Abstract
Amphiphilic polymers are increasingly applied in the detergent-free isolation and functional studies of membrane proteins. However, the carboxylate group present in the structure of many popular variants, such as styrene-maleic acid (SMA) copolymers, brings limitations in terms of polymer sensitivity to precipitation at acidic pH or in the presence of divalent metal cations. Herein, we addressed this problem by replacing carboxylate with the more acidic sulfonate groups. To this end, we synthesized a library of amphiphilic poly[styrene-co-(sodium 4-styrene sulfonate)] copolymers (termed SSS), differing in their molecular weight and overall polarity. Using model cell membranes (Jurkat), we identified two copolymer compositions (SSS-L30 and SSS-L36) that solubilized membranes to an extent similar to SMA. Interestingly, the density gradient ultracentrifugation/SDS-PAGE/Western blotting analysis of cell lysates revealed a distribution of studied membrane proteins in the gradient fractions that was different than for SMA-solubilized membranes. Importantly, unlike SMA, the SSS copolymers remained soluble at low pH and in the presence of Mg2+ ions. Additionally, the solubilization of DMPC liposomes by the lead materials was studied by turbidimetry, DLS, SEC, and high-resolution NMR, revealing, for SSS-L36, the formation of stable particles (nanodiscs), facilitated by the direct hydrophobic interaction of the copolymer phenyls with lipid acyl chains.
Keywords: amphiphilic copolymer, sulfonated polystyrene, membrane protein, solubilization, cell membrane
1. Introduction
Membrane proteins are critical components of biological systems and represent a significant group of targets for pharmacological intervention [1]. However, structural and functional studies on these proteins present a significant challenge due to their destabilization outside their native lipid bilayer environment [2–4]. Detergents have traditionally been employed to isolate membrane proteins; however, this method removes the protein from its surrounding lipids, potentially affecting its structure, dynamics and function [5]. Recently, a detergent-free solubilization approach for membrane proteins was introduced that employs low-molecular weight (MW) styrene-maleic acid (SMA) copolymers [6]. The use of SMA copolymers results in the formation of nanodisc-shaped particles that contain membrane proteins embedded in a native-like lipid environment [3,6,7]. The use of SMA and other amphiphilic (co)polymers has facilitated the successful reconstitution, isolation, and characterization of various proteins [8–17], and the technology has found uses in related applications such as antibacterial vaccination [18] or bacterial toxin neutralization [19].
Nevertheless, the SMA copolymers are associated with some limitations hindering their use in certain applications. For instance, the most widely applied SMA variants are produced through free-radical polymerization [20], leading to their high dispersity (in terms of MW) [21,22], which may negatively influence the membrane solubilization efficiency and the obtained nanodisc homogeneity [21,23]. Another significant limitation of SMA stems from the presence of carboxylate in its structure since the acid-base behavior of this group severely limits SMA solubility at acidic pH [22]. In addition, the carboxylate groups easily bind with divalent cations such as Ca2+ and Mg2+, which makes the copolymer susceptible to precipitation under conditions necessary for the solubilization and function of certain proteins [22,24,25]. To address these limitations, different strategies were employed, including synthesizing better-defined SMA copolymers [21,26–29], and derivatizing the original SMA structure [30–40]. Furthermore, numerous other amphiphilic copolymers for membrane protein isolation have been introduced in recent years, including diisobutylene/maleic acid copolymers (DIBMA) and their derivatives [39,41–45], butyl methacrylate copolymers with methacroylcholine chloride (PMA) [46] or methacrylic acid (BMAA) [47], alkylamine-modified poly(acrylic acid) (APAA) [48], acrylic acid copolymers with styrene (AASTY) [23,49] or substituted styrenes (R-SAA) [38], modified inulin [13,50,51], methylstilbene/maleic acid copolymers (STMA) [52], cycloalkylamine- or arylamine-modified poly(acrylic acid) (CyclAPol, ArylAPol) [53,54], and hydrophobe-containing polypeptoids [55]. In addition, amphiphilic, non-polymeric, low-MW macromolecules have been recently also applied [56,57].
Even though SMA derivatives and new types of amphiphilic (co)polymers successfully addressed some of the shortcomings of the original SMA, the search for new polymeric materials usable in cell membrane solubilization still continues. This is motivated by various remaining challenges, such as the hydrolytic lability of some SMA derivatives [40], undesired polymer interactions with the solubilized membranes and proteins [37,58–62], the need for nanodiscs of controlled size and narrow size distribution [63], and also by the general desire to better understand the process of nanodisc formation [3,38]. Expanding the toolkit of polymers suitable for the disintegration of biological membranes and biochemical studies on native membrane proteins is expected to eventually allow a broader array of targets to be tackled under a range of different conditions. The availability of a variety of nanodisc-forming polymers would enable approaches to overcome other limitations such as the effects of polymers on the lamellar phase behavior of encased lipids and the reconstitution of a mixture of lipids and creating membrane domains.
Various parameters influence the (co)polymer performance in membrane solubilization assays. These mainly include MW, MW distribution (dispersity), polymer microstructure/topology, and balanced amphiphilicity [17,28,47,61,64]. With rare exceptions [31,47], a great majority of (co)polymers shown as effective in cell membrane solubilization are of low MW (i.e., several thousands). The underlying reason is currently unclear, with the potential mismatch between the typical nanodisc dimensions and high-MW polymer length suggested as a possible explanation [22]. Recently, we have proposed that a low MW may enable water-solubility of more hydrophobic (co)polymers [47], allowing the material to strike the right balance between hydrophilic and hydrophobic units, which is a prerequisite for the successful membrane solubilization [23,47]. The notion that in high-dispersity copolymers, such as the standard SMA, some polymeric fractions are considerably more efficient in membrane solubilization and nanodisc stabilization inevitably highlights the importance of controlling the copolymer dispersity (MW distribution) [23,26,61]. Indeed, the significance of this parameter is becoming increasingly realized, with controlled polymerization methods applied in multiple recent studies to assume MW control [21,23,28,38,43,47,61]. Finally, it was found that the distribution of hydrophobic/hydrophilic monomeric units within the copolymer chain is also a significant factor, with excessive “blockiness” being detrimental to the application [21,26].
When designing a new (co)polymeric system for membrane solubilization, most of the aforementioned factors can be addressed by using an appropriate polymerization or post-polymerization modification method. Nevertheless, it is practically impossible to a priori predict the copolymer composition (the ratio of hydrophobic and hydrophilic units) affording the best performance in solubilization assays. For this reason, recent studies have often opted for synthesizing a broader library of materials with incremental variations in the key parameters that is then screened using model membranes or liposomes [21,38,47,54]. In principle, two approaches can be used to obtain the targeted set of amphiphilic copolymers: direct copolymerization of selected monomers (or monomeric unit precursors) [47] or post-polymerization modification (polymer analogous reaction) of a suitable polymeric precursor [54]. While the first option appears straightforward, it can be complicated by various factors, such as unfavorable copolymerization parameters leading to compositional drift or the necessity to optimize polymerization conditions for polymerization mixtures of widely different compositions [61]. In addition, it may not be always easy to retain the pre-determined MW and dispersity for each targeted copolymer composition, which complicates evaluating the impact these parameters have on membrane solubilization. On the other hand, the post-polymerization modification approach allows for using well-defined starting polymeric materials as precursors to which substituents modulating the overall copolymer polarity are introduced. This enables creating copolymer libraries where important parameters such as polymeric backbone MW and dispersity are fixed, facilitating the structure-property-performance correlation and thus streamlining the identification of lead materials. A possible disadvantage of the post-modification approach may be the comparatively narrower range of achievable copolymeric compositions since bulky or charged substituents may not allow for a high degree of modification due to steric factors or Coulombic repulsion, respectively. Nevertheless, depending on the system, high degrees of modification may not be necessary to achieve the amphiphilic materials used in cell membrane solubilization.
In this study, we attempted at circumventing some of the limitations of the contemporary polymer designs used in cell membrane solubilization. Firstly, we desired to replace the typically used carboxylic groups with sulfonate groups. Carboxylate-based (co)polymers, such as SMA, are weak polyelectrolytes for which the backbone hydrophobicity easily prevails when the charge is lost upon carboxylate protonation at lower pH values, leading to aggregation and macroscopic precipitation [65]. On the other hand, sulfonate-based (co)polymers tend to behave as strong polyelectrolytes, with their sulfonate groups staying dissociated in a wide pH range, which potentially removes the (co)polymer sensitivity to lower pH [22]. Secondly, we focused on preparing well-defined copolymers, which enables the evaluation of the polymer MW impact on the membrane solubilization process. To this end, we employed a post-polymerization modification strategy to create a small library of well-defined amphiphilic poly[styrene-co-(sodium 4-styrene sulfonate)] copolymers (termed SSS) differing in their MW and composition characterized by their degree of sulfonation (DS). Employing a biologically relevant cell model (human T cell line Jurkat), we screened the library for copolymers suitable for cell membrane solubilization, and we further studied the lead materials in the solubilization of DMPC liposomes using DLS, SEC, high-resolution NMR and turbidity measurements. NMR data revealed the direct hydrophobic interactions between the copolymer aromatic groups and acyl chains of lipids. The SEC and DLS data, along with NMR results, confirm the formation of stable polymer-lipid particles (nanodiscs).
2. Results and Discussion
2.1. Synthesis of SSS copolymers
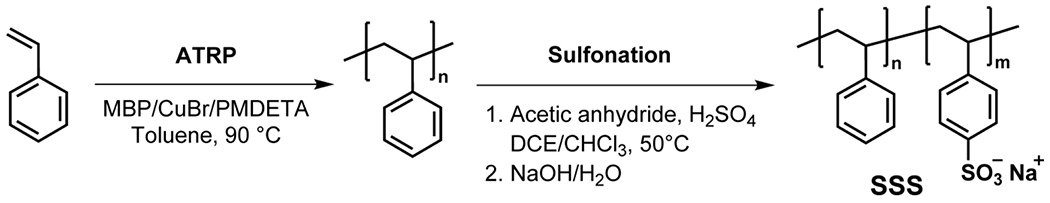
SSS copolymers were synthesized using a straightforward two-step protocol depicted in Scheme 1. First, two parent polystyrenes were synthesized via atom transfer radical polymerization (ATRP) using methyl 2-bromopropionate (MBP) as an initiator, CuBr as a catalyst, 1,1,4,7,7-pentamethyldiethylenetriamine (PMDETA) as a ligand, and toluene as a solvent at 90 °C. The results of these polymerizations are collected in Table S1. An SEC analysis of the prepared polystyrenes showed that they had very low dispersity (Ð = 1.12) and markedly different molecular weights of approximately 3 800 and 12 600 (Figure 1). Gravimetry was used for determining the monomer conversion for the high-MW polystyrene, while for the low-MW polystyrene, the conversion was calculated from the 1H NMR spectrum of the crude polymerization mixture (see Figure S1) as losses upon polymer precipitation made the gravimetric analysis inaccurate. Figure S2 provides a representative 1H NMR spectrum of isolated polystyrene.
Scheme 1.
Two-step synthesis of SSS copolymers.
Figure 1.
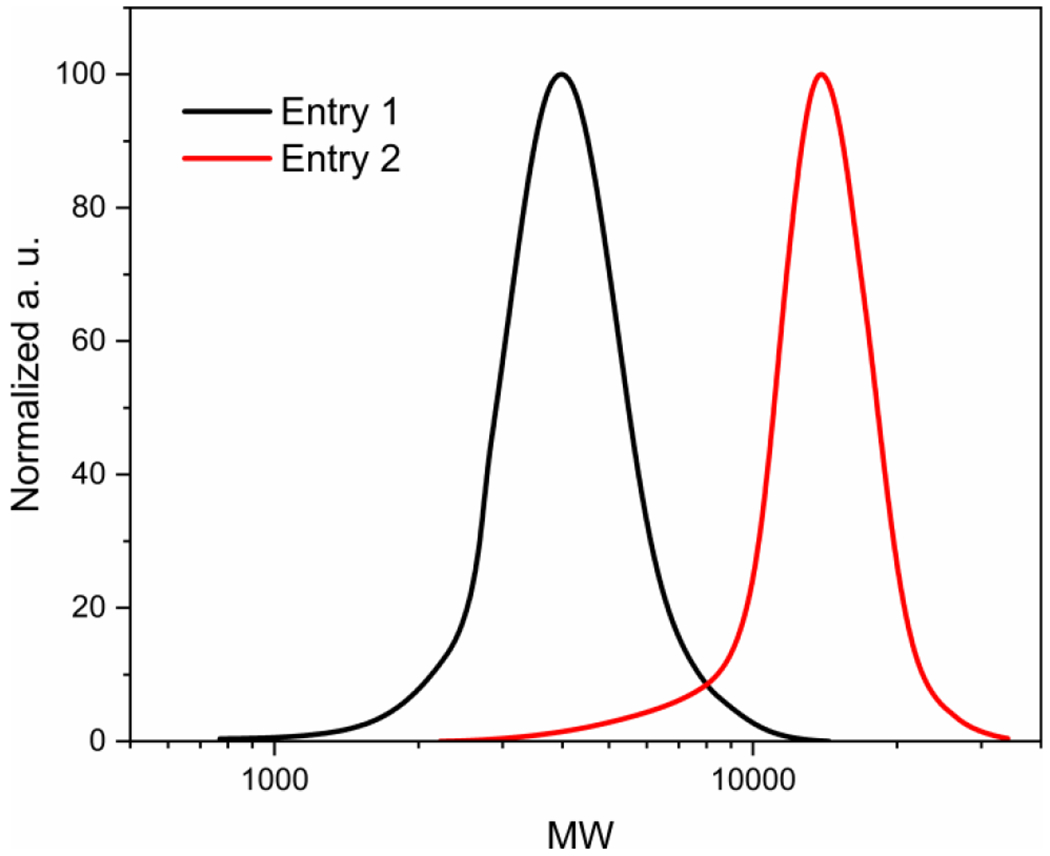
MW distributions of the parent polystyrenes used in the subsequent sulfonation step to prepare SSS copolymers (distributions correspond to the polymers shown in Table S1).
In a second step, we sulfonated the synthesized polystyrenes (in the para-position of phenyl rings) to different DS through a mild sulfonation method using acetyl sulfate, generated in situ by the reaction of sulfuric acid with an excess of acetic anhydride, in the mixture of chloroform and 1,2-dichloroethane (DCE) [66]. This approach is advantageous as it helps avoiding polymer degradation and sulfone formation that could lead to crosslinking reactions [67]. DS of the starting polymer was modulated by changing the H2SO4/styrene unit ratio and/or the reaction time. The obtained copolymers were subsequently neutralized with aqueous NaOH, dialyzed, and isolated by freeze drying. A typical 1H NMR spectrum of the final SSS copolymer is provided in Figure S3. The synthesized SSS copolymers were assigned codes corresponding to the MW of the parent polystyrene, i.e. “L” for the low-MW and “H” for the high-MW polystyrene (entries 1 and 2, Table S1, respectively), and to the DS value (in %) as determined by elemental analysis.
As can be seen from the sulfonation results displayed in Table 1, the parent polystyrenes can be readily sulfonated to the DS of approximately 25 % under the mildest conditions employed (H2SO4/styrene = 0.5, 3 h), with the achieved DS increasing only moderately when the H2SO4/styrene ratio is increased to 1 and the reaction time is prolonged, ultimately reaching approximately 35 % for both low-MW and high-MW polystyrenes. In one case (SSS-H38), we tried to apply more forcing conditions by increasing the H2SO4/styrene ratio to 2 and substantially prolonging the reaction time to 24 h. However, the DS was increased only marginally to 38 %. These observations indicate that the sulfonate groups already present on the polymeric backbone progressively hinder the introduction of additional sulfonate groups, which appears to be typical for polystyrene sulfonations [68]. Whatever is the reason for such behavior, we assume that this repulsion effectively directs the sulfonating agent to the less-sulfonated regions of the polymeric chain during the sulfonation reaction, maintaining the uniform distribution of sulfonate groups along the polystyrene chain. All the SSS copolymers were soluble under the conditions used in the cell membrane solubilization assays (0.02 M Tris-HCl buffer of pH 8.2, containing 0.1 M NaCl), only the lowest-DS samples (SSS-L26 and SSS-H24) had to be briefly heated to 90°C to achieve stable, clear solutions. This behavior suggests that the used sulfonation conditions afford copolymers of well-balanced amphiphilicity (an appropriate ratio of styrene and styrene sulfonate units), i.e., soluble in an aqueous buffer but still sufficiently hydrophobic.
Table 1.
Partial sulfonation of polystyrene to prepare the library of SSS copolymers.a
| Copolymerb | Mn (parent polystyrene) | H2SO4/styrene units | Time (h) | DS (%)c |
|---|---|---|---|---|
| SSS-L26 | 3 800 | 0.5 | 3 | 26 |
| SSS-L30 | 3 800 | 1 | 3 | 30 |
| SSS-L36 | 3 800 | 1 | 7 | 36 |
| SSS-H24 | 12 600 | 0.5 | 3 | 24 |
| SSS-H27 | 12 600 | 1 | 3 | 27 |
| SSS-H35 | 12 600 | 1 | 7 | 35 |
| SSS-H38 | 12 600 | 2 | 24 | 38 |
Standard reaction conditions: Ac2O/H2SO4 = 2, Ac2O/DCE = 1:1 (v/v), solvent: DCE/CHCl3, 50 °C.
Coding reflects the MW of the parent polystyrene (L – low, H – high) and the DS value.
Determined by elemental analysis.
2.2. Biochemical studies
2.2.1. Copolymer library screening using cell membranes
As in our previous study [47], we screened the synthesized SSS copolymers with respect to their ability to solubilize a biologically relevant model, cell membranes isolated from human T-cell line Jurkat. For this purpose, we dissolved all the copolymers in the previously specified buffer to achieve 1% (w/V) concentration. First, we briefly examined the effect of the copolymers on whole cells. Visual inspection revealed that all the copolymers lysed Jurkat T cells after 30 min exposure. We also note that all the SSS copolymers rapidly lysed human erythrocytes, showing a markedly different behavior when compared to the BMAA copolymers, screened in our previous study, that were completely inactive in this respect [47]. This observation indicates that the comparatively more rigid phenyl groups, present in the structure of both SSS and SMA, are more effective in attacking the cholesterol-rich erythrocyte membranes as compared to butyls in BMAA [69]. Subsequently, we proceeded to test the solubilization of isolated Jurkat T-cell membranes as a cleaner system with limited interference with cytoplasmic and nuclear biopolymers. As shown in Figure 2, only the SSS-L30 and SSS-L36 copolymers solubilized cell membrane proteins in an extent comparable to SMA, as determined by an SDS-PAGE/Western blotting analysis of the supernatant vs. sediment, detecting three typical membrane proteins (CD5, LCK, LAT). The ineffectiveness of the H-series copolymers points to a similar behavior as observed for SMA and most other copolymers used in this application where the low-MW variants show substantially higher solubilization power [70]. Furthermore, the negative result obtained for SSS-L26 highlights the importance of well-balanced amphiphilicity where subtle changes to the ratio of hydrophilic and hydrophobic groups can have a pronounced impact on copolymer performance [71].
Figure 2.
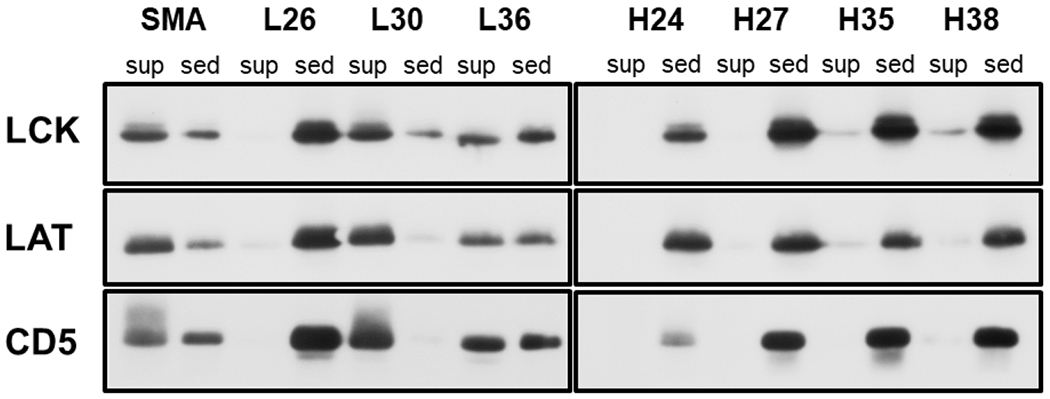
SDS PAGE/Western blot analysis of Jurkat T-cell membranes solubilized by the SSS copolymers. The cell membranes were treated with the lysis buffer containing 1% of a copolymer, and the insoluble material was removed by centrifugation. Three typical membrane proteins (CD5, LCK, LAT) were detected by immunostaining in the sediments and supernatants of the copolymer-treated membrane samples. Only the relevant parts of the blots are shown, corresponding to the area around the MW of the respective proteins (55 kDa for LCK, 45 kDa for LAT and 65 kDa for CD5). Sed – sediment, sup – supernatant.
Biological membranes are known to be spatially heterogeneous. Various micro/nano domains differing in their protein and lipid composition may also differ in their sensitivity to detergent- or copolymer-mediated disintegration. The best-known type of such microdomains are the so-called membrane rafts, enriched in cholesterol, lipids with saturated fatty acids, and lipid-modified membrane proteins [72]. Our previous study demonstrated that the SMA copolymer (used here as a comparative standard) disintegrates most of the Jurkat cell membrane into small nanodiscs [73] while membrane raft proteins are mostly present in larger SMA-resistant membrane fragments (SRMs). The raft-derived SRMs can be isolated by density gradient ultracentrifugation of the disintegrated membranes. Therefore, we subjected the membrane lysates obtained by using the SSS-L30 and SSS-L36 copolymers (together with SMA as a positive control) to this treatment, and then examined by SDS PAGE, followed by Western blotting, the distribution of four membrane markers (CD59, LCK, LAT, CD5) differing in their affinity to lipid raft microdomains (Figure 3).
Figure 3.
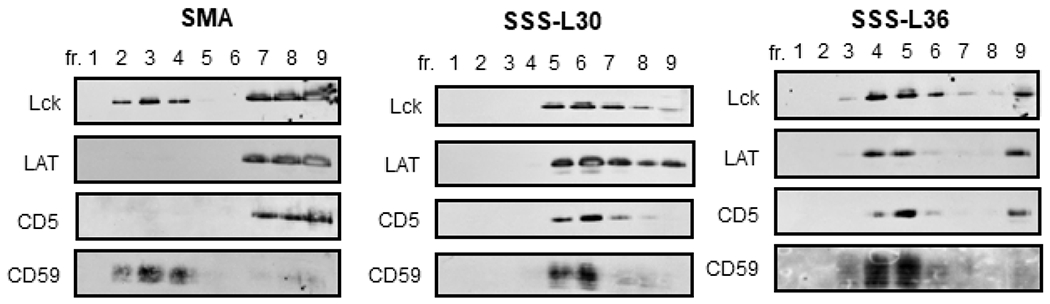
Distribution of Jurkat cell membrane proteins in the density gradient ultracentrifugation. Jurkat cell membranes were solubilized by the copolymers, the lysates were fractionated by density gradient ultracentrifugation, and the indicated proteins in the fractions were detected by Western blotting. The fractions are numbered from the top of the gradient. Only the relevant parts of the blots are shown, corresponding to the area around the MW of the respective proteins (55 kDa for LCK, 45 kDa for LAT, 65 kDa for CD5, and 20 kDa for CD59).
For the SMA-solubilized sample, the presence of membrane raft markers LCK (a palmitoylated and myristoylated protein tyrosine kinase) and CD59 (a GPI-anchored glycoprotein) indicated that the top (buoyant) gradient fractions contained copolymer-resistant membrane fragments (CRMs). The bottom fractions of the gradient then included the transmembrane protein CD5 (a non-raft marker), presumably solubilized within small membrane nanodiscs, and another potential membrane raft protein, the palmitoylated transmembrane adaptor protein LAT (Figure 3). These results are consistent with our previous study [73]. A somewhat different pattern was observed in gradient fractions of membranes solubilized by the SSS copolymers. Interestingly, the distribution of the LAT and CD5 membrane proteins was similar to that of LCK. This indicates that these proteins reside in larger and buoyant membrane fragments. Nevertheless, it is currently unclear what is their relationship to membrane rafts, if any. Furthermore, it should be noted that the CRMs are probably similar to the previously described detergent-resistant membrane fragments (DRMs), the relationship of which to native membrane rafts has been a subject of ongoing literature debate [72].
2.2.2. Resistance of SSS copolymers to low pH and divalent metal ions
One of the expected advantages of the SSS copolymers is their strong electrolyte character, potentially mitigating the major drawback of SMA and similar copolymers that lies in their sensitivity to acidic environment. As documented by the turbidimetric data shown in Table 2, both SSS-L30 and SSS-L36 remained soluble at pH 4 while SMA, used as a control, precipitated under these conditions. Furthermore, we evaluated the sensitivity of the SSS copolymers to biologically relevant concentrations of Ca2+ or Mg2+ ions. While the behavior toward Ca2+ ions was found to be comparable to that of SMA, the SSS copolymers did not precipitate in the presence of 10 mM Mg2+ ions, with the SSS-L30 copolymer tolerating even 100 mM Mg2+. Collectively, these results suggest that the use of SSS copolymers may be advantageous in the scenarios where low pH or the presence of divalent cations is required for the stability or function of the isolated membrane protein or where the divalent ions play an important role in bioassays [22,24].
Table 2.
Turbidimetric study of the resistance of the selected SSS copolymers to the presence of divalent metal ions and to acidic pH.a
| Copolymer | A490 |
||||
|---|---|---|---|---|---|
| pH 4 | Ca2+ (10 mM) | Ca2+ (100 mM) | Mg2+ (10 mM) | Mg2+ (100 mM) | |
| SSS-L30 | 0 | 0.08 | >3 | 0.09 | 0.12 |
| SSS-L36 | 0 | 0.26 | >3 | 0 | >3 |
| SMA (control) | >3 | 0.04 | >3 | >3 | >3 |
The experiments with Ca2+ and Mg2+ were conducted at the standard buffer solution pH of 8.2.
2.2.3. Solubilization of DMPC liposomes by SSS copolymers
To obtain additional insights into the lipid bilayer solubilization by SSS copolymers, we applied the two lead materials identified in the Jurkat membrane solubilization assay, SSS-L30 and SSS-L36, in the solubilization of DMPC liposomes. As can be seen from the turbidimetry measurements shown in Figure 4, SSS-L36 was considerably more effective in solubilizing DMPC liposomes than SSS-L30 that led to only a small decrease in turbidity. Considering the membrane solubilization data presented above, these results highlight the possible mismatch between the outcomes obtained with different models (i.e., crude cell membranes vs. artificial liposomes) used for assessing the solubilization performance of various (co)polymer types [21]. We also note that the efficacy of DMPC solubilization by polymers at 25 °C and 37 °C was not significantly different (Figure 4A, B). The DLS profile of the 1:1 w/w SSS-L36:DMPC sample showed particles with 6±2 nm hydrodynamic radius (Figure S4). However, in the DLS spectra of the 1:1 w/w SSS-L30:DMPC (1 mg/mL) sample, a range of different size particles were observed (Figure S4).
Figure 4.
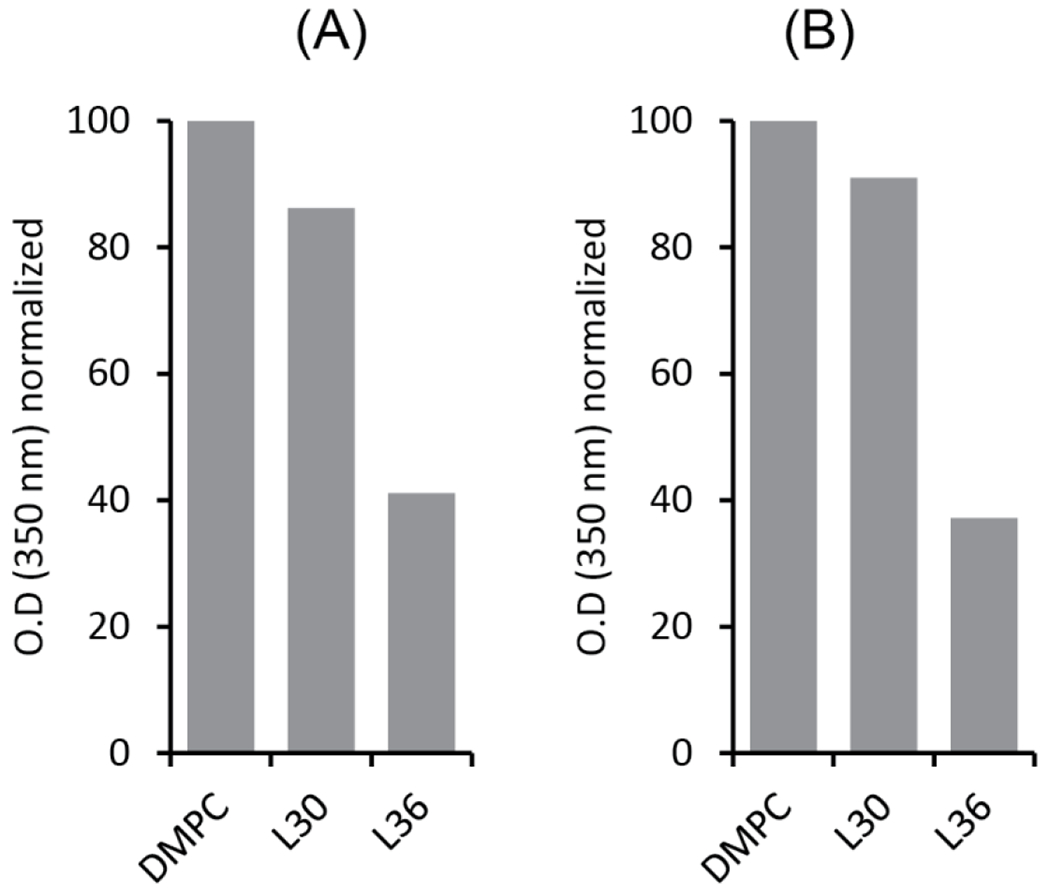
Turbidimetric analysis to follow DMPC liposomes solubilization using SSS-L30 and SSS-L36 copolymers at SSS:DMPC ratio of 1:1 (w/w). The O.D. was measured for samples under shaking at two different temperatures, 25 °C (A) and 37 °C (B). The labels L30 and L36 indicate DMPC liposomes solubilized by SSS-L30 and SSS-L36, respectively.
The SSS-L36:DMPC sample was further characterized by SEC and 1H NMR. Two major SEC elution peaks (P1 and P2) were detected at 214/254 nm (Figure 5A, B). The size of the formed particles was studied by DLS. The DLS profile of the SSS-L36:DMPC sample showed particles with a 5.6±1 nm hydrodynamic radius (Figures 5C and S4). Although both peaks showed similar DLS profiles, the DLS profile of P1 only showed the best autocorrelation plot (Figure S4). The 1H NMR spectra of the SSS-L36:DMPC sample showed peaks corresponding to DMPC lipids in the aliphatic region (0.5 to 5.4 ppm) and polymer-specific peaks in the aromatic region (6.55 – 7.95 ppm) (Figures 5D and S5). The early elution of the polymer-DMPC complex in SEC (8 – 13 mL), and the presence of peaks from both the polymer and lipids in the 1H NMR spectrum, suggest good stability of the SSS-L36-DMPC complex on the SEC column. To probe the intermolecular interactions between the polymer and lipid, a 2D 1H/1H NOESY NMR experiment was carried out (Figure 6). Both 1D 1H and 2D NOESY spectra (and the projections) exhibit a combination of narrow and broad spectral lines from the polymer-lipid particles. Narrow lines are observed for the acyl chains of lipid molecules (0.5 to 5.4 ppm) and low intensity broad lines are observed for the aromatic protons (line-width = ~150 Hz) from polymer molecules (6 to 8 ppm) (Figures 5D and 6). Overall, the observation of motionally-averaged isotropic chemical shifts indicates the small size of polymer-lipid particles that undergo fast-tumbling on the NMR time scale, which is in good agreement with the DLS data. The broad aromatic peaks observed from polymer molecules suggest a slow motion for the phenyl groups, which is likely due to their strong interaction with the hydrophobic acyl chains of lipids (Figures 5D and 6). This is further supported by the 2D 1H/1H NOESY spectrum that showed cross-peaks between polymer aromatic ring protons and protons from the acyl chains/methyl groups and the quaternary ammonium or choline methyl groups (γCH3) from DMPC lipids (Figure 6). This observation of intermolecular cross-peaks also confirms the formation of stable polymer-DMPC particles in which the polymer molecules constitute the belt surrounding the DMPC lipid bilayer [74,75]. The observed cross-peaks between protons from DMPC acyl chains and polymer aliphatic protons (0.6 to 1.6 ppm) and polymer aromatic protons (6 to 8 ppm) are not symmetric in S/N with respect to the diagonal of the 2D 1H/1H NOESY spectrum. This is mainly due to the fast spin-spin relaxation (i.e., short T2) of aromatic protons as indicated by the broad and low S/N peaks from aromatic protons. This is also the reason for the absence of the cross-peaks above the diagonal corresponding to the interaction between choline methyl group protons of DMPC and polymer aromatic protons.
Figure 5.
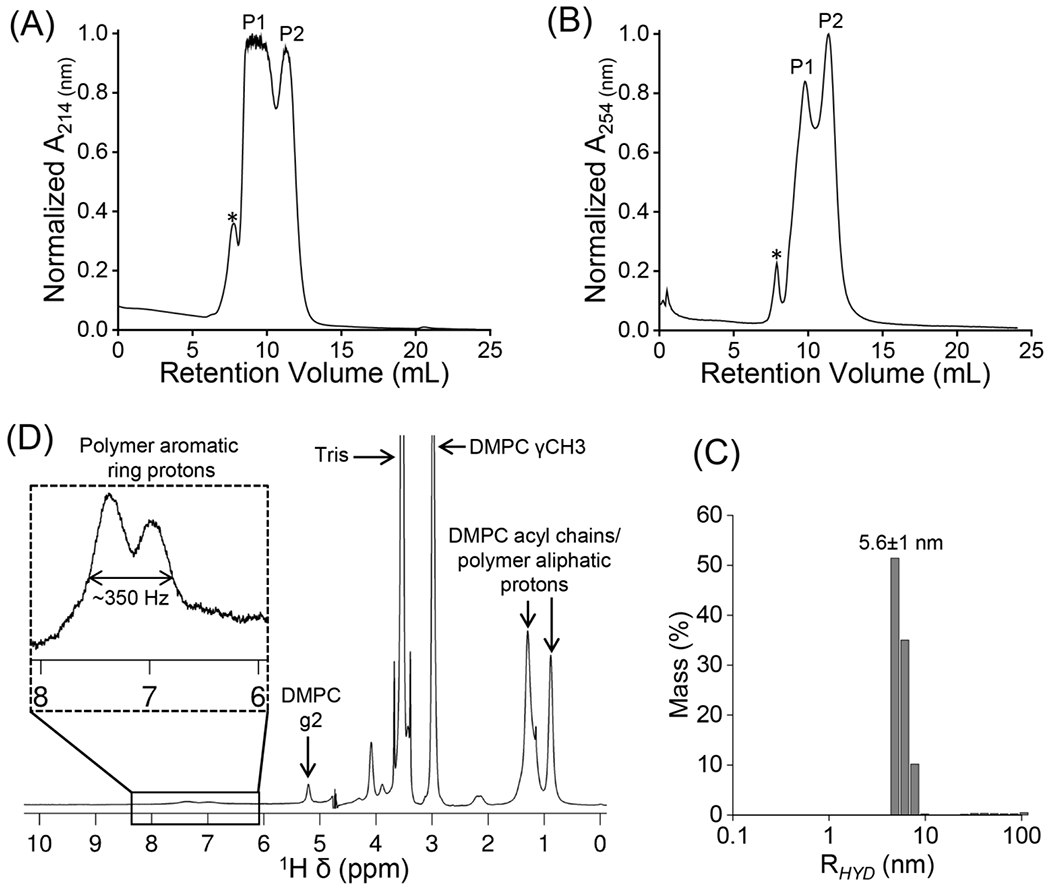
Characterization of SSS-L36:DMPC (1:1 w/w) particles. (A, B) SEC chromatograms recorded at 214 and 254 nm. The two major elution peaks are labelled PI, P2; * indicates an uncharacterized peak, likely to be from assemblies larger in size than that of PI and P2. (C) DLS profile of PI from SEC. (D) 1D 1H NMR spectrum of P1 from SEC. For clarity, the aromatic peaks which are at the noise level are expanded. The major peaks corresponding to polymer, lipids and buffer are labelled with assignments.
Figure 6.
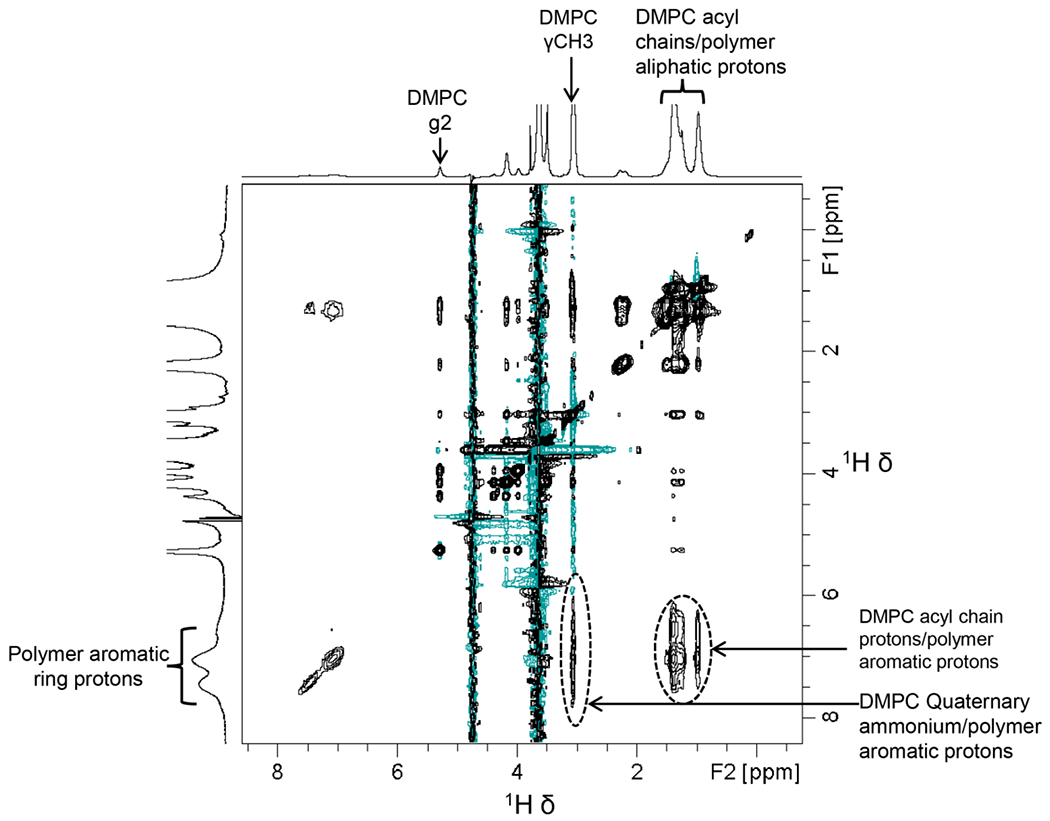
High-resolution two-dimensional 1H/1H NOESY NMR spectrum mapping of SSS-L36-DMPC interactions. The cross-peaks corresponding to polymer-lipid intermolecular interactions are labelled. F1 and F2 are the indirect and direct frequency dimensions, respectively. The cross-peaks not labelled are from intramolecular NOE interactions.
In this study, we did not specifically probe the shape of the formed polymer-lipid particles. Nevertheless, the good size homogeneity of the particles shown by the SEC/DLS measurements, together with the direct observation of the polymer-lipid interaction by NMR, support the assumption that homogeneous polymer nanodiscs are formed upon the DMPC solubilization by SSS copolymers. Indeed, this process may be facilitated by the high uniformity of SSS copolymers, in terms of composition and MW, provided by the used synthetic protocol.
3. Conclusions
In conclusion, a small library of well-defined amphiphilic SSS copolymers, differing in their MW and composition, was synthesized by a two-step procedure combining ATRP and sulfonation under mild conditions. We used a biologically relevant model of Jurkat T-cell membranes to screen the library, identifying two SSS copolymers (SSS-L30 and SSS-L36) that solubilized the membranes to a similar extent as the standard SMA copolymer. However, the analysis of the membrane lysates by density gradient ultracentrifugation followed by SDS PAGE/Western blotting indicated that the distribution of the LAT and CD5 membrane proteins in the gradient fractions was different from that obtained when SMA was used for membrane solubilization. This indicates that the SSS copolymers probably produce comparatively larger membrane fragments, the nature, heterogeneity and composition of which should be the subject of future studies, possibly identifying novel types of membrane micro/nanodomains compositionally and functionally different from membrane rafts. Importantly, in contrast to SMA, the sulfonate group-based SSS copolymers remained soluble at low pH and in the presence of Mg2+ ions, highlighting the importance of the charged group structure. Finally, we confirmed that one of the lead copolymers identified in the membrane solubilization assay (SSS-L36) efficiently dissolved DMPC liposomes to form stable polymer-lipid particles (probably nanodiscs) as revealed by SEC, DLS, and NMR experiments. We expect these nanodiscs to be useful for the reconstitution and high-resolution structural studies of membrane proteins.
In summary, this study establishes well-defined sulfonated polystyrenes as a new class of copolymers applicable to the solubilization of cell membranes and isolation of membrane proteins, expanding thus the current polymeric toolbox. We note that due to the commercial availability of extremely well-defined (Ð ≈ 1.05) polystyrenes in a wide range of MWs (SEC standards), close-to-monodisperse SSS copolymers should be easily accessible, possibly serving in future studies as a unique model for studying the impact of polymer MW on the process of nanodisc formation.
Supplementary Material
Highlights.
A library of sulfonated polystyrenes differing in MW and polarity was synthesized
Polymers effective in solubilizing model cell membranes were identified
The lead polymer forms stable particles (nanodiscs) from model DMPC liposomes
Polymers remain soluble at low pH and in the presence of Mg2+ ions
Acknowledgments
This work was supported by the Czech Science Foundation (Grant 19-04047S). The research in the Ramamoorthy lab is supported by NIH (R35 GM139572 to A.R.).
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Vaclav Horejsi reports financial support was provided by Czech Science Foundation. Ayyalusamy Ramamoorthy reports financial support was provided by National Institutes of Health. Vladimir Raus reports financial support was provided by Czech Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Experimental details, materials, and methods; additional NMR and DLS results.
Conflict of Interest
The authors declare no competing financial interest.
Data Availability Statement
Data are available in Supporting Information or on request from the authors.
References
- [1].Overington JP, Al-Lazikani B, Hopkins AL, How many drug targets are there?, Nat. Rev. Drug Discovery 5 (12) (2006) 993–996. 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- [2].Sligar SG, Denisov IG, Nanodiscs: A toolkit for membrane protein science, Protein Science 30 (2) (2021) 297–315. 10.1002/pro.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Overduin M, Esmaili M, Memtein: The fundamental unit of membrane-protein structure and function, Chem. Phys. Lipids 218 (2019) 73–84. 10.1016/j.chemphyslip.2018.11.008. [DOI] [PubMed] [Google Scholar]
- [4].Ravula T, Hardin NZ, Ramamoorthy A, Polymer nanodiscs: Advantages and limitations, Chem. Phys. Lipids 219 (2019) 45–49. 10.1016/j.chemphyslip.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zoonens M, Comer J, Masscheleyn S, Pebay-Peyroula E, Chipot C, Miroux B, Dehez F, Dangerous Liaisons between Detergents and Membrane Proteins. The Case of Mitochondrial Uncoupling Protein 2, J. Am. Chem. Soc 135 (40) (2013) 15174–15182. 10.1021/ja407424v. [DOI] [PubMed] [Google Scholar]
- [6].Knowles TJ, Finka R, Smith C, Lin Y-P, Dafforn T, Overduin M, Membrane Proteins Solubilized Intact in Lipid Containing Nanoparticles Bounded by Styrene Maleic Acid Copolymer, J. Am. Chem. Soc 131 (22) (2009) 7484–7485. 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- [7].Dörr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schäfer M, van Walree CA, Killian JA, The styrene–maleic acid copolymer: a versatile tool in membrane research, Eur. Biophys. J 45 (1) (2016) 3–21. 10.1007/s00249-015-1093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Barniol-Xicota M, Verhelst SHL, Stable and Functional Rhomboid Proteases in Lipid Nanodiscs by Using Diisobutylene/Maleic Acid Copolymers, J. Am. Chem. Soc 140 (44) (2018) 14557–14561. 10.1021/jacs.8b08441. [DOI] [PubMed] [Google Scholar]
- [9].Sun C, Benlekbir S, Venkatakrishnan P, Wang Y, Hong S, Hosier J, Tajkhorshid E, Rubinstein JL, Gennis RB, Structure of the alternative complex III in a supercomplex with cytochrome oxidase, Nature 557 (7703) (2018) 123–126. 10.1038/s41586-018-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Swainsbury DJK, Scheidelaar S, van Grondelle R, Killian JA, Jones MR, Bacterial Reaction Centers Purified with Styrene Maleic Acid Copolymer Retain Native Membrane Functional Properties and Display Enhanced Stability, Angew. Chem. Int. Ed 53 (44) (2014) 11803–11807. 10.1002/anie.201406412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bada Juarez JF, Harper AJ, Judge PJ, Tonge SR, Watts A, From polymer chemistry to structural biology: The development of SMA and related amphipathic polymers for membrane protein extraction and solubilisation, Chem. Phys. Lipids 221 (2019) 167–175. 10.1016/j.chemphyslip.2019.03.008. [DOI] [PubMed] [Google Scholar]
- [12].Krishnarjuna B, Im S-C, Ravula T, Marte J, Auchus RJ, Ramamoorthy A, Non-Ionic Inulin-Based Polymer Nanodiscs Enable Functional Reconstitution of a Redox Complex Composed of Oppositely Charged CYP450 and CPR in a Lipid Bilayer Membrane, Anal. Chem 94 (34) (2022) 11908–11915. 10.1021/acs.analchem.2c02489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Krishnarjuna B, Ravula T, Ramamoorthy A, Detergent-free isolation of CYP450-reductase’s FMN-binding domain in E. coli lipid-nanodiscs using a charge-free polymer, Chem. Commun 58 (31) (2022) 4913–4916. 10.1039/D1CC07193A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shu S, Mi W, Regulatory mechanisms of lipopolysaccharide synthesis in Escherichia coli, Nat. Commun 13 (1) (2022) 4576. 10.1038/s41467-022-32277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Swainsbury DJK, Hawkings FR, Martin EC, Musiat S, Salisbury JH, Jackson PJ, Farmer DA, Johnson MP, Siebert CA, Hitchcock A, Hunter CN, Cryo-EM structure of the four-subunit Rhodobacter sphaeroides cytochrome bc(1) complex in styrene maleic acid nanodiscs, Proc. Natl. Acad. Sci. U. S. A 120 (12) (2023) e2217922120. 10.1073/pnas.2217922120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Harant K, Čajka T, Angelisová P, Pokorná J, Hořejší V, Composition of raft-like cell membrane microdomains resistant to styrene-maleic acid copolymer (SMA) solubilization, Biophys. Chem 296 (2023) 106989. 10.1016/j.bpc.2023.106989. [DOI] [PubMed] [Google Scholar]
- [17].Krishnarjuna B, Ramamoorthy A, Detergent-Free Isolation of Membrane Proteins and Strategies to Study Them in a Near-Native Membrane Environment, Biomolecules 12 (8) (2022) 1076. 10.3390/biom12081076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Noh I, Guo Z, Zhou J, Gao W, Fang RH, Zhang L, Cellular Nanodiscs Made from Bacterial Outer Membrane as a Platform for Antibacterial Vaccination, ACS Nano 17 (2) (2023) 1120–1127. 10.1021/acsnano.2c08360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sun L, Wang D, Noh I, Fang RH, Gao W, Zhang L, Synthesis of Erythrocyte Nanodiscs for Bacterial Toxin Neutralization, Angew. Chem. Int. Ed n/a (n/a) (2023) e202301566. 10.1002/anie.202301566. [DOI] [PubMed] [Google Scholar]
- [20].Klumperman B, Mechanistic considerations on styrene–maleic anhydride copolymerization reactions, Polym. Chem 1 (5) (2010) 558–562. 10.1039/B9PY00341J. [DOI] [Google Scholar]
- [21].Smith AAA, Autzen HE, Laursen T, Wu V, Yen M, Hall A, Hansen SD, Cheng Y, Xu T, Controlling Styrene Maleic Acid Lipid Particles through RAFT, Biomacromolecules 18 (11) (2017) 3706–3713. 10.1021/acs.biomac.7b01136. [DOI] [PubMed] [Google Scholar]
- [22].Stroud Z, Hall SCL, Dafforn TR, Purification of membrane proteins free from conventional detergents: SMA, new polymers, new opportunities and new insights, Methods 147 (2018) 106–117. 10.1016/j.ymeth.2018.03.011. [DOI] [PubMed] [Google Scholar]
- [23].Smith AAA, Autzen HE, Faust B, Mann JL, Muir BW, Howard S, Postma A, Spakowitz AJ, Cheng Y, Appel EA, Lipid Nanodiscs via Ordered Copolymers, Chem 6 (10) (2020) 2782–2795. 10.1016/j.chempr.2020.08.004. [DOI] [Google Scholar]
- [24].Sawczyc H, Heit S, Watts A, A comparative characterisation of commercially available lipid-polymer nanoparticles formed from model membranes, Eur. Biophys. J 52 (1) (2023) 39–51. 10.1007/s00249-023-01632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bariwal J, Ma H, Altenberg GA, Liang H, Nanodiscs: a versatile nanocarrier platform for cancer diagnosis and treatment, Chem. Soc. Rev 51 (5) (2022) 1702–1728. 10.1039/D1CS01074C. [DOI] [PubMed] [Google Scholar]
- [26].Cunningham RD, Kopf AH, Elenbaas BOW, Staal BBP, Pfukwa R, Killian JA, Klumperman B, Iterative RAFT-Mediated Copolymerization of Styrene and Maleic Anhydride toward Sequence- and Length-Controlled Copolymers and Their Applications for Solubilizing Lipid Membranes, Biomacromolecules 21 (8) (2020) 3287–3300. 10.1021/acs.biomac.0c00736. [DOI] [PubMed] [Google Scholar]
- [27].Domínguez Pardo JJ, Koorengevel MC, Uwugiaren N, Weijers J, Kopf AH, Jahn H, van Walree CA, van Steenbergen MJ, Killian JA, Membrane Solubilization by Styrene-Maleic Acid Copolymers: Delineating the Role of Polymer Length, Biophys. J 115 (1) (2018) 129–138. 10.1016/j.bpj.2018.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hall SCL, Tognoloni C, Price GJ, Klumperman B, Edler KJ, Dafforn TR, Arnold T, Influence of Poly(styrene-co-maleic acid) Copolymer Structure on the Properties and Self-Assembly of SMALP Nanodiscs, Biomