ABSTRACT
Bifidobacterium is the dominant genus, particularly in the intestinal tract niche of healthy breast-fed infants, and many of these strains have been proven to elicit positive effects on infant development. In addition to its effective antimicrobial activity against detrimental microorganisms, it helps to improve the intestinal microbiota balance. The isolation and identification of bacteriocins from Bifidobacterium have been limited since the mid-1980s, leading to an underestimation of its ability for bacteriocin production. Here, we employed a silicon-based search strategy to mine 354 putative bacteriocin gene clusters (BGCs), most of which have never been reported, from the genomes of 759 Bifidobacterium strains distributed across 9 species. Consistent with previous reports, most Bifidobacterium strains did not carry or carry only a single BGC; however, Bifidobacterium longum subsp. infantis, in contrast to other Bifidobacterium species, carried numerous BGCs, including lanthipeptides, lasso peptides, thiopeptides, and class IId bacteriocins. The antimicrobial activity of the crude bacteriocins and transcription analysis confirmed its potential for bacteriocin biosynthesis. Additionally, we investigated the association of bacteriocins with the phylogenetic positions of their homologs from other genera and niches. In conclusion, this study re-examines a few Bifidobacterium species traditionally regarded as a poor source of bacteriocins. These bacteriocin genes impart a competitive advantage to Bifidobacterium in colonizing the infant intestinal tract.
IMPORTANCE
Development of the human gut microbiota commences from birth, with bifidobacteria being among the first colonizers of the newborn intestinal tract and dominating it for a considerable period. To date, the genetic basis for the successful adaptation of bifidobacteria to this particular niche remains unclear since studies have mainly focused on glycoside hydrolase and adhesion-related genes. Bacteriocins are competitive factors that help producers maintain colonization advantages without destroying the niche balance; however, they have rarely been reported in Bifidobacterium. The advancement in sequencing methods and bacteriocin databases enables the use of a silicon-based search strategy for the comprehensive and rapid re-evaluation of the bacteriocin distribution of Bifidobacterium. Our study revealed that B. infantis carries abundant bacteriocin biosynthetic gene clusters for the first time, presenting new evidence regarding the competitive interactions of Bifidobacterium in the infant intestinal tract.
KEYWORDS: genome mining, bacteriocin gene clusters, Bifidobacterium, B. longum subsp. infantis, antimicrobial activity, RT-qPCR, niche competition
INTRODUCTION
Bacteriocins are antimicrobial peptides synthesized by bacterial ribosomes that can inhibit the growth of other related or unrelated microorganisms (1). Bacteriocins are widespread across the human intestinal tract and are crucial in determining the composition of gut microbiota (2, 3). They help producers compete in their specific ecological niches, maintain a balance in microbial populations (4, 5), and actively participate in host defense against potential gastrointestinal pathogens (6). Many bacteriocins have been isolated and identified from the gut microbiota and are roughly divided into three classes based on molecular weight, post-translational modification, chemical structure, thermal stability, and other properties (3, 7). Class I bacteriocins are post-translationally modified peptides (RiPPs), which are further subdivided into lanthipeptides, lasso peptides, sactipeptides, autoinducing peptides (AIPs), thiopeptides, and other subgroups according to the comprehensive nomenclature of RiPPs (8). Lanthipeptides are characterized by the β-thioether cross-linked bis-amino acids lanthionine (Lan) and methyllanthionine (MeLan). Class I lanthipeptides are characterized by two proteins that individually catalyze dehydration (LanB) and cyclization (LanC). Class II lanthipeptides biosynthesis involves a single protein with both dehydration and cyclization domains (LanM). Class III and class IV lanthipeptides are characterized by distinct phosphorylation-mediated dehydration processes (9). Lasso peptides are named for their topology resembling a threaded lasso or a slipknot, rendering them resistant to heat, chemicals, and proteases (10). Sactipeptides are distinguished by radical S-adenosylmethionine (rSAM) enzymes that form intramolecular thioether linkage between a donor Cys sulfur and the alpha carbon of another acceptor amino acid (11). AIPs are peptide signals modulating the expression of virulence factors via the Agr quorum-sensing system, which comprises response regulator AgrA, histidine kinase AgrC, and protease AgrB that cleaves AgrD (precursor peptide) (12). Thiopeptides are highly modified macrocyclic peptides with a six-membered heterocycle grafted on the peptidic backbone being central to pyridine/piperidine/dehydropiperidine, and the enzymes that are necessary for primary macrocyclization, such as YcaO, ThiF, dehydrogenase, dehydratase, and macrocyclase, are generally conserved. Small (<10 kDa), heat-stable class II bacteriocins are subdivided into class IIa peptides (containing the YGNGV motif), class IIb peptides (two-peptide bacteriocins), class IIc peptides (circular bacteriocins), and class IId peptides (unmodified, linear, non-pediocin-like, and single-peptide bacteriocins) based on their structure and function (3). Class III bacteriocins are large (>30 kDa), heat-labile proteins that have not been extensively characterized (3).
Bifidobacterium is among the first bacteria that colonize and inhabit the human intestinal tract for life. Certain species, such as B. longum subsp. infantis (B. infantis) and Bifidobacterium breve, are more common in the infant gut microbiome. Other species co-exist in the intestinal tract of infants and adults, including B. longum subsp. longum (B. longum), Bifidobacterium adolescentis, and Bifidobacterium pseudocatenulatum (13). Bifidobacterium is the dominant genus in the intestinal tract of healthy breast-fed infants, and its relative abundance appears to be negatively correlated with several disease states (14). Members of the Bifidobacterium genus are widely recognized as beneficial health-promoting bacteria due to their extensive prebiotic functions. However, the bacteriocin-producing capability of bifidobacteria has been underestimated despite their inhibition of detrimental microbial growth (15). Typically, their antibacterial activity is attributed to the inhibition of organic acids and a decrease in pH (16). Compared to the extensively reported bacteriocin-producing lactic acid bacteria (LAB) (17), such as Lactobacillus and Enterococcus, only a few strains of bacteriocin-producing Bifidobacterium have been discovered, with scarce information on the genes or structure of the bacteriocins (18). The above mining of bacteriocin from bifidobacteria is based on antimicrobial phenotype to screen, which is labor- and time-intensive.
With the rapid advances in sequencing technologies, numerous bacteriocins have been identified and characterized by laborious screening methods and biochemical characterization; in silico screening strategy based on genomics and bioinformatics has become a more efficient means of bacteriocin mining. Genome mining tools, such as the bacteriocin mining tool BAGEL (19) and the secondary metabolite mining tool antiSMASH (20), exploit the conserved domains of genetic elements in bacteriocin gene clusters (BGCs), enabling the easy identification of new compounds in microorganisms (21 – 23). Despite these advancements, the BGCs in the Bifidobacterium genus were rarely mined in a recent study (24), possibly due to the lack of sufficiently comprehensive data for analysis. Equipped with the available genome data of the Bifidobacterium genus, we addressed these shortcomings by evaluating the bacteriocin biosynthesis potential of the Bifidobacterium genus. Through genome mining, we characterized valuable phenotypic traits associated with antimicrobial activity and explored the potential associations among Bifidobacterium, bacteriocins, and niches.
RESULTS
Most Bifidobacterium species are poor sources of bacteriocins
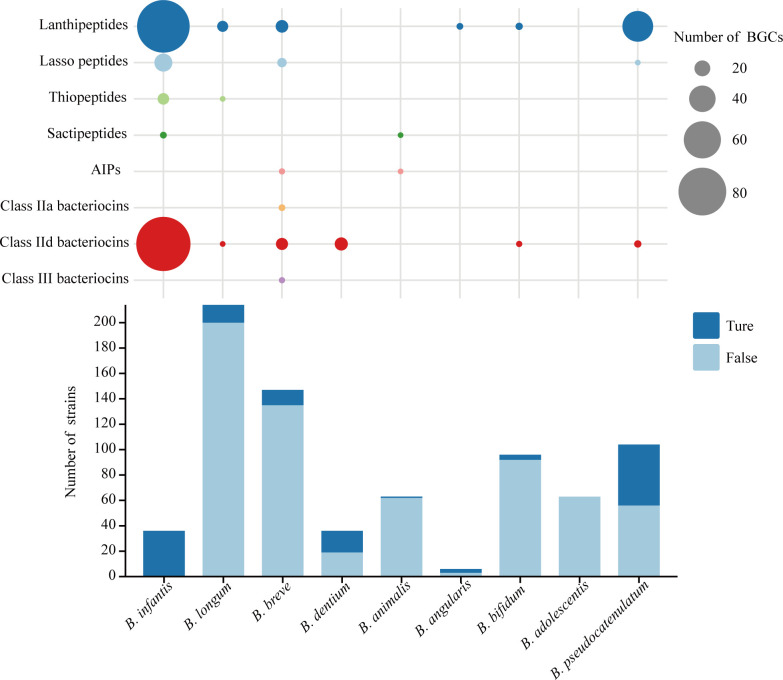
We identified 354 putative BGCs from 133 Bifidobacterium strains (Table S3). Based on the gene organization and homology of their structural and biosynthetic genes, these BGCs were classified into three classes harboring eight subgroups, namely, lanthipeptides, lasso peptides, thiopeptides, sactipeptides, AIPs, class IIa bacteriocins, class IId bacteriocins, and class III bacteriocins (Fig. 1). Gene clusters with the same structural composition and order under a subgroup of BGCs were grouped into a “type.” The situation of each Bifidobacterium species carrying BGCs was diverse. No BGC was found in B. adolescentis, and only one Bifidobacterium animalis strain carried BGCs among strains of the species. Three of the six Bifidobacterium angulatum strains possessed one type of class II lanthipeptide gene cluster, and 15 Bifidobacterium dentium strains possessed one type of class II BGC. A total of 48 B. pseudocatenulatum strains harbored three types of BGC, primarily (~90%) concentrated in the undetermined class IV lanthipeptide gene cluster. Thirty-eight BGCs encoding lanthipeptides, lasso peptides, AIPs, and class II and III bacteriocins were mined from 12 B. breve strains.
