gp41 subunits, which are non-covalently associated (6 – 8). In the infected cell, Env is synthesized in the rough endoplasmic reticulum, where signal peptide cleavage, trimerization, and high-mannose glycan addition occur (9 – 12). The resulting gp160 Env precursor traffics to the cell surface via two pathways (13). In the canonical secretory pathway, Env is transported through the Golgi compartment, where it is cleaved into gp120 and gp41 subunits and further decorated by complex N-linked glycans (9 – 13). This mature (cleaved) Env trimer is selectively incorporated into budding virions. In an alternate pathway, uncleaved gp160 bypasses the Golgi apparatus to reach the cell surface, but this immature Env is largely excluded from virions (13).
Based on single-molecule fluorescence resonance energy transfer (smFRET) studies, the flexible HIV-1 Env protomers in the virion trimer spontaneously sample at least three conformations (States 1–3) (14). During the process of virus entry, the pretriggered (State-1) Env sequentially engages the receptors, CD4 and CCR5/CXCR4 (6, 7, 15 – 17), triggering a cascade of conformational changes in the metastable, high-potential-energy Env trimer. Initially, CD4 binding transforms the State-1 Env to a default intermediate conformation (State 2) and then to the full CD4-bound State 3 (14). Upon binding the CCR5 or CXCR4 coreceptor, the State-3 prehairpin intermediate undergoes rearrangements in gp41 (18 – 20). The hydrophobic fusion peptide at the N-terminus of gp41 is translocated close to and inserts into the target cell membrane. Further rearrangements of the gp41 ectodomain lead to the formation of an energetically favorable six-helix bundle (21 – 23). This process drives the fusion of the viral and target cell membranes and allows virus entry (24, 25).
HIV-1 is a persistent virus and has evolved multiple mechanisms by which Env avoids host antibodies: Env diversity among HIV-1 strains, extensive glycosylation, and conformational lability (26 – 30). The vast majority of antibodies elicited against Env during natural HIV-1 infection fail to neutralize primary viruses (31 – 33). These poorly neutralizing antibodies (pNAbs) are directed against non-functional Envs: uncleaved gp160, which is conformationally flexible; shed gp120 and the resulting gp41 “stumps”; and Env trimers that spontaneously sample more open State 2/3 conformations (31 – 37). As pNAbs do not efficiently recognize the cleaved pretriggered (State-1) Env, they do not inhibit virus infection. After several years of HIV-1 infection, ~10% of infected individuals generate broadly neutralizing antibodies (bNAbs) (38 – 42). Most bNAbs show a preference for binding State 1, often by evolving distinctive features that allow them to bind glycans and/or access recessed, conserved protein epitopes on the trimer (3 – 5, 14, 26 – 28, 43 – 46).
A successful HIV-1 vaccine likely needs to elicit bNAbs that recognize the pretriggered (State-1) Env and therefore should include immunogens that closely mimic this conformation. Soluble stabilized (sgp140 SOSIP.664) Env trimers have been designed that retain known bNAb epitopes (47 – 50). Structures of these sgp140 SOSIP.664 Env trimers complexed with bNAbs have provided detailed information on the targeted epitopes (29, 30, 46, 51 – 56). Although many strategies to stabilize these soluble Env trimers have been employed, bNAbs have not been consistently elicited in animals or humans (30, 57 – 66). Studies have suggested that differences in conformation and glycosylation exist between sgp140 SOSIP.664 Env trimers and the State-1 Env in membranes (47, 67 – 77), which could influence immunogen efficacy.
Efforts to design more accurate mimics of the functional State-1 Env trimer would benefit from detailed structural information. Stabilizing and enriching the pretriggered (State-1) conformation of membrane Env trimers could assist efforts to characterize the structure of this metastable state and evaluate its immunogenicity. Env changes that increase the resistance of membrane HIV-1 Env trimers to denaturation or functional inactivation by exposure to heat or cold have been reported (78 – 90). In several cases, these increases in Env trimer stability were associated with decreases in triggerability by soluble CD4 (sCD4) or CD4-mimetic compounds (CD4mcs), phenotypes indicative of State-1 stabilization (82 – 93). Some of the Env changes that stabilize the pretriggered membrane Env trimer are located near the trimer axis of structurally well-characterized sgp140 SOSIP.664 Envs (85 – 88). However, stabilizing alterations are not always interchangeable between membrane Envs and sgp140 SOSIP.664 trimers, possibly due to differences in conformation or post-translational modifications between these Envs (47, 65, 67 – 77). Therefore, for the present, efforts to stabilize the pretriggered (State-1) conformation of the membrane Env must proceed empirically.
Here, we present evidence that alterations in either the carbohydrate or the protein components of the HIV-1 Env trimer can modulate the stability of the pretriggered (State-1) Env conformation. We identify changes in specific gp120 glycans (Fig. 1A) or amino acid residues in the gp41 FPPR (Fig. 1B) that increase or decrease the stability of the pretriggered state. In some cases, the State-1-stabilizing phenotypes are additive. These State-1-stabilizing changes in Env can also compensate for Env changes that destabilize the pretriggered conformation. Our results provide insights into the mechanisms by which the functional HIV-1 Env trimer maintains the pretriggered (State-1) conformation, and identify State-1-stabilizing Env changes potentially useful for structural and immunogenicity studies.
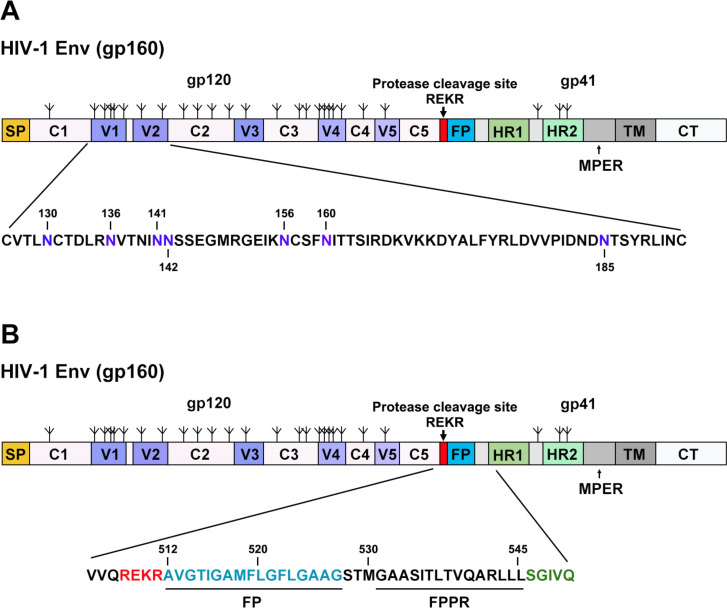
Fig 1.
HIV-1AD8 Env regions altered in this study. Schematic representations of the HIV-1AD8 Env are shown, with gp120 and gp41 subunits designated. The proteolytic cleavage site [(508)REKR(511)] between gp120 and gp41 is colored red. The gp120 variable regions (V1–V5) and conserved regions (C1–C5) are indicated. The following elements are labeled: SP, signal peptide; FP, fusion peptide; HR1 and HR2, heptad repeat regions 1 and 2; MPER, membrane-proximal external region; TM, transmembrane region; CT, cytoplasmic tail. The N-linked glycosylation sites are depicted above the Env diagram. The sequences in V1 and V2 (A) and the Env region surrounding the gp120-gp41 cleavage site (B) are shown. The HIV-1AD8 Env amino acids are numbered according to standard nomenclature, referring to the prototypic HXB2 sequence (94). In A, the N-linked glycosylation sites in the V1/V2 region, several of which are modified in this study, are colored purple, and are numbered.
RESULTS
Effect of gp120 V1/V2 insertions on HIV-1 Env conformational state
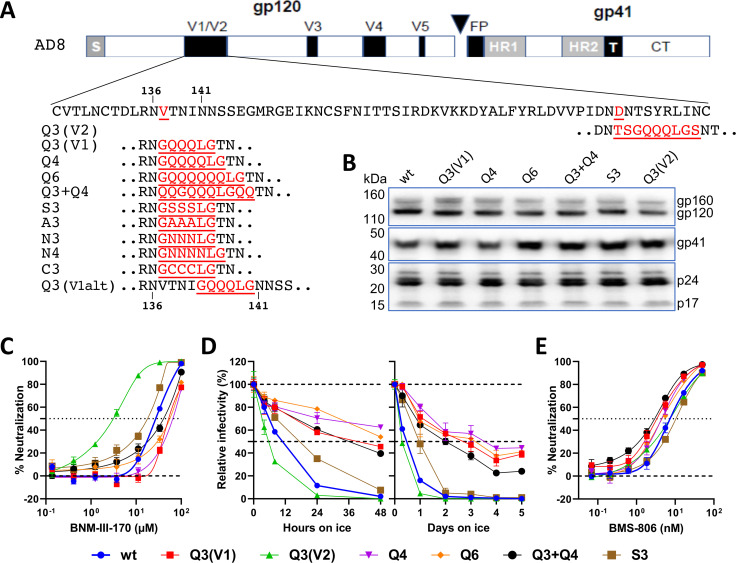
The introduction of sequence tags into HIV-1 Envs has allowed smFRET labeling and monitoring of Env conformational states (14, 76, 95). During the course of our studies of Env conformation, we introduced the Q3 tag into HIV-1AD8 gp120 V1 and V2 regions that tolerate large insertions in natural HIV-1 variants (96) (Fig. 2A). Compared with the wild-type (wt) HIV-1AD8 Env, the Q3(V1) and Q3(V2) Envs were processed and incorporated into virions efficiently (Fig. 2B). The Q3(V1) and Q3(V2) Envs supported virus infection only slightly less efficiently than the wt Env (Table 1).
Fig 2.
Phenotypes of V1/V2 insertion mutants. (A) The sequences of the HIV-1AD8 V1/V2 insertion mutants are shown. The gp120-gp41 cleavage site is designated with a triangle. S, signal peptide; FP, fusion peptide; HR1 and HR2, heptad repeat regions 1 and 2; T, transmembrane region; CT, cytoplasmic tail. The Asn 136 and Asn 141 glycosylation sites are shown. (B) Pseudovirus particles were prepared from transfected 293T cells, lysed, and analyzed by western blotting. The gp160, gp120, and gp41 Envs and the p24 CA (capsid) and p17 MA (matrix) proteins are shown. (C) The inhibition of the infectivity of recombinant viruses pseudotyped by the indicated wild-type (wt) or mutant HIV-1AD8 Envs by the CD4mc BNM-III-170 is shown. (D) Recombinant viruses with wt or mutant Envs were incubated at 0°C for up to 2 days (left panel) or 5 days (right panel), and their infectivity on TZM-bl cells was measured. The infectivity relative to a control virus not incubated at 0°C is reported. (E) The inhibition of the infectivity of recombinant viruses with the wt or mutant Envs by BMS-806 is shown. In C–E, the means and SDs of triplicate measurements are shown. The experiments were repeated with comparable results.
TABLE 1.
Phenotypes of HIV-1AD8 Env variants with V1/V2 insertions
| Env | Virion Env a | Infectivity b | Resistance/sensitivity compared to wt AD8 Env c | |||
|---|---|---|---|---|---|---|
| Cold | BNM-III-170 | 19b | 17b | |||
| wt AD8 | +++ | +++ | ● | ● | ● | ● |
| Q3(V1) | +++ | ++ | RR | R | ● | ● |
| Q3(V2) | +++ | ++ | S | S | ||
| Q4 | +++ | ++ | RR | R | ||
| Q6 | +++ | ++ | RR | R | ||
| Q3 + Q4 | +++ | ++ | RR | Slight R | ||
| S3 | +++ | +++ | ● | ● or slight S | ||
| A3 | +++ | RR | ||||
| N3 | + | R | ||||
| N4 | +++ | RR | ||||
| C3 | – | ND | ||||
| Q3(V1alt) | +++ | S | S | |||
The levels of Env on virus particles produced by transfection of 293T cells were estimated by pelleting and lysing the virus particles, followed by western blot analysis. +++, 75%–125% wt level; ++, 25%–74% wt level; +, 5%–24% wt level; –, <5% wt level.
The infectivity of recombinant viruses pseudotyped by the indicated Env was measured on TZM-bl target cells. +++, 75%–125% wt infectivity; ++, 25%–74% wt infectivity; +, 5%–24% wt infectivity; –, <5% wt infectivity.
The sensitivity/resistance of recombinant viruses pseudotyped by the indicated Env to cold exposure (0°C), the CD4mc BNM-III-170, and the 19b and 17b pNAbs, relative to that of the wt HIV-1AD8, is reported. ●, wt level; R indicates resistance relative to the wt virus; S indicates sensitivity relative to the wt virus. An increase in the number of R or S symbols indicates a greater level of resistance or sensitivity, respectively, compared to the wild-type virus level. The values in the table that were not determined are designated ND or are left blank. The data shown are representative of results obtained in at least two independent experiments.
The Q3(V1) and Q3(V2) Envs exhibited opposite phenotypes related to Env conformational state. The Q3(V1) Env was more resistant to a CD4mc, BNM-III-170, and exposure to cold (0°C) temperature than wt Env (Fig. 2C and D; Table 1). Viruses with the Q3(V1) and wt Envs were neutralized equivalently by the PGT145 and PGT151 bNAbs and were equally resistant to inhibition by the 19b and 17b pNAbs (Table 1 and data not shown). Thus, the Q3(V1) Env exhibits phenotypes associated with a relative stabilization of the pretriggered (State-1) conformation (87 – 92). By contrast, the Q3(V2) Env was more sensitive than the wt Env to BNM-III-170 and cold, phenotypes indicative of destabilization of State 1 (87 – 92).
