ABSTRACT
The asexual sporulation of filamentous fungi is an important mechanism for their reproduction, survival, and pathogenicity. In Aspergillus and several filamentous fungi, BrlA, AbaA, and WetA are the key elements of a central regulatory pathway controlling conidiation, and MedA is a developmental modifier that regulates temporal expression of central regulatory genes; however, their roles are largely unknown in nematode-trapping (NT) fungi. Arthrobotrys oligospora is a representative NT fungus, which can capture nematodes by producing adhesive networks (traps). Here, we characterized the function of AoMedA and three central developmental regulators (AoBrlA, AoAbaA, and AoWetA) in A. oligospora by gene disruption, phenotypic comparison, and multi-omics analyses, as these regulators are required for conidiation and play divergent roles in mycelial development, trap formation, lipid droplet accumulation, vacuole assembly, and secondary metabolism. A combined analysis of phenotypic traits and transcriptome showed that AoMedA and AoWetA are involved in the regulation of peroxisome, endocytosis, and autophagy. Moreover, yeast one-hybrid analysis showed that AoBrlA can regulate AoMedA, AoAbaA, and AoWetA, whereas AoMedA and AoAbaA can regulate AoWetA. Our results highlight the important roles of AoMedA, AoBrlA, AoAbaA, and AoWetA in conidiation, mycelia development, trap formation, and pathogenicity of A. oligospora and provide a basis for elucidating the relationship between conidiation and trap formation of NT fungi.
IMPORTANCE
Conidiation is the most common reproductive mode for many filamentous fungi and plays an essential role in the pathogenicity of fungal pathogens. Nematode-trapping (NT) fungi are a special group of filamentous fungi owing to their innate abilities to capture and digest nematodes by producing traps (trapping devices). Sporulation plays an important role in the growth and reproduction of NT fungi, and conidia are the basic components of biocontrol reagents for controlling diseases caused by plant-parasitic nematodes. Arthrobotrys oligospora is a well-known NT fungus and is a routinely used model fungus for probing the interaction between fungi and nematodes. In this study, the functions of four key regulators (AoMedA, AoBrlA, AoAbaA, and AoWetA) involved in conidiation were characterized in A. oligospora. A complex interaction between AoMedA and three central regulators was noted; these regulators are required for conidiation and trap formation and play a pleiotropic role in multiple intracellular activities. Our study first revealed the role of AoMedA and three central regulators in conidiation, trap formation, and pathogenicity of A. oligospora, which contributed to elucidating the regulatory mechanism of conidiation in NT fungi and helped in developing effective reagents for biocontrol of nematodes.
KEYWORDS: Arthrobotrys oligospora, developmental regulator, conidiation, trap formation, secondary metabolism
INTRODUCTION
Filamentous fungi are organisms with diversified reproduction strategies, with most of their lifecycle being spent on asexual reproduction (1). Asexual spores are critical in the lifecycle of most filamentous fungi (2), and conidia are the main type of asexual spores, which occur after a period of vegetative growth when the specialized aerial hyphae differentiate into conidia (3). Conidiation in Aspergillus species, particularly in the model organism Aspergillus nidulans, has been studied extensively. Conidiation of filamentous fungi involves many aspects, including spatial and temporal regulation of gene expression, specialized cell differentiation, intracellular/intercellular communication, and response to environmental factors (4). BrlA activates a central regulatory pathway to control the temporal and spatial expression of conidiation-specific genes (5), and BrlA, AbaA, and WetA are the key core regulatory proteins, and MedA is identified as a temporal modifier of the expression of these core conidiation proteins (6).
The central development pathway consists of BrlA, AbaA, and WetA which are indispensable for sporulation in the model fungus A. nidulans and other Aspergillus species (7). MedA expression begins after the induction of conidiation and persists throughout the asexual cycle (8). It is a developmental modifier necessary for correct conidial morphogenesis through spatial and temporal regulation of brlA and abaA expression (9). The regulatory sequence for central regulatory components is BrlA→ AbaA→ WetA, and they play a crucial role in asexual development. Central regulatory components are functionally conserved in conidiogenesis in A. nidulans (10), A. fumigatus (11), Penicillium decumbens (12), and P. digitatum (13). In plant-pathogenic fungus Magnaporthe grisea (syn. M. oryzae), ACR1 is the ortholog of MedA, which is required for conidiophore architecture and pathogenicity and infection-related morphogenesis (14, 15). Recently, the role of central regulatory components was also revealed in several entomopathogenic fungi. For example, BrlA and AbaA are important regulators of conidiation, insect pathogenicity, and dimorphism transformation in Beauveria bassiana (16), and WetA is dispensable for conidiation as well as conidial maturation and virulence (17). In addition, as conidia are diffusive propagules, they are essential in disease transmission and are also effective components of fungal insecticides; hence, conidia have vital significance in pathogenicity (18).
Most fungal lifestyle transitions are complex. Among pathogenic fungi, nematode-trapping (NT) fungi are unique as their hyphae can form ingenious structures (traps) to capture nematodes when sensing prey, and the formation of traps is the key indicator of their lifestyle transition from saprophytes to predators; thus, they are a good model for studying the pathogenesis and adaptation mechanism of fungi (19, 20). Arthrobotrys oligospora is a typical NT fungus that can complete its reproduction asexually by producing abundant conidia and adhesive three-dimensional networks for nematode predation; therefore, it has been widely used in studying the interactions between fungi and nematodes (21, 22). Recent studies have shown that several signaling proteins, including regulators of G-protein (23, 24), mitogen-activated protein kinases (25 – 28), and small GTPases (29 – 31), are involved in conidiation in A. oligospora. In addition, autophagy-related proteins (32, 33) and peroxisome biogenesis proteins (34, 35) play an important role in the interaction between fungi and nematodes and regulating conidiation. In our previous study, two velvet proteins VosA and VelB involved in conidiation were identified in A. oligospora. VelB is essential for conidiation, trap formation, and pathogenicity, whereas VosA plays a minor role in the regulation of conidial germination and heat shock stress (36). Conidia play a key role in the virulence of pathogenic fungi, for example, in destructive hemibiotrophic phytopathogen M. oryzae, when its conidia are attached to the host surface, they can form appressorium, causing rice blast disease (37). However, the role of most sporulation-related genes is still unknown in NT fungi.
In this study, we investigated the homologous proteins MedA (AoMedA), BrlA (AoBrlA), AbaA (AoAbaA), and WetA (AoWetA) in A. oligospora via phenotypic comparison and multi-omics approaches. Our results showed that AomedA and three central regulatory genes (AobrlA, AoabaA, and AowetA) are required for conidiation, and they play divergent roles in trap formation, lipid droplet (LD) accumulation, autophagy, peroxisome, vacuole assembly, and secondary metabolism. We also detected the interaction between AomedA and three central regulatory genes using a yeast one-hybrid (Y1H) assay. In addition, the role and potential regulation of these sporulation-related genes were investigated by transcriptomic and metabonomic analyses.
RESULTS
Bioinformatic analysis of AoMedA, AoBrlA, AoAbaA, and AoWetA
Orthologs of MedA, BrlA, AbaA, and WetA were retrieved from A. oligospora based on the homologous sequences of A. nidulans. The partial sequence properties of these proteins are summarized in Table 1. The orthologs of MedA, BrlA, AbaA, and WetA were divided into different evolutionary branches (Fig. S1A), and they were highly homologous to orthologs derived from other NT fungi; for example, AoBrlA shared a high degree of sequence similarity with NT fungi Arthrobotrys flagrans (91.9%), Drechslerella brochopaga (60.7%), and Dactylellina haptotyla (77%), and it had a middle degree of similarity with A. nidulans (58.2%) (Fig. S1B).
TABLE 1.
Partial sequence properties of AoMedA and three central developmental regulators in A. oligospora
| Gene | Open reading frame (bp) | Introns | Amino acid residues | Isoelectric point | Molecular weight (kDa) |
|---|---|---|---|---|---|
| AomedA | 2018 | 2 | 608 | 9.16 | 67.14 |
| AobrlA | 966 | 0 | 321 | 7.34 | 35.39 |
| AoabaA | 1921 | 2 | 588 | 5.50 | 67.49 |
| AowetA | 2415 | 0 | 804 | 5.93 | 86.80 |
AoMedA, AoBrlA, AoAbaA, and AoWetA are required for conidiation
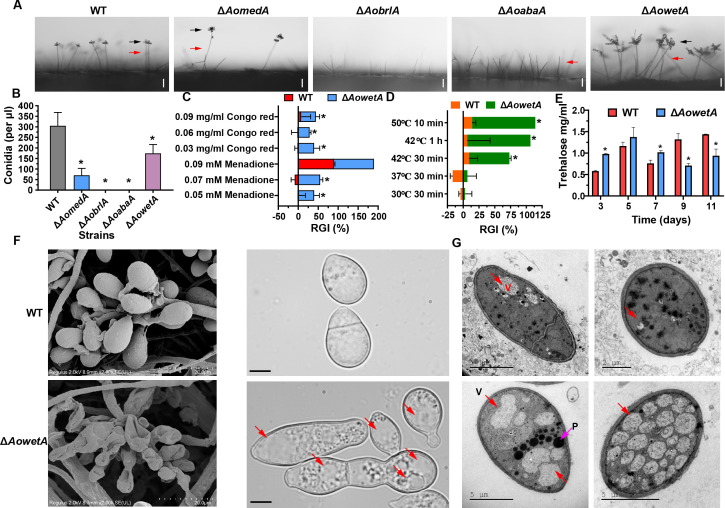
Three mutants of each gene (AomedA, AobrlA, AoabaA, and AowetA) were generated, as described in the Materials and methods, and were then verified using PCR and Southern blot analyses (Fig. S2) using paired primers (Table 2). As independent mutant strains of each gene showed similar phenotypic traits, a single mutant from each gene was randomly selected for subsequent study. Deletion of AomedA resulted in a reduction in conidiophores and conidia yield when being compared with those in the wild-type (WT) strain (Fig. 1A and B). Moreover, the ΔAobrlA and ΔAoabaA mutants completely lost the ability to produce conidia and the ΔAobrlA mutant did not form conidiophores, whereas ΔAoabaA mutant could form conidiophores but not conidia (Fig. 1A and B). In particular, the ΔAowetA mutant produced deformed conidia (Fig. 1A), and the number of conidia by ΔAowetA mutant decreased compared with WT strain (Fig. 1B). Next, the stress response of conidia of WT and ΔAowetA mutant strains was tested with Congo red and menadione, and it was observed that the relative growth inhibition rate (RGI) of the ΔAowetA mutant increased remarkably under 0.05–0.07 mM menadione and 0.03–0.09 mg/mL Congo red compared with WT strain (Fig. 1C). Similarly, when the conidia of ΔAowetA mutant was treated at 42°C for 30 min, the germination ability of conidia was inhibited remarkably and approximately 89% and 36% of conidia had germinated in the WT and ΔAowetA, respectively. Moreover, the conidia of ΔAowetA mutant lost the ability to germinate when treated at 42°C for 1 h, whereas 70% conidia of WT strain could germinate (Fig. 1D). The trehalose content increased in ΔAowetA mutant from 3 to 7 d, whereas it decreased considerably from 9 to 11 d compared with the WT strain (Fig. 1E). In addition, conidia of WT and ΔAowetA mutant strains were observed by scanning electron microscopy (SEM), and it was observed that the conidia of ΔAowetA mutant attached on conidiophore had shrunk and the conidia of ΔAowetA mutant were abnormal (Fig. 1F). Furthermore, a high number of vacuoles and peroxisome-like structures were observed in the conidia of the ΔAowetA mutant by transmission electron microscopy (TEM) (Fig. 1G).
TABLE 2.
Primers used for genetic manipulation
| Primers | Sequence (5′−3′) | Description |
|---|---|---|
| AomedA_ZF | GTAACGCCAGGGTTTTCCCAGTCACGACGGATACCCACTTGACGACCCA | Amplify the AomedA gene 5′ flank |
| AomedA_ZR | ATCCACTTAACGTTACTGAAATCTCCAACGTAAGACCGGCTTCAGCGT | |
| AobrlA_ZF | GTAACGCCAGGGTTTTCCCAGTCACGACGGAAGGATGTGCGTGGCTCTA | Amplify the AobrlA gene 5′ flank |
| AobrlA_ZR | ATCCACTTAACGTTACTGAAATCTCCAACGGAGCTGGCATTTGTTTCCG | |
| AoabaA_ZF | GTAACGCCAGGGTTTTCCCAGTCACGACGCAACACTGCAAGGCTTCGTA | Amplify the AoabaA gene 5′ flank |
| AoabaA_ZR | ATCCACTTAACGTTACTGAAATCTCCAACAAGCTTCCTCAACCTCGTCA | |
| AowetA_ZF | GTAACGCCAGGGTTTTCCCAGTCACGACGCACTTCTTCACCCATCGGCT | Amplify the AowetA gene 5′ flank |
| AowetA_ZR | ATCCACTTAACGTTACTGAAATCTCCAACGATTCCGCGGAGACAGAGAG | |
| AomedA_YF | CTCCTTCAATATCATCTTCTGTCTCCGACTCGCGGTGTACTGTTTTCCA | Amplify the AomedA gene 3′ flank |
| AomedA_YR | GCGGATAACAATTTCACACAGGAAACAGCCGAGCCTCAGATCAGACGAAA | |
| AobrlA_YF | CTCCTTCAATATCATCTTCTGTCTCCGACTTATGGGTACGGGGGTCTGG | Amplify the AobrlA gene 3′ flank |
| AobrlA_YR | GCGGATAACAATTTCACACAGGAAACAGCGAGTTTGCGCTGCCAATTCA | |
| AoabaA_YF | CTCCTTCAATATCATCTTCTGTCTCCGACCCAGTCACATCCAGGTGTTG | Amplify the AoabaA gene 3′ flank |
| AoabaA_YR | GCGGATAACAATTTCACACAGGAAACAGCCCGTATCGACATTTGTGGTG | |
| AowetA_YF | CTCCTTCAATATCATCTTCTGTCTCCGACCGACGTTGAGGGGTTTGGAT | Amplify the AowetA gene 3′ flank |
| AowetA_YR | GCGGATAACAATTTCACACAGGAAACAGCAAAATCCCGCCTTTCACCGA | |
| Hph_F | GTCGGAGACAGAAGATGATATTGAAGGAGC | Amplify the hph cassette |
| Hph_R | GTTGGAGATTTCAGTAACGTTAAGTGGAT | |
| AomedA_F | TCCTCCACCATTCGATTATCAG | Verify the transformants |
| AomedA_R | GACAAGTGGCAAGATAGGTA | |
| AobrlA_F | CGGAAACAAATGCCAGCTCC | Verify the transformants |
| AobrlA_R | CCAGACCCCCGTACCCATAA | |
| AoabaA_F | CAACACTGCAAGGCTTCGTA | Verify the transformants |
| AoabaA_R | CCGTATCGACATTTGTGGTG | |
| AowetA_F | CTCTCTGTCTCCGCGGAATC | Verify the transformants |
| AowetA_R | ATCCAAACCCCTCAACGTCG | |
| AomedA_TZF | GTTTGAAGACCGTTCCACCA | Make Southern blotting probe |
| AomedA_TZR | TGCTCAGACTCAGATGGACG | |
| AobrlA_TZF | AGCCTGCCCAGTAATATGTATCG | Make Southern blotting probe |
| AobrlA_TZR | GCTGGCATTTGTTTCCGAAGA | |
| AoabaA_TZF | TCTAAGACACGGGCTGCAAG | Make Southern blotting probe |
| AoabaA_TZR | GGTGGTGTTTGTAAAGGTCGC | |
| AowetA_TZF | TTTGCCTCGAGTCGGTTCTC | Make Southern blotting probe |
| AowetA_TZR | TGCTAAAACCGCGTTCAAAGG |
Fig 1.
Comparison of conidiophores and conidia between WT and mutant strains. (A) Conidiophores and conidia of WT and mutant strains were observed under a light microscope. Bar = 10 µm. Red arrows indicate conidiophores, and black arrows indicate conidia. (B) Comparison of conidia yields. (C) Stress tolerance of conidia of WT and ΔAowetA mutant strains to chemical reagents. (D) Stress tolerance of conidia of WT and ΔAowetA mutant strains to heat shock. (E) Comparison of trehalose content. (F) Observation of conidiophores and conidia of WT and ΔAowetA mutant strains using a scanning electron microscope (SEM). Red arrows indicated vacuole. (G) Observation of ultrastructure of conidia of WT and ΔAowetA mutant strains under a transmission electron microscope. Red arrows indicate vacuoles and pink arrow indicates peroxisome-like structures. Asterisk (B–E) indicates a significant difference between mutant and WT strain (Tukey’s HSD, P < 0.05).
AoWetA had complex interactions with AoMedA, AoBrlA, and AoAbaA
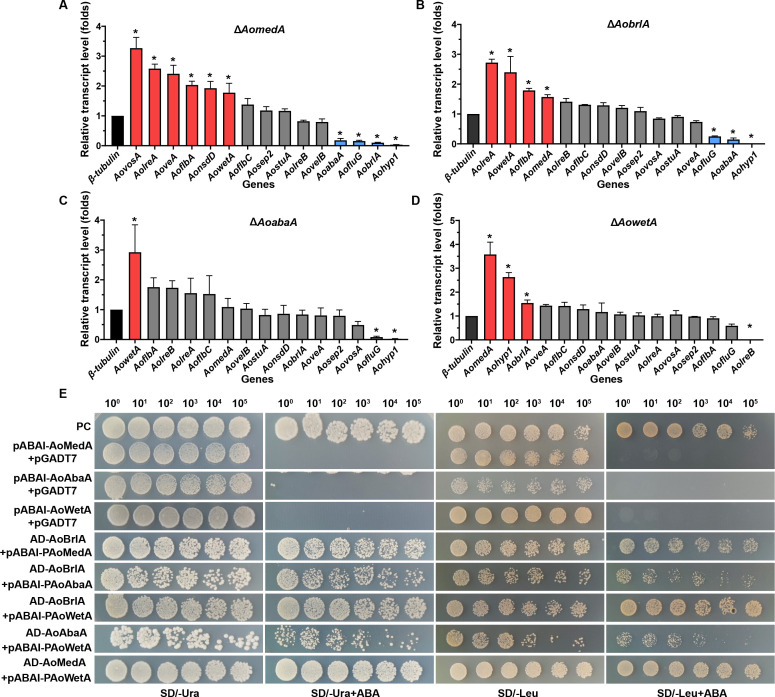
The transcriptional level of sporulation-related genes in WT and mutant strains was determined by real-time quantitative PCR (RT-qPCR) analysis. Deletion of AomedA led to considerably increased transcription levels of AovosA, AolreA, AoveA, AoflbA, AonsdD, and AowetA, whereas AoabaA, AofluG, AobrlA, and Aohyp1 were considerably downregulated (Fig. 2A). In ΔAobrlA mutant, the transcripts of AolreA, AowetA, AoflbA, and AomedA were upregulated, whereas AofluG, AoabaA, and Aohyp1 were downregulated (Fig. 2B). In ΔAoabaA mutant, only the AowetA gene was upregulated but AofluG and Aohyp1 were downregulated (Fig. 2C). In ΔAowetA mutant, AomedA, Aohyp1, and AobrlA were considerably upregulated, whereas AolreB was considerably downregulated (Fig. 2D). Y1H assay showed that AoBrlA can physically bind to the promoter region of AoMedA, AoAbaA, and AoWetA; meanwhile, both AoMedA and AoAbaA can bind to AoWetA (Fig. 2E).
Fig 2.
Comparison of relative transcription levels (RTLs) of sporulation-related genes between WT and mutant strains and yeast one-hybrid assay. (A–D) Comparison of RTLs of sporulation-related genes in ΔAomedA (A), AobrlA (B), AoabaA (C), and ΔAowetA mutants (D) versus WT strain. Error bars: SD from three replicates. Asterisk (A–D) indicates a significant difference between mutant and WT strain (Tukey’s HSD, P < 0.05). (E) Yeast one-hybrid assay of AoMedA, AoBrlA, AoAbaA, and AoWetA. pGADT7-Rec-p53/p53-AbAi as positive control and pABAI-AoMedA, pABAI-AoAbaA, and pABAI-AoWetA plus pGADT7 as negative controls. Yeast transformants were diluted in 0.9% NaCl, and serially diluted five times with equal volume for obtaining 100, 10−1, 10−2, 10−3, 10−4, and 10−5 dilutions.
AoMedA and three regulators play divergent roles in trap formation and pathogenicity
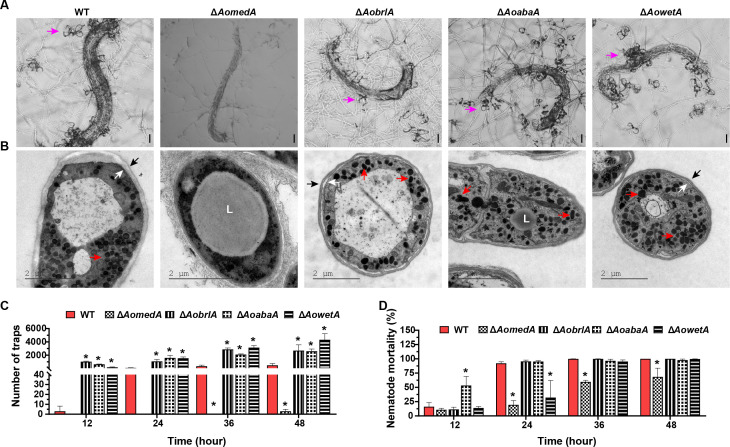
The ΔAomedA mutant lost the ability to form traps, and nematode mortality reduced remarkably (Fig. 3A), but the mycelium of the ΔAomedA mutant could still penetrate and decompose nematodes. Furthermore, many LDs were observed in the mycelia of ΔAomedA mutant after induction with nematodes, whereas electron-dense bodies (EDs) disappeared (Fig. 3B). The deletion of AobrlA, AoabaA, and AowetA genes resulted in increased trap formation than WT strain, and at 12 h post-induction (hpi), the average number of traps formed by ΔAobrlA and ΔAoabaA strains was 1021 and 627, respectively, which was higher than that of WT strain (179 traps), and the nematode mortality (53.36%) of ΔAoabaA strains increased considerably, whereas that of WT strain was 16.13%. At 24, 36, and 48 hpi, the number of traps formed by ΔAobrlA, ΔAoabaA, and ΔAowetA strains was also considerably more than that of the WT strain, whereas nematode mortality was not different from WT strain, except for the ΔAowetA mutant, which showed remarkable reduction in nematode mortality at 24 hpi (Fig. 3C and D). Furthermore, the ultrastructures of trap cells were observed by TEM, and the traps of WT, ΔAobrlA, ΔAoabaA, and ΔAowetA mutants were filled with EDs and separation of the plasma membrane and cell wall occurred in the trap cells of the three mutant strains (Fig. 3B).
Fig 3.
Comparison of trap formation and pathogenicity between WT and mutant strains. (A) Observation of trap formation and captured nematodes using a light microscope. Bar = 50 µm. Pink arrows indicate traps. (B) Observation of ultrastructure of traps using TEM. Blank arrows indicate cell wall, white arrows indicate cell membrane, red arrows indicate electron-dense bodies, and L indicates lipid droplets. (C) Comparison of the number of traps. (D) Comparison of nematode mortality. Error bars: SD from three replicates. Asterisk (C and D) indicates a significant difference between mutant and WT strain (Tukey’s HSD, P < 0.05).