Fig 1.
BGCs distribution of the Bifidobacterium. The bar plot shows the number of Bifidobacterium strains with/without putative BGCs; the bubble plot shows the number of different groups of BGCs carried by the Bifidobacterium species.
Abundant and diverse BGCs were mined in B. infantis
In contrast to the aforementioned Bifidobacterium species exhibiting a low BGC distribution frequency, B. infantis has great potential to produce a wide variety of bacteriocins. A total of 36 B. infantis isolates harbored five subgroups of 220 BGCs, and the occurrence of putative BGCs was 100%. Moreover, except class II and III bacteriocins found in two B. breve strains and three AIPs found in one B. breve strain and one B. animalis strain, B. infantis harbored all the BGCs found in other Bifidobacterium species. We constructed the phylogenetic tree of homologous sequences of various bacteriocins and found that most bacteriocins from B. infantis were native and were only distributed in the branches of Bifidobacterium. As an additional note, we tested four other strains (CECT7210, 157F, JCM11660, and KCTC5934) marked as B. infantis in NCBI; the results revealed that they did not harbor any BGC. The average nucleotide identity (ANI) analysis found that they were clustered with B. longum subsp. longum (Fig. S2). Therefore, we did not include them in our study.
The following sections describe the characteristics of various BGCs classified by bacteriocin groups in B. infantis in detail. The precursor peptide sequences carried by other Bifidobacterium species are also displayed.
Lanthipeptides
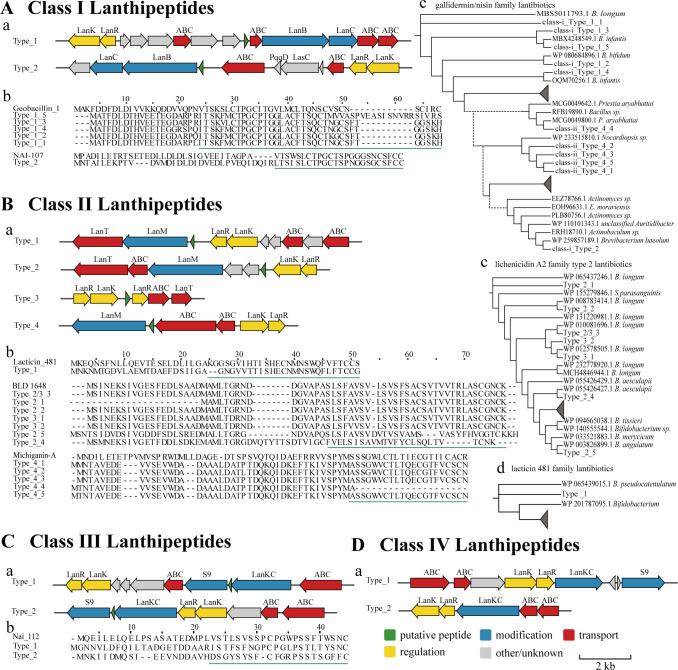
In this study, 104 putative lanthipeptide gene clusters were discovered in B. infantis, which was also the largest subgroup. Among these, two types of class I lanthipeptide gene clusters were distributed across the genomes of the 15 B. infantis strains. According to the results of basic local alignment search tool (BLAST) in the nonredundant (nr) protein database, all annotated precursor peptides of class I lanthipeptides belonged to the gallidermin/nisin family lantibiotics (Fig. 2Ac). Five different precursor peptide sequences were obtained from the first gene cluster and were found to cluster with the other lantibiotics from Bifidobacterium on the phylogenetic tree differing only at a few amino acid positions (Fig. 2Ab). Alignment of the precursor peptides with identified bacteriocins revealed higher similarity with the geobacillin I sequence from Geobacillus thermodenitrificans (25). Three B. infantis (FHuNCS6M8, FJND16M4, and FLNSY3M1) and two Bifidobacterium bifidum (FGZ3I2M1 and FGZ6I2M1) strains had class I lanthipeptide gene clusters similar to the first type; however, they lacked precursor peptide genes. After excluding the sequencing factor via reverse transcription-quantitative PCR (RT-qPCR) analysis, we assumed that the precursor genes of these strains could have been lost due to genetic drift; therefore, these strains were excluded. Similarly, two B. dentium strains (FNXHL9M3 and FNXHL9M2) were disregarded due to the absence of precursor genes. In the second cluster, the lanthipeptide gene cluster was adjacent to the lasso peptide gene cluster; hence, we suspect that they share the two-component regulatory system genes, LanR and LanK. One precursor peptide was obtained from the second gene cluster, which did not cluster with the specific genus. Its core peptide sequence shared 83.3% identity with the NAI-107 (also known as microbisporicin or 107,891) produced by Microbispora sp. (26, 27).
Fig 2.
BGC mining results of class I–IV lanthipeptides (A–D) containing bacteriocin biosynthetic gene clusters (A), multiple sequence alignment (MSA) results of putative lantibiotic prepeptides with known bacteriocin (B), and the phylogenetic tree of homologous sequences in the NCBI (C and D). The green underline indicates the core peptide sequence in the MSAs.
We identified four types of class II lanthipeptide gene clusters from 29 B. infantis strains (Fig. 2B). The first type was homologous to the class II lanthipeptide lacticin 481 (also known as lactococcin DR) gene cluster of Lactococcus lactis (28). The second cluster had been reported to encode BLD 1648 (based on the BAGEL4 database) in B. longum DJO10A (2, 29). The third cluster, mined using BAGEL4, encoded a similar precursor peptide as the second cluster but lacked the important modification gene lanM. Although nearly half of these strains encoded other class II lanthipeptides, we could not determine the influence of the absence of lanM in the gene cluster on the normal activity of the lantibiotic. BLAST results identified that these compounds belong to lichenicidin A2 family type 2 lantibiotics. The fourth type was found exclusively in B. infantis and shared homology with five similar precursor peptides annotated as gallidermin/nisin family lantibiotics, based on BLAST results. When compared with known bacteriocins in the BAGEL4 database, they exhibited the highest similarity to michiganin-A, obtained from Clavibacter michiganensis subsp. michiganensis (30). B. infantis strain M203F0227 lacked this precursor gene by annotation; however, we confirmed its presence by RT-qPCR.
Two types of class III lanthipeptide gene clusters were distributed across the genomes of 20 B. infantis strains, as well as 5 B. pseudocatenulatum strains, 1 B. brevis strain, and 1 B. bifidum strain (Fig. 2C). The precursor peptide from the first gene cluster was homologous to the class III lanthipeptide NAI-112 produced by Actinoplanes spp. when aligned against the sequences in the BAGEL4 database (31, 32). No known bacteriocins homologous to the second precursor peptide were identified, suggesting it may be a novel class III lanthipeptide. Additionally, we found two types of class IV lanthipeptide gene clusters, the first was identified in 3 B. infantis, and the second was identified in 46 B. pseudocatenulatum strains (Fig. 2D). We were unable to identify the potential precursor peptides using BLAST.
Lasso peptides
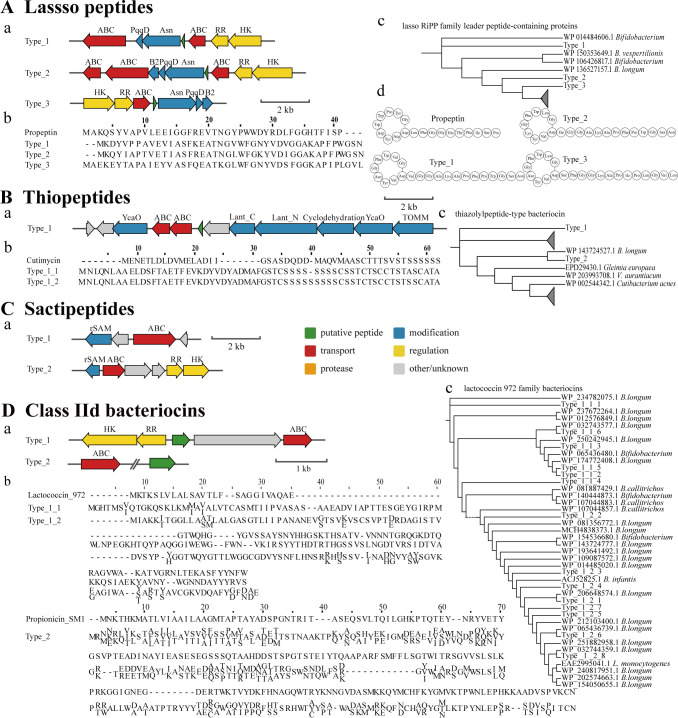
A phylogenetic tree was constructed for the lasso RiPP family leader peptide-containing protein sequences from NCBI (https://www.ncbi.nlm.nih.gov/), and the three precursor peptides were clustered with other lasso peptides from Bifidobacterium (Fig. 3Ad). The identified lasso peptides exhibited higher similarities to the propeptin sequence from Microbispora sp. SNA-115 (33, 34). We predicted the cyclization structure of core peptides based on the structure of propeptin (Fig. 3Ac).