As the known HIV-1 alterations that stabilize State 1 are fewer than State-1-destabilizing Env changes, we sought to understand the basis of the Q3(V1) Env phenotypes. We varied the length and specific amino acid residues in the V1 insert (Fig. 2A). With one exception, the S3 Env, the Env mutants with various V1 inserts were similar to the Q3(V1) Env with respect to resistance to BNM-III-170 and cold (Fig. 2C and D; Table 1). The sensitivities of the S3 Env to BNM-III-170 and cold were closer to those of the wt HIV-1AD8 Env (Fig. 2C and D; Table 1). The V1 insertions exerted minimal effects on virus sensitivity to BMS-806 (Fig. 2E).
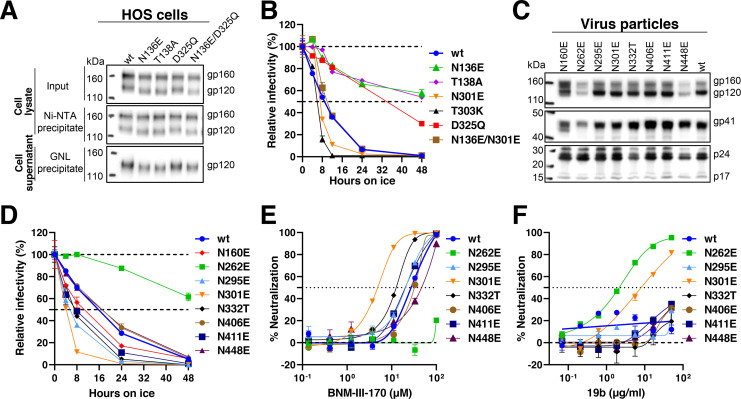
We noticed that the serine insertions in the S3 Env restored the Asn-X-(Thr/Ser) sequon for the Asn 136 glycosylation site that was altered in Q3(V1) and the other V1 insertion mutants. To test whether the absence of the carbohydrate chain modifying Asn 136 resulted in the observed cold- and CD4mc-resistant phenotypes, we studied the N136E and the T138A HIV-1AD8 Env mutants, both of which lack this sequon. The faster migration of the N136E and T138A gp120 and gp160 bands on SDS-polyacrylamide gels relative to that of the corresponding wt HIV-1AD8 Envs (Fig. 3A) indicates that Asn 136 on the wt Env is modified by carbohydrate. The N136E and the T138A Env mutants lacking the Asn 136 glycan both exhibited cold-resistant phenotypes similar to those of Q3(V1) (Fig. 3B). The sensitivity of the N136E and T138A mutants to BNM-III-170 was similar to that of the wt HIV-1AD8 Env (Table 2). Thus, the loss of the glycan at Asn 136 in the gp120 V1 region results in cold-resistant phenotypes comparable to those associated with the V1 insertions.
Fig 3.
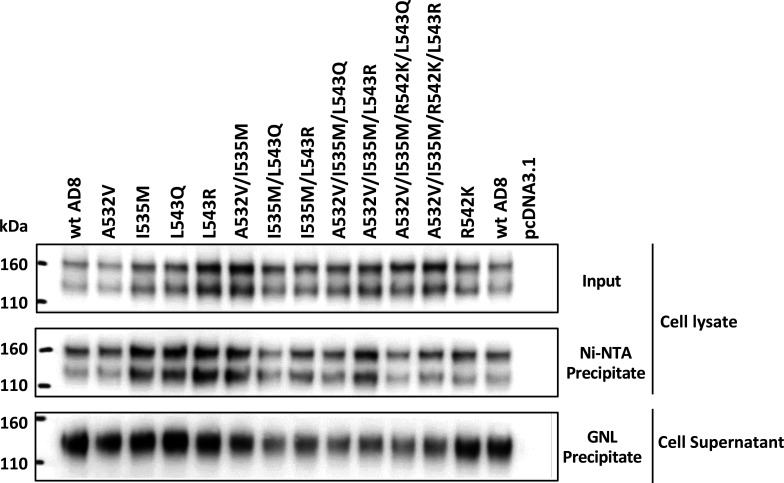
Phenotypes of Env glycosylation mutants. (A) The processing and subunit association of the wt and indicated HIV-1AD8 Env mutants in transfected HOS cells were evaluated. The two upper panels show the cell lysates, and the lower panel shows the cell supernatants. The upper panel shows the total input Envs in the cell lysates. The middle panel shows the Env proteins precipitated by Ni-NTA beads, using the His6 tags at the carboxyl terminus of the gp41 glycoprotein. The lower panel shows the gp120 in the cell supernatants, which was precipitated with Galanthus nivalis lectin (GNL)-beads. (B and D) Recombinant viruses with the indicated wt or mutant Envs were incubated at 0°C for the indicated times, and their infectivity on TZM-bl cells was measured. The infectivity relative to a control virus not incubated at 0°C is reported. (C) Virus particles were prepared from transfected 293T cells, lysed, and analyzed by western blotting. (E and F) Recombinant viruses pseudotyped with the indicated wt or mutant Envs were incubated with the indicated concentrations of BNM-III-170 (E) or the 19b pNAb (F) for 1 h at 37°C. The virus preparations were then incubated with TZM-bl cells, and the infectivity was measured 48 h later. In B and D–F, the means and SDs of triplicate measurements are shown. The experiments were repeated with comparable results.
TABLE 2.
Phenotypes of HIV-1AD8 Envs with altered sites of N-linked glycosylation
| Env | Virion Env a | Infectivity b | Resistance/sensitivity compared to wt AD8 Env c | ||
|---|---|---|---|---|---|
| Cold | BNM-III-170 | 19b | |||
| N130E | +++ | S | S | ||
| N136E | +++ | RR | ● | ||
| T138A | +++ | RR | ● | ||
| S143D | +++ | R | ● | ||
| S158E | – | ND | ND | ||
| N160E | d | + | ● | ||
| N262E | + | + | RR | RR | SS |
| N295E | +++ | +++ | S | ● | ● |
| N301E | +++ | ++ | SS | SS | S |
| T303K | ++ | SS | |||
| D325Q | +++ | R | ● | ||
| R327Q | +++ | ||||
| N332T | +++ | ++ | S | S | |
| N406E | +++ | +++ | ● | ● | ● |
| N411E | +++ | ++ | S | ● | ● |
| N448E | + | +++ | ● | ● | ● |
| N136E/D325Q | +++ | RR | R | ||
| N130E/T138A | +++ | R | ● | ||
| T138A/S143E | ++ | R | S | ||
| T138A/S158E | – | ND | ND | ||
| Q3(V1)/N301E | ++ | ||||
| Q3(V1)/T303K | ++ | ||||
| N136E/N301E | ● | ||||
The levels of Env on virus particles produced by transfection of 293T cells were estimated by pelleting and lysing the virus particles, followed by western blot analysis. +++, 75%–125% wt level; ++, 25%–74% wt level; +, 5%–24% wt level; –, <5% wt level.
The infectivity of recombinant viruses pseudotyped by the indicated Env was measured on TZM-bl target cells. +++, 75%–125% wt infectivity; ++, 25%–74% wt infectivity; +, 5%–24% wt infectivity; –, <5% wt infectivity.
The sensitivity/resistance of recombinant viruses pseudotyped by the indicated Env to cold exposure (0°C), the CD4mc BNM-III-170, and the 19b and 17b pNAbs, relative to that of the wt HIV-1AD8, is reported. ●, wt level; R indicates resistance relative to the wt virus; S indicates sensitivity relative to the wt virus. An increase in the number of R or S symbols indicates a greater level of resistance or sensitivity, respectively, compared to the wild-type virus level. The values in the table that were not determined are designated ND or are left blank. The data shown are representative of results obtained in at least two independent experiments.
The virion Envs for the N160E mutant exhibit decreases in the ratio of mature gp120 to uncleaved (gp160) Env.
Effect of removal of other gp120 N-linked glycans on Env conformation
To evaluate whether the removal of gp120 N-linked glycans other than the Asn 136 glycan might also influence the conformational state of the functional Env trimer, we examined the cold sensitivity of viruses with HIV-1AD8 Envs missing particular glycans in the general vicinity of the V1 loop (Fig. 3; Table 2). These Env mutants were processed and incorporated into virions efficiently, with the exception of the slowly processed N160E mutant and the poorly incorporated N262E and N448E mutants (Fig. 3C). Several Env mutants (N295E, N301E, N332T, and N411E) were more sensitive than the wt HIV-1AD8 to cold inactivation (Fig. 3D). The most cold-sensitive Env mutant, N301E, was also more sensitive than wt Env to inhibition by BNM-III-170 and neutralization by the 19b pNAb (Fig. 3E and F; Table 2). The T303K Env mutant exhibited phenotypes similar to those of the N301E mutant, supporting the conclusion that loss of the N-linked glycan at Asn 301 destabilizes the pretriggered (State-1) conformation (Table 2). Our results suggest that some gp120 glycans, particularly those modifying Asn 301 and Asn 332, contribute to the maintenance of the State-1 Env conformation.
Phenotypes of Env combination mutants
We asked whether the State-1-stabilizing phenotype associated with the removal of the Asn 136 glycan depended on the presence of other glycans in the vicinity of residue 136. The identification of candidate glycans is complicated by the lack of a detailed State-1 Env structure and by the existence of smFRET data raising the possibility that the gp120 V1 region undergoes substantial shifts in position as a result of the transition from State 1 to State 2 (14, 76). Cognizant of the potential differences from State 1, we evaluated currently available HIV-1 Env trimer structures and identified three gp120 glycans at Asn 130, Asn 141, and Asn 156 that might be spatially proximal to Asn 136. We measured the sensitivity to cold and BNM-III-170 of mutant viruses with these glycans removed, individually or in combination with the removal of the Asn 136 glycan (Table 2). Viruses with the N130E change, which eliminates the Asn 130 N-linked glycan, infected target cells efficiently and were more sensitive to cold and BNM-III-170 than viruses with the wt HIV-1AD8 Env. These phenotypes are consistent with mild destabilization of State 1. Compared with the wt HIV-1AD8 Env, the N130E/T138A Env mutant was more cold resistant but was inhibited by BNM-III-170 equivalently. Thus, the loss of the glycan at Asn 136 can stabilize the State-1 conformation even when Asn 130 is not glycosylated and can compensate for the mild State 1-destabilizing effects of the loss of the Asn 130 glycan. More complex relationships between the Asn 136 and Asn 141 glycans were observed (Table 2). The S143D change, which results in the loss of Asn 141 glycosylation, resulted in mild resistance to cold relative to the wt HIV-1AD8 Env. Based on the phenotypes of the T138A/S143D mutant, the loss of the Asn 136 glycan exhibited little or no State-1-stabilizing effects in the absence of the Asn 141 glycan. Thus, the phenotypic effects of glycan removal at Asn 136 can be influenced by the presence or absence of the adjacent V1 glycan at Asn 141. The S158E change, which results in the removal of the Asn 156 glycan, did not support virus infection and was therefore not evaluated further.
During the course of our study of HIV-1 Env variants, we found that compared with the wt HIV-1AD8 Env, the D325Q mutant was cold resistant (Fig. 3B). To evaluate whether the D325Q change would contribute additively to the cold resistance of Envs lacking the glycan at Asn 136, we tested the N136E/D325Q and Q4/D325Q mutants. These combination mutants exhibited a level of cold resistance greater than or equal to that of either of the parental Env mutants (Fig. 4A; Table 2). Alteration of the nearby Arg 327 residue did not significantly alter the cold resistance of the Q4 Env (Fig. 4A). The N136E/D325Q mutant was also more resistant to BNM-III-170 than either the N136E or D325Q mutants (Table 2). These results indicate that some of the State-1-stabilizing phenotypes associated with the removal of the Asn 136 glycan and the D325Q change are additive.
Fig 4.
Viral phenotypes of Envs with combinations of glycosylation changes. (A–D) Recombinant viruses pseudotyped with the indicated Env variants were incubated at 0°C for the period of time shown, and their infectivity on TZM-bl cells was measured. The infectivity relative to a control virus not incubated at 0°C is reported. (E and F) Recombinant viruses pseudotyped with the indicated Envs were incubated with BNM-III-170 (E) or BMS-806 (F) for 1 h at 37°C. Viruses were then incubated with TZM-bl cells for 48 h, and the infectivity was measured. The means and SDs of triplicate measurements are reported. The experiments were repeated with comparable results.
The results above suggest that the loss of the Asn 136 glycan can decrease the mild State 1-destabilizing effects of the removal of the adjacent glycan at Asn 130. To evaluate whether the State-1-stabilizing phenotypes associated with the removal of the Asn 136 glycan could compensate for State-1 destabilization resulting from the loss of other glycans, we evaluated the phenotypes of mutants with either Q3(V1) or N136E changes combined with the destabilizing changes N295E, N301E, N332T, or N411E. In each case, the Envs with State-1-destabilizing changes exhibited increased resistance to cold inactivation as a result of the Q3(V1) or N136E changes (Fig. 3B and 4B through D; Table 2). The addition of the Q3(V1) change also rendered the N332T Env more resistant to BNM-III-170 (Fig. 4E). These changes individually or in combination minimally affected sensitivity to BMS-806 (Fig. 4F). These results indicate that State-1 stabilization resulting from V1 changes including the loss of the Asn 136 glycan can reduce the State-1-destabilizing effects of the removal of other gp120 glycans.
Mechanisms of altered cold sensitivity of Env glycan mutants
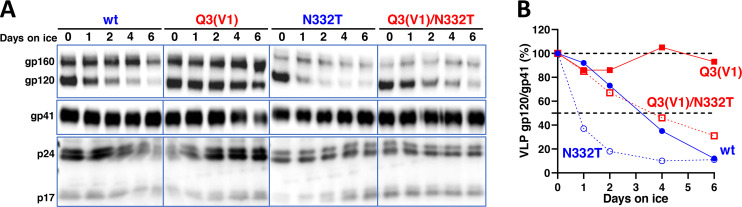
The cold inactivation of multimeric protein complexes often results from the damaging effects of ice formation in intersubunit or interprotomer interfaces (97 – 99). We examined the effect of the individual and combined Q3(V1) and N332T changes on the integrity of Env trimers on the surface of virus particles. After 6 days of incubation on ice, nearly all of the gp120 glycoprotein was lost from virus particles with the wt HIV-1AD8 Env (Fig. 5). Virus particles with the N332T Env shed gp120 even faster than virus particles with the wt HIV-1AD8 Env. By contrast, the Q3(V1) Env on virus particles was relatively stable during the 6-day incubation on ice. The stability of the Q3(V1)/N332T Env on virus particles incubated at 0°C was similar to that of the wt Env. Thus, the integrity of the viral Env trimers correlated with Env functional stability after cold exposure. The results indicate that the State-1-stabilizing change in the gp120 V1 region and the State-1-destabilizing removal of the Asn 332 glycan can exert their phenotypes when combined in a single Env.
Fig 5.
Stability of viral Envs on ice. Virus particles with the indicated Envs were prepared from the supernatants of transfected 293T cells and incubated at 0°C for the period of time shown. The viruses were then pelleted, lysed, and analyzed by western blotting (A). The gp120/gp41 ratio (%) on the virus particles at each time point relative to that on a control virus not incubated at 0°C is reported (B). The results of a typical experiment are shown. The experiment was repeated with comparable results.
Phenotypes of HIV-1 Env mutants with changes near the gp120-gp41 cleavage site
The flexible gp160 Env precursor samples multiple conformations; proteolytic maturation in the Golgi stabilizes State 1, decreasing the spontaneous exposure of pNAb epitopes (34 – 36, 45, 90, 95). To understand better the contribution of Env cleavage to the maintenance of a pretriggered (State-1) Env conformation, we studied the phenotypes of a panel of HIV-1AD8 Env mutants with changes in gp120 and gp41 amino acid residues near the cleavage site. Although a few gp120 residues were studied, most of the changes involved the gp41 fusion peptide, FPPR, and the N-terminal portion of the heptad repeat (HR1) region (HR1N; Fig. 1B). As these regions contribute to Env function (100 – 103), which we wished to preserve to allow more thorough evaluation of Env conformational states in viral assays, we employed a conservative mutagenesis strategy. The changes introduced were based either on natural polymorphisms in HIV-1 strains or on variants selected for efficient replication from populations of mutagenized viruses (96, 104). As we anticipated that some of the phenotypes relevant to State-1 stabilization/destabilization could be subtle in this panel of conservatively altered Env mutants, the phenotypes were evaluated quantitatively in different contexts. Env alterations with interesting and consistent phenotypes were studied in combination.
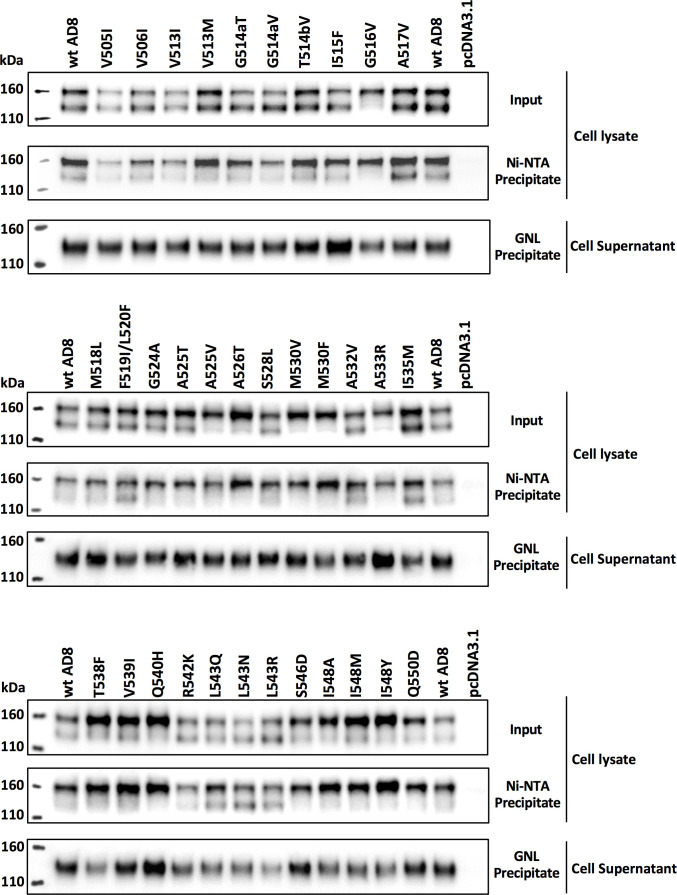
We evaluated the processing and subunit association of the Env mutants in human osteosarcoma (HOS) cells, in which gp120-gp41 cleavage is efficient (Fig. 6 and 7). Compared with the wt HIV-1AD8 Env, several mutants with changes in the FPPR (I535M, T538F, V539I, R542K, L543Q, L543N, L543R, and I548M) exhibited stronger subunit association, as measured by the ratio of cell-associated:shed gp120. With the exception of the T538F mutant, all of these Env mutants with strong subunit association were processed efficiently, in some cases significantly more efficiently than wt Env. We also examined the strength of the gp120 association with the Env trimer solubilized in NP-40 detergent solutions; in this assay, Ni-NTA beads precipitate the solubilized Env by capturing the His6 tags at the gp41 carboxyl terminus (45, 88). When the precipitated gp120/gp160 ratio was normalized to the gp120/gp160 ratio in the input lysate, the Envs with stronger subunit association in the cell membrane did not generally exhibit significant increases in the Ni-NTA precipitation assay relative to wt Env (Fig. 7). Thus, some Envs with alterations in the gp41 FPPR exhibit stronger subunit association when anchored in the cell membrane, but this phenotype is less evident when the Envs are solubilized in detergent. None of the Env mutants with alterations outside the FPPR exhibited this interesting combination of phenotypes.
Fig 6.
Effects of HIV-1AD8 Env FPPR changes on processing, subunit association, and gp120-trimer association. HOS cells transiently expressing the wt HIV-1AD8 Env and the indicated mutants with C-terminal His6 tags were harvested 72 h after transfection. The cells were lysed, and fractions of the cell lysates were reserved as Input samples. The remainder of the cell lysates was incubated at room temperature for 1.5 h with Ni-NTA beads. The cell supernatants were incubated at room temperature for 1.5 h with GNL beads. Total cell lysates (Input), Ni-NTA precipitates, and GNL precipitates were western blotted with a goat anti-gp120 antibody. HOS cells transfected with the empty pcDNA3.1 vector serve as a negative control. The GNL precipitates from the cell supernatants reflect the level of gp120 shed from the cell-associated Env and were used to calculate subunit association. The Ni-NTA precipitates from the cell lysates measure the relative levels of gp160 and gp120 precipitated in the presence of NP-40 detergent and were used to calculate gp120-trimer association. The results of a typical experiment are shown.
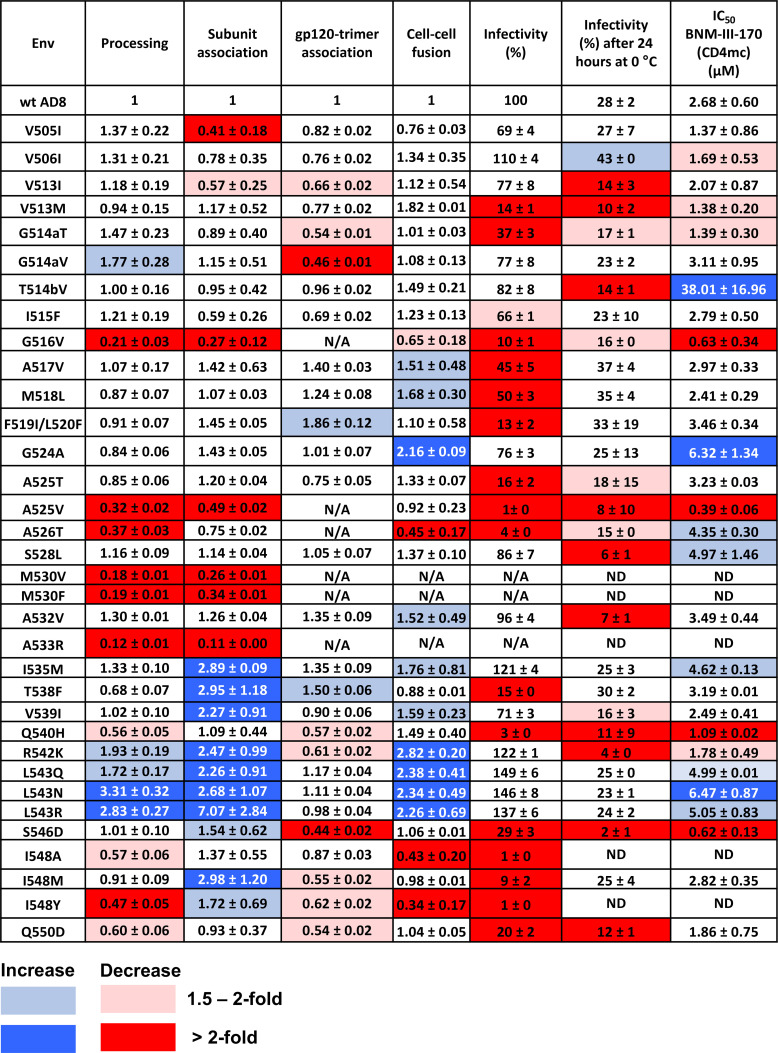
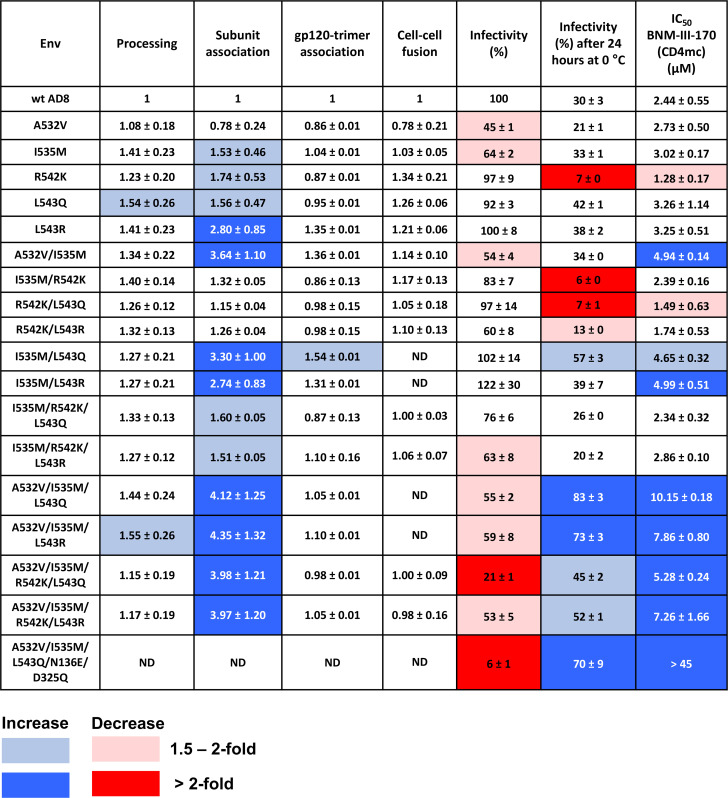
Fig 7.
Phenotypes of HIV-1AD8 Env variants with changes near the gp120-gp41 cleavage site. Env processing, subunit association, gp120-trimer association, cell-cell fusion, and infectivity are shown for each HIV-1AD8 Env mutant, normalized to those values observed for the wt HIV-1AD8 Env. Cold sensitivity represents the pseudovirus infectivity after 24 h of incubation at 0°C, relative to that of the untreated virus. The 50% inhibitory concentrations (IC50 values) of the CD4-mimetic compound, BNM-III-170, are reported in µM. N/A, not available; ND, not determined. Values are colored according to the key, based on their fold increase or decrease compared to those of the wt HIV-1AD8 Env. The results shown are the means and SDs derived from at least two independent experiments.
The function of the Env mutants was assessed by evaluating their ability to mediate cell-cell fusion and pseudovirus infectivity (Fig. 7). Except for three Env mutants (M530V, M530F, and A533R) with severe defects in gp160 processing, all the mutants supported some level of cell-cell fusion. The capacity of several Env mutants to fuse cells was preserved better than their ability to support virus entry. This relationship has been previously observed with HIV-1 Env mutants and indicates that the function of virion Envs is more labile than that of the newly synthesized cell-surface Envs (87, 105). Of note, several of the Env mutants (I535M, R542K, L543Q, L543N, and L543R) with increases in subunit association mediated cell-cell fusion and pseudovirus entry more efficiently than the wt Env. These observations imply that the effects of these FPPR changes on Env trimer stability apply to functional Envs on the cell surface and virion.
The sensitivity of the Env mutant pseudotypes to inhibition by a CD4mc, BNM-III-170, and to incubation at 0°C was examined (Fig. 7). Compared with wt HIV-1AD8, the T514bV, G524A, and L543N mutants were significantly more resistant to BNM-III-170. Lower levels of resistance to BNM-III-170 were observed for the A526T, S528L, I535M, L543Q, and L543R Envs. The R542K mutant was more sensitive to BNM-III-170 than the wt Env. Therefore, with the exception of this R542K mutant, all the FPPR Envs with increases in subunit association also exhibited a higher level of resistance to the CD4mc. Viruses with the wt Env and all of the replication-competent Env mutants in our panel lost more than half of their infectivity after 24 h of incubation on ice (Fig. 7). In summary, our screen identified functional Env variants with gp41 FPPR changes that exhibit tighter association of the Env subunits and decreased triggerability by a CD4mc. This combination of phenotypes was not observed for the Env mutants with changes in the gp41 fusion peptide.
In addition to the R542K mutant, three other mutants (V513M, G514aT, and S546D) exhibited phenotypes indicative of increased triggerability, with increased sensitivity to cold inactivation and to inhibition by BNM-III-170 (Fig. 7). These residues are located in the fusion peptide (Val 513 and Gly 514a), FPPR (Arg 542), and at the junction of the FPPR and HR1 regions of gp41 (Ser 546). Thus, particular changes in Env residues located throughout the gp41 N-terminal regions can apparently destabilize State 1.
Phenotypes of Env mutants with combinations of changes in the gp41 FPPR
As several individual amino acid changes in the gp41 FPPR resulted in phenotypes that potentially reflect modest increases in Env stability, we examined the phenotypes of Envs with combinations of these changes (Fig. 8 and 9). We also included the A532V change in these combinations. The processing, subunit association, gp120-trimer association, and functional assays were performed in parallel to allow the ranking of the Env mutants. Combinations of three Env changes resulted in more pronounced phenotypes than the single or double changes, suggesting the additivity of the Env alterations. Notably, the combination mutants (A532V/I535M/L543Q, A532V/I535M/L543N, and A532V/I535M/L543R) with the highest levels of subunit association and BNM-III-170 resistance also exhibited the best ability to withstand incubation on ice. Therefore, these functional Envs exhibit strong phenotypes indicative of State-1 stabilization (87 – 92). The A532V/I535M/L543Q and A532V/I535M/L543N mutants exhibited phenotypes suggestive of nearly equivalent degrees of State-1 stabilization; both levels of State-1 stability appeared to be greater than that of the A532V/I535M/L543R mutant (Fig. 9 and data not shown). The slight decrease in virus infectivity observed for the A532V/I535M/L543Q, A532V/I535M/L543N, and A532V/I535M/L543R pseudotypes may be a consequence of their lower triggerability.
Fig 8.
Effects of combinations of FPPR changes on HIV-1AD8 Env processing, subunit association, and gp120-trimer association. HOS cells transiently expressing the indicated wt and mutant HIV-1AD8 Env variants were processed as described in the Fig. 6 legend. The results of a typical experiment are shown.
Fig 9.
Phenotypes of HIV-1AD8 Env FPPR combination mutants. The phenotypes of Envs with combined changes and selected Env mutants with individual amino acid changes in the FPPR were compared to those of the wild-type HIV-1AD8 Env. Values are colored according to the key, based on their fold increase or decrease compared to those of the wt HIV-1AD8 Env. The results shown are the means and SDs derived from two independent experiments.
Although the phenotype of the individual A532V Env mutant differed minimally from that of wt Env, the inclusion of the A532V change in combination with other FPPR changes (A532V/I535M, A532V/I535M/L543Q, and A532V/I535M/L543R) increased Env subunit association and virus resistance to cold and BNM-III-170 (Fig. 9). This enhancement of State-1-associated phenotypes supports the inclusion of the A532V change in Envs in which FPPR alterations help to stabilize the pretriggered conformation.
The R542K alteration in the FPPR, which individually results in State-1-destabilizing phenotypes, was combined with one or more of the FPPR changes with State-1-stabilizing phenotypes (Fig. 9). Env combination mutants with the R542K change were generally more sensitive to cold and BNM-III-170 than their matched counterparts without the R542K change. Conversely, the combined State-1-stabilizing changes reduced or nullified the R542K phenotypes of increased virus sensitivity to inactivation by cold and the CD4mc. Thus, State-1 stability can be positively or negatively adjusted by particular FPPR changes in an additive way.
We identified one instance where an individual Env change, R542K, that strengthens subunit association did not contribute to this phenotype when combined with other Env changes stabilizing subunit association. This contrasts with the additivity of subunit association phenotypes observed for FPPR residues 535 and 543.
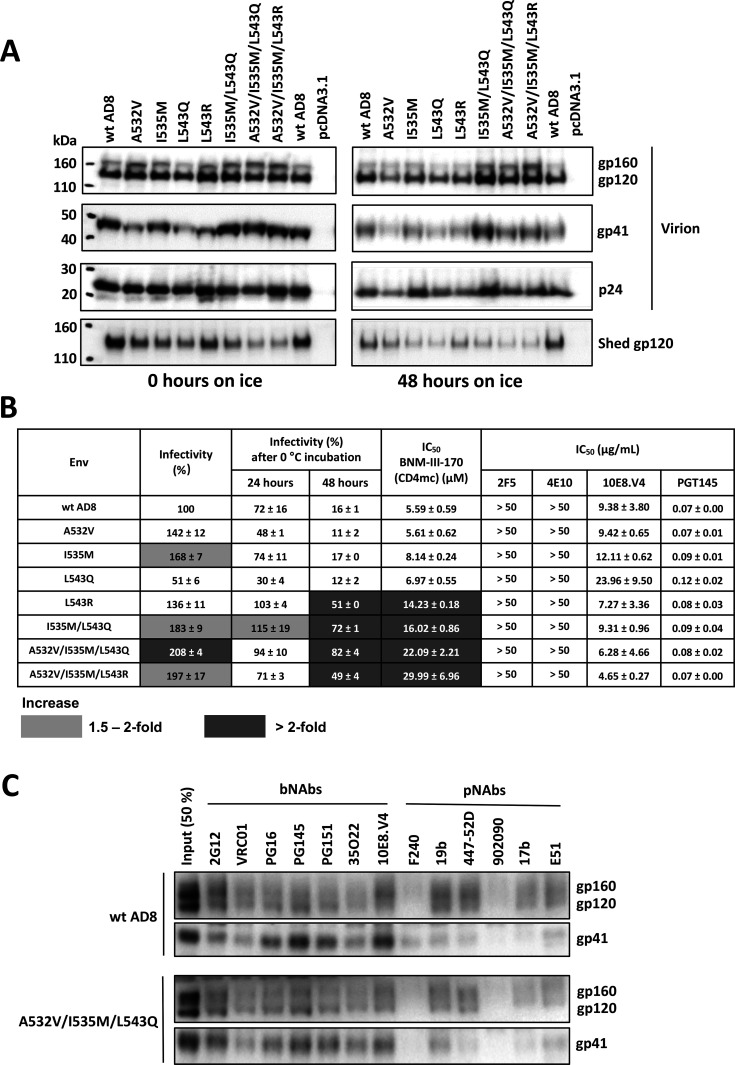
Effects of FPPR changes in a proviral context
We introduced selected changes in the FPPR into the Env encoded by an infectious NL4-3.AD8 Bam provirus (90). Viruses produced transiently by transfection of 293T cells with these infectious molecular clones (IMCs) were examined by western blotting. All the viruses exhibited efficient Env cleavage; the A532V/I535M/L543Q and A532V/I535M/L543R combination mutants shed gp120 less than the wt HIV-1AD8 Env or the Env mutants with fewer FPPR changes (Fig. 10A, left panels). After a 48 h incubation on ice, the combination mutants maintained higher levels of gp120 on the virions compared with the wt Env or Envs with individual FPPR changes (Fig. 10A, right panels). The viruses with FPPR changes were infectious on TZM-bl cells, and the viruses with the combinations of FPPR changes retained infectivity after 48 h on ice better than the viruses with the wt HIV-1AD8 Env (Fig. 10B). The combined FPPR Env mutants were more resistant to BNM-III-170 inhibition than wt HIV-1AD8. Minimal differences were seen in the sensitivity to antibody neutralization between the FPPR Env mutants and wt HIV-1AD8; the L543Q mutant virus was slightly more resistant to the 10E8 MPER bNAb than wt HIV-1AD8. We also examined the effects of the A532V/I535M/L543Q changes on the antigenicity of the AD8 Bam Env on virus particles. Other than a decrease in the low level of F240 recognition of gp41 from which gp120 has been shed, the antigenicity of the A532V/I535M/L543Q Bam Env did not significantly differ from that of the wt AD8 Bam Env (Fig. 10C). These results are consistent with stabilization of a pretriggered (State-1) conformation for the I535M/L543Q, A532V/I535M/L543Q, and A532V/I535M/L543R Env mutants.
Fig 10.
Analysis of HIV-1AD8 Env FPPR mutants expressed from an infectious HIV-1 proviral clone. (A) HEK 293T cells were transfected with plasmids containing an infectious NL4-3 provirus with the envs expressing the indicated wild-type (wt) or mutant AD8 Bam Envs. The Bam changes (S752F I756F) restore the natural HIV-1AD8 Env sequence near the AD8/HXBc2 junction and reduce the clipping of the gp41 cytoplasmic tail by the viral protease (90). Seventy-two hours later, the cell supernatants were collected, filtered (0.45 µm), and aliquoted. The virus-containing supernatants were incubated on ice for 0 or 48 h, after which they were centrifuged at 14,000 × g for 1 h at 4°C. The pellets were resuspended in 1× PBS and represent the virion samples. The supernatants were incubated with GNL beads for 1.5 h at room temperature; the GNL precipitates represent the shed gp120. Virion and shed gp120 samples were western blotted with a goat anti-gp120 antibody. (B) The infectivity of the virions with the indicated HIV-1AD8 Env variants without treatment or after 24 and 48 h of incubation on ice was measured on TZM-bl cells. The values are normalized to that of the untreated wt AD8 Bam virions. The fold increases observed for the Env mutants relative to the values for the wt AD8 Bam virus under the same treatment conditions are colored according to the key. The inhibition of virus infectivity after 1 h incubation at 37°C with the CD4mc, BNM-III-170, or antibodies is reported. The 50% inhibitory concentrations (IC50 values) are reported in µM for BNM-III-170 and in µg/mL for the antibodies. Note that all the viruses were resistant to the 2F5 and 4E10 bNAbs against the gp41 membrane-proximal external region (MPER). The results shown are the means and SDs derived from two independent experiments. Inhibition by BNM-III-170 is colored according to the key. (C) Purified virus particles were incubated with a panel of broadly neutralizing antibodies (bNAbs) and poorly neutralizing antibodies (pNAbs) for 1 h at room temperature. The virus-antibody mixture was washed with 1× PBS and centrifuged. The virus-antibody pellet was lysed and precipitated with Protein A-agarose beads for 1 h at 4°C. The beads were washed three times and western blotted with a goat anti-gp120 antibody and the 4E10 anti-gp41 antibody.
Env mutants with combined State-1-stabilizing changes in the gp120 V1 glycan and gp41 FPPR
To examine whether the State-1-stabilizing changes in the gp120 V1 glycan and the gp41 FPPR could have additive phenotypes, we made and tested the A532V/I535M/L543Q/N136E/D325Q mutant. Although this combination mutant exhibited relatively low infectivity, its resistance to cold and BNM-III-170 was at least as great as that of any of the other Env mutants tested in this study (Fig. 9). These results indicate that the State-1-stabilizing changes in gp120 and gp41 are compatible.
DISCUSSION
Although the pretriggered (State-1) HIV-1 Env conformation is an important target for virus entry inhibitors and antibodies, its plasticity and dependence on membrane anchorage have created challenges for its detailed characterization (30, 76, 90, 106 – 108). Stabilizing State-1 Env could provide a means to overcome these challenges. Here, we show that changes in two dissimilar Env components, gp120 carbohydrates, and the gp41 FPPR, can positively or negatively influence the stability of the pretriggered Env conformation.
Our attention was drawn to the gp120 V1 variable loop when the insertion of a Q3 tag resulted in a virus that was more resistant to cold and to a CD4mc than the parental HIV-1AD8. These phenotypes are an indicator of State-1 stabilization (87 – 92), prompting us to explore the basis of this effect. The relative degree of cold resistance of viruses with diverse V1 insertions indicated that the insert length and amino acid sequence exerted little effect on the ability of the functional Env to withstand exposure to 0°C. Removal of the glycan at Asn 136 explained the observed cold-resistant phenotypes. Although the exact position varies among HIV-1 strains, sequons for N-linked glycosylation are nearly always present in this stretch of the V1 region (96). In a previous study (86), alterations of V1 that included an N136S change were found to increase the temperature of HIV-1 inactivation, enhance sensitivity to the PGT145 bNAb, which recognizes the trimer apex (109), and confer resistance to pNAb neutralization; stabilization of the State-1 Env conformation could account for these phenotypes. How might the loss of the Asn 136 glycan stabilize State 1? This glycan is not resolved in HIV-1AD8 Env cryo-electron microscopy (cryo-EM) maps (110), suggesting that the carbohydrate chain is poorly ordered and likely exposed to the solvent. Indeed, crystal structures show that the trimannosyl core modifying an adjacent residue, Asn 137, in the sgp140 SOSIP.664 trimer derived from HIV-1BG505 is located at the periphery of the protomeric arms of the trimer and extends directly away from the trimer axis (51, 53). A similarly positioned, flexible Asn 136 glycan extending into the solvent could create stress on the V1/V2 structure and destabilize the pretriggered conformation (Fig. 11A). Consistent with a model in which a bulky V1 destabilizes State 1, when the Asn 136 glycan was retained, a Q3 insertion in V1 resulted in increased cold sensitivity [see Q3(V1 alt) in Table 1]. We also found that the removal of the glycan modifying the nearby V1 residue Asn 141 slightly reduced cold sensitivity but nullified the State-1-stabilizing effects associated with the loss of the Asn 136 glycan. By contrast, the State-1-stabilizing phenotypes of Asn 136 glycan removal were not dependent on the other nearby V1 glycan at Asn 130. These results are consistent with a contribution of a bulky distal V1 structure to the destabilization of the State-1 conformation and underscore the unique nature of the Asn 136 glycan in the context of the HIV-1AD8 Env.
Fig 11.
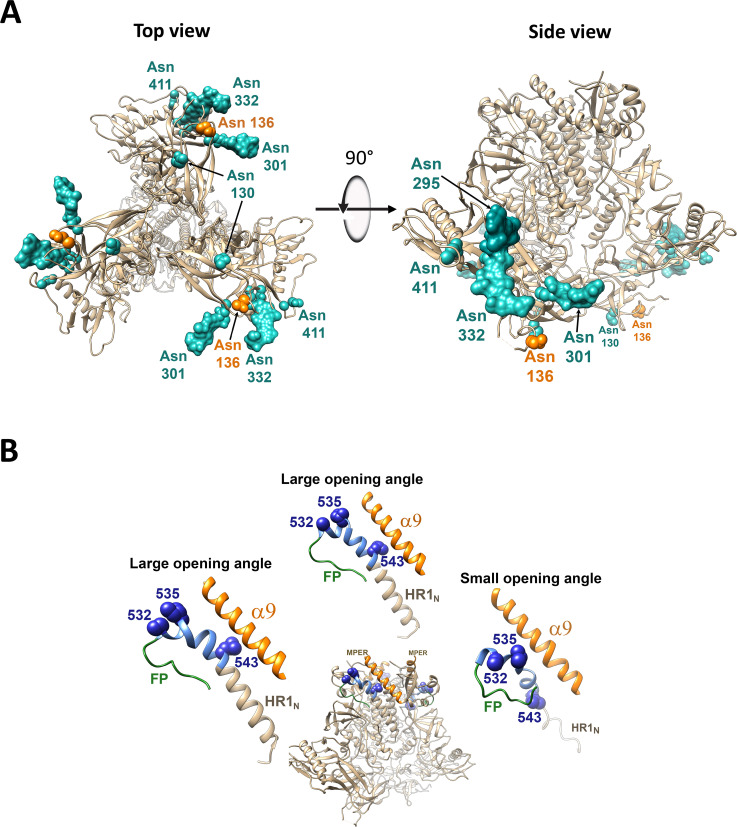
Location of Env changes in current Env structural models. (A) A cryo-EM structure of the AE2.1 variant of the HIV-1AD8 Env trimer (110) is shown from the perspective of the target cell (left panel) or as a side view, with the viral membrane at the top of the figure (right panel). Although the detailed structure of the State-1 conformation of the membrane Env trimer is unknown, the structure of the AE2.1 Env trimer, which has been suggested to represent a default intermediate (State-2-like) conformation (110), is informative. Specific glycosylated Asn residues studied herein are indicated. Well-defined, stable densities in the cryo-EM map allowed modeling of the peptide-proximal glycans at Asn 295, Asn 301, and Asn 332, whereas the glycans associated with Asn 130, Asn 136, and Asn 411 were not resolved (110). Removal of the gp120 glycans in cyan resulted in State-1-destabilizing phenotypes. Where resolved, these glycans project into the interprotomer spaces of the Env trimer. Removal of the Asn 136 glycan (orange) resulted in the phenotypes associated with State-1 stabilization. The poorly resolved glycan at Asn 136 presumably projects from the gp120 V1 loop into the solvent. (B) A side view of the AE2.1 variant of the HIV-1AD8 trimer is shown, with the MPER and viral membrane at the top of the figure. The fusion peptide (FP; green), FPPR (blue), and α9 helices (orange) are highlighted. The structures of these regions in all three protomers of the asymmetric AE2.1 Env trimer are shown. In the asymmetric AE2.1 Env trimer, two opening angles between the protomers are >120°, and one opening angle is <120° (110). The FPPR residues that are implicated in State-1 stabilization are shown as blue Corey-Pauling-Koltun (CPK) models. Note that the conformation of the FPPR differs in the two protomers associated with the larger opening angles compared with that in the protomer associated with the smaller opening angle. The as-yet-known structure of the State-1 Env may hypothetically consist of a symmetric trimer. In such a model, the FPPR could contribute to State-1 stabilization and Env trimer symmetry through interactions with the α9 helices and/or MPERs of the adjacent protomers. FPPR interactions with α9 or MPER could also potentially influence the conformations of the adjacent HR1N regions. The HR1N region is helical in the protomers associated with the larger opening angles, whereas HR1N is a random coil in the protomer associated with the smaller opening angle. Thus, variation in the interactions of the FPPR and HR1N regions with other Env elements can potentially influence trimer symmetry.
In contrast to the State-1-stabilizing Asn 136 phenotype, removal of glycans at Asn residues 130, 301, and 332 resulted in phenotypes indicative of destabilization of State 1. These carbohydrate chains project from V1 and from the base of the gp120 V3 region into the interprotomer space between the trimer arms (Fig. 11A). If trimer symmetry is important for maintaining a State-1 Env conformation, filling the interprotomer space with glycans could help to resist changes in the interprotomer angles leading to trimer asymmetry. Furthermore, loss of the Asn 301 and Asn 332 glycans may alter the V3 conformation, which can disrupt Env trimer integrity (111). Indeed, previous studies have shown that the removal of glycans at Asn 301 and Asn 332 increases HIV-1 sensitivity to V3-directed pNAbs (111 – 114) as well as to soluble CD4 and some pNAbs against the CD4-binding site (115, 116).
The State-1-destabilizing phenotypes resulting from glycan removal at Asn 130, Asn 301, or Asn 332 were suppressed by the loss of the glycan at Asn 136. In some cases, phenotypes related to stability or instability of the HIV-1 Env pretriggered conformation have been shown to be expressed additively even when the Env determinants of the phenotypes are located distantly on the Env trimer (88, 89). Here, we observed a similar counterbalancing of State-1-stabilizing and -destabilizing phenotypes; in this case, both were determined by glycan removal.
As a second line of inquiry, we investigated the contribution of HIV-1 Env proteolytic cleavage to Env conformation. We evaluated the phenotypes of HIV-1AD8 Env mutants with individual or combined changes in sequences near the cleavage site. As we wished to use the resistance of virus infectivity to a CD4mc or to cold exposure as an indicator of State-1 stabilization, we employed a conservative mutagenesis strategy, focusing on polymorphic changes in natural HIV-1 strains and on Env variants detected in HIV-1 populations replicating in tissue culture (96, 104).
Despite our conservative strategy, several of the mutants exhibited defects that compromised their ability to support cell-cell fusion and virus entry. Changes in the highly conserved Met 530 and Ala 533 resulted in severe defects in gp160 processing and function, consistent with the predicted role of this region in folding the gp41 subunit (53, 117). Other mutants (G516V, A525V, A526T, T538F, Q540H, I548A, I548Y, and Q550D) showed reductions in cleavage efficiency that correlated with decreases in pseudovirus infectivity; cell-cell fusion mediated by these mutants was attenuated to a lesser degree, as expected for inefficiently folded Envs with decreases in the stability of the functional trimer (105). Finally, changes in residues 546, 548, and 550 at the FPPR-HR1N junction resulted in Envs that were processed, exhibited good subunit association, and mediated cell-cell fusion but were deficient in supporting pseudovirus infection. As above, specific deficiencies in the ability to maintain functional virion Env trimers underly these phenotypes.
Most of the Env mutants in the gp41 FP and FPPR retained a detectable level of pseudovirus infectivity. This allowed us to evaluate their sensitivity to a CD4mc, BNM-III-170, and to cold inactivation, two indicators of HIV-1 Env conformational state (87 – 89). Individual changes in HIV-1AD8 FPPR residues Ile 535 and Leu 543, converting them to common HIV-1 polymorphic variants, resulted in modest decreases in sensitivity to BNM-III-170, unusually strong subunit association and good proteolytic processing. These individual mutants (I535M, L543Q, L543N, and L543R) resisted cold inactivation only slightly better than the wt HIV-1AD8 Env when measured in careful side-by-side assays (Fig. 9). Thus, for the individual FPPR mutants, the degree of State-1 stabilization is mild at best. However, when introduced in combination, changes in FPPR residues 532, 535, and 543 resulted in strong virus resistance to exposure to both cold and a CD4mc, clearcut signatures of State-1 stabilization (87 – 89). The A532V change, which exhibited minimal evidence of State-1 stabilization on its own, intensified the State-1-associated phenotypes of the I535M change. Of interest, some natural HIV-1 variants have Env residues at 535 and 543 associated with greater State-1 stability (96), likely contributing to some of the variation in Env triggerability among primary virus strains (118 – 120). We also note that alteration of these FPPR residues has been included in efforts to stabilize both soluble gp140 SOSIP.664 trimers and membrane Envs (63, 65, 66, 80, 81, 85). Apparently, FPPR changes can benefit the integrity of Env trimers in multiple contexts.
The increases in State-1 stabilization and subunit association of the FPPR mutants were not recapitulated in the gp120-trimer association assay. This result contrasts with increases in gp120-trimer association observed for other State-1-stabilizing Env changes (88). Qualitative differences in the mechanisms of State-1 stabilization may account for these observations; for example, the phenotypes of the FPPR variants may be more dependent on membrane-proximal Env elements that are disrupted by the detergent solubilization of Env in the gp120-trimer association assay. Recent cryo-EM structures of HIV-1 Env trimers indicate that solubilization may result in the asymmetrical opening of the trimer, with allosterically coupled transformations of the gp41 FPPR and HR1N regions (110). The proximity of the gp41 FPPR to the cleavage site and its association with Env conformational modulation supports the contribution of the FPPR to State-1 stabilization following Env cleavage. Finally, quantitative differences in the degree to which State 1 is stabilized may also account for the distinct outcomes of Env variants in the gp120-trimer association assay.
How do FPPR changes modulate the stability of the State-1 Env conformation? In available Env trimer structures (51 – 53, 56, 73, 107), the FPPR is located near the α9 helix and MPER of the adjacent protomer. Of interest, the FPPR residues (535 and 543) implicated in State 1 stabilization exhibit significant differences in orientation among the asymmetric protomers of Envs solubilized in styrene-maleic acid lipid nanoparticles (110) (Fig. 11B). As these asymmetric Env trimers are thought to represent an early, State-2-like conformation, such FPPR movement may reflect changing interactions (e.g., with the α9 helix or MPER) involved in the release of State 1 and the promotion of State 2. Modulation of Env trimer symmetry may represent a common mechanism whereby FPPR and glycan changes influence State 1-to-State 2 transitions. A complete understanding of this mechanism will require detailed structures of State-1 Envs.
Our study also identified gp41 changes (R542K and S546D) that increased HIV-1 sensitivity to cold inactivation and to inhibition by the CD4mc, phenotypes indicative of State-1 destabilization (87 – 92). These results are consistent with the contribution of the FPPR to the maintenance of the pretriggered Env conformation, as also suggested by recent studies (121, 122). The phenotypes of the R542K Env are noteworthy. Although the R542K Env exhibits strong subunit association and increased cell-cell fusion like the I535M and Leu 543 mutants, its sensitivity to cold and BNM-III-170 clearly suggest a higher triggerability. Thus, the increased subunit association of the membrane Env trimer and stabilization of the pretriggered (State-1) Env conformation are separable consequences of FPPR modification.
This study provides a set of Env changes that additively contribute to the stabilization of the pretriggered (State-1) conformation. Such Env modifications may be useful in reducing Env conformational heterogeneity for studies of State-1 structure and immunogenicity.
MATERIALS AND METHODS
HIV-1 Env mutants
The wt HIV-1AD8 env cloned in the pSVIIIenv expression plasmid was used as a template to construct HIV-1 Env mutants in this study. The signal peptide/N-terminus (residues 1–33) and the cytoplasmic tail C-terminus (residues 751–856) of this Env are derived from the HIV-1HXBc2 Env (88, 89). Changes were introduced by using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies). All the Envs contain a His6 tag at the carboxyl terminus. For the Env mutants expressed in IMCs, site-directed changes were introduced into the pNL4-3.AD8 Bam IMC (90) by using the Q5 high-fidelity DNA polymerase (New England Biolabs) and One Shot Stbl3 chemically competent Escherichia coli (Invitrogen), following the manufacturer’s protocols. All the mutations were confirmed by DNA sequencing.
Antibodies
pNAbs (19b, 17b, and E51) and bNAbs (2F5, 4E10, 10E8.v4, and PGT145) against the HIV-1 Env were obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH.
Cell lines
HEK293T, TZM-bl, and HOS cells (ATCC) were cultured in Dulbecco-modified Eagle medium supplemented with 10% fetal bovine serum and 100 µg/mL penicillin-streptomycin (Life Technologies). Cf2Th-CD4/CCR5 cells were cultured in the same medium supplemented with 400 µg/mL G418.
Env expression
To evaluate Env processing, subunit association, and gp120-trimer association, 3 × 105 HOS cells were seeded in six-well plates. After 24 h of incubation, they were transfected with plasmids encoding His6-tagged Env variants and Tat at a ratio of 8:1, using the Lipofectamine 3,000 transfection reagent (Life Technologies) according to the manufacturer’s instructions. Seventy-two hours after transfection, the supernatants were collected and incubated with Galanthus Nivalis Lectin (GNL)-agarose beads (Vector Laboratories) for 1.5 h at room temperature. Beads were washed three times with 1× phosphate-buffered saline (PBS) containing 0.1% NP-40 and processed for western blotting with a goat anti-gp120 antibody (Invitrogen). The cells were lysed in PBS buffer containing 1.0% NP-40 and protease inhibitor (Sigma-Aldrich). Clarified lysates were harvested and separated into two portions. One portion was used for the “Input” sample. The other was incubated with Ni-NTA beads at room temperature for 1.5 h. Then the beads were washed, boiled, and analyzed by western blotting with 1:2,500 goat anti-gp120 antibody (Invitrogen) and 1:2,500 HRP-conjugated rabbit anti-goat antibody (Invitrogen). The intensities of the gp120 and gp160 bands from unsaturated western blots were quantified by using ImageJ software. The Env processing index was calculated by dividing gp120 by gp160 in the Input samples. The subunit association was calculated by dividing gp120 in the Input samples by the gp120 in the GNL precipitates. The gp120-trimer association index was calculated by dividing the gp120:gp160 ratio in the Ni-NTA precipitates by the gp120:gp160 ratio in the Input samples. The processing, subunit association, and gp120-trimer association indices of the Env mutants were normalized to those of the wt HIV-1AD8 Env.
Cell-cell fusion assay
For the alpha-complementation assay measuring cell-cell fusion, 2 × 104 COS-1 effector cells were seeded in black-and-white 96-well plates and then cotransfected with plasmids expressing ⍺-gal, Env variants, and Tat at a 1:1:0.125 ratio, using Effectene transfection reagent (Qiagen) following the manufacturer’s protocol. At the same time, Cf2Th-CD4/CCR5 cells target cells in 6-well plates were cotransfected with a plasmid expressing ω-gal using Effectene transfection reagent. Forty-eight hours after transfection, target cells were detached and resuspended in medium. The medium was aspirated from the effector cells, and target cell suspensions in 50-µL volumes were added to the effector cells (one target-cell well provides sufficient cells for 50 effector-cell wells). Plates were spun at 500 × g for 3 min and then incubated at 37°C in 5% CO2 for 8 h. Medium was removed, and cells were lysed in Tropix lysis buffer (Thermo Fisher Scientific). The β-galactosidase activity in the cell lysates was measured using the Galacto-Star Reaction Buffer Diluent with Galacto-Star Substrate (Thermo Fisher Scientific), according to the manufacturer’s instruction.
Virus infectivity
To produce pseudoviruses, HEK293T cells were cotransfected with the Env-expressing pSVIIIenv plasmid, a Tat-encoding plasmid, and the luciferase-encoding pNL4-3.Luc.R-E- vector (NIH HIV Reagent Program) at a 1:1:3 ratio using polyethyleneimine (PEI, Polysciences). To prepare replication-competent viruses, HEK293T cells were transfected with the pNL4-3.Env.Bam IMC plasmid using PEI. The medium was replaced 6–8 h after transfection. The cell supernatants containing pseudoviruses or infectious viruses were harvested 72 h later and centrifuged (3,500 rpm for 5 min), aliquoted, and either used directly to measure pseudovirus and infectious virus infectivity or stored at −80°C until use.
To compare the infectivity of the variants, freshly prepared pseudoviruses and infectious viruses were serially diluted in 96-well plates and incubated with TZM-bl cells in the presence of 20 µg/mL DEAE-dextran. After 48 h of incubation, TZM-bl cells were lysed, and the luciferase activity was measured using a luminometer.
Virus sensitivity to cold inactivation
To evaluate virus sensitivity to cold inactivation, pseudoviruses and infectious viruses were incubated on ice for different lengths of time, and virus infectivity was subsequently measured, as described above.
Virus inhibition/neutralization
The inhibitors to be tested (antibodies, BNM-III-170, or BMS-806) were serially diluted in triplicate wells of 96-well plates. Then approximately 100–200 TCID50 (50% tissue culture infectious dose) of either pseudoviruses or infectious viruses was added and incubated at 37°C for 1 h. Subsequently, approximately 2 × 104 TZM-bl cells with 20 µg/mL DEAE-dextran in the medium were added to each well, and the mixture was incubated at 37°C/5%CO2 for 48 h. Then luciferase activity was measured, as described above. The concentrations of antibodies and other inhibitors that inhibit 50% of infection (the IC50 values) were determined by fitting the data in four-parameter dose-response curves using GraphPad Prism 9.
Analysis of Env on virus particles
Approximately 1 mL of the pseudovirus or infectious virus suspension in the supernatants of expressing cells was filtered (0.45 µm) and incubated on ice for different periods of time. Then the virus suspensions were centrifuged at 14,000 × g for 1 h at 4°C. The pelleted virus particles were resuspended in 1× PBS. Equal volumes of the virus suspensions were then analyzed by western blotting. Western blots were developed with 1:2,500 goat anti-gp120 polyclonal antibody (Invitrogen), 1:2,500 4E10 anti-gp41 antibody, and 1:5,000 rabbit polyclonal antibody against Gag p55/p24/p17 (Abcam). The HRP-conjugated secondary antibodies were 1:2,500 rabbit anti-goat antibody (Invitrogen), 1:2,500 goat anti-human antibody (Invitrogen), and 1:5,000 goat anti-rabbit antibody (Sigma-Aldrich), respectively. The supernatants containing shed gp120 were collected and incubated with GNL-agarose beads for 1.5 h at room temperature. Beads were washed three times with 1× PBS/0.1% NP-40 and processed for western blotting with a goat anti-gp120 antibody (Invitrogen), as described above.
Antigenicity of Env on virus particles
HEK293T cells were transfected with the pNL4-3.AD8 Bam IMCs encoding the wt and mutant Envs using PEI. Seventy-two hours later, the cell supernatants were collected, filtered (0.45 µm), and centrifuged at 100,000 × g for 1 h at 4°C. Virus pellets were resuspended in 1× PBS, and 100 µL aliquots were incubated with a panel of antibodies at 10 µg/mL concentration for 1 h at room temperature. One milliliter of chilled 1× PBS was added, and samples were centrifuged at 14,000 × g for 1 h at 4°C. The pellets were lysed in 100 µL chilled 1× PBS/0.5% NP-40/protease inhibitor cocktail. Lysates were rotated during incubation with Protein A-agarose beads for 1 h at 4°C and washed with chilled 1× PBS/0.1% NP-40 three times. The beads were processed for western blotting. To prepare the Input (50%) sample, 25 µL purified virus was mixed with 1 mL chilled 1× PBS and centrifuged at 14,000 × g for 1 h at 4°C; the pellet was processed for western blotting.
ACKNOWLEDGMENTS
We thank Ms. Elizabeth Carpelan for manuscript preparation. Antibodies against HIV-1 were kindly supplied by John C. Kappes (University of Alabama at Birmingham), Dennis Burton (Scripps), Peter Kwong and John Mascola (Vaccine Research Center NIH), Barton Haynes (Duke University), Hermann Katinger (Polymun), James Robinson (Tulane University), and Marshall Posner (Mount Sinai Medical Center). We thank the NIH HIV Reagent Program for providing additional reagents.
This work was supported by grants from the National Institutes of Health (grants AI 145547, AI 124982, AI 150471, AI 129017, and AI 164562), a grant from Gilead Sciences, and a gift from the late William F. McCarty-Cooper.
Z.Z., Q.W., and H.T.N. performed the experiments. H.-C.C., T.-J.C., and A.B.S. III synthesized the CD4mc, BNM-III-170. Z.Z., Q.W., H.T.N., and J.G.S. interpreted the data and wrote the paper. All authors contributed to manuscript editing and revision.
We declare no conflicts of interest.
Contributor Information
Joseph G. Sodroski, Email: joseph_sodroski@dfci.harvard.edu.
Viviana Simon, Icahn School of Medicine at Mount Sinai, New York, New York, USA .
REFERENCES
- 1. Gray GE, Laher F, Lazarus E, Ensoli B, Corey L. 2016. Approaches to preventative and therapeutic HIV vaccines. Curr Opin Virol 17:104–109. doi: 10.1016/j.coviro.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Corey L, Gilbert PB, Tomaras GD, Haynes BF, Pantaleo G, Fauci AS. 2015. Immune correlates of vaccine protection against HIV-1 acquisition. Sci Transl Med 7:310rv7. doi: 10.1126/scitranslmed.aac7732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haynes BF, Wiehe K, Borrow P, Saunders KO, Korber B, Wagh K, McMichael AJ, Kelsoe G, Hahn BH, Alt F, Shaw GM. 2023. Strategies for HIV-1 vaccines that induce broadly neutralizing antibodies. Nat Rev Immunol 23:142–158. doi: 10.1038/s41577-022-00753-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seabright GE, Doores KJ, Burton DR, Crispin M. 2019. Protein and glycan mimicry in HIV vaccine design. J Mol Biol 431:2223–2247. doi: 10.1016/j.jmb.2019.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andrabi R, Bhiman JN, Burton DR. 2018. Strategies for a multi-stage neutralizing antibody-based HIV vaccine. Curr Opin Immunol 53:143–151. doi: 10.1016/j.coi.2018.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and Immunogens. Science 280:1884–1888. doi: 10.1126/science.280.5371.1884 [DOI] [PubMed] [Google Scholar]
- 7. Chen B. 2019. Molecular mechanism of HIV-1 entry. Trends Microbiol 27:878–891. doi: 10.1016/j.tim.2019.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harrison SC. 2015. Viral membrane fusion. Virology 479–480:498–507. doi: 10.1016/j.virol.2015.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Willey RL, Bonifacino JS, Potts BJ, Martin MA, Klausner RD. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci U S A 85:9580–9584. doi: 10.1073/pnas.85.24.9580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Earl PL, Moss B, Doms RW. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol 65:2047–2055. doi: 10.1128/JVI.65.4.2047-2055.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pal R, Hoke GM, Sarngadharan MG. 1989. Role of oligosaccharides in the processing and maturation of envelope glycoproteins of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 86:3384–3388. doi: 10.1073/pnas.86.9.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dewar RL, Vasudevachari MB, Natarajan V, Salzman NP. 1989. Biosynthesis and processing of human immunodeficiency virus type 1 envelope glycoproteins: effects of monensin on glycosylation and transport. J Virol 63:2452–2456. doi: 10.1128/JVI.63.6.2452-2456.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang S, Nguyen HT, Ding H, Wang J, Zou S, Liu L, Guha D, Gabuzda D, Ho DD, Kappes JC, Sodroski J. 2021. Dual pathways of human immunodeficiency virus type 1 envelope glycoprotein trafficking modulate the selective exclusion of uncleaved oligomers from virions. J Virol 95:e01369-20. doi: 10.1128/JVI.01369-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183. doi: 10.1038/384179a0 [DOI] [PubMed] [Google Scholar]
- 16. Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187. doi: 10.1038/384184a0 [DOI] [PubMed] [Google Scholar]
- 17. Khasnis MD, Halkidis K, Bhardwaj A, Root MJ. 2016. Receptor activation of HIV-1 Env leads to asymmetric exposure of the gp41 trimer. PLoS Pathog 12:e1006098. doi: 10.1371/journal.ppat.1006098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Furuta RA, Wild CT, Weng Y, Weiss CD. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol 5:276–279. doi: 10.1038/nsb0498-276 [DOI] [PubMed] [Google Scholar]
- 19. Koshiba T, Chan DC. 2003. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J Biol Chem 278:7573–7579. doi: 10.1074/jbc.M211154200 [DOI] [PubMed] [Google Scholar]
- 20. He Y, Vassell R, Zaitseva M, Nguyen N, Yang Z, Weng Y, Weiss CD. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J Virol 77:1666–1671. doi: 10.1128/jvi.77.3.1666-1671.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chan DC, Fass D, Berger JM, Kim PS. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273. doi: 10.1016/s0092-8674(00)80205-6 [DOI] [PubMed] [Google Scholar]
- 22. Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430. doi: 10.1038/387426a0 [DOI] [PubMed] [Google Scholar]
- 23. Lu M, Blacklow SC, Kim PS. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol 2:1075–1082. doi: 10.1038/nsb1295-1075 [DOI] [PubMed] [Google Scholar]
- 24. Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol 151:413–423. doi: 10.1083/jcb.151.2.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilen CB, Tilton JC, Doms RW. 2012. Molecular mechanisms of HIV entry. Adv Exp Med Biol 726:223–242. doi: 10.1007/978-1-4614-0980-9_10 [DOI] [PubMed] [Google Scholar]
- 26. Bonsignori M, Liao HX, Gao F, Williams WB, Alam SM, Montefiori DC, Haynes BF. 2017. Antibody-virus co-evolution in HIV infection: paths for HIV vaccine development. Immunol Rev 275:145–160. doi: 10.1111/imr.12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kwong PD, Mascola JR. 2018. HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity 48:855–871. doi: 10.1016/j.immuni.2018.04.029 [DOI] [PubMed] [Google Scholar]
- 28. Sok D, Burton DR. 2018. Recent progress in broadly neutralizing antibodies to HIV. Nat Immunol 19:1179–1188. doi: 10.1038/s41590-018-0235-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stewart-Jones GBE, Soto C, Lemmin T, Chuang G-Y, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens A-J, Bylund T, Choi CW, Davison JR, Georgiev IS, Joyce MG, Kwon YD, Pancera M, Taft J, Yang Y, Zhang B, Shivatare SS, Shivatare VS, Lee C-CD, Wu C-Y, Bewley CA, Burton DR, Koff WC, Connors M, Crispin M, Baxa U, Korber BT, Wong C-H, Mascola JR, Kwong PD. 2016. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165:813–826. doi: 10.1016/j.cell.2016.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ward AB, Wilson IA. 2017. The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol Rev 275:21–32. doi: 10.1111/imr.12507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470 [DOI] [PubMed] [Google Scholar]
- 32. Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol 77:10557–10565. doi: 10.1128/jvi.77.19.10557-10565.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moore PL, Ranchobe N, Lambson BE, Gray ES, Cave E, Abrahams MR, Bandawe G, Mlisana K, Abdool Karim SS, Williamson C, Morris L, Study C, NCfHAV I. 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog 5:e1000598. doi: 10.1371/journal.ppat.1000598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haim H, Salas I, Sodroski J. 2013. Proteolytic processing of the human immunodeficiency virus envelope glycoprotein precursor decreases conformational flexibility. J Virol 87:1884–1889. doi: 10.1128/JVI.02765-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pancera M, Wyatt R. 2005. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332:145–156. doi: 10.1016/j.virol.2004.10.042 [DOI] [PubMed] [Google Scholar]
- 36. Chakrabarti BK, Pancera M, Phogat S, O’Dell S, McKee K, Guenaga J, Robinson J, Mascola J, Wyatt RT. 2011. HIV type 1 Env precursor cleavage state affects recognition by both neutralizing and nonneutralizing gp41 antibodies. AIDS Res Hum Retroviruses 27:877–887. doi: 10.1089/AID.2010.0281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moore JP, McKeating JA, Weiss RA, Sattentau QJ. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139–1142. doi: 10.1126/science.2251501 [DOI] [PubMed] [Google Scholar]
- 38. Gray ES, Taylor N, Wycuff D, Moore PL, Tomaras GD, Wibmer CK, Puren A, DeCamp A, Gilbert PB, Wood B, Montefiori DC, Binley JM, Shaw GM, Haynes BF, Mascola JR, Morris L. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J Virol 83:8925–8937. doi: 10.1128/JVI.00758-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, Stamatatos L. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol 83:757–769. doi: 10.1128/JVI.02036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6:e1001028. doi: 10.1371/journal.ppat.1001028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L, Team CS. 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol 85:4828–4840. doi: 10.1128/JVI.00198-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. 2014. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28:163–169. doi: 10.1097/QAD.0000000000000106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guttman M, Cupo A, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. 2015. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun 6:6144. doi: 10.1038/ncomms7144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haim H, Salas I, McGee K, Eichelberger N, Winter E, Pacheco B, Sodroski J. 2013. Modeling virus- and antibody-specific factors to predict human immunodeficiency virus neutralization efficiency. Cell Host Microbe 14:547–558. doi: 10.1016/j.chom.2013.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zou S, Zhang S, Gaffney A, Ding H, Lu M, Grover JR, Farrell M, Nguyen HT, Zhao C, Anang S, Zhao M, Mohammadi M, Blanchard SC, Abrams C, Madani N, Mothes W, Kappes JC, Smith AB 3rd, Sodroski J. 2020. Long-acting BMS-378806 analogues stabilize the state-1 conformation of the human immunodeficiency virus type 1 envelope glycoproteins. J Virol 94:e00148-20. doi: 10.1128/JVI.00148-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McCoy LE, Burton DR. 2017. Identification and specificity of broadly neutralizing antibodies against HIV. Immunol Rev 275:11–20. doi: 10.1111/imr.12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sanders Rogier W., Derking R, Cupo A, Julien J-P, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, Golabek M, de los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP, Trkola A. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sanders R.W, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76:8875–8889. doi: 10.1128/jvi.76.17.8875-8889.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sanders R.W, Schiffner L, Master A, Kajumo F, Guo Y, Dragic T, Moore JP, Binley JM. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J Virol 74:5091–5100. doi: 10.1128/jvi.74.11.5091-5100.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schülke N, Vesanen MS, Sanders RW, Zhu P, Lu M, Anselma DJ, Villa AR, Parren PWHI, Binley JM, Roux KH, Maddon PJ, Moore JP, Olson WC. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol 76:7760–7776. doi: 10.1128/jvi.76.15.7760-7776.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lyumkis D, Julien J-P, de Val N, Cupo A, Potter CS, Klasse P-J, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang G-Y, Ofek G, Stewart-Jones GBE, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Peña AT, Cupo A, Julien J-P, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. 2014. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40:669–680. doi: 10.1016/j.immuni.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang G-Y, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Peña AT, Derking R, Klasse P-J, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515:138–142. doi: 10.1038/nature13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rantalainen K, Berndsen ZT, Antanasijevic A, Schiffner T, Zhang X, Lee W-H, Torres JL, Zhang L, Irimia A, Copps J, Zhou KH, Kwon YD, Law WH, Schramm CA, Verardi R, Krebs SJ, Kwong PD, Doria-Rose NA, Wilson IA, Zwick MB, Yates JR III, Schief WR, Ward AB. 2020. HIV-1 envelope and MPER antibody structures in lipid assemblies. Cell Reports 31:107583. doi: 10.1016/j.celrep.2020.107583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, Eroshkin AM, Guenaga J, Kaushik K, Kulp DW, Liu J, McCoy LE, Oom AL, Ozorowski G, Post KW, Sharma SK, Steichen JM, de Taeye SW, Tokatlian T, Torrents de la Peña A, Butera ST, LaBranche CC, Montefiori DC, Silvestri G, Wilson IA, Irvine DJ, Sanders RW, Schief WR, Ward AB, Wyatt RT, Barouch DH, Crotty S, Burton DR. 2017. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity 46:1073–1088. doi: 10.1016/j.immuni.2017.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Torrents de la Peña A, de Taeye SW, Sliepen K, LaBranche CC, Burger JA, Schermer EE, Montefiori DC, Moore JP, Klasse PJ, Sanders RW. 2018. Immunogenicity in rabbits of HIV-1 SOSIP trimers from clades A, B, and C, given individually, sequentially, or in combination. J Virol 92:e01957-17. doi: 10.1128/JVI.01957-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dubrovskaya V, Tran K, Ozorowski G, Guenaga J, Wilson R, Bale S, Cottrell CA, Turner HL, Seabright G, O’Dell S, Torres JL, Yang L, Feng Y, Leaman DP, Vázquez Bernat N, Liban T, Louder M, McKee K, Bailer RT, Movsesyan A, Doria-Rose NA, Pancera M, Karlsson Hedestam GB, Zwick MB, Crispin M, Mascola JR, Ward AB, Wyatt RT. 2019. Vaccination with glycan-modified HIV NFL envelope trimer-liposomes elicits broadly neutralizing antibodies to multiple sites of vulnerability. Immunity 51:915–929. doi: 10.1016/j.immuni.2019.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xu K, Acharya P, Kong R, Cheng C, Chuang G-Y, Liu K, Louder MK, O’Dell S, Rawi R, Sastry M, Shen C-H, Zhang B, Zhou T, Asokan M, Bailer RT, Chambers M, Chen X, Choi CW, Dandey VP, Doria-Rose NA, Druz A, Eng ET, Farney SK, Foulds KE, Geng H, Georgiev IS, Gorman J, Hill KR, Jafari AJ, Kwon YD, Lai Y-T, Lemmin T, McKee K, Ohr TY, Ou L, Peng D, Rowshan AP, Sheng Z, Todd J-P, Tsybovsky Y, Viox EG, Wang Y, Wei H, Yang Y, Zhou AF, Chen R, Yang L, Scorpio DG, McDermott AB, Shapiro L, Carragher B, Potter CS, Mascola JR, Kwong PD. 2018. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat Med 24:857–867. doi: 10.1038/s41591-018-0042-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramírez M, Back JW, Koff WC, Julien J-P, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 vaccines. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. doi: 10.1126/science.aac4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Klasse PJ, LaBranche CC, Ketas TJ, Ozorowski G, Cupo A, Pugach P, Ringe RP, Golabek M, van MJ, Guttman M, Lee KK, Wilson IA, Butera ST, Ward AB, Montefiori DC, Sanders RW, Moore JP. 2016. Sequential and simultaneous immunization of rabbits with HIV-1. PLoS Pathog 12:e1005864. doi: 10.1371/journal.ppat.1005864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. de Taeye SW, Ozorowski G, Torrents de la Peña A, Guttman M, Julien J-P, van den Kerkhof TLGM, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, LaBranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW. 2015. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163:1702–1715. doi: 10.1016/j.cell.2015.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Feng Y, Tran K, Bale S, Kumar S, Guenaga J, Wilson R, de Val N, Arendt H, DeStefano J, Ward AB, Wyatt RT, Trkola A. 2016. Thermostability of well-ordered HIV spikes correlates with the elicitation of autologous tier 2 neutralizing antibodies. PLoS Pathog 12:e1005767. doi: 10.1371/journal.ppat.1005767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Torrents de la Peña A, Sanders RW. 2018. Stabilizing HIV-1 envelope glycoprotein trimers to induce neutralizing antibodies. Retrovirology 15:63. doi: 10.1186/s12977-018-0445-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Del Moral-Sánchez I, Russell RA, Schermer EE, Cottrell CA, Allen JD, Torrents de la Peña A, LaBranche CC, Kumar S, Crispin M, Ward AB, Montefiori DC, Sattentau QJ, Sliepen K, Sanders RW. 2022. High thermostability improves neutralizing antibody responses induced by native-like HIV-1 envelope trimers. NPJ Vaccines 7:27. doi: 10.1038/s41541-022-00446-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Alsahafi N, Debbeche O, Sodroski J, Finzi A. 2015. Effects of the I559P gp41 change on the conformation and function of the human immunodeficiency virus (HIV-1) membrane envelope glycoprotein trimer. PLoS One 10:e0122111. doi: 10.1371/journal.pone.0122111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Alsahafi Nirmin, Anand SP, Castillo-Menendez L, Verly MM, Medjahed H, Prévost J, Herschhorn A, Richard J, Schön A, Melillo B, Freire E, Smith AB III, Sodroski J, Finzi A, Simon V. 2018. SOSIP changes affect human immunodeficiency virus type 1 envelope glycoprotein conformation and CD4 engagement. J Virol 92:e01080–18. doi: 10.1128/JVI.01080-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Castillo-Menendez LR, Nguyen HT, Sodroski J. 2019. Conformational differences between functional human immunodeficiency virus envelope glycoprotein trimers and stabilized soluble trimers. J Virol 93:e01709-18. doi: 10.1128/JVI.01709-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nguyen HT, Alsahafi N, Finzi A, Sodroski JG. 2019. Effects of the SOS (A501C/T605C) and DS (I201C/A433C) disulfide bonds on HIV-1 membrane envelope glycoprotein conformation and function. J Virol 93:e00304-19. doi: 10.1128/JVI.00304-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Go EP, Herschhorn A, Gu C, Castillo-Menendez L, Zhang S, Mao Y, Chen H, Ding H, Wakefield JK, Hua D, Liao HX, Kappes JC, Sodroski J, Desaire H. 2015. Comparative analysis of the glycosylation profiles of membrane-anchored HIV-1 envelope glycoprotein trimers and soluble gp140. J Virol 89:8245–8257. doi: 10.1128/JVI.00628-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cao L, Pauthner M, Andrabi R, Rantalainen K, Berndsen Z, Diedrich JK, Menis S, Sok D, Bastidas R, Park S-KR, Delahunty CM, He L, Guenaga J, Wyatt RT, Schief WR, Ward AB, Yates JR 3rd, Burton DR, Paulson JC. 2018. Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Nat Commun 9:3693. doi: 10.1038/s41467-018-06121-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Torrents de la Peña A, Rantalainen K, Cottrell CA, Allen JD, van Gils MJ, Torres JL, Crispin M, Sanders RW, Ward AB. 2019. Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics. PLoS Pathog 15:e1007920. doi: 10.1371/journal.ppat.1007920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Struwe WB, Chertova E, Allen JD, Seabright GE, Watanabe Y, Harvey DJ, Medina-Ramirez M, Roser JD, Smith R, Westcott D, Keele BF, Bess JW, Sanders RW, Lifson JD, Moore JP, Crispin M. 2018. Site-specific glycosylation of virion-derived HIV-1 Env is mimicked by a soluble trimeric immunogen. Cell Rep 24:1958–1966. doi: 10.1016/j.celrep.2018.07.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Castillo-Menendez LR, Witt K, Espy N, Princiotto A, Madani N, Pacheco B, Finzi A, Sodroski J. 2018. Comparison of uncleaved and mature human immunodeficiency virus membrane envelope glycoprotein trimers. J Virol 92:e00277-18. doi: 10.1128/JVI.00277-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B, Zhou T, Reichard N, Wang K, Grover JR, Carman BP, Gardner MR, Nikić-Spiegel I, Sugawara A, Arthos J, Lemke EA, Smith AB III, Farzan M, Abrams C, Munro JB, McDermott AB, Finzi A, Kwong PD, Blanchard SC, Sodroski JG, Mothes W. 2019. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568:415–419. doi: 10.1038/s41586-019-1101-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mangala Prasad V, Leaman DP, Lovendahl KN, Croft JT, Benhaim MA, Hodge EA, Zwick MB, Lee KK. 2022. Cryo-ET of Env on intact HIV virions reveals structural variation and positioning on the gag lattice. Cell 185:641–653. doi: 10.1016/j.cell.2022.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Klasse PJ, McKeating JA, Schutten M, Reitz MS, Robert-Guroff M. 1993. An immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 (HXB2-Env:Ala 582(-->Thr)) decreases viral neutralization by monoclonal antibodies to the CD4-binding site. Virology 196:332–337. doi: 10.1006/viro.1993.1484 [DOI] [PubMed] [Google Scholar]
- 79. Thali M, Charles M, Furman C, Cavacini L, Posner M, Robinson J, Sodroski J. 1994. Resistance to neutralization by broadly reactive antibodies to the human immunodeficiency virus type 1 gp120 glycoprotein conferred by a gp41 amino acid change. J Virol 68:674–680. doi: 10.1128/jvi.68.2.674-680.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dey AK, David KB, Klasse PJ, Moore JP. 2007. Specific amino acids in the N-terminus of the gp41 ectodomain contribute to the stabilization of a soluble, cleaved gp140 envelope glycoprotein from human immunodeficiency virus type 1. Virology 360:199–208. doi: 10.1016/j.virol.2006.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dey AK, David KB, Ray N, Ketas TJ, Klasse PJ, Doms RW, Moore JP. 2008. N-terminal substitutions in HIV-1 Gp41 reduce the expression of non-trimeric envelope glycoproteins on the virus. Virology 372:187–200. doi: 10.1016/j.virol.2007.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kassa A, Finzi A, Pancera M, Courter JR, Smith AB III, Sodroski J. 2009. Identification of a human immunodeficiency virus type 1 envelope glycoprotein variant resistant to cold inactivation. J Virol 83:4476–4488. doi: 10.1128/JVI.02110-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kassa A, Madani N, Schön A, Haim H, Finzi A, Xiang S-H, Wang L, Princiotto A, Pancera M, Courter J, Smith AB III, Freire E, Kwong PD, Sodroski J. 2009. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J Virol 83:8364–8378. doi: 10.1128/JVI.00594-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. McGee K, Haim H, Korioth-Schmitz B, Espy N, Javanbakht H, Letvin N, Sodroski J. 2014. The selection of low envelope glycoprotein reactivity to soluble CD4 and cold during simian-human immunodeficiency virus infection of rhesus macaques. J Virol 88:21–40. doi: 10.1128/JVI.01558-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Leaman DP, Zwick MB. 2013. Increased functional stability and homogeneity of viral envelope spikes through directed evolution. PLoS Pathog 9:e1003184. doi: 10.1371/journal.ppat.1003184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gift SK, Leaman DP, Zhang L, Kim AS, Zwick MB. 2017. Functional stability of HIV-1 envelope trimer affects accessibility to broadly neutralizing antibodies at its apex. J Virol 91:e01216-17. doi: 10.1128/JVI.01216-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pacheco B, Alsahafi N, Debbeche O, Prévost J, Ding S, Chapleau J-P, Herschhorn A, Madani N, Princiotto A, Melillo B, Gu C, Zeng X, Mao Y, Smith AB III, Sodroski J, Finzi A, Silvestri G. 2017. Residues in the gp41 ectodomain regulate HIV-1 envelope glycoprotein conformational transitions induced by gp120-directed inhibitors. J Virol 91:e02219–16. doi: 10.1128/JVI.02219-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Nguyen HT, Qualizza A, Anang S, Zhao M, Zou S, Zhou R, Wang Q, Zhang S, Deshpande A, Ding H, Chiu T-J, Smith AB III, Kappes JC, Sodroski JG, Simon V. 2022. Functional and highly cross-linkable HIV-1 envelope glycoproteins enriched in a pretriggered conformation. J Virol 96. doi: 10.1128/jvi.01668-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wang Q, Esnault F, Zhao M, Chiu T-J, Smith AB III, Nguyen HT, Sodroski JG, Simon V. 2022. Global increases in human immunodeficiency virus neutralization sensitivity due to alterations in the membrane-proximal external region of the envelope glycoprotein can be minimized by distant state 1-stabilizing changes. J Virol 96. doi: 10.1128/jvi.01878-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nguyen HT, Wang Q, Anang S, Sodroski JG. 2023. Characterization of the human immunodeficiency virus (HIV-1) envelope glycoprotein conformational states on infectious virus particles. J Virol 97:e0185722. doi: 10.1128/jvi.01857-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Herschhorn A, Ma X, Gu C, Ventura JD, Castillo-Menendez L, Melillo B, Terry DS, Smith AB 3rd, Blanchard SC, Munro JB, Mothes W, Finzi A, Sodroski J. 2016. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. mBio 7:e01598-16. doi: 10.1128/mBio.01598-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Herschhorn A, Gu C, Moraca F, Ma X, Farrell M, Smith AB 3rd, Pancera M, Kwong PD, Schön A, Freire E, Abrams C, Blanchard SC, Mothes W, Sodroski JG. 2017. The beta20-beta21 of gp120 is a regulatory switch for HIV-1 Env conformational transitions. Nat Commun 8:1049. doi: 10.1038/s41467-017-01119-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Melillo B, Liang S, Park J, Schön A, Courter JR, LaLonde JM, Wendler DJ, Princiotto AM, Seaman MS, Freire E, Sodroski J, Madani N, Hendrickson WA, Smith AB 3rd. 2016. Small-molecule CD4-mimics: structure-based optimization of HIV-1 entry inhibition. ACS Med Chem Lett 7:330–334. doi: 10.1021/acsmedchemlett.5b00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Korber BT, Foley BT, Kuiken CL, Pillai SK, Sodroski JG. 1998. Numbering positions in HIV relative to HXB2CG. Available from: https://hfv.lanl.gov/content/sequence/HIV/COMPENDIUM/1998/III/HXB2.pdf
- 95. Lu M, Ma X, Reichard N, Terry DS, Arthos J, Smith AB III, Sodroski JG, Blanchard SC, Mothes W, Simon V. 2020. Shedding-resistant HIV-1 envelope glycoproteins adopt downstream conformations that remain responsive to conformation-preferring ligands. J Virol 94:e00597–20. doi: 10.1128/JVI.00597-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, Mullins J, Rambaut A, Wolinsky S, Korber B (ed). 2014. HIV sequence compendium 2014. Los Alamos National Laboratory, Los Alamos, NM. LA-UR-14-26717. [Google Scholar]
- 97. Privalov PL. 1990. Cold denaturation of proteins. Crit Rev Biochem Mol Biol 25:281–305. doi: 10.3109/10409239009090612 [DOI] [PubMed] [Google Scholar]
- 98. Tsai CJ, Maizel JV, Nussinov R. 2002. The hydrophobic effect: a new insight from cold denaturation and a two-state water structure. Crit Rev Biochem Mol Biol 37:55–69. doi: 10.1080/10409230290771456 [DOI] [PubMed] [Google Scholar]
- 99. Lopez CF, Darst RK, Rossky PJ. 2008. Mechanistic elements of protein cold denaturation. J Phys Chem B 112:5961–5967. doi: 10.1021/jp075928t [DOI] [PubMed] [Google Scholar]
- 100. Kowalski M, Potz J, Basiripour L, Dorfman T, Goh WC, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351–1355. doi: 10.1126/science.3629244 [DOI] [PubMed] [Google Scholar]
- 101. Freed EO, Myers DJ, Risser R. 1990. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc Natl Acad Sci U S A 87:4650–4654. doi: 10.1073/pnas.87.12.4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Delahunty MD, Rhee I, Freed EO, Bonifacino JS. 1996. Mutational analysis of the fusion peptide of the human immunodeficiency virus type 1: identification of critical glycine residues. Virology 218:94–102. doi: 10.1006/viro.1996.0169 [DOI] [PubMed] [Google Scholar]
- 103. Bellamy-McIntyre AK, Lay C-S, Baaör S, Maerz AL, Talbo GH, Drummer HE, Poumbourios P. 2007. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J Biol Chem 282:23104–23116. doi: 10.1074/jbc.M703485200 [DOI] [PubMed] [Google Scholar]
- 104. Haddox HK, Dingens AS, Bloom JD. 2016. Experimental estimation of the effects of all amino-acid mutations to HIV’s envelope protein on viral replication in cell culture. PLoS Pathog 12:e1006114. doi: 10.1371/journal.ppat.1006114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Finzi A, Xiang SH, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. 2010. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Molecular Cell 37:656–667. doi: 10.1016/j.molcel.2010.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang Q, Finzi A, Sodroski J. 2020. The conformational states of the HIV-1 envelope glycoproteins. Trends Microbiol 28:655–667. doi: 10.1016/j.tim.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhang S, Wang K, Wang WL, Nguyen HT, Chen S, Lu M, Go EP, Ding H, Steinbock RT, Desaire H, Kappes JC, Sodroski J, Mao Y, Simon V. 2021. Asymmetric structures and conformational plasticity of the uncleaved full-length human immunodeficiency virus envelope glycoprotein trimer. J Virol 95. doi: 10.1128/JVI.00529-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Salimi H, Johnson J, Flores MG, Zhang MS, O’Malley Y, Houtman JC, Schlievert PM, Haim H. 2020. The lipid membrane of HIV-1 stabilizes the viral envelope glycoproteins and modulates their sensitivity to antibody neutralization. J Biol Chem 295:348–362. doi: 10.1074/jbc.RA119.009481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Lee JH, Andrabi R, Su CY, Yasmeen A, Julien JP, Kong L, Wu NC, McBride R, Sok D, Pauthner M, Cottrell CA, Nieusma T, Blattner C, Paulson JC, Klasse PJ, Wilson IA, Burton DR, Ward AB. 2017. A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic beta-hairpin structure. Immunity 46:690–702. doi: 10.1016/j.immuni.2017.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wang K, Zhang S, Go EP, Ding H, Wang WL, Nguyen HT, Kappes JC, Desaire H, Sodroski J, Mao Y. 2023. Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles. Commun Biol 6:535. doi: 10.1038/s42003-023-04916-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Xiang SH, Finzi A, Pacheco B, Alexander K, Yuan W, Rizzuto C, Huang CC, Kwong PD, Sodroski J. 2010. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J Virol 84:3147–3161. doi: 10.1128/JVI.02587-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wang Y, O’Dell S, Turner HL, Chiang C-I, Lei L, Guenaga J, Wilson R, Martinez-Murillo P, Doria-Rose N, Ward AB, Mascola JR, Wyatt RT, Karlsson Hedestam GB, Li Y. 2017. HIV-1 cross-reactive primary virus neutralizing antibody response elicited by immunization in nonhuman primates. J Virol 91:e00910-17. doi: 10.1128/JVI.00910-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Powell RLR, Totrov M, Itri V, Liu X, Fox A, Zolla-Pazner S. 2017. Plasticity and epitope exposure of the HIV-1 envelope trimer. J Virol 91:e00410-17. doi: 10.1128/JVI.00410-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zolla-Pazner S, Cohen SS, Boyd D, Kong XP, Seaman M, Nussenzweig M, Klein F, Overbaugh J, Totrov M. 2016. Structure/function studies involving the V3 region of the HIV-1 envelope delineate multiple factors that affect neutralization sensitivity. J Virol 90:636–649. doi: 10.1128/JVI.01645-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Binley JM, Ban Y-EA, Crooks ET, Eggink D, Osawa K, Schief WR, Sanders RW. 2010. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J Virol 84:5637–5655. doi: 10.1128/JVI.00105-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Gach JS, Quendler H, Tong T, Narayan KM, Du SX, Whalen RG, Binley JM, Forthal DN, Poignard P, Zwick MB. 2013. A human antibody to the CD4 binding site of gp120 capable of highly potent but sporadic cross clade neutralization of primary HIV-1. PLoS One 8:e72054. doi: 10.1371/journal.pone.0072054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kumar S, Sarkar A, Pugach P, Sanders RW, Moore JP, Ward AB, Wilson IA. 2019. Capturing the inherent structural dynamics of the HIV-1 envelope glycoprotein fusion peptide. Nat Commun 10:763. doi: 10.1038/s41467-019-08738-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB 3rd, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope Glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog 7:e1002101. doi: 10.1371/journal.ppat.1002101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84:1439–1452. doi: 10.1128/JVI.02108-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Montefiori DC, Roederer M, Morris L, Seaman MS. 2018. Neutralization tiers of HIV-1. Curr Opin HIV AIDS 13:128–136. doi: 10.1097/COH.0000000000000442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Rojas Chávez RA, Boyt D, Schwery N, Han C, Wu L, Haim H. 2022. Commonly elicited antibodies against the base of the HIV-1 Env trimer guide the population-level evolution of a structure-regulating region in gp41. J Virol 96:e0040622. doi: 10.1128/jvi.00406-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lu W, Chen S, Yu J, Behrens R, Wiggins J, Sherer N, Liu S-L, Xiong Y, Xiang S-H, Wu L, Kirchhoff F. 2019. The polar region of the HIV-1 envelope protein determines viral fusion and infectivity by stabilizing the gp120-gp41 association. J Virol 93:e02128–18. doi: 10.1128/JVI.02128-18 [DOI] [PMC free article] [PubMed] [Google Scholar]