AoMedA and three regulators play pleiotropic roles in mycelial growth, hyphal septum, cell nucleus, and LD accumulation
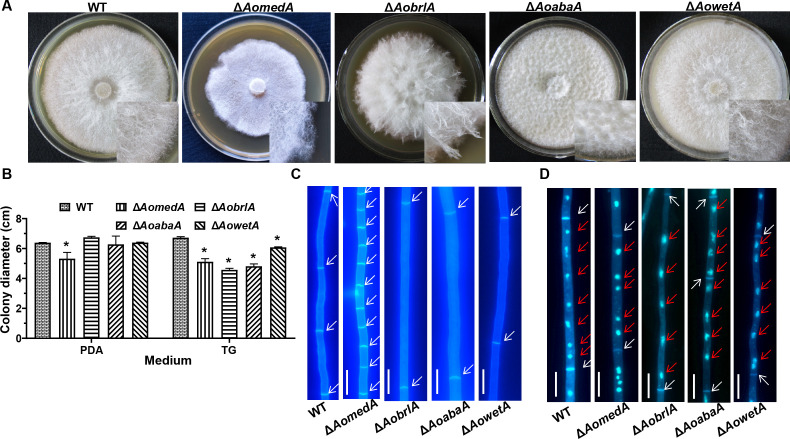
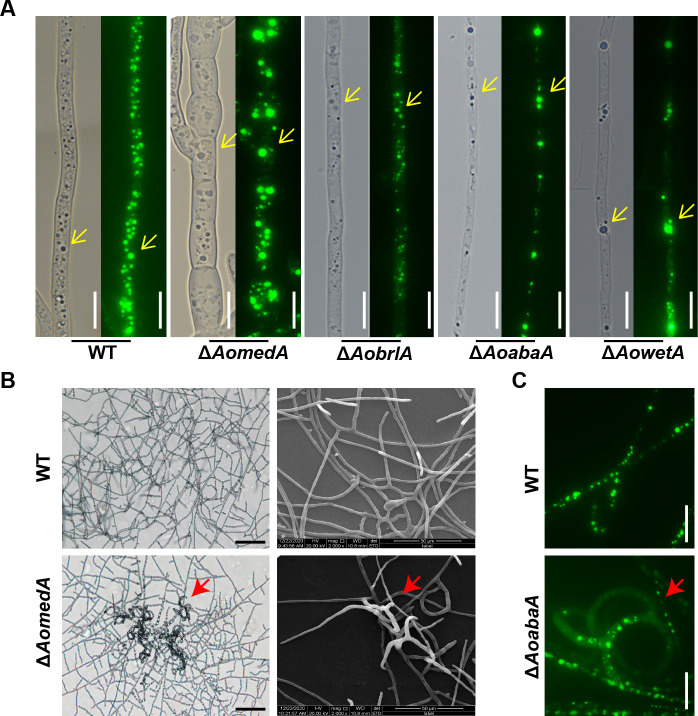
The colony and mycelial morphology of WT and mutant strains were observed in tryptone–yeast extract–glucose (TYGA) medium. Compared with the WT strain, the aerial hyphae of the ΔAomedA mutant were sparse, whereas the colony grew intensively; the aerial hyphae of ΔAobrlA mutant were extremely thriving, but the aerial hyphae of ΔAoabaA mutant became very compact, whereas the growth of aerial hyphae of ΔAowetA mutant was comparable with WT strain (Fig. 4A). Moreover, the radial growth of ΔAomedA mutant was remarkably slower on potato dextrose gar (PDA) or tryptone–glucose (TG) media, and the growth of ΔAobrlA, ΔAoabaA, and ΔAowetA mutants was mired on TG medium, whereas the growth was consistent with WT strain on PDA medium (Fig. 4B). The hyphal septum was observed using calcofluor white (CFW) dye (Fig. 4C), more septa were observed in ΔAomedA mutant, and the average length of mycelial cells of ΔAomedA mutant was considerably shorter, whereas the mycelial length of ΔAobrlA and ΔAoabaA was considerably longer than that of WT strain (Fig. S3A). In addition, mycelial nuclei were visualized by staining with 4′,6-diamino-2-phenylindole (DAPI) (Fig. 4D), the number of nuclei in WT strain was 7.0 per cell, and the number of nuclei in ΔAomedA, ΔAobrlA, ΔAoabaA, and ΔAowetA mutants was 5.0, 4.0, 4.0, and 6.0, respectively (Fig. S3B). When mycelial samples were stained with boron–dipyrromethene (BODIPY) (Fig. 5A), the fluorescence intensity of LDs in ΔAobrlA mutant was less than that of WT and the other three mutant strains. In addition, the volume of LDs in ΔAomedA mutant was higher than that of WT, and the number of LDs in ΔAoabaA, ΔAobrlA, and ΔAowetA was less than the WT strain (Fig. S3C). In addition, the mycelia of ΔAomedA and ΔAoabaA mutants formed trap-like structures when they were incubated on water agar (WA) plates (Fig. 5B and C).
Fig 4.
Comparison of mycelial growth, septa, and nuclei between WT and mutant strains. (A) Colony morphology of WT and mutant strains incubated on TYGA medium for 5 d. (B) Comparison of colony diameters of WT and mutant strains incubated on PDA and TG medium for 5 d. Error bars: SD from three replicates. Asterisk indicates a significant difference between mutant and WT strain (Tukey’s HSD, P < 0.05). (C) Mycelial cells were stained with CFW dye. White arrows indicate cell septa of the hyphae. (D) The nuclei of mycelial cells were stained using CFW and DAPI. White arrows indicate the cell septa of hyphae, and red arrows indicate nuclei. Bar = 10 µm.
Fig 5.
Comparison of the lipid droplets between WT and mutant strains. (A) Lipid droplets were stained by BIODIPY and observed using a light microscope (left panel) and fluorescence electron microscope (right panel), respectively. Yellow arrows indicate lipid droplets. (B) Observation of mycelial morphology of WT and ΔAomedA mutant strains by light microscopy (left panel) and SEM (right panel). The red arrow indicates the trap-like structure produced by ΔAomedA mutant. (C) Observation of mycelial morphology of WT and ΔAoabaA mutant strains by fluorescence electron microscopy. The red arrow indicates the trap-like structure produced by ΔAomedA (B) and ΔAoabaA (C) mutants. Bar (A and C) =10 µm.
AoMedA and three regulators play crucial roles in vacuole assembly
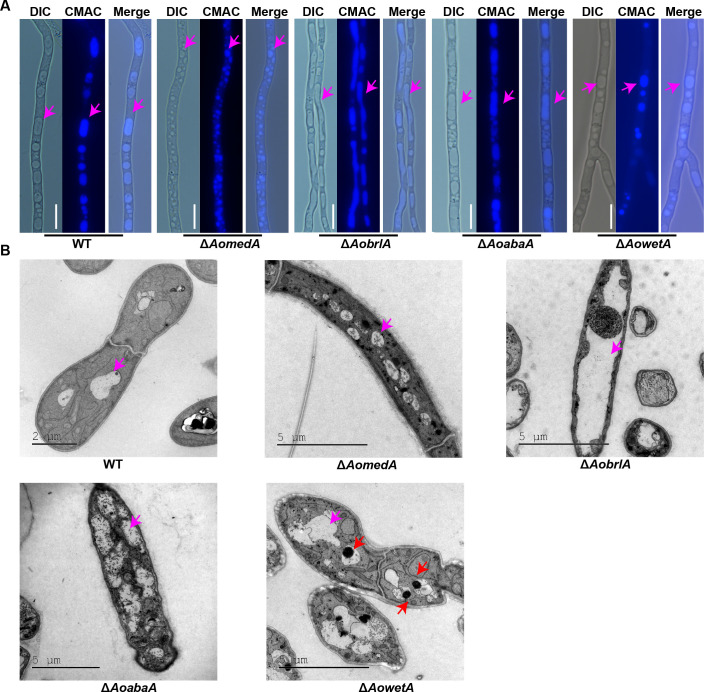
The vacuoles in the mycelia of WT and mutant strains were observed by 7-amino-4-chloromethylcoumarin (CMAC) staining, and the vacuoles in the WT strain were usually regular round and long oval, whereas the vacuoles were small and fragmented in the hyphal cells of ΔAomedA mutant and were irregular, elongated, and almost occupied the whole mycelial cell in ΔAobrlA mutant, and the vacuoles in ΔAoabaA strains mostly existed in the form of long ellipses. Although there was no difference in vacuole morphology between WT and ΔAowetA mutant strains, several pexophagy-like structures were observed in most vacuoles of ΔAowetA mutant (Fig. 6A). Furthermore, similar results of vacuoles were observed in the mycelia of WT and mutants by TEM (Fig. 6B).
Fig 6.
Comparison of vacuole morphology and ultrastructure of hyphae between WT and mutant strains. (A) Observation of vacuole morphology stained using 7-amino-4-chloromethylcoumarin (CMAC). Pink arrows indicate vacuoles. Bar = 10 µm. (B) Observation of ultrastructure of WT and mutant strains by TEM. Pink arrows indicate vacuoles, and red arrows indicate pexophagy-like structures.
AoMedA and AoWetA participate in the regulation of autophagy and peroxisome
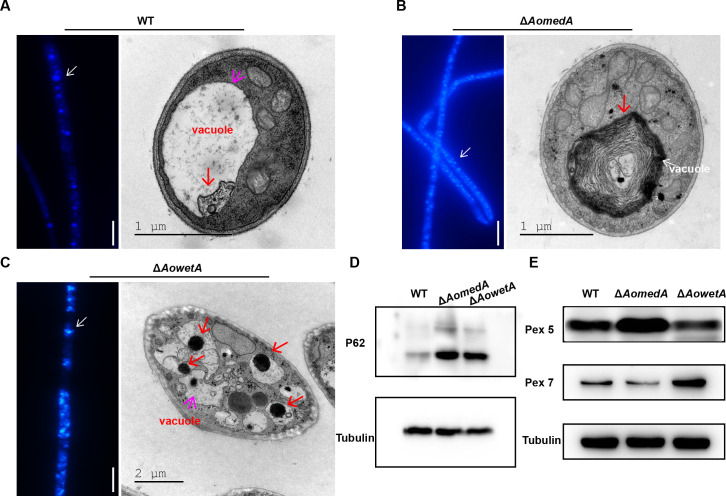
Mycelial samples of WT, ΔAomedA, and ΔAowetA strains were stained with monodansylcadaverine (MDC), and the ΔAomedA and ΔAowetA mutants had higher fluorescence intensities than that of the WT strain (Fig. 7A through C; Fig. S3D). Furthermore, more autophagosome-like structures were observed in the two mutants than WT strain (Fig. 7A through C). The mycelia of WT, ΔAomedA, and ΔAowetA mutant strains were collected and lysed to obtain proteins for Western blot analysis with p62 antibody (autophagosome marker). The results showed that autophagy in ΔAomedA and ΔAowetA mutants was considerably higher than that in the WT strain (Fig. 7D). In addition, the results of Western blot analysis with Pex5 and Pex7 antibody (peroxisome marker) showed that Pex5 in ΔAomedA and Pex7 in ΔAowetA mutants were considerably higher than that in WT strain, respectively (Fig. 7E).
Fig 7.
Detection of autophagy in WT and mutants (ΔAomedA and ΔAowetA). (A–C). Observation of autophagic vesicles of WT strain (A), ΔAomedA (B), and ΔAowetA (C) mutants stained by MDC (left panel) and observed by TEM (right panel). (D) Western blot analysis of autophagy in WT and mutant strains. Anti-SQSTM1/p62 antibody was used as an autophagosome marker and tubulin as the control. (E) Western blot analysis of peroxisome in WT and mutant strains. Anti-PEX5/PER3 antibody and anti-PEX7 antibody were used as peroxisome markers and tubulin as the control.
Analysis of transcriptome profiles of the WT, ΔAomedA, and ΔAowetA mutant strains
To further explore the regulation mechanism of conidiation in A. oligospora, mixed samples of hyphae and conidia of WT and mutant (ΔAomedA and ΔAowetA) strains were collected for transcriptome analysis, and sequencing statistics showed that the genes in each group were expressed efficiently (Table S1). Principal components analysis (PCA) showed that the three repeats of each sample shared a high degree of similarity (Fig. S4A). The accuracy of the transcriptomic data were confirmed by RT-qPCR of genes associated with endocytosis, phagosome, lipid metabolism, cell growth, and peroxisome (Fig. S4B and C).
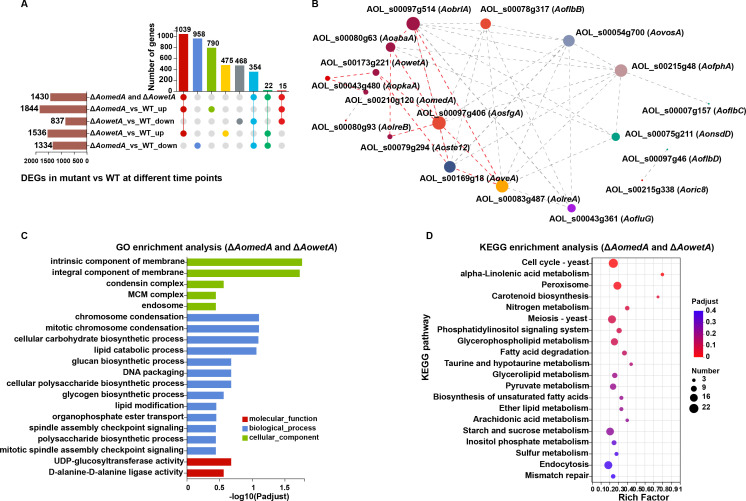
There were 1,844 upregulated differentially expressed genes (DEGs) and 1,334 downregulated DEGs in ΔAomedA mutant versus WT strain, there were 1,536 upregulated DEGs and 837 downregulated DEGs in ΔAowetA mutant versus WT strain, and 1,430 DEGs were shared in the ΔAomedA and ΔAowetA mutants, of which 1,039 were upregulated and 354 genes were downregulated in both groups (Fig. 8A). There were 17 sporulation-related DEGs in both groups. In gene co-expression analysis, deletion of AomedA and AowetA revealed that AobrlA played a central role in conidiation, which connected with AosfgA and AoabaA. AosfgA was linked with AowetA, AowetA was linked with AopkaA, and AopkaA was linked with AomedA; these genes were linked with other genes and constituted a complex network to regulate conidiation (Fig. 8B).
Fig 8.
Comparison of DEGs in WT and mutants (ΔAomedA and ΔAowetA). (A) UpSet plot analysis of DEGs between ΔAomedA/ΔAowetA mutant versus WT strain. The bar chart on the bottom left represents the number of the up/downregulated DEGs. The dotted line at the bottom right shows the number of DEGs in the different groups. (B) Visual representation of gene expression correlation of sporulation-related genes. In the figure, each node represents a gene/transcript and the connection between nodes represents the correlation between gene/transcript expression. The size of nodes indicates the transcript levels of genes/transcripts, and indicates the transcript correlation between different genes/transcripts. Node size is positively correlated with transcription level of gene/transcript. (C) Gene Ontology (GO) enrichment analysis of DEGs of ΔAomedA and ΔAowetA mutants versus WT strain. (D) Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis of ΔAomedA and ΔAowetA mutants versus WT strain.
Gene Ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) enrichment analyses were performed for these 1,430 DEGs, and GO enrichment analysis showed that intrinsic component of membrane and integral component of membrane were considerably enriched in cellular component; chromosome condensation, mitotic chromosome condensation, cellular carbohydrate biosynthetic process, and lipid catabolic process were considerably enriched in biological process; and two categories UDP-glucosyltransferase activity and D-alanine-D-alanine ligase activity were enriched in molecular function (Fig. 8C). Unlike these, upregulated DEGs in ΔAomedA mutant versus WT strain were also considerably enriched in transmembrane transport, transport, establishment of localization, localization, and obsolete oxidation–reduction process, whereas downregulated DEGs were enriched in organic acid metabolic process, cellular amino acid metabolic process, small molecule biosynthetic process, and organic acid biosynthetic process (Fig. S5). The upregulated DEGs of ΔAowetA mutant versus WT strain were also considerably enriched in transmembrane transport and membrane, whereas downregulated DEGs were enriched in catalytic activity, organic substance metabolic process, cellular process, and nitrogen compound metabolic process (Fig. S6).
KEGG enrichment analysis showed that genes of cell cycle, peroxisome, meiosis, glycerophospholipid metabolism, pyruvate metabolism, starch and sucrose metabolism, and endocytosis were enriched (Fig. 8D). Upregulated DEGs of ΔAomedA mutant versus WT strain were considerably enriched in linoleic acid metabolism, longevity regulating pathway, phagosome, cyanoamino acid metabolism, and fructose and mannose metabolism, and downregulated DEGs were enriched in DNA replication; homologous recombination; aminoacyl-tRNA biosynthesis; lysine biosynthesis; cysteine and methionine metabolism; nucleotide excision repair; valine, leucine, and isoleucine biosynthesis; glycine, serine, and threonine metabolism; and protein processing in the endoplasmic reticulum (Fig. S5). The interaction network analysis of AoMedA protein showed that AoMedA can regulate endocytosis, phagosome, lipid metabolism, cell growth, and peroxisome (Fig. S7A). Yeast two-hybrid (Y2H) assay showed that AoMedA can interact with AOL_s00054g434 (Fig. S7B), which encodes the transcription factor Atf21. Upregulated DEGs of ΔAowetA mutant versus WT strain were considerably enriched in ABC transporters and valine, leucine, and isoleucine degradation, whereas ribosome biogenesis in eukaryotes, aminoacyl-tRNA biosynthesis, ubiquinone and other terpenoid–quinone biosynthesis, DNA replication, and glycosaminoglycan degradation were enriched in downregulated DEGs (Fig. S6).
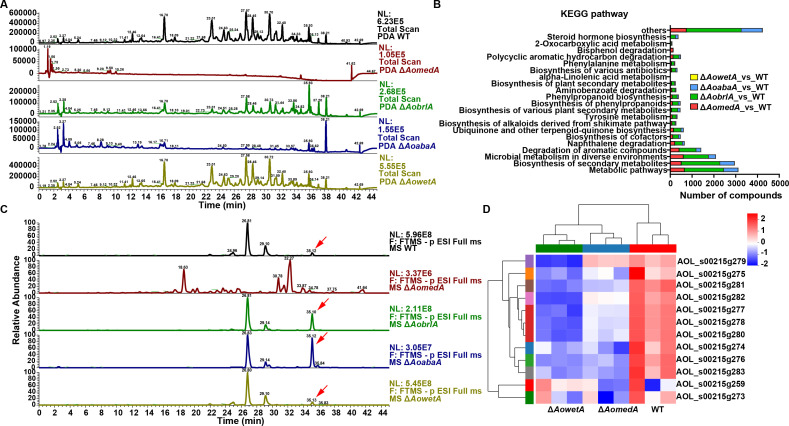
AoMedA and three regulators play a different role in secondary metabolism
Crude extracts of WT and mutant strains were detected by ultra-performance liquid chromatography (UPLC)–mass spectrometry (MS), and the chromatogram analysis showed that the peak values and secondary metabolites of ΔAomedA and ΔAoabaA mutants decreased considerably compared with the WT strain. The peak value of ΔAobrlA mutant decreased considerably to 12 min from 23–38 min, whereas the peak value of ΔAowetA mutant exhibited no difference from WT strain (Fig. 9A). Volcanic map analyses showed that more downregulated compounds were detected in the mutants ΔAomedA, ΔAoabaA, and ΔAobrlA compared with WT strain (Fig. S8A through D). KEGG pathway analysis showed that metabolic pathways, biosynthesis of secondary metabolites, microbial metabolism in diverse environments, degradation of aromatic compounds, and biosynthesis of cofactors were considerably enriched in the three mutant strains ΔAomedA, ΔAoabaA, and ΔAobrlA. In addition, alpha-linolenic acid metabolism and bisphenol degradation were particularly enriched in ΔAomedA mutant; 2-oxocarboxylic acid metabolism was enriched in ΔAobrlA, ΔAoabaA, and ΔAowetA mutants; and steroid hormone biosynthesis was enriched in ΔAobrlA and ΔAoabaA mutants (Fig. 9B).
Fig 9.
Comparison of secondary metabolism between WT and mutant strains. (A) Comparison of UPLC-MS profiles of WT and mutant strains. (B) Comparison of KEGG pathways of mutant versus WT strains. (C) Detection of the peak of arthrobotrisins in the chromatogram. The red arrow indicates the peak of arthrobotrisins. (D) Heatmap shows the relative transcript levels of genes associated with the biosynthesis of arthrobotrisins in WT, ΔAomedA, and ΔAomedA mutant strains.
The peak of arthrobotrisins was analyzed in WT and mutant strains, which was detected at 35 min, and the mass spectra were 139, 393, and 429 m/z (Fig. S8E). The content of arthrobotrisins in ΔAobrlA and ΔAoabaA mutants was less than that of WT strain, and no arthrobotrisins were detected in ΔAomedA mutant (Fig. 9C). Transcriptional level of the 215 g gene cluster associated with the biosynthesis of arthrobotrisins was analyzed, and most of these genes were downregulated in ΔAomedA and ΔAowetA mutants versus WT strain (Fig. 9D).
DISCUSSION
Asexual sporulation (conidiation) is the most common reproductive mode for many filamentous fungi, and the number of genes involved in conidiation has been identified, particularly, a FluG-mediated conidiation signaling pathway has been proposed for several filamentous fungi (38). In this study, we characterized the developmental regulator AoMedA and three core regulatory proteins AoBrlA, AoAbaA, and AoWetA in A. oligospora as they are crucial for conidiation and trap formation and play pleiotropic roles in mycelial development, LD accumulation, autophagy, vacuole assembly, and secondary metabolism.
The crucial role of MedA and three core regulatory proteins in conidiation has been revealed in several filamentous fungi. Mutations in the developmental modifier MedA resulted in frequent reinitiation of secondary conidiophores in A. nidulans (39). Similarly, deletion of AomedA caused the formation of secondary conidiophores in A. oligospora, and the conidia yield of ΔAomedA mutant decreased remarkably compared with that in the WT strain, which is consistent with A. fumigatus (40), whereas ΔAomedA mutant produced more mature conidia with a septum, possibly associated with high expression of AovosA and AowetA, as VosA and WetA are indispensable for conidia maturation (11). ΔAobrlA and ΔAoabaA strains completely lost the ability to produce conidia, and the results were consistent with A. nidulans (41), A. oryzae (42), B. bassiana (16), and Metarhizium robertsii (43). Based on our results, AobrlA is essential for conidiophore development and AoabaA is required for the formation of conidia, but there was no effect on conidia in ΔbrlA mutant of Monascus ruber M7 (44) and ΔbrlA mutant of Neurospora crassa (45). ΔAowetA mutant produced conidia, but the conidial yields decreased, which is similar to that observed in M. robertsii (18), and many conidia were immature with no septum, as observed in Fusarium graminearum (46). In particular, the morphology of the conidia of ΔAowetA mutant was acutely deformed and the cytoplasm of the conidia was filled with vacuoles. In addition, ΔAowetA strains were more sensitive to chemical stress reagents (Congo red and menadione) and high temperature, as also observed in B. bassiana (17) and P. digitatum (13). Moreover, the trehalose content in ΔAowetA strains changed with the culture times, which was different from B. bassiana (17), Aspergillus flavus (47), and A. fumigatus (11). Taken together, AomedA, AobrlA, AoabaA, and AowetA play a conserved and distinct role in conidiation in A. oligospora and other filamentous fungi.
At the transcriptional level, AomedA, AobrlA, and AoabaA had negative feedback regulation with AowetA. AobrlA had negative feedback regulation with AomedA but had positive regulation with AoabaA. Y1H assay revealed that AoBrlA regulated AoMedA, AoAbaA, and AoWetA and AoMedA and AoAbaA regulated AoWetA. The regulation relationship of AoBrlA, AoAbaA, and AoWetA was the same as in M. robertsii (18), whereas the regulation relationship between AoMedA and AoBrlA was different from MedA and BrlA in Penicillum chrysogenum; the deletion of brlA had no effect on medA (48), and the regulation relationship of AoBrlA, AoAbaA, and AoWetA was different from BrlA, AbaA, and WetA in A. nidulans (41) and P. digitatum (13) at the transcript level. The inactivation of brlA inhibits the expression of abaA and wetA, whereas the inactivation of abaA inhibits the expression of wetA. Therefore, the regulation relationships of AomedA, AobrlA, AoabaA, and AowetA were varied in fungal species, which implied that asexual sporulation is very complex in filamentous fungi and could be affected by various factors.
Conidiation is usually closely related to the pathogenicity of fungi (49). In this study, AomedA, AobrlA, AoabaA, and AowetA as the core regulatory genes in conidiation also had important roles in trap formation and pathogenicity. In ΔAomedA mutant, trap formation was completely abolished; therefore, the pathogenicity of ΔAomedA mutant decreased remarkably compared with the WT strain, which is similar to studies on A. fumigatus (40), Ustilago maydis (50), and M. grisea (15, 51). Conversely, ΔAobrlA, ΔAoabaA, and ΔAowetA mutants produced more traps than the WT strain, the pathogenicity of ΔAoabaA mutant increased remarkably at 12 hpi, and the pathogenicity of ΔAowetA mutant decreased remarkably at 24 hpi, whereas their pathogenicity had no difference from WT strain at 36 and 48 hpi. In M. robertsii (43) and B. bassiana (16), brlA and abaA strains had no conidia and reduced colonization capacity to host, and in F. graminearum, AbaA and WetA were indispensable for conidiation while not for virulence (46, 52). Thus, the orthologs of MedA, BrlA, AbaA, and WetA play a varied role in the pathogenicity of different fungal species and are crucial for trap formation in A. oligospora.
A previous study indicated that ΔmedA mutant had deficient mycelial growth in U. maydis (50). In this study, the deletion of AomedA caused a reduction in mycelial growth and the number of nuclei but increased hyphal septa, LD accumulation, and autophagy, particularly, the deletion of AomedA impaired vacuole assembly. In contrast, ΔAobrlA, ΔAoabaA, and ΔAowetA mutants had no significant difference in PDA compared with the WT strain, but their growth was decelerated on TG medium, and the average length of mycelia cells of ΔAobrlA and ΔAoabaA strains increased. In contrast to our results, BrlA, AbaA, and WetA are essential for conidiation but not for mycelial growth in B. bassiana (16) and F. graminearum (46). In addition, the deletion of AobrlA and AoabaA resulted in a reduction in hyphal septa, number of nuclei, and LD accumulation, and the vacuole became bigger and was filled with hyphal cells. Similarly, the deletion of AowetA reduced the number of nuclei and LD accumulation. Moreover, ΔAomedA and ΔAoabaA mutants formed a trap-like structure, and ΔAomedA and ΔAowetA mutants had a high autophagic level compared with the WT strain. Therefore, AoMedA and three core regulatory proteins play an important role in mycelial growth, nuclei, LD accumulation, vacuole assembly, and autophagy, whereas their roles are varied in A. oligospora.