Fig 3.
BGC mining results of lasso peptides (A), thiopeptides (B), sactipeptides(C), and class IId bacteriocins (D) containing bacteriocin biosynthetic gene clusters (A), multiple sequence alignments (MSAs) of putative precursor peptides with known bacteriocins (B), the phylogenetic tree of homologous sequences in the NCBI (C), and predicted structures based on identified bacteriocins (D). The green underline indicates the core peptide sequence in the MSAs. Considering that multiple sequences are only different at several amino acid sites, MSA uses the consensus results to show in D.
Thiopeptides
Over 100 thiopeptides have been isolated and characterized heretofore; however, none have been isolated from Bifidobacterium before this study (35). We found one type of putative thiopeptide BGC, which most likely encodes a series D thiopeptide in 12 B. infantis strains and 1 B. longum strain (Fig. 3B). Although, none of the thiopeptides in the bacteriocin databases were found to be similar to the precursor peptides in this Bifidobacterium strain, we did identify a precursor peptide and a thiopeptide from Cutibacterium acnes, called cutimycin, located on a subunit in the phylogenetic tree of the thiazolylpeptide-type bacteriocin sequences (36).
Sactipeptides
We identified two types of putative sactipeptide gene clusters in the genomes of three B. infantis strains and one B. animalis strain; however, no precursor peptides were predicted (Fig. 3C). Unlike the genomes of the three B. infantis strains, B.animalis FJSNJLH3M11 genomes encoded histidine kinase and response regulators besides encoding rSAM enzymes and ABC transporter. This is the only reported putative sactipeptide gene cluster from Bifidobacterium, although this finding requires further investigation.
Class IId bacteriocins
Lactococcin 972 (Lcn972) is a member of the class IId bacteriocins (37). We found one or two gene clusters encoding Lcn972 analogs in all 36 B. infantis strains. This type of gene cluster encodes a precursor peptide, an ABC transporter protein, and a two-component regulatory system, portraying a simple genetic organization. The precursor peptides were clustered in the phylogenetic tree of Lcn972 together with sequences from other B. longum strains (Fig. 3Dc). The second type of precursor peptide cluster also shared homology with Lmo2776, a bacteriocin produced by L. monocytogenes (38).
Propionicin SM1-like genes were mined using BAGEL4 in 59 genomes distributed among 30 B. infantis, 15 B. dentium, 8 B. brevis, 4 B. pseudocatenulatum, and 2 B. bifidum strains (39). Some gene clusters included only precursor peptides, without ABC transporter proteins (Fig. 3E). The overall similarity of amino acid sequences between these precursor peptides and propionicin SM1 was 26.1%–50.5%, although they were annotated as DUF2599 domain-containing proteins in the nr protein database.
BGC horizontal transfer exists in B. breve and B. animalis strains
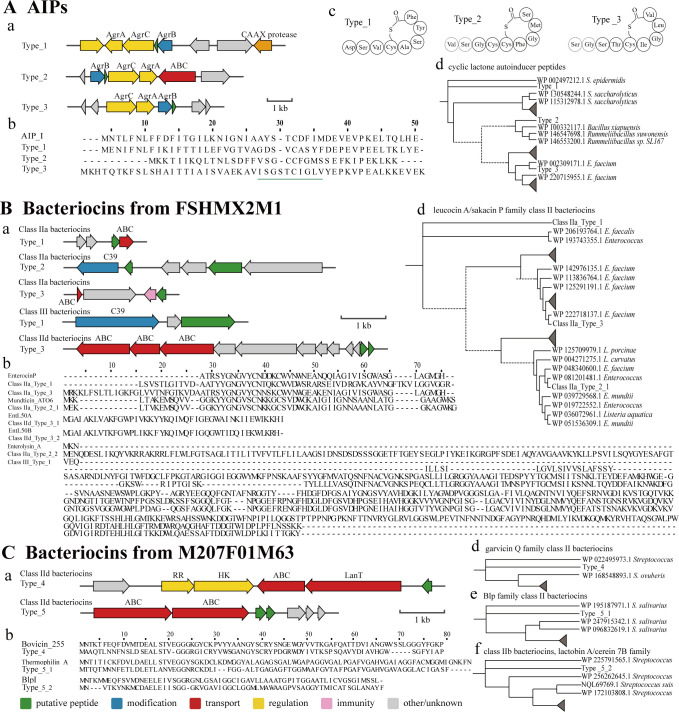
Three bacteriocins previously associated with other species were identified in B. breve and B. animalis strains. A core AIP from Staphylococcus epidermidis (40) was identified in B. animalis FJSNJLH3M1 (Fig. 4A type_1); additionally, EntL50A and EntL50B from Enterococcus faecium L50 (41) were identified in B. breve FSHMX2M1 (Fig. 4B class IId type_3). Additionally, some putative bacteriocins were identified in the above strains and B. breve M207F01M63 that do not exist in other Bifidobacterium. Most of these bacteriocins are in a cluster with those bacteriocins from, e.g., Enterococcus, Staphylococcus, and Streptococcus in their respective phylogenetic tree.
Fig 4.
BGC mining results of AIPs (A), B. brevis FSHMX2M1 (B), and B. brevis M207F01M63 (C) containing bacteriocin biosynthetic gene clusters (A), MSAs of putative precursor peptides with known bacteriocins (B), predicted structures based on identified bacteriocins (C), and the phylogenetic tree of homologous sequences in the NCBI (D–F). The green underline indicates the core peptide sequence in the MSAs.
Although AIPs did not exist in B. infantis, three putative gene clusters were found in the genomes of one B. animalis strain and one B. breve strain (Fig. 4A). One of the AIPs in B. breve was homologous to the AIP in Enterococcus faecalis. Based on the similarity to the S. epidermidis, the AIP structure of B. animalis was shown in Fig. 4Ac (40). The structure of the other two AIPs from B. breve still needed experimental verification.
Seven putative bacteriocins encoding Class II and Class III bacteriocins were identified in five gene clusters of FSHMX2M1 (Fig. 4B). The precursor peptides encoded by the first and third gene cluster were similar to the class IIa bacteriocin enterocin P produced by E. faecium (48.3% and 56.3%, respectively) (42). Two putative bacteriocins were identified in the second cluster. One of the bacteriocins, located upstream, had 94.8% sequence similarity with the class IIa bacteriocin mundticin ATO6 produced by Enterococcus mundtii ATO6 (43). The other bacteriocin, located downstream, showed 29.8% sequence similarity with enterolysin A. Additionally, the bacteriocin encoded in the fourth gene cluster was similar to enterolysin A with 20.20% similarity (44, 45). The fifth gene cluster carried bacteriocins consistent with class IIb bacteriocin Enterocin L50 (46).
Two class II bacteriocin gene clusters were identified in B. brevis M207F01M63 (Fig. 4C). The first precursor peptide showed 60.8% sequence similarity to the class IId bacteriocin Bovicin 255 produced by Streptococcus gallolyticus LRC0255 (47). This peptide and Streptococcus-derived peptides were clustered in the same cluster on the phylogenetic tree of the garvicin Q family class II bacteriocin. The second gene cluster carried two hypothetical class II bacteriocin genes, which were located downstream of two ABC transporter genes and showed sequence similarity to thermophilin A (68.2%) and BlpI (60.8%) (48, 49). The first precursor peptide clustered with a peptide from Streptococcus in the phylogenetic tree of Blp family class II bacteriocins. The second precursor peptide was grouped with peptides from Streptococcus in the phylogenetic tree of the lactobin A/cerein 7B family.
B. infantis strains and their crude bacteriocins inhibit the growth of common competitive bacteria in the infant intestinal tract
Owing to the abundance of bacteriocin synthesis genes harbored by B. infantis, we tested the antimicrobial activity of 29 B. infantis strains against 9 bacteria that were common in infant intestines. As shown in Table 1, all 29 B. infantis strains exhibited broad antimicrobial activity against both Gram-negative bacteria and Gram-positive bacteria, except for the Lactobacillus. During testing, all crude bacteriocins lost their activity against Gram-negative bacteria (Table 2). Most crude bacteriocins showed strong inhibitory activity against E. faecalis and L. monocytogenes. Considering that each B. infantis strain produces several bacteriocins and other antimicrobial substances may also be present in the crude bacteriocins, the target bacteria of specific bacteriocin need to be validated later after isolation and purification.
TABLE 1.
The antimicrobial activity of B. infantis strains a
| Zone size | E. faecalis | S. aureus | B. adolescens | B. bifidum | L. plantarum | L. rhamnosus | L. monocytogenes | K. pneumoniae | E. coli | S. enterica |
|---|---|---|---|---|---|---|---|---|---|---|
| FGZ19I2M3 | + | + | + | + | − | − | ++ | + | ++ | ++ |
| FGZ23I1M7 | ++ | ++ | ++ | ++ | − | − | +++ | + | ++ | +++ |
| FHeNJZ3M1 | ++ | ++ | + | ++ | − | − | +++ | ++ | +++ | +++ |
| FHNFQ45M2 | + | + | ++ | +++ | − | − | +++ | + | +++ | +++ |
| FHuNCS6M8 | ++ | ++ | + | ++ | − | − | +++ | ++ | +++ | +++ |
| FJND2M2 | + | + | + | +++ | − | − | +++ | + | +++ | +++ |
| FJND16M4 | ++ | ++ | ++ | ++ | − | − | +++ | ++ | +++ | +++ |
| FJSYZ1M3 | ++ | ++ | + | ++ | − | − | +++ | + | +++ | +++ |
| HeNJZ8M1 | +++ | ++ | ++ | ++ | − | − | +++ | ++ | +++ | +++ |
| JSSZ7M7 | ++ | ++ | + | ++ | − | − | +++ | ++ | +++ | +++ |
| JSWX3M1 | ++ | ++ | ++ | +++ | − | − | +++ | ++ | +++ | +++ |
| JSWX6M2 | + | + | + | ++ | − | − | ++ | + | +++ | ++ |
| JSWX25_6 | + | + | + | + | − | − | ++ | + | +++ | +++ |
| JSWX25M6 | + | + | + | +++ | − | − | +++ | ++ | +++ | +++ |
| SDZC2M4 | + | + | + | ++ | − | − | +++ | + | +++ | +++ |
| FGZ17I1M1 | ++ | ++ | + | ++ | − | − | +++ | + | +++ | +++ |
| FGZ19I1M3 | + | + | + | ++ | − | − | ++ | + | +++ | +++ |
| FGZ23I1M2 | + | + | + | ++ | − | − | ++ | + | +++ | +++ |
| FHeJZ44M8 | ++ | ++ | ++ | ++ | − | − | +++ | ++ | +++ | +++ |
| M203F0227 | + | + | + | ++ | − | − | ++ | ++ | +++ | +++ |
| FBJTZ1M6 | ++ | ++ | ++ | ++ | − | − | +++ | + | +++ | ++ |
| FHBSJZ9M1 | ++ | ++ | ++ | + | − | − | +++ | + | ++ | + |
| FHBSJZ50M1 | ++ | ++ | ++ | ++ | − | − | +++ | ++ | +++ | + |
| FHeBSJZ1M1 | ++ | ++ | ++ | ++ | − | − | +++ | + | +++ | + |
| FHNFQ4M11 | ++ | ++ | ++ | ++ | − | − | +++ | + | ++ | + |
| FJSWXI2MIM1 | ++ | ++ | ++ | ++ | − | − | +++ | ++ | +++ | + |
| FJSWXI4MIM1 | + | ++ | ++ | + | − | − | ++ | + | ++ | + |
| FLNSY3M1 | ++ | ++ | ++ | + | − | − | +++ | ++ | +++ | + |
| M207F01M51 | ++ | ++ | ++ | + | − | − | +++ | + | ++ | + |
Zone size = diameter of the zone − diameter of the spot in millimeters. – means no antimicrobial activity; + means 0–10 mm; ++ means 10–20 mm; +++ means >20 mm.
TABLE 2.
The antimicrobial activity of crude bacteriocins from B. infantis strains a
| Zone size | E. faecalis | S. aureus | B. adolescens | B. bifidum | L. plantarum | L. rhamnosus | L. monocytogenes | K. pneumoniae | E. coli | S. enterica |
|---|---|---|---|---|---|---|---|---|---|---|
| FGZ19I2M3 | ++ | ++ | − | ++ | − | − | +++ | − | − | − |
| FGZ23I1M7 | ++ | ++ | − | − | − | − | +++ | − | − | − |
| FHeNJZ3M1 | ++ | ++ | − | − | − | − | +++ | − | − | − |
| FHNFQ45M2 | + | ++ | − | − | − | − | +++ | − | − | − |
| FHuNCS6M8 | +++ | ++ | − | − | − | − | +++ | − | − | − |
| FJND2M2 | +++ | ++ | − | − | − | − | +++ | − | − | − |
| FJND16M4 | +++ | + | − | ++ | − | − | ++ | − | − | − |
| FJSYZ1M3 | ++ | ++ | + | ++ | − | − | ++ | − | − | − |
| HeNJZ8M1 | ++ | + | − | − | − | − | +++ | − | − | − |
| JSSZ7M7 | +++ | ++ | − | − | − | − | +++ | − | − | − |
| JSWX3M1 | +++ | ++ | +++ | +++ | − | − | +++ | − | − | − |
| JSWX6M2 | ++ | ++ | − | − | − | − | ++ | − | − | − |
| JSWX25_6 | ++ | ++ | − | − | − | − | +++ | − | − | − |
| JSWX25M6 | +++ | ++ | − | − | − | − | ++ | − | − | − |
| SDZC2M4 | +++ | + | ++ | +++ | − | − | ++ | − | − | − |
| FGZ17I1M1 | ++ | ++ | − | − | − | − | +++ | − | − | − |
| FGZ19I1M3 | ++ | + | − | − | − | − | +++ | − | − | − |
| FGZ23I1M2 | ++ | + | − | − | − | − | +++ | − | − | − |
| FHeJZ44M8 | +++ | ++ | − | +++ | − | − | +++ | − | − | − |
| M203F0227 | +++ | ++ | − | + | − | − | ++ | − | − | − |
| FBJTZ1M6 | +++ | ++ | +++ | ++ | − | − | +++ | − | − | − |
| FHBSJZ9M1 | +++ | + | − | + | − | − | +++ | − | − | − |
| FHBSJZ50M1 | ++ | + | + | − | − | − | ++ | − | − | − |
| FHeBSJZ1M1 | ++ | + | + | − | − | − | + | − | − | − |
| FHNFQ4M11 | ++ | + | − | ++ | − | − | +++ | − | − | − |
| FJSWXI2MIM1 | ++ | + | − | − | − | − | ++ | − | − | − |
| FJSWXI4MIM1 | +++ | + | ++ | + | − | − | ++ | − | − | − |
| FLNSY3M1 | +++ | + | − | ++ | − | − | ++ | − | − | − |
| M207F01M51 | ++ | + | − | ++ | − | − | ++ | − | − | − |
Zone size = diameter of the zone − diameter of Oxford cup in millimeters. – means no antimicrobial activity; + means 0–5 mm; ++ means 5–10 mm; +++ means 10–15 mm.
The levels of BGCs transcription varied among the confirmed clusters
All confirmed BGCs were transcribed in the deMan Rogosa Sharpe (MRS) liquid medium at different levels (Fig. S1). Among the 29 Bifidobacterium strains, the genes with the least difference in transcription levels were lan-i-T_2, lan-iii-T_1, lan-iii-T_2, lan-ii-T_1, and lasA, whereas the transcription levels of lan-ii-T_4, lcn972-T_1_1, and lcn972-T_1_2 were quite different. During the analysis, we found that many precursor peptide genes had low transcription intensity in the liquid medium, which complicated the screening of bacteriocin-producing Bifidobacterium strains using the fermentation supernatant and purification of bacteriocin (50). Using previously mentioned primers, we excluded a few BGCs that lacked the precursor gene and confirmed the BGCs whose precursor genes were disregarded due to sequencing/assembly reasons. These primers may facilitate the efficient screening of Bifidobacterium strains carrying bacteriocin synthesis genes.
DISCUSSION
The first putative Bifidobacterium-associated bacteriocin was bifidin, isolated from B. bifidum NCDC 1452 in the mid-1980s (51). However, the research progress on bacteriocins involving Bifidobacterium has been slow since then. To date, only nine putative Bifidobacterium-associated bacteriocins have been identified, one in B. lactis Bb-12 (52), two in B. bifidum (53, 54), three in B. longum (29, 55), one each in B. thermophilum RBL67 (56), B. animals BB04 (57), and B. infantis BCRC14602 strains (51); however, most of these bacteriocins have not been strictly characterized or sequenced. Bifidocin B, bifidin I, and BLD 1648 are the only Bifidobacterium-associated bacteriocins whose sequences have been partially elucidated. The mining of the above bacteriocin mainly depends on the inhibitory effects exerted by the producer on the indicator bacteria and is evaluated using agar plates or direct tests. These in vitro methods can be used to discover new bacteriocins and evaluate their diverse antimicrobial abilities. However, the antimicrobial ability detected by these methods is affected by nutritional and biological factors, such as medium composition, stress conditions, and the presence of target bacteria, which may result in false-negative results (18). Numerous bacterial whole-genome sequence data and BGC mining tools have made the identification of hypothetical BGCs using in silico analysis efficient and convenient (21, 22). In this study, we found that the proportion of BGCs harbored by each of Bifidobacterium species was either low or that of a single type, except for B. infantis, explaining the scarce detection of bifidobacterial bacteriocins previously (18). B. infantis stands out as a potential bacteriocin producer and harbors abundant BGCs that can serve as its distinct species marker.
In certain cases, complete gene clusters lacking evident bacteriocin precursor genes were identified, which predominantly belonged to class IV lanthipeptides and sactipeptides. Excluding low-quality genome assembly, the reasons for “unidentified” are as follows: (i) the precursor peptide is encoded elsewhere in the genome, (ii) the precursor gene is lost due to genetic drift, (iii) rSAM is not involved in sactipeptide biosynthesis (58), and (iv) scarce class IV lanthipeptides- and sactipeptide-associated data are used for alignment. The disadvantage of bacteriocin mining in silico analysis is that it mainly depends on the similarity with the previously discovered BGCs, indicating the possible oversight of novel BGCs. To date, all class IV lanthipeptides are analogs of venezuelin from Streptomyces venezuelae (59), and only eight sactipeptides have been identified (11, 60). The number of described and characterized bacteriocins of this class is relatively small compared to that of other classes; to that effect, only two class IV lanthipeptides and six sactipeptides have been included in the BAGEL database. Zheng et al. also reported a relatively low incidence of sactipeptides (61). Therefore, we speculate that there may be novel class IV lanthipeptides and sactipeptides in Bifidobacterium (62). Histidine kinase and response regulator genes were identified in the sactipeptide gene cluster from B. animalis FJSNJLH3M11 indicating that a two-component regulatory system was subjected to sactipeptide, which had never been reported in other known sactipeptide genes.
BGCs are important genetic determinants of competitive fitness in a given niche and are frequently associated with mobile genetic elements. Competitive growth experiments in a simulated fecal environment showed that B. longum DJO10A outcompeted both E. coli and C. difficile more effectively than B. longum DJO10A-JH1 (lantibiotic operon deletion strain) (50). This indicates that bacteriocin-encoding genes play an important role in intestinal niche competition. B. infantis has the potential to produce a tremendous diversity of bacteriocins. It mainly exists in the intestinal tract of healthy breast-fed infants and dominates the stool microbiota for a period (63). Compared to the adult intestinal microbiome, the infant microbiome is less dense yet more diverse because of the neonatal diet of human milk. It can be assumed that dominant bacteria in the infant intestinal tract compete more effectively than the other bacteria. The homologous sequences of bacteriocins in the NCBI database can be used to understand the associations between bacteriocins and niches. Considering the comprehensiveness of bacteriocins carried by B. infantis, we suspect B. infantis to be the main source of BGCs in Bifidobacterium. One of the reasons that B. infantis rather than other Bifidobacterium species carrying many BGCs may be the metabolic costs. Although there are direct benefits to the host, the expression of BGCs brings more burden on the cell (64). This genetic material is transformed from a precursor peptide to a mature bacteriocin by modification, cleavage, and transport, which requires the secretion and production of numerous secondary metabolites, and the cell energy level may consequently be reduced (65). These processes require adaptive mutations or changes in nutrient supply to optimize the production of compounds (66). B. infantis has a tremendous number of genes involved in the metabolism of complex carbohydrates and is the only member of the Bifidobacterium genus that can digest all human milk oligosaccharide (HMO) structures (67). Previous studies indicate that their excellent utilization of HMOs is also the main colonization mechanism of B. infantis in this niche. The abundance of BGCs in the HMO-friendly B. breve, another dominant Bifidobacterium species in the infant intestinal tract, was second to that in B. infantis. Conversely, in some “adult-type” Bifidobacterium species (13), such as B. adolescentis, BGCs were either absent or scarce. These “adult-type” Bifidobacterium species lack the advantage of saccharolytic metabolism in the infant intestinal niche, while bacteriocin gene expression imposes an additional metabolic burden. It is a trade-off for other “adult-type” Bifidobacterium species to acquire these BGCs, as B. infantis and B.breve produce enough bacteriocins to eliminate targeted competitors. On the contrary, we hypothesized that the bacteria targeted by these bacteriocins could be potentially threatening competitors of Bifidobacterium in the infant intestinal niche. In the complex intestinal niche of adults, these BGCs are not sufficient to provide robust competition to the host and thus not prevalent in the “adult-type” Bifidobacterium species.
Several other Bifidobacterium strains carried bacteriocins with homology similar to bacteriocins from Staphylococcus, Enterococcus, and Streptococcus. The distribution of similar BGCs among different species indicates that horizontal gene transfer occurs actively among species. These three genera are also some of the first microorganisms to colonize the infant intestinal tract, accounting for a high abundance. Many studies have reported their rich production of bacteriocins (68 – 70). Bacteriocins are generally thought to exhibit inhibitory activities against bacterial strains that are closely related to the bacteriocin-producing bacteria strain. After searching the antimicrobial activity of those most similar bacteriocins to that from Bifidobacterium as reference (Table S4), many similar bacteriocins could inhibit Enterococcus and Staphylococcus. The bacteriocin bifidin I has strong anti-Listeria activity (51) and is homologous to propionicin SM1 which is widely expressed in the test B. infantis. This may partially explain the strong antibacterial activity of crude bacteriocins against L. monocytogenes. When comes to crude bacteriocins of test B. infantis, antimicrobial activities against Gram-negative bacteria are lost. In addition to the fact that other non-proteinaceous antimicrobial substances were produced by the strains, there can be two potential explanations: (i) lower pH values exert a dual effect both by promoting the production of bacteriocins (71) and by enhancing their antibacterial effects (55); additionally, (ii) compared with agar culture, the expression of bacteriocin precursor genes in broth culture was obviously repressed (29). Some research has reported the antibacterial activity of crude extracts from other Bifidobacterium species against E. faecalis, L. monocytogenes, and S. aureus (18).We speculate that B. infantis facilitates the predominance of Bifidobacterium in the infant intestinal tract by harboring BGCs that inhibit the growth of competing microorganisms. This study provides novel evidence regarding the niche competition of Bifidobacterium within the infant intestinal tract that needs to be corroborated by further experimentation.
MATERIALS AND METHODS
Genome sequences and mining for BGCs
The details of the genome sequences of 759 strains belonging to B. longum subsp. infantis (n = 36), B. longum subsp. longum (n = 210), B. breve (n = 146), B. dentium (n = 36), B. animalis (n = 63), B. angulatum (n = 6), B. bifidum (n = 96), B. adolescentis (n = 63), and B. pseudocatenulatum (n = 103) are presented in Table S1. Most of these sequences were previously determined by our institution and the rest were retrieved from public data. The pairwise ANI (https://github.com/widdowquinn/pyani/) was performed, and values were calculated and visualized to distinguish B. longum subsp. infantis and B. longum subsp. longum.
The bioinformatics tools, antiSMASH6 (20) and BAGEL4 (19), were used to mine putative BGCs, and the results (regions and areas of interest from antiSMASH6 and BAGEL4, respectively) were manually categorized. Bacteriocins were classified according to Garcia-Gutierrez et al. and Cotter et al. (3, 7). The classification of specific class I bacteriocin subgroups of gene clusters was based on the presence of highly conserved enzymes. Gene clusters with the same structural composition and order under a subgroup of BGCs were grouped into a “type.” To determine the location of precursor peptides in a particular type, all identified precursor peptides were taken into account (via comparison with the respective constructed bacteriocin databases); additionally, all proteins encoded in the gene clusters were examined using BLAST in the nr protein database to search for potential precursor peptides that may have been omitted in bacteriocin databases. A discrepancy of two or more amino acids between the identified and previously discovered bacteriocins was considered novel (72). Peptide sequences were aligned using MUSCLE (73) and visualized using Jalview (74). The similarity of pairwise sequence alignment was derived via Needle (https://www.ebi.ac.uk/Tools/psa/emboss_needle/). After the putative bacteriocins were described using BLAST, they were aligned with all sequences with the same description in Identical Protein Groups of NCBI, and phylogenetic analyses were constructed by IQ-TREE (75). The resulting nexus tree files were exported to iTOL for graphical adjustment (76). Most branches of the phylogenetic trees were folded for a better illustration of the evolutionary distance between the target sequence and other sequences (which may have otherwise led to more than 1,000 branches); however, this did not modify the topological structure of the phylogenetic tree.
Evaluation of the antimicrobial activity of B. infantis strains and their crude bacteriocins
The antimicrobial activities of 29 B. infantis strains from our team among 36 B. infantis strains were tested (Table 1). All tested strains were deposited in the Culture Collection of Food Microorganisms at Jiangnan University. Each tested strain was cultivated anaerobically for three generations in MRS liquid medium with 0.05% cysteine hydrochloride at 37°C for 24 h. After centrifugation (Eppendorf 5424R centrifuge, Hamburg, Germany) for cell removal, the supernatant was incubated with 80% saturated ammonium sulfate at 4°C overnight to obtain crude bacteriocin, which was dissolved in phosphate buffer (pH 5.5) to achieve a 20-fold concentration after desalting and then filtered through a sterilized 0.22-µm nitrocellulose membrane.
The antimicrobial activity of B. infantis was determined using a colony overlay assay with modifications (77). Briefly, strains were inoculated as a 3 µL spot on MRS agar plates (1.5% agar) at 37°C for 24 h under anaerobic conditions. Subsequently, cover the plate with 0.8% soft agar containing 107 CFU/mL indicator strain and incubated under appropriate conditions for 24 h. Enterococcus faecalis ATCC19433, Staphylococcus aureus CGMCC1.1861, Lactobacillus plantarum LABCC IMAU60055, Lactobacillus rhamnosus ATCC7469, B. adolescens CGMCC 12190, B. bifidum CICC6071, Listeria monocytogenes ATCC19114, Klebsiella pneumoniae JNGMMA3, Escherichia coli CICC10413, and Salmonella enterica CICC21482 were included as indicator strains as they are commonly associated with the infant intestinal tract. Antimicrobial activity was evaluated based on the diameter of the inhibition zone around the spot.
The antimicrobial activity of the crude bacteriocin was determined using the Oxford cup method with modifications (78). The bottom of the plate was covered with 1.5% sterile agar, and then, 0.8% soft agar containing 107 CFU/mL of the indicator strain was overlaid on the plates. The Oxford cups were placed on the agar surface and lightly pressed. Next, 200 µL of crude bacteriocin was poured into the cups. MRS liquid medium was used as a control. After 4 h of storage at 4°C, the plates were incubated under appropriate conditions at 37°C for 24 h. The antimicrobial activity of crude bacteriocins was evaluated based on the diameter of the inhibition zone from the edge of the cups. All experiments were performed in three independent replicates.
Transcription analysis
B. infantis strains were collected after 24 h of culture by centrifugation at 2,500 g for 10 min at 4°C. RNA extraction was performed using the Vazyme Bacteria RNA Extraction Kit (R403-01, Vazyme Biotech Co., Ltd., Nanjing, China). RNA integrity was analyzed by electrophoresis on a 1% agarose gel. Total RNA samples were reverse transcribed into cDNA using the Vazyme HiScript III-RT SuperMix for qPCR (R323-01). Each strain was performed in three independent biological replicates.
qPCR was used to detect the bacteriocin gene expression. The Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/Primer-Blast/) was used to construct qPCR primers targeting bacteriocin genes in the conserved region and checked the specificity of primers (Table S2). Using the 23S rRNA gene as an internal reference gene (79), qPCR was performed in a 10-µL mixture containing 5 µL of 2× iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, USA), 0.5 µL of each primer (10 nM), 1 µL of cDNA template, and 3 µL of deionized distilled water under the following conditions on a CFX-Connected Real-Time PCR System (Bio-Rad, Hercules, CA, USA): polymerase activation at 95°C for 30 s followed by 39 cycles, each at 95°C for 5 s and 60°C for 30 s. The amplification specificity was confirmed by melt curve analysis under the following conditions: 65°C, 5 s; 95°C, 0.5°C. The relative expression was calculated by the 2−△△CT method (80). Each replicate was performed in triplicate.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (No. 32072228, 32021005, 31820103010), 111project (BP0719028), National Key Research and Development Program of China (2022YFF1100203), and the Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province.
Authors have no conflict of interest to disclose.
Contributor Information
Wenwei Lu, Email: luwenwei@jiangnan.edu.cn.
Danilo Ercolini, Universita degli Studi di Napoli Federico II, Naples, Italy .
DATA AVAILABILITY
All sequence data of Bifidobacterium strains have been deposited in the NCBI GenBank database. The accession numbers for all individual genomes used in this study (including 129 genomes downloaded from NCBI) are presented in Table S1.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.00979-23.
Fig. S1 Transcription analysis of BGCs of the 36 B. infantis strains. The X-axis is the target gene whose specific name can be found in Table S2; the gray bottom indicates that the gene is not tested in the strain.
Fig. S2 Heatmap and dendrogram of ANIb values of 59 B. longum genomes. Sequences of 40 genomes marked as B. longum subsp. infantis (11 of which were downloaded from NCBI) and 19 genomes marked as B. longum subsp. longum (all downloaded from NCBI) were distinguished by pairwise ANIb values. ANIb, average nucleotide identity blast. The red color code refers to the strains that may be incorrectly marked based on clustering results.
Table S1 Specific information of 763 Bifidobacterium genomes.
Table S2 Primers designed and used for transcription analysis.
Table S3 Specific distribution of different types of BGCs in the Bifidobacterium.
Information summary table of bacteriocins and their most similar bacteriocins.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Alvarez-Sieiro P, Montalbán-López M, Mu D, Kuipers OP. 2016. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol 100:2939–2951. doi: 10.1007/s00253-016-7343-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walsh CJ, Guinane CM, Hill C, Ross RP, O’Toole PW, Cotter PD. 2015. In silico identification of bacteriocin gene clusters in the gastrointestinal tract, based on the human microbiome project's reference genome database. BMC Microbiol 15:183. doi: 10.1186/s12866-015-0515-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garcia-Gutierrez E, Mayer MJ, Cotter PD, Narbad A. 2019. Gut microbiota as a source of novel antimicrobials. Gut Microbes 10:1–21. doi: 10.1080/19490976.2018.1455790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH. 2015. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526:719–722. doi: 10.1038/nature15524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu J, Ma L, Nie Y, Chen J, Zheng W, Wang X, Xie C, Zheng Z, Wang Z, Yang T, Shi M, Chen L, Hou Q, Niu Y, Xu X, Zhu Y, Zhang Y, Wei H, Yan X. 2018. A microbiota-derived bacteriocin targets the host to confer diarrhea resistance in early-weaned piglets. Cell Host Microbe 24:817–832. doi: 10.1016/j.chom.2018.11.006 [DOI] [PubMed] [Google Scholar]
- 6. Corr SC, Li Y, Riedel CU, O’Toole PW, Hill C, Gahan CGM. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A 104:7617–7621. doi: 10.1073/pnas.0700440104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cotter PD, Ross RP, Hill C. 2013. Bacteriocins - a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. doi: 10.1038/nrmicro2937 [DOI] [PubMed] [Google Scholar]
- 8. Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. doi: 10.1039/c2np20085f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Repka LM, Chekan JR, Nair SK, van der Donk WA. 2017. Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem Rev 117:5457–5520. doi: 10.1021/acs.chemrev.6b00591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hegemann JD, Zimmermann M, Xie X, Marahiel MA. 2015. Lasso peptides: an intriguing class of bacterial natural products. Acc Chem Res 48:1909–1919. doi: 10.1021/acs.accounts.5b00156 [DOI] [PubMed] [Google Scholar]
- 11. Chen Y, Wang J, Li G, Yang Y, Ding W. 2021. Current advancements in sactipeptide natural products. Front Chem 9:595991. doi: 10.3389/fchem.2021.595991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu S, Liu J, Liu C, Yang A, Qiao J. 2020. Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell Mol Life Sci 77:1319–1343. doi: 10.1007/s00018-019-03326-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arboleya S, Watkins C, Stanton C, Ross RP. 2016. Gut bifidobacteria populations in human health and aging. Front Microbiol 7:1204. doi: 10.3389/fmicb.2016.01204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhuang L, Chen HH, Zhang S, Zhuang JH, Li QP, Feng ZC. 2019. Intestinal microbiota in early life and its implications on childhood health. Genom Proteom Bioinform 17:13–25. doi: 10.1016/j.gpb.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lim HJ, Shin HS. 2020. Antimicrobial and immunomodulatory effects of Bifidobacterium strains: a review. J Microbiol Biotechnol 30:1793–1800. doi: 10.4014/jmb.2007.07046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Devika NT, Raman K. 2019. Deciphering the metabolic capabilities of bifidobacteria using genome-scale metabolic models. Sci Rep 9:18222. doi: 10.1038/s41598-019-54696-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LMT. 2018. Functions and emerging applications of bacteriocins. Curr Opin Biotechnol 49:23–28. doi: 10.1016/j.copbio.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martinez FAC, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP. 2013. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol Adv 31:482–488. doi: 10.1016/j.biotechadv.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 19. van Heel AJ, de Jong A, Song C, Viel JH, Kok J, Kuipers OP. 2018. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res 46:W278–W281. doi: 10.1093/nar/gky383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, Weber T. 2021. AntiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49:W29–W35. doi: 10.1093/nar/gkab335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao X, Kuipers OP. 2016. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genomics 17:882. doi: 10.1186/s12864-016-3224-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu LW, Hao TT, Xie ZJ, Horsman GP, Chen YH. 2016. Genome mining unveils widespread natural product biosynthetic capacity in human oral microbe Streptococcus mutans. Sci Rep 6:37479. doi: 10.1038/srep37479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu YY, Teng KL, Huang FQ, Xia TQ, Zhang JQ, Wang TW, Zhong J. 2022. High-throughput discovery of novel lanthipeptides and producers by metagenomic mining of isolates population (MMIP) from Chinese spicy cabbage. Food Res Int 154:110991. doi: 10.1016/j.foodres.2022.110991 [DOI] [PubMed] [Google Scholar]
- 24. Darvishi N, Fard NA, Sadrnia M. 2021. Genomic and proteomic comparisons of bacteriocins in probiotic species Lactobacillus and Bifidobacterium and inhibitory ability of Escherichia coli MG 1655. Biotechnol Rep (Amst) 31:e00654. doi: 10.1016/j.btre.2021.e00654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garg N, Tang W, Goto Y, Nair SK, van der Donk WA. 2012. Lantibiotics from Geobacillus thermodenitrificans. Proc Natl Acad Sci U S A 109:5241–5246. doi: 10.1073/pnas.1116815109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sandiford SK. 2019. Current developments in lantibiotic discovery for treating Clostridium difficile infection. Expert Opin Drug Discov 14:71–79. doi: 10.1080/17460441.2019.1549032 [DOI] [PubMed] [Google Scholar]
- 27. Jabés D, Brunati C, Candiani G, Riva S, Romanó G, Donadio S. 2011. Efficacy of the new lantibiotic NAI-107 in experimental infections induced by multidrug-resistant Gram-positive pathogens. Antimicrob Agents Chemother 55:1671–1676. doi: 10.1128/AAC.01288-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Piard JC, Delorme F, Giraffa G, Commissaire J, Desmazeaud M. 1990. Evidence for a bacteriocin produced by Lactococcus lactis CNRZ 481. Neth Milk Dairy J 44:143–158. [Google Scholar]
- 29. Lee J-H, Li X, O’Sullivan DJ. 2011. Transcription analysis of a lantibiotic gene cluster from Bifidobacterium longum DJO10A. Appl Environ Microbiol 77:5879–5887. doi: 10.1128/AEM.00571-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holtsmark I, Mantzilas D, Eijsink VGH, Brurberg MB. 2006. Purification, characterization, and gene sequence of michiganin A, an actagardine-like lantibiotic produced by the tomato pathogen Clavibacter michiganensis subsp michiganensis. Appl Environ Microbiol 72:5814–5821. doi: 10.1128/AEM.00639-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iorio M, Sasso O, Maffioli SI, Bertorelli R, Monciardini P, Sosio M, Bonezzi F, Summa M, Brunati C, Bordoni R, Corti G, Tarozzo G, Piomelli D, Reggiani A, Donadio S. 2014. A glycosylated, labionin-containing lanthipeptide with marked antinociceptive activity. ACS Chem Biol 9:398–404. doi: 10.1021/cb400692w [DOI] [PubMed] [Google Scholar]
- 32. Krawczyk JM, Völler GH, Krawczyk B, Kretz J, Brönstrup M, Süssmuth RD. 2013. Heterologous expression and engineering studies of labyrinthopeptins, class III lantibiotics from Actinomadura namibiensis. Chem Biol 20:111–122. doi: 10.1016/j.chembiol.2012.10.023 [DOI] [PubMed] [Google Scholar]
- 33. Esumi Y, Suzuki Y, Itoh Y, Uramoto M, Kimura K, Goto M, Yoshihama M, Ichikawa T. 2002. Propeptin, a new inhibitor of prolyl endopeptidase produced by Microbispora - II. determination of chemical structure. J Antibiot 55:296–300. doi: 10.7164/antibiotics.55.296 [DOI] [PubMed] [Google Scholar]
- 34. Kimura KI, Kanou F, Takahashi H, Esumi Y, Uramoto M, Yoshihama M. 1997. Propeptin, a new inhibitor of prolyl endopeptidase produced by Microbispora .1. fermentation, isolation and biological properties. J Antibiot 50:373–378. doi: 10.7164/antibiotics.50.373 [DOI] [PubMed] [Google Scholar]
- 35. Shen X, Mustafa M, Chen Y, Cao Y, Gao J. 2019. Natural thiopeptides as a privileged scaffold for drug discovery and therapeutic development. Med Chem Res 28:1063–1098. doi: 10.1007/s00044-019-02361-1 [DOI] [Google Scholar]
- 36. Claesen J, Spagnolo JB, Ramos SF, Kurita KL, Byrd AL, Aksenov AA, Melnik AV, Wong WR, Wang S, Hernandez RD, Donia MS, Dorrestein PC, Kong HH, Segre JA, Linington RG, Fischbach MA, Lemon KP. 2020. A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci Transl Med 12:eaay5445. doi: 10.1126/scitranslmed.aay5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martínez B, Böttiger T, Schneider T, Rodríguez A, Sahl H-G, Wiedemann I. 2008. Specific interaction of the unmodified bacteriocin lactococcin 972 with the cell wall precursor lipid II. Appl Environ Microbiol 74:4666–4670. doi: 10.1128/AEM.00092-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rolhion N, Chassaing B, Nahori M-A, de Bodt J, Moura A, Lecuit M, Dussurget O, Bérard M, Marzorati M, Fehlner-Peach H, Littman DR, Gewirtz AT, Van de Wiele T, Cossart P. 2019. A Listeria monocytogenes bacteriocin can target the commensal Prevotella copri and modulate intestinal infection. Cell Host Microbe 26:691–701. doi: 10.1016/j.chom.2019.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miescher S, Stierli MP, Teuber M, Meile L. 2000. Propionicin SM1, a bacteriocin from Propionibacterium jensenii DF1: isolation and characterization of the protein and its gene. Syst Appl Microbiol 23:174–184. doi: 10.1016/S0723-2020(00)80002-8 [DOI] [PubMed] [Google Scholar]
- 40. Otto M, Süssmuth R, Jung G, Götz F. 1998. Structure of the pheromone peptide of the Staphylococcus epidermidis agr system. FEBS Lett 424:89–94. doi: 10.1016/s0014-5793(98)00145-8 [DOI] [PubMed] [Google Scholar]
- 41. Cintas LM, Casaus P, Holo H, Hernandez PE, Nes IF, Håvarstein LS. 1998. Enterocins L50A and L50B, two novel bacteriocins from Enterococcus faecium L50, are related to staphylococcal hemolysins. J Bacteriol 180:1988–1994. doi: 10.1128/JB.180.8.1988-1994.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cintas LM, Casaus P, Håvarstein LS, Hernández PE, Nes IF. 1997. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl Environ Microbiol 63:4321–4330. doi: 10.1128/aem.63.11.4321-4330.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kawamoto S, Shima J, Sato R, Eguchi T, Ohmomo S, Shibato J, Horikoshi N, Takeshita K, Sameshima T. 2002. Biochemical and genetic characterization of mundticin KS, an antilisterial peptide produced by Enterococcus mundtii NFRI 7393. Appl Environ Microbiol 68:3830–3840. doi: 10.1128/AEM.68.8.3830-3840.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nigutova K, Morovsky M, Pristas P, Teather RM, Holo H, Javorsky P. 2007. Production of enterolysin A by rumen Enterococcus faecalis strain and occurrence of enlA homologues among ruminal Gram-positive cocci. J Appl Microbiol 102:563–569. doi: 10.1111/j.1365-2672.2006.03068.x [DOI] [PubMed] [Google Scholar]
- 45. Nilsen T, Nes IF, Holo H. 2003. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl Environ Microbiol 69:2975–2984. doi: 10.1128/AEM.69.5.2975-2984.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Basanta A, Sánchez J, Gómez-Sala B, Herranz C, Hernández PE, Cintas LM. 2008. Antimicrobial activity of Enterococcus faecium L50, a strain producing enterocins L50 (L50A and L50B), P and Q, against beer-spoilage lactic acid bacteria in broth, wort (hopped and unhopped), and alcoholic and non-alcoholic lager beers. Int J Food Microbiol 125:293–307. doi: 10.1016/j.ijfoodmicro.2008.04.011 [DOI] [PubMed] [Google Scholar]
- 47. Whitford MF, McPherson MA, Forster RJ, Teather RM. 2001. Identification of bacteriocin-like inhibitors from rumen Streptococcus spp. and isolation and characterization of bovicin 255. Appl Environ Microbiol 67:569–574. doi: 10.1128/AEM.67.2.569-574.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marciset O, Jeronimus-Stratingh MC, Mollet B, Poolman B. 1997. Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor*. J Biol Chem 272:14277–14284. doi: 10.1074/jbc.272.22.14277 [DOI] [PubMed] [Google Scholar]
- 49. Wholey W-Y, Abu-Khdeir M, Yu EA, Siddiqui S, Esimai O, Dawid S. 2019. Characterization of the competitive pneumocin peptides of Streptococcus pneumoniae. Front Cell Infect Microbiol 9:55. doi: 10.3389/fcimb.2019.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee J-H, Karamychev VN, Kozyavkin SA, Mills D, Pavlov AR, Pavlova NV, Polouchine NN, Richardson PM, Shakhova VV, Slesarev AI, Weimer B, O’Sullivan DJ. 2008. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9:247. doi: 10.1186/1471-2164-9-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cheikhyoussef A, Cheikhyoussef N, Chen HQ, Zhao JX, Tang J, Zhang H, Chen W. 2010. Bifidin I - A new bacteriocin produced by Bifidobacterium infantis BCRC 14602: purification and partial amino acid sequence. Food Control 21:746–753. doi: 10.1016/j.foodcont.2009.11.003 [DOI] [Google Scholar]
- 52. Saleh F, El-Sayed E. 2004. “Isolation and characterization of bacteriocins produced by Bifidobacterium lactis BB-12 and Bifidobacterium longum BB-46” 9th Egyptian conference for dairy science and technology; International Agriculture Centre, Cairo, Egypt: [Google Scholar]
- 53. Yildirim Z, Johnson MG. 1998. Characterization and antimicrobial spectrum of bifidocin B, a bacteriocin produced by Bifidobacterium bifidum NCFB 1454. J Food Prot 61:47–51. doi: 10.4315/0362-028x-61.1.47 [DOI] [PubMed] [Google Scholar]
- 54. Anand SK, Srinivasan RA, Rao LK. 1984. Antimicrobial activity associated with Bifidobacterium bifidum-I. Cultured Dairy Prod J 2:6–7. [Google Scholar]
- 55. KH K, HJ S, YH P, TS L. 1989. Studies on the antibacterial substances produced by lactic acid bacteria: purification and some properties of antibacterial substance “Bifilong” produced by Bifidobacterium longum. Korean Dairy Sci 57–11:204–216. [Google Scholar]
- 56. Zihler A, Gagnon M, Chassard C, Hegland A, Stevens MJA, Braegger CP, Lacroix C. 2010. Unexpected consequences of administering bacteriocinogenic probiotic strains for Salmonella populations, revealed by an in vitro colonic model of the child gut. Microbiology 156:3342–3353. doi: 10.1099/mic.0.042036-0 [DOI] [PubMed] [Google Scholar]
- 57. Liu GR, Ren L, Song ZQ, Wang CT, Sun BG. 2015. Purification and characteristics of bifidocin A, a novel bacteriocin produced by Bifidobacterium animals BB04 from centenarians' intestine. Food Control 50:889–895. [Google Scholar]
- 58. Hudson GA, Burkhart BJ, DiCaprio AJ, Schwalen CJ, Kille B, Pogorelov TV, Mitchell DA. 2019. Bioinformatic mapping of radical SAM-dependent RiPPs identifies new C alpha, C beta, and C gamma-linked thioether-containing peptides. J Am Chem Soc 141:8228–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Q, Doroghazi JR, Zhao X, Walker MC, van der Donk WA, Elliot MA. 2015. Expanded natural product diversity revealed by analysis of lanthipeptide-like gene clusters in Actinobacteria. Appl Environ Microbiol 81:4339–4350. doi: 10.1128/AEM.00635-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wambui J, Stevens MJA, Sieber S, Cernela N, Perreten V, Stephan R. 2021. Targeted genome mining reveals the psychrophilic Clostridium estertheticum complex as a potential source for novel bacteriocins, including cesin A and estercticin A. Front Microbiol 12:801467. doi: 10.3389/fmicb.2021.801467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zheng J, Gänzle MG, Lin XB, Ruan L, Sun M. 2015. Diversity and dynamics of bacteriocins from human microbiome. Environ Microbiol 17:2133–2143. doi: 10.1111/1462-2920.12662 [DOI] [PubMed] [Google Scholar]
- 62. Goto Y, Li B, Claesen J, Shi Y, Bibb MJ, van der Donk WA. 2010. Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol 8:e1000339. doi: 10.1371/journal.pbio.1000339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MA, Mills DA, Stephensen CB. 2014. Stool microbiota and vaccine responses of infants. Pediatrics 134:E362–E372. doi: 10.1542/peds.2013-3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Krauss S, Harbig TA, Rapp J, Schaefle T, Franz-Wachtel M, Reetz L, Elsherbini AMA, Macek B, Grond S, Link H, Nieselt K, Krismer B, Peschel A, Heilbronner S. 2023. Horizontal transfer of bacteriocin biosynthesis genes requires metabolic adaptation to improve compound production and cellular fitness. Microbiol Spectr 11:e0317622. doi: 10.1128/spectrum.03176-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Blanchard AE, Liao C, Lu T. 2016. An ecological understanding of quorum sensing-controlled bacteriocin synthesis. Cel Mol Bioeng 9:443–454. doi: 10.1007/s12195-016-0447-6 [DOI] [Google Scholar]
- 66. Huo L, Hug JJ, Fu C, Bian X, Zhang Y, Müller R. 2019. Heterologous expression of bacterial natural product biosynthetic pathways. Nat Prod Rep 36:1412–1436. doi: 10.1039/c8np00091c [DOI] [PubMed] [Google Scholar]
- 67. Underwood MA, German JB, Lebrilla CB, Mills DA. 2015. Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr Res 77:229–235. doi: 10.1038/pr.2014.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. de Freire Bastos M do C, Miceli de Farias F, Carlin Fagundes P, Varella Coelho ML. 2020. Staphylococcins: an update on antimicrobial peptides produced by staphylococci and their diverse potential applications. Appl Microbiol Biotechnol 104:10339–10368. doi: 10.1007/s00253-020-10946-9 [DOI] [PubMed] [Google Scholar]
- 69. Almeida-Santos AC, Novais C, Peixe L, Freitas AR. 2021. Enterococcus spp. as a producer and target of bacteriocins: a double-edged sword in the antimicrobial resistance crisis context. Antibiotics (Basel) 10:1215. doi: 10.3390/antibiotics10101215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. O’Shea EF, Cotter PD, Stanton C, Ross RP, Hill C. 2012. Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: bacteriocins and conjugated linoleic acid. Int J Food Microbiol 152:189–205. doi: 10.1016/j.ijfoodmicro.2011.05.025 [DOI] [PubMed] [Google Scholar]
- 71. Zhang J, Caiyin Q, Feng WJ, Zhao XL, Qiao B, Zhao GR, Qiao JJ. 2016. Enhance nisin yield via improving acid-tolerant capability of Lactococcus lactis F44. Sci Rep 6:27973. doi: 10.1038/srep27973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Angelopoulou A, Warda AK, O’Connor PM, Stockdale SR, Shkoporov AN, Field D, Draper LA, Stanton C, Hill C, Ross RP. 2020. Diverse bacteriocins produced by strains from the human milk microbiota. Front Microbiol 11:788. doi: 10.3389/fmicb.2020.00788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tejero-Sariñena S, Barlow J, Costabile A, Gibson GR, Rowland I. 2012. In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: evidence for the effects of organic acids. Anaerobe 18:530–538. doi: 10.1016/j.anaerobe.2012.08.004 [DOI] [PubMed] [Google Scholar]
- 78. Bian X, Evivie SE, Muhammad Z, Luo GW, Liang HZ, Wang NN, Huo GC. 2016. In vitro assessment of the antimicrobial potentials of Lactobacillus helveticus strains isolated from traditional cheese in Sinkiang China against food-borne pathogens. Food Funct 7:789–797. doi: 10.1039/c5fo01041a [DOI] [PubMed] [Google Scholar]
- 79. Kurakawa T, Ogata K, Tsuji H, Kado Y, Takahashi T, Kida Y, Ito M, Okada N, Nomoto K. 2015. Establishment of a sensitive system for analysis of human vaginal microbiota on the basis of rRNA-targeted reverse transcription-quantitative PCR. J Microbiol Methods 111:93–104. doi: 10.1016/j.mimet.2015.01.021 [DOI] [PubMed] [Google Scholar]
- 80. Ma Q, Pei Z, Fang Z, Wang H, Zhu J, Lee Y-K, Zhang H, Zhao J, Lu W, Chen W. 2021. Evaluation of tetracycline resistance and determination of the tentative microbiological cutoff values in lactic acid bacterial species. Microorganisms 9:2128. doi: 10.3390/microorganisms9102128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Transcription analysis of BGCs of the 36 B. infantis strains. The X-axis is the target gene whose specific name can be found in Table S2; the gray bottom indicates that the gene is not tested in the strain.
Fig. S2 Heatmap and dendrogram of ANIb values of 59 B. longum genomes. Sequences of 40 genomes marked as B. longum subsp. infantis (11 of which were downloaded from NCBI) and 19 genomes marked as B. longum subsp. longum (all downloaded from NCBI) were distinguished by pairwise ANIb values. ANIb, average nucleotide identity blast. The red color code refers to the strains that may be incorrectly marked based on clustering results.
Table S1 Specific information of 763 Bifidobacterium genomes.
Table S2 Primers designed and used for transcription analysis.
Table S3 Specific distribution of different types of BGCs in the Bifidobacterium.
Information summary table of bacteriocins and their most similar bacteriocins.
Data Availability Statement
All sequence data of Bifidobacterium strains have been deposited in the NCBI GenBank database. The accession numbers for all individual genomes used in this study (including 129 genomes downloaded from NCBI) are presented in Table S1.