Transcriptome analysis is considered a robust method to study differential gene expression in organisms under different sets of conditions (53). Here, RNA-Seq was performed to analyze differential gene expression caused by the deletion of sporulation-related genes. We focused on common DEGs of ΔAomedA and ΔAowetA mutants compared with WT strain and observed that several pathways, such as lipid catabolic and modification process, cell cycle, peroxisome, nitrogen metabolism, meiosis, glycerophospholipid metabolism, pyruvate metabolism, and endocytosis, were enriched. Lipid catabolic and modification process may be associated with the altered LD accumulation in ΔAomedA and ΔAowetA strains. Similarly, peroxisomes are associated with EDs, which contain existing trap cells (54 – 56). In our previous studies, several peroxisome biogenesis genes were identified in A. oligospora, and they were indispensable for conidiation and trap formation (34, 35). Here, the deletion of AomedA caused the loss of trap formation. In addition, nitrogen metabolism and endocytosis may be associated with autophagy (56, 57), and the deletion of AomedA and AowetA genes facilitated autophagy. A combination of transcriptome analysis and Y2H verification revealed that AoMedA can interact with AOL_s00054g434 that encodes the transcription factor Atf21; Atf21 participates in meiosis, osmotic pressure reaction, and sporulation in fission yeast (58), but the function of Atf21 has not been revealed in filamentous fungi. Therefore, many enriched pathways in transcriptome coincide well with the phenotypic features of ΔAomedA and ΔAowetA mutants.
Secondary metabolites produced by NT fungi, which act as chemoattractants, were studied to determine the interaction between nematodes and NT fungi and arthrobotrisins, specific compounds produced by A. oligospora and other NT fungi that can impair trap formation (59, 60). In this study, AoMedA, AoBrlA, and AoAbaA played an important role in the biosynthesis of arthrobotrisins and other secondary metabolites; volcanic map analysis showed that a high number of compounds were downregulated in mutant strains. There was no significant difference in the content of secondary metabolites between the ΔAowetA mutant and WT strain, whereas WetA has an important role in secondary metabolites in Aspergillus species (61, 62). Overall, AoMedA, AoBrlA, and AoAbaA play a vital role in the secondary metabolism of A. oligospora, and AoMedA is required for the biosynthesis of arthrobotrisins.
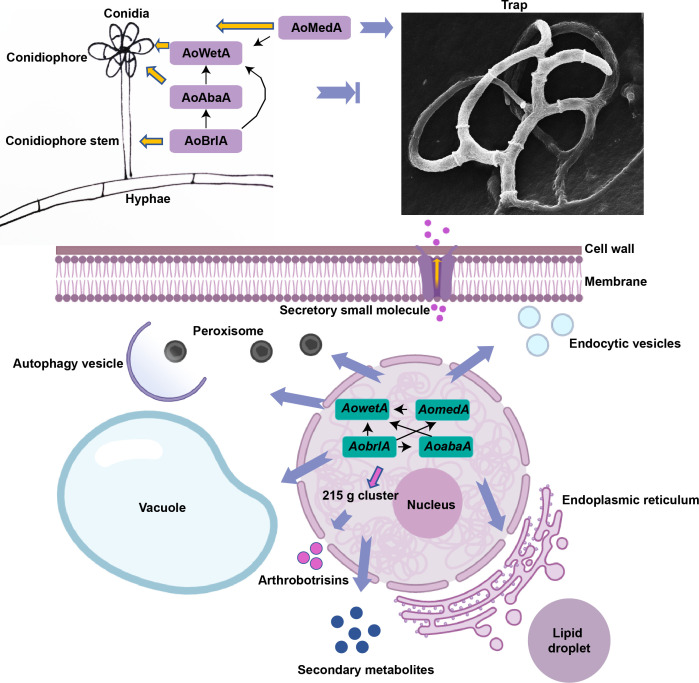
In this study, the roles of four key conidiation genes AomedA, AobrlA, AoabaA, and AowetA were characterized in A. oligospora, and these genes were essential for conidiation and trap formation and had pleiotropic roles in mycelial development, trap formation, LD accumulation, vacuole assembly, and secondary metabolism (Fig. 10). Based on Y1H and RT-qPCR analyses, there are complex interactions between AomedA and three central regulatory genes. AobrlA regulates AomedA, AoabaA, and AowetA and AomedA and AoabaA regulate AowetA; these genes are involved in multiple intracellular events, such as the number of septa and nuclei, lipid metabolism, vacuole assembly, autophagy, peroxisome, endocytosis, and secondary metabolism. In conidiation, AoBrlA is required for the development of conidiophores. AoAbaA is indispensable for forming conidia and AoMedA and AoWetA are necessary for conidia morphology and conidia yield. Interestingly, AoMedA is indispensable for trap formation, whereas three central regulators play a negative role in trap formation. In summary, our study first elaborated the functions and regulatory mechanism of AomedA and three central regulatory genes in mycelial growth, development, and differentiation of the NT fungus A. oligospora, provided a broad basis for elucidating the molecular mechanism of conidiation, and outlined the regulatory relationship between conidiation and trap formation in NT fungi.
Fig 10.
proposed model for the regulation of AoMedA, AoBrlA, AoAbaA, and AoWetA in A. oligospora. In this model, complex interactions between AomedA and three central regulatory genes (AobrlA, AoabaA, and AowetA) are illustrated; they are required for conidiation and trap formation and play a varied role in cell septa, nuclei, lipid metabolism, vacuole assembly, autophagy, endocytosis, and secondary metabolism.
Materials and Methods
Strains and culture conditions
A. oligospora (ATCC24927) WT and derived mutant strains were incubated on PDA, TG, TYGA, and corn dextrose with yeast extract (CMY) at 28°C for determining mycelial growth and conidia (63). Saccharomyces cerevisiae (FY834) and Escherichia coli strain DH5α were used for constructing the knockout vectors, as previously described (64). Caenorhabditis elegans (strain N2) was cultured on an oatmeal medium at 26°C and used for trap induction (27).
Sequence analysis of AoMedA, AoBrlA, AoAbaA, and AoWetA
The orthologous AomedA (AOL_s00210g120), AobrlA (AOL_s00097g514), AoabaA (AOL_s00080g63), and AowetA (AOL_s00173g221) were identified from A. oligospora using the orthologs of A. nidulans as a query. Their orthologous sequences from different fungi were blasted, and the biochemical properties of proteins were analyzed by the ProtParam tool (https://web.expasy.org/protparam/). DNAman software package (Version 5.2.2) was used to align different sequences and analyze their similarity, and a neighbor-joining tree was constructed by the MEGA 5 software package (65).
Deletion of AomedA, AobrlA, AoabaA, and AowetA
The disruption of AomedA, AobrlA, AoabaA, and AowetA was performed using a modified yeast cloning procedure, as previously described (66, 67). A. oligospora genome DNA was used as a template for amplifying the 5′- and 3′-flanking sequences of these genes, and the hph cassette was amplified from pCSN44, the paired primers listed in Table 2. The plasmid of pRS426 was used to construct the corresponding knockout vectors pRS426-AomedA-hph, pRS426-AobrlA-hph, pRS426-AoabaA-hph, and pRS426-AowetA-hph. The disruption sequences were transformed into A. oligospora protoplasts, as previously described (54). Positive transformants were confirmed via PCR and Southern blotting, as described previously (54, 68).
Analysis of mycelial growth, conidiation, and stress tolerance
WT and mutant strains were cultured on PDA and TG medium for 5 d for observing mycelial growth, colony morphology, and aerial hyphae, and the colony diameters were recorded at 24-h intervals. To observe the morphology of cell nuclei, LDs, and vacuole, 10 µg/mL of CFW (Sigma Aldrich), DAPI (Sigma-Aldrich), BODIPY (Sigma-Aldrich), and CMAC (Sigma-Aldrich) dyes were used to stain the mycelia (69, 70). The mycelia were examined by TEM.
To analyze sporulation, CMY medium was used to cultivate the fungal strains, and the mycelium was transferred to the slide for observing the conidiophore structures by light microscopy and SEM (71). Meanwhile, the conidia were washed with sterile water; hyphae were filtered for collecting conidia and observed via light microscopy and TEM; conidia were stained with CFW and observed by fluorescence electron microscopy.
The stress response of fungal strains to chemical reagents was performed, as described previously (64). In addition, to induce temperature stress, plates coated with 10,000 conidia were incubated on TG medium at 28, 30, 37, and 42°C for 30 min, 42°C for 1 h, and 50°C for 10 min and cultured at 28°C until 24 h and at 28°C for 30 min as control. RGI values of the strains were calculated, as previously described (72, 73).
Analysis of trap formation and pathogenicity
Mycelial discs (6 mm) of WT and mutant strains were incubated on a WA plate at 28°C for 4 d. Approximately 200 nematodes were added to each plate for trap induction. The number of traps and captured nematodes were counted and imuntaaged at 12-h intervals, and the ultrastructure of the trap cells was observed by TEM.
Observation of hyphal autophagy and western blot analysis
WT and mutant strains were inoculated in TYGA for 5 d, then mycelial samples were stained with MDC (100 µg/mL) for 30 min at 37°C, and observed by fluorescence electron microscopy.
WT and mutant strains were inoculated in potato dextrose liquid medium for 3 d at 28°C and 180 rpm, hyphae were collected, total protein was extracted by liquid nitrogen grinding using radioimmunoprecipitation assay lysis buffer, the concentration of total proteins was calculated using a bicinchoninic acid (BCA) protein assay reagent (Beijing Dingguochangsheng Biotechnology, China), and 50 µg of proteins was loaded into each well for Western blot analysis with anti-SQSTM1/p62, anti-Pex5/PER3, and anti-Pex7 antibodies (Abcam, Cambridge, UK), which diluted 1,000 times with primary antibody dilution buffer, and β-tubulin (Beyotime, Shanghai, China) as control. Antibody binding was visualized using an ECL Plus Western blotting detection reagent (Amersham Biosciences) after binding to a horseradish peroxidase-conjugated secondary antibody (74).
Transcriptome sequencing and RT-qPCR analyses
The WT, ΔAomedA, and ΔAowetA mutant strains were cultured on CMY medium with cellophane at 28°C for 5 d, and the mycelia were collected and three independent biological replicates were used for each sample. The samples were sent to Shanghai Meiji Biological Company (Shanghai, China) for RNA sequencing, and the RNA-Seq data were analyzed through the OmicShare online platform (www.majorbio.com). DEGs were identified based on the thresholds of | log2 ratio | ≥ 1 and adjusted P < 0.05.
The Axygen kit procedure (Axygen Biotech Company, Hangzhou, China) was used for total RNA extraction of WT and mutant strains, which were cultured on CMY at 28°C for 5 d. To verify the accuracy of transcriptome data, several genes associated with endocytosis, phagosome, lipid metabolism, cell growth, and peroxisome were selected, and their transcript levels were determined by RT-qPCR analysis, as previously described (23). All primers used for RT-qPCR are listed in Table 3. The relative transcription level (RTL) of each gene was calculated as the ratio of the transcription level between mutant and WT strain according to the 2 −ΔΔCt method (75), and the β-tubulin gene (AOL_s00076g640) was used as an internal standard.
TABLE 3.
Primers used for RT-qPCR analysis
| Primers | Sequence (5′−3′) | Primers | Sequence (5′−3′) |
|---|---|---|---|
| 54g700 F | CAAACCACCCACCACCAAAT | 215g893 F | ATACCGCCAACACCCTCTAC |
| 54g700 R | GGATGGACAGGAGAAGGACC | 215g893 R | AACCATCTTCATCTCGGCCT |
| 83g487 F | TTCTCTTCGTCCCAAGCCAC | 83g25 F | AGCTCCCGAAACGAGTCTAA |
| 83g487 F | ACCGGTTCGAGTGGAGTCTA | 83g25 R | ATTGATCATGTGATTATCCT |
| 169g18 F | AAGCTACACCCAATCAACGC | 80g93 F | CCAGGGTCGTCAGTATCTT |
| 169g18 R | TTGCGATGCTGACGATCTTG | 80g93 R | CAGCATCTTCCAGGTCAA |
| 215g516 F | TTCAAACGCAGCTCCTTCAC | 54g811 F | ATTCCGCAACTTCTCCCTCA |
| 215g516 R | AAGCGGGTTGACAGATGAGA | 54g811 R | GGCATGTTTGGATTCTGGGG |
| 75g211 F | ATTACGGCCGCCTAGTAGTC | 80g63 F | AACTTTATGCGCCTTGTCGT |
| 75g211 R | CTCGTTTGGACCTGGTTGTG | 80g63 R | TTGGCTAGGTGGTCTGTACG |
| 173g221 F | TTACATGCCACCCCAAGTCC | 43g361 F | GATTCCAGTCCCGTGAATTC |
| 173g221 R | CAATTGCAACTGCGTCCACA | 43g361 R | GCTAAGGAGAGGATGGGCAT |
| 7g157 F | CTCTCCGGCAAAGACAATCG | 97g514 F | TTGAGGCCTCGATCCGTAGA |
| 7g157 R | GTCGACTGAGGATAGTAGCT | 97g514 R | AGGTAGATGGCGCTGTTACG |
| 6g570 F | GCGGATCCAACATGAAGCTT | 210g120 F | TCCGGCCCAATGATTCAGAA |
| 6g570 R | GGTTGACAACTGGGATGCTG | 210g120 R | AGATCGCAGGAACATGGTGA-3 |
| 97g317 F | GAATGTGAGGGTGGCGAAGA | 54g737 F | CATCTCTTCGCCAACCCACC |
| 97g317 R | TAGCACGAACCCATGAACGG | 54g737 R | TTCTTTGTCGGCTCATCGGG |
| 110g119 F | GCCTGCGTCGTTAGTGTTTT | 4g362 F | CTTCCTTGCCCTTCACCCAA |
| 110g119 R | ACCCTTGCCTGCTGAAGAAT | 4g362 R | TTCTTGAGGCTACCGTCGTG |
| 193g4 F | ACCGGCAATCAACCTGTCTT | 79g361 F | GCGAACTACCCAACAGACCA |
| 193g4 R | CGCAAGCTCTCCGATCTCAT | 79g361 R | TGACCTTGTAACGCCGGATT |
| 79g276 F | GGATGTGGGCCGGTATTAGG | 83g431 F | ATTGGATCCGGACATGTGCTT |
| 79g276 R | CTACAACTGCGACCGTCCTT | 83g431 R | GCCATTTGCGCTTTGAAGGA |
| 112g52 F | CTGCCGGTCCTCACTTCAAC | 215g205 F | GCCTTCGAGAATGCCAATGT |
| 112g52 R | CAACAACAGTTCGTCCGAGG | 215g205 R | CCGCGGGTTCTCAAGAGTAG |
| 75g201 F | TTCCCTGCAATCAATTCGGC | 78g492 F | ATCCAGCTGTTCCATTCCCG |
| 75g201 R | GCTTCATACCCAACAAACCCG | 78g492 R | ACGGCTTTGGTATTGGCTTC |
| 210g359 F | AACCTGGCCTCGTCAAGAAG | 97g208 F | CCCCCGCATAAGGACATTCA |
| 210g359 R | GTGGGTGTAGGTGCGAAGAG | 97g208 R | GTAGAAGCGCCACATCAAGC |
| 54g368 F | AAGTTCGTCCACGCCAAGAA | 97g207 F | ATCCTCCAACAAGACGTCGC |
| 54g368 R | CCAACTGGCGCTACAGGTTT | 97g207 R | TCTCCTCCGACCGTTTTTCG |
| 78g71 F | ACCACTCCCAATTACCGCAG | 117g35 F | CTTCGCTCAAATGCTCGTCG |
| 78g71 R | TCGGTGTGATCCTCGCTCTA | 117g35 R | TAGAAGCACCCGTAACCCCT |
| 80g44 F | TTCGCGGATGGTGGAGATTT | 215g336 F | TCTCCGTCGCACCCCTATTA |
| 80g44 R | GCAAAACCAAAGTCGGCGAT | 215g336 R | TCTCCGTCGCACCCCTATTA |
| 83g233 F | TTCGGTCAAAAGGCGCAATG | 97g364 F | CAAATTCCCAGCCGGCAAAA |
| 83g233 R | GCTAACGATAAGGTCCCGCA | 97g364 R | CGGCGAATAAGACTGGCTCT |
| 78g27 F | ACAGGCGGTGTACAGTCTTC | 83g260 F | AGGGTGGTGAGCGAAGATTG |
| 78g27 R | CGAATTTCCCGAGCGAACTG | 83g260 R | GGAACCGTCTCGTTTTTGGC |
| 43g730 F | ATACCTTCTTGCTCACCCCG | 83g512 F | ATCCGCCAATTCAAACGCAC |
| 43g730 R | GCTTCGTTGAACTTGTCGGC | 83g512 R | TCTCTGAACAATGGACCGGC |
| 173g374 F | TCATCAACAAGACCGGCGAA | 4g606 F | GTCGCCAGCTACACAAATGC |
| 173g374 R | GGCTTCCTTTGGTGTCATGG | 4g606 R | AAGCGTGTGAGATCAGTCGT |
| 54g687 F | CACCAAACCTATGTCAACGGC | 4g235 F | ACTTTTGGCTCTGGACCTCG |
| 54g687 R | GCCTCGCTCAACTTACCACT | 4g235 R | CCTGGGCAGCTTGTTCGATA |
| 4g133 F | ACTCGTGACCAACTTACCCG | 6g253 F | AAGTTTAGGGATCGGTGCGG |
| 4g133 R | CTAGCAACGATCAGACGGGG | 6g253 R | CCTCAGGGTCGCTATCATCG |
| 6g411 F | TCACAGATAACTACGGCGGC | 109g23 F | CATTCAGCACACCGGTTCTC |
| 6g411 R | CTGTTTCCCCAGCATATCCCA | 109g23 R | CGAGCGGTCTGGATGTCAAT |
| 78g58 F | GTTCCTTCGCAGTTCCCTCA | 81g328 F | AGAGCGGAAACCTAGACATCA |
| 78g58 R | ACCGGTAGCTTGTTGGTTGA | 81g328 R | TTAGGTTGCCTTCGGTTCGG |
| 75g198 F | TCAAACCTGCCGACGAACTA | 75g194 F | TCACCGCGCAATCAGTCTAA |
| 75g198 R | GGGTGGCCTGTTCTAACTCG | 75g194 R | GAAGGGGAACGGTTCTTGGA |
| 6g248 F | GATCCCGAGCTCCAATACCC | 75g120 F | GTCATGGACGTTTCGGGGAT |
| 6g248 R | AATGCAAGTCCTTTCCAGCG | 75g120 R | TTGTGGAATGTTAGCCGGGA |
| 215g255 F | TCCACCCGTATCCCCAAGAT | 173g67 F | CCCAGCTAGACCCTCCATTG |
| 215g255 R | GAGAGAGGAGCAACGTCGAG | 173g67 R | AATCTCCCTGGTCAACTCGC |
| 78g38 F | ACAGACAATGTTCGAGCGGT | 78g181 F | ACAAAGTCGCCGTCAGAGAG |
| 78g38 R | CTTCCGAGAGCTGACCCATT | 78g181 R | CCGTCAGTTCGTTGGGAGTT |
| 188g58 F | CCGTCTTCCCAGTTCTCGTC | 4g330 F | TTCGAAGTCGTCATCTCCCG |
| 188g58 R | AGCCATCATTGATACCGCGT | 4g330 R | TCGCTGTCTTTTTCTCGGGG |
| 6g247 F | CCAGTGCGAAGACAGGACAT | 43g133 F | GCAACAAATCGGCCTAACGG |
| 6g247 R | GCACAACCGTCCTTGTTTGT | 43g133 R | CTGTTCGCCCAGCTAGTGAT |
| 78g47 F | AATGTTACCGCAGCTCCCTC | 78g126 F | GGCGTTCCAAAATCAGTTCCA |
| 78g47 R | TGACCGTAAGCATGCTCGAA | 78g126 R | GTCTCGCCGGTTTCACTAGG |
| 54g446 F | GTAGCCTTCTATCGCGGTGC | 76g160 F | GCGATGTGGCTGAATAACGG |
| 54g446 R | TGCCTCTTTCCATCGTCCAC | 76g160 R | ACAACCGAATCTCCACCACC |
| 159g2 F | TCACCGGACAGAGGGTTTTG | 54g253 F | AGAAATACAAGGGGACGGCG |
| 159g2 R | CAAGACTTCCGCCATCTCGT | 54g253 R | GGGACTCCGTGACAATAGCA |
| 215g358 F | CATATGCCAAGGAAGCCGGT | 43g20 F | TTGGACCGCACATACTAGCC |
| 215g358 R | TTAGGTTGGCGGTTTGAGGT | 43g20 R | CGTCTTATTCTCTGCGCGTG |
| 188g92 F | CCGCCTTTCATCCCTCGAAT | 54g312 F | GACAAGACCAAGGCCGAAGT |
| 188g92 R | TGCGAATCCTTCTTTGTCCCA | 54g312 R | CCGGTGCAGGAGTTGATGTA |
| 76g585 F | ATCCTCAAACCTTCCCTCGC | 76g229 F | AGCGATGGGAATGTGGTTGA |
| 76g585 R | ACCGCTTCTGATCTCTGTGG | 76g229 R | GTACTTCGCGGAACAAACCC |
| 97g406 F | CTCCAACAATGAGGCCGAGA | 97g406 F | GGGCTGCGGACAAGGATAAA |
| 76g640 F | CCACCTTCGTCGGTAACTC | 76g640 R | TCGTCCATACCCTCACCAG |
Y1H assay
The association of a transcription factor with a ~1,000 bp DNA fragment (putative promoter region of a gene) was assayed by Y1H according to the Matchmaker Gold Y1H Library Screening System User Manual (Clontech, USA). The promoter region with a ~1,000 bp DNA fragment (AomedA, AoabaA, and AowetA) was cloned into the pAbAi vector. The plasmids (pAbAi-AoMedA, pAbAi-AoAbaA, and pAbAi-AoWetA) were linearized and cloned into S. cerevisiae Y1HGold cells (TaKaRa, Dalian, China) as bait plasmids. Then, the cDNA sequence of AomedA, AobrlA, and AoabaA was cloned into a pGADT7 vector. The recombinant plasmids pGADT7-AoMedA, pGADT7-AoBrlA, and pGADT7-AoAbaA were further transformed into the Y1H1baitGold (pAbAi-AoMedA, pAbAi-AoAbaA, and pAbAi-AoWetA) strain. The transformed cells were plated on an SD/-Ura and SD/-Leu agar medium with 600 ng/mL aureobasidin A to identify the interactions between them. SD/-Ura and SD/-Leu agar medium without aureobasidin A was used as a control; pGADT7-Rec-p53/p53-AbAi as a positive control; and pABAI-AoMedA, pABAI-AoAbaA, and pABAI-AoWetA plus pGADT7 as negative controls (76). All primers used for the Y1H assay are listed in Table 4.
TABLE 4.
Primers used for Y1H and Y2H assays
| Primers | Sequence (5’−3’) | Description |
|---|---|---|
| AD-AoMedA F | ATATGGCCATGGAGGCCAGTATGGCAGCCTCCTACACCAA | Connected to pGADT7 vector |
| AD-AoMedA R | TATCGATGCCCACCCGGGTGGGACATCGAGTAGGTGTTGT | |
| AD-AoBrlA F | ATATGGCCATGGAGGCCAGTATGTCCCATCATAACATTCA | |
| AD-AoBrlA R | TATCGATGCCCACCCGGGTGATTTTCAAGCTTGATCCCCG | |
| AD-AoAbaA F | ATATGGCCATGGAGGCCAGTATGAGGAGCAGAACAAAGTC | |
| AD-AoAbaA R | TATCGATGCCCACCCGGGTGCTGCATATTGCCCATCCCAC | |
| ABI-AoMedA F | TTGAATTCGAGCTCGGTACCGATGAGAGATGGATGGTTTG | Connected to pABAI vector |
| ABI-AoMedA R | TCGACAGATCCCCGGGTACCGTTGCTGAGAGCCTGGGTGT | |
| ABI-AoAbaA F | TTGAATTCGAGCTCGGTACCCCGCATCGAGCAGAGAATTT | |
| ABI-AoAbaA R | TCGACAGATCCCCGGGTACCATTTGGGGATGATGTATATC | |
| ABI-AoWetA F | TTGAATTCGAGCTCGGTACCTAGGAAATTTCGCTGCATCG | |
| ABI-AoWetA R | TCGACAGATCCCCGGGTACCTTGAACTCAAGTTGCCCGGA | |
| AD-AoMedA F | ATATGGCCATGGAGGCCAGTATGGCAGCCTCCTACACCAA | Connected to pGADT7 vector |
| AD-AoMedA R | TATCGATGCCCACCCGGGTGGGACATCGAGTAGGTGTTGT | |
| BD-54g434 F | TGCATATGGCCATGGAGGCCATGGCTGCCGTTGCAAAACA | Connected to pGBKT7 vector |
| BD-54g434 R | GCAGGTCGACGGATCCCCGGGTCGTAAGCAGGTGGAGATG |
Y2H assay
The cDNA of A. oligospora was obtained as aforementioned, and then the cDNA sequences of AomedA and AOL_s00054g434 were amplified using the paired primers (Table 4). The cDNA sequence of AomedA was cloned into pGADT-7, and the cDNA sequence of AOL_s00054g434 was cloned into pGBKT-7. The Y2H assay was performed as previously described (64).
UPLC-MS analysis assay
WT and mutant strains were inoculated into PD broth at 28°C and 180 rpm for 6 d, and the fermentation broth was then extracted using ethyl acetate and dried under vacuum (64, 69). The samples were dissolved in methanol and analyzed by LC-MS using the Thermo Scientific Dionex Ultimate 3000 UHPLC system with a Thermo high-resolution Q Exactive focus mass spectrometer (Thermo, Bremen, Germany). Compounds Discoverer 3.0 software (Thermo Fisher Scientific, CA, USA) was used for untargeted metabolomics analysis.
Statistical analysis
Prism 8.0 (GraphPad Software, San Diego, CA, USA) was used as the tool for statistical analysis, with one-way ANOVA followed by Tukey’s honestly significant difference test being performed with P < 0.05 considered as statistically significant. All experiments were repeated three times.
ACKNOWLEDGMENTS
We are grateful to the Microbial Library of the Germplasm Bank of Wild Species from Southwest China for preserving and providing experimental strains, and to Guo Ying-qi (Kunming Institute of `Zoology, Chinese Academy of Sciences) for her help with taking and analyzing TEM images.
Funding for this study was provided by the National Natural Science Foundation of China (grant 31960556), the Special Fund of the Yunnan University "double first-class" Construction, and Applied Basic Research Foundation of Yunnan Province (grant nos. 202201BC070004, 202001BB050004).
K.-Q.Z. and J.Y. conceived and designed the study. N.B. performed the experiments. N.B., M.X., Q.L., Y.Z., and X.Y. analyzed the data. N.B. and J.Y. contributed to manuscript preparation and revision. All authors read and approved the final manuscript.
We declare that we have no conflicts of interests.
Contributor Information
Jinkui Yang, Email: jinkui960@ynu.edu.cn.
Yvonne Nygård, Chalmers University of Technology, Gothenburg, Sweden .
DATA AVAILABILITY
The data that support the findings of this study are included in the paper and the associated Supplementary Materials. All the RNA-seq data of this study have been deposited in the Gene Expression Omnibus database under accession number GSE230299.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.00983-23.
Figure S1-S8; Table S1.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Nieuwenhuis BPS, James TY. 2016. The frequency of sex in fungi. Philos Trans R Soc Lond B Biol Sci 371:20150540. doi: 10.1098/rstb.2015.0540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jung B, Kim S, Lee J. 2014. Microcycle conidiation in filamentous fungi. Mycobiology 42:1–5. doi: 10.5941/MYCO.2014.42.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yuan BH, Li H, Liu L, Du XH. 2021. Successful induction and recognition of conidiation, conidial germination and chlamydospore formation in pure culture of Morchella. Fungal Biol 125:285–293. doi: 10.1016/j.funbio.2020.11.005 [DOI] [PubMed] [Google Scholar]
- 4. Park HS, Yu JH. 2012. genetic control of asexual sporulation in filamentous fungi. Curr Opin Microbiol 15:669–677. doi: 10.1016/j.mib.2012.09.006 [DOI] [PubMed] [Google Scholar]
- 5. Mirabito PM, Adams TH, Timberlake WE. 1989. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell 57:859–868. doi: 10.1016/0092-8674(89)90800-3 [DOI] [PubMed] [Google Scholar]
- 6. Al Abdallah Q, Choe SI, Campoli P, Baptista S, Gravelat FN, Lee MJ, Sheppard DC. 2012. A conserved C-terminal domain of the Aspergillus fumigatus developmental regulator meda is required for nuclear localization, adhesion and virulence. PLoS One 7:e49959. doi: 10.1371/journal.pone.0049959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krijgsheld P, Bleichrodt R, van Veluw GJ, Wang F, Müller WH, Dijksterhuis J, Wösten HAB. 2013. Development in Aspergillus. Stud Mycol 74:1–29. doi: 10.3114/sim0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Etxebeste O, Garzia A, Espeso EA, Ugalde U. 2010. Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol 18:569–576. doi: 10.1016/j.tim.2010.09.007 [DOI] [PubMed] [Google Scholar]
- 9. Prade RA, Timberlake WE. 1993. The Aspergillus Nidulans brlA regulatory locus consists of overlapping transcription units that are individually required for conidiophore development. EMBO J 12:2439–2447. doi: 10.1002/j.1460-2075.1993.tb05898.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adams TH, Wieser JK, Yu JH. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol Mol Biol Rev 62:35–54. doi: 10.1128/MMBR.62.1.35-54.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tao L, Yu JH. 2011. Abaa and WetA govern distinct stages of Aspergillus fumigatus development. Microbiology (Reading) 157:313–326. doi: 10.1099/mic.0.044271-0 [DOI] [PubMed] [Google Scholar]
- 12. Qin Y, Bao L, Gao M, Chen M, Lei Y, Liu G, Qu Y. 2013. Penicillium decumbens BrlA extensively regulates secondary metabolism and functionally associates with the expression of cellulase genes. Appl Microbiol Biotechnol 97:10453–10467. doi: 10.1007/s00253-013-5273-3 [DOI] [PubMed] [Google Scholar]
- 13. Wang M, Sun X, Zhu C, Xu Q, Ruan R, Yu D, Li H. 2015. Pdbrla, PdabaA and PdwetA control distinct stages of conidiogenesis in Penicillium digitatum. Res Microbiol 166:56–65. doi: 10.1016/j.resmic.2014.12.003 [DOI] [PubMed] [Google Scholar]
- 14. Ohara T, Inoue I, Namiki F, Kunoh H, Tsuge T. 2004. Ren1 is required for development of microconidia and macroconidia, but not of chlamydospores, in the plant pathogenic fungus Fusarium oxysporum. Genetics 166:113–124. doi: 10.1534/genetics.166.1.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lau GW, Hamer JE. 1998. Acropetal: a genetic locus required for conidiophore architecture and pathogenicity in the rice blast fungus. Fungal Genet Biol 24:228–239. doi: 10.1006/fgbi.1998.1053 [DOI] [PubMed] [Google Scholar]
- 16. Zhang AX, Mouhoumed AZ, Tong SM, Ying SH, Feng MG. 2019. BrlA and AbaA govern virulence-required dimorphic switch, conidiation, and pathogenicity in a fungal insect pathogen. mSystems 4:e00140-19. doi: 10.1128/mSystems.00140-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li F, Shi HQ, Ying SH, Feng MG. 2015. WetA and VosA are distinct regulators of conidiation capacity, conidial quality, and biological control potential of a fungal insect pathogen. Appl Microbiol Biotechnol 99:10069–10081. doi: 10.1007/s00253-015-6823-7 [DOI] [PubMed] [Google Scholar]
- 18. Zeng G, Chen X, Zhang X, Zhang Q, Xu C, Mi W, Guo N, Zhao H, You Y, Dryburgh F-J, Bidochka MJ, St Leger RJ, Zhang L, Fang W. 2017. Genome-wide identification of pathogenicity, conidiation and colony sectorization genes in Metarhizium Robertsii. Environ Microbiol 19:3896–3908. doi: 10.1111/1462-2920.13777 [DOI] [PubMed] [Google Scholar]
- 19. Ji X, Yu Z, Yang J, Xu J, Zhang Y, Liu S, Zou C, Li J, Liang L, Zhang KQ. 2020. Expansion of adhesion genes drives pathogenic adaptation of nematode-trapping fungi. iScience 23:101057. doi: 10.1016/j.isci.2020.101057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu MC, Li XM, Zhao N, Yang L, Zhang KQ, Yang JK. 2022. Regulatory mechanism of trap formation in the nematode-trapping fungi. JoF 8:406. doi: 10.3390/jof8040406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang J, Wang L, Ji X, Feng Y, Li X, Zou C, Xu J, Ren Y, Mi Q, Wu J, Liu S, Liu Y, Huang X, Wang H, Niu X, Li J, Liang L, Luo Y, Ji K, Zhou W, Yu Z, Li G, Liu Y, Li L, Qiao M, Feng L, Zhang K-Q, Andrianopoulos A. 2011. Genomic and Proteomic analyses of the fungus Arthrobotrys oligospora provide insights into Nematode-trap formation. PLoS Pathog 7:e1002179. doi: 10.1371/journal.ppat.1002179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang C-T, Vidal-Diez de Ulzurrun G, Gonçalves AP, Lin H-C, Chang C-W, Huang T-Y, Chen S-A, Lai C-K, Tsai IJ, Schroeder FC, Stajich JE, Hsueh Y-P. 2020. Natural diversity in the predatory behavior facilitates the establishment of a robust model strain for nematode-trapping fungi. Proc Natl Acad Sci U S A 117:6762–6770. doi: 10.1073/pnas.1919726117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bai N, Zhang G, Wang W, Feng H, Yang X, Zheng Y, Yang L, Xie M, Zhang KQ, Yang J. 2022. Ric8 acts as a regulator of G-protein signalling required for nematode-trapping lifecycle of Arthrobotrys oligospora. Environ Microbiol 24:1714–1730. doi: 10.1111/1462-2920.15735 [DOI] [PubMed] [Google Scholar]
- 24. Ma N, Zhao Y, Wang Y, Yang L, Li D, Yang J, Jiang K, Zhang K-Q, Yang J. 2021. Functional analysis of seven regulators of G protein signaling (RGSs) in the nematode-trapping fungus Arthrobotrys oligospora. Virulence 12:1825–1840. doi: 10.1080/21505594.2021.1948667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen SA, Lin HC, Schroeder FC, Hsueh YP. 2021. Prey sensing and response in a nematode-trapping fungus is governed by the MAPK pheromone response pathway. Genetics 217:iyaa008. doi: 10.1093/genetics/iyaa008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xie M, Bai N, Yang J, Jiang K, Zhou D, Zhao Y, Li D, Niu X, Zhang K-Q, Yang J. 2019. Protein kinase Ime2 is required for mycelial growth, conidiation, osmoregulation, and pathogenicity in nematode-trapping fungus Arthrobotrys Oligospora. Front Microbiol 10:3065. doi: 10.3389/fmicb.2019.03065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xie M, Yang J, Jiang K, Bai N, Zhu M, Zhu Y, Zhang KQ, Yang J. 2021. AoBck1 and AoMkk1 are necessary to maintain cell wall integrity, vegetative growth, conidiation, stress resistance, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front. Microbiol 12:649582. doi: 10.3389/fmicb.2021.649582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhen Z, Xing X, Xie M, Yang L, Yang X, Zheng Y, Chen Y, Ma N, Li Q, Zhang KQ, Yang J. 2018. MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genet Biol 116:42–50. doi: 10.1016/j.fgb.2018.04.011 [DOI] [PubMed] [Google Scholar]
- 29. Yang L, Li X, Xie M, Bai N, Yang J, Jiang K, Zhang KQ, Yang J. 2021. Pleiotropic roles of Ras GTPases in the nematode-trapping fungus Arthrobotrys oligospora identified through multi-Omics analyses. iScience 24:102820. doi: 10.1016/j.isci.2021.102820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang L, Li X, Bai N, Yang X, Zhang K-Q, Yang J. 2022. Transcriptomic analysis reveals that Rho GTPases regulate trap development and lifestyle transition of the nematode-trapping fungus Arthrobotrys oligospora. Microbiol Spectr 10:e0175921. doi: 10.1128/spectrum.01759-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma Y, Yang X, Xie M, Zhang G, Yang L, Bai N, Zhao Y, Li D, Zhang KQ, Yang J. 2020. The Arf-GAP AoGlo3 regulates conidiation, endocytosis, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Fungal Genet Biol 138:103352. doi: 10.1016/j.fgb.2020.103352 [DOI] [PubMed] [Google Scholar]
- 32. Zhou D, Zhu Y, Bai N, Xie M, Zhang KQ, Yang J. 2021. Aolatg1 and Aolatg13 regulate autophagy and play different roles in conidiation, trap formation, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Front Cell Infect Microbiol 11:824407. doi: 10.3389/fcimb.2021.824407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li X, Zhu M, Liu Y, Yang L, Yang J. 2023. Aoatg11 and Aoatg33 are indispensable for mitophagy, and contribute to conidiation, the stress response, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Microbiol Res 266:127252. doi: 10.1016/j.micres.2022.127252 [DOI] [PubMed] [Google Scholar]
- 34. Liu Q, Li D, Jiang K, Zhang KQ, Yang J. 2022. Aopex1 and Aopex6 are required for mycelial growth, conidiation, stress response, fatty acid utilization, and trap formation in Arthrobotrys oligospora. Microbiol Spectr 10:e0027522. doi: 10.1128/spectrum.00275-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Q, Li D, Bai N, Zhu Y, Yang J. 2023. Peroxin Pex14/17 is required for trap formation, and plays pleiotropic roles in mycelial development, stress response, and secondary metabolism in Arthrobotrys oligospora. mSphere 8:e0001223. doi: 10.1128/msphere.00012-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang G, Zheng Y, Ma Y, Yang L, Xie M, Zhou D, Niu X, Zhang KQ, Yang J. 2019. The velvet proteins vosa and velb play different roles in conidiation, trap formation, and pathogenicity in the nematode-trapping fungus Arthrobotrys Oligospora Front Microbiol 10:1917. doi: 10.3389/fmicb.2019.01917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lim YJ, Lee YH. 2020. F-box only and CUE proteins are crucial ubiquitination-associated components for conidiation and pathogenicity in the rice blast fungus, Magnaporthe oryzae. Fungal Genet Biol 144:103473. doi: 10.1016/j.fgb.2020.103473 [DOI] [PubMed] [Google Scholar]
- 38. Li HX, Lu ZM, Zhu Q, Gong JS, Geng Y, Shi JS, Xu ZH, Ma YH. 2017. Comparative transcriptomic and proteomic analyses reveal a flug-mediated signaling pathway relating to Asexual sporulation of Antrodia Camphorata. Proteomics 17. doi: 10.1002/pmic.201700256 [DOI] [PubMed] [Google Scholar]
- 39. Busby TM, Miller KY, Miller BL. 1996. Suppression and enhancement of the Aspergillus nidulans medusa mutation by altered dosage of the bristle and stunted genes. Genetics 143:155–163. doi: 10.1093/genetics/143.1.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gravelat FN, Ejzykowicz DE, Chiang LY, Chabot JC, Urb M, Macdonald KD, al-Bader N, Filler SG, Sheppard DC. 2010. Aspergillus fumigatus meda governs adherence, host cell interactions and virulence. Cell Microbiol 12:473–488. doi: 10.1111/j.1462-5822.2009.01408.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boylan MT, Mirabito PM, Willett CE, Zimmerman CR, Timberlake WE. 1987. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol Cell Biol 7:3113–3118. doi: 10.1128/mcb.7.9.3113-3118.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamada O, Lee BR, Gomi K, Iimura Y. 1999. Cloning and functional analysis of the Aspergillus oryzae conidiation regulator gene brlA by its disruption and Misscheduled expression. J Biosci Bioeng 87:424–429. doi: 10.1016/s1389-1723(99)80089-9 [DOI] [PubMed] [Google Scholar]
- 43. Zhang JG, Xu SY, Ying SH, Feng MG. 2022. Roles of BrlA and AbaA in mediating asexual and insect pathogenic Lifecycles of Metarhizium Robertsii. J Fungi 8:1110. doi: 10.3390/jof8101110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jia L, Yu JH, Chen F, Chen W. 2021. Characterization of the asexual developmental genes brlA and wetA in Monascus ruber M7. Fungal Genet Biol 151:103564. doi: 10.1016/j.fgb.2021.103564 [DOI] [PubMed] [Google Scholar]
- 45. Boni AC, Ambrósio DL, Cupertino FB, Montenegro-Montero A, Virgilio S, Freitas FZ, Corrocher FA, Gonçalves RD, Yang A, Weirauch MT, Hughes TR, Larrondo LF, Bertolini MC. 2018. Neurospora crassa developmental control mediated by the FLB-3 transcription factor. Fungal Biol 122:570–582. doi: 10.1016/j.funbio.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 46. Son H, Kim MG, Min K, Lim JY, Choi GJ, Kim JC, Chae SK, Lee YW. 2014. Weta is required for conidiogenesis and conidium maturation in the Ascomycete fungus Fusarium graminearum. Eukaryot Cell 13:87–98. doi: 10.1128/EC.00220-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu MY, Mead ME, Kim SC, Rokas A, Yu JH. 2017. Weta bridges cellular and chemical development in Aspergillus flavus. PLoS One 12:e0179571. doi: 10.1371/journal.pone.0179571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sigl C, Haas H, Specht T, Pfaller K, Kürnsteiner H, Zadra I. 2011. Among developmental regulators, StuA but not BrlA is essential for penicillin V production in Penicillium chrysogenum. Appl Environ Microbiol 77:972–982. doi: 10.1128/AEM.01557-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim KS, Lee YH. 2012. Gene expression profiling during conidiation in the rice blast pathogen Magnaporthe oryzae. PLoS One 7:e43202. doi: 10.1371/journal.pone.0043202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chacko N, Gold S.. 2012. Deletion of the ustilago maydis ortholog of the Aspergillus sporulation regulator medA affects mating and virulence through pheromone response. Fungal Genet Biol 49(6):426–432. [DOI] [PubMed] [Google Scholar]
- 51. Nishimura M, Hayashi N, Jwa NS, Lau GW, Hamer JE, Hasebe A. 2000. Insertion of the LINE retrotransposon MGL causes a conidiophore pattern mutation in Magnaporthe grisea. Mol Plant Microbe Interact 13:892–894. doi: 10.1094/MPMI.2000.13.8.892 [DOI] [PubMed] [Google Scholar]
- 52. Son H, Kim MG, Min K, Seo YS, Lim JY, Choi GJ, Kim JC, Chae SK, Lee YW. 2013. AbaA regulates Conidiogenesis in the Ascomycete fungus Fusarium graminearum. PLoS One 8:e72915. doi: 10.1371/journal.pone.0072915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pandit R, Patel R, Patel N, Bhatt V, Joshi C, Singh PK, Kunjadia A. 2017. RNA-Seq reveals the molecular mechanism of trapping and killing of root-knot nematodes by nematode-trapping fungi. World J Microbiol Biotechnol 33:65. doi: 10.1007/s11274-017-2232-7 [DOI] [PubMed] [Google Scholar]
- 54. Xie M, Ma N, Bai N, Yang L, Yang X, Zhang KQ, Yang J. 2022. PKC-Swi6 signaling regulates asexual development, cell wall integrity, stress response, and lifestyle transition in the nematode-trapping fungus Arthrobotrys oligospora . Sci China Life Sci 65:2455–2471. doi: 10.1007/s11427-022-2118-0 [DOI] [PubMed] [Google Scholar]
- 55. Veenhuis M, Van Wijk C, Wyss U, Nordbring-Hertz B, Harder W. 1989. Significance of electron dense microbodies in trap cells of the nematophagous fungus Arthrobotrys oligospora. Antonie Van Leeuwenhoek 56:251–261. doi: 10.1007/BF00418937 [DOI] [PubMed] [Google Scholar]
- 56. Zhou D, Zhu Y, Bai N, Yang L, Xie M, Yang J, Zhu M, Zhang KQ, Yang J. 2022. AoATG5 plays pleiotropic roles in vegetative growth, cell nucleus development, conidiation, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Sci China Life Sci 65:412–425. doi: 10.1007/s11427-020-1913-9 [DOI] [PubMed] [Google Scholar]
- 57. Birgisdottir ÅB, Johansen T. 2020. Autophagy and endocytosis - interconnections and interdependencies. J Cell Sci 133:10. doi: 10.1242/jcs.228114 [DOI] [PubMed] [Google Scholar]
- 58. Morita T, Yamada T, Yamada S, Matsumoto K, Ohta K. 2011. Fission yeast ATF/CREB family protein Atf21 plays important roles in production of normal spores. Genes Cells 16:217–230. doi: 10.1111/j.1365-2443.2010.01480.x [DOI] [PubMed] [Google Scholar]
- 59. Yu X, Hu X, Pop M, Wernet N, Kirschhöfer F, Brenner-Weiß G, Keller J, Bunzel M, Fischer R. 2021. Fatal attraction of Caenorhabditis elegans to predatory fungi through 6-methyl-salicylic acid. Nat Commun 12. doi: 10.1038/s41467-021-25535-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. He Z-Q, Tan J-L, Li N, Zhang H-X, Chen Y-H, Wang L-J, Zhang K-Q, Niu X-M. 2019. Sesquiterpenyl epoxy-cyclohexenoids and their signaling functions in nematode-trapping fungus Arthrobotrys Oligospora. J Agric Food Chem 67:13061–13072. doi: 10.1021/acs.jafc.9b04968 [DOI] [PubMed] [Google Scholar]
- 61. Wu MY, Mead ME, Lee MK, Ostrem Loss EM, Kim SC, Rokas A, Yu JH. 2018. Systematic dissection of the evolutionarily conserved weta developmental regulator across a genus of filamentous fungi. mBio 9:e01130-18. doi: 10.1128/mBio.01130-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu MY, Mead ME, Lee MK, Neuhaus GF, Adpressa DA, Martien JI, Son YE, Moon H, Amador-Noguez D, Han KH, Rokas A, Loesgen S, Yu JH, Park HS. 2021. Transcriptomic, protein-DNA interaction, and metabolomic studies of vosa, velb, and weta in Aspergillus Nidulans asexual spores. mBio 12:e03128-20. doi: 10.1128/mBio.03128-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang X, Ma N, Yang L, Zheng Y, Zhen Z, Li Q, Xie M, Li J, Zhang KQ, Yang J. 2018. Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl Microbiol Biotechnol 102:4601–4613. doi: 10.1007/s00253-018-8929-1 [DOI] [PubMed] [Google Scholar]
- 64. Bai N, Xie M, Liu Q, Wang W, Liu Y, Yang J. 2023. AoSte12 is required for mycelial development, conidiation, trap morphogenesis, and secondary metabolism by regulating hyphal fusion in nematode-trapping fungus Arthrobotrys Oligospora. Microbiol Spectr 11:e0395722. doi: 10.1128/spectrum.03957-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. Mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tunlid A, Ahman J, Oliver RP. 1999. Transformation of the nematode-trapping fungus Arthrobotrys oligospora. FEMS Microbiol Lett 173:111–116. doi: 10.1111/j.1574-6968.1999.tb13491.x [DOI] [PubMed] [Google Scholar]
- 67. Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. 2006. A high-throughput gene knockout procedure for neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103:10352–10357. doi: 10.1073/pnas.0601456103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jiang KX, Liu QQ, Bai N, Zhu MC, Zhang KQ, Yang JK. 2022. AoSsk1, a response regulator required for mycelial growth and development, stress responses, trap formation, and the secondary metabolism in Arthrobotrys oligospora. J Fungi (Basel) 8:260. doi: 10.3390/jof8030260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhu MC, Zhao N, Liu YK, Li XM, Zhen ZY, Zheng YQ, Zhang KQ, Yang JK. 2022. The cAMP-PKA signalling pathway regulates hyphal growth, conidiation, trap morphogenesis, stress tolerance, and autophagy in Arthrobotrys oligospora. Environ Microbiol 24:6524–6538. doi: 10.1111/1462-2920.16253 [DOI] [PubMed] [Google Scholar]
- 70. Yang J, Wang W, Liu Y, Xie M, Yang J. 2023. The MADS-box transcription factor AoRlmA is involved in the regulation of Mycelium development, conidiation, cell-wall integrity, stress response, and trap formation of Arthrobotrys oligospora. Microbiol Res 268:127299. doi: 10.1016/j.micres.2022.127299 [DOI] [PubMed] [Google Scholar]
- 71. Wang W, Zhao Y, Bai N, Zhang K-Q, Yang J, Ianiri G. 2022. AMPK is involved in regulating the utilization of carbon sources, conidiation, pathogenicity, and stress response of the nematode-trapping fungus Arthrobotrys Oligospora. Microbiol Spectr 10. doi: 10.1128/spectrum.02225-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu J, Wang ZK, Sun HH, Ying SH, Feng MG. 2017. Characterization of the Hog1 MAPK pathway in the Entomopathogenic fungus Beauveria bassiana. Environ Microbiol 19:1808–1821. doi: 10.1111/1462-2920.13671 [DOI] [PubMed] [Google Scholar]
- 73. Pang MY, Lin HY, Hou J, Feng MG, Ying SH. 2022. Different contributions of the peroxisomal import protein Pex5 and Pex7 to development, stress response and virulence of insect fungal pathogen Beauveria bassiana . J Appl Microbiol 132:509–519. doi: 10.1111/jam.15216 [DOI] [PubMed] [Google Scholar]
- 74. Madamanchi NR, Runge MS. 2001. Western blotting. Methods Mol Med 51:245–256. doi: 10.1385/1-59259-087-X:245 [DOI] [PubMed] [Google Scholar]
- 75. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔδCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 76. Dey B, Thukral S, Krishnan S, Chakrobarty M, Gupta S, Manghani C, Rani V. 2012. DNA-protein interactions: methods for detection and analysis. Mol Cell Biochem 365:279–299. doi: 10.1007/s11010-012-1269-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1-S8; Table S1.
Data Availability Statement
The data that support the findings of this study are included in the paper and the associated Supplementary Materials. All the RNA-seq data of this study have been deposited in the Gene Expression Omnibus database under accession number GSE230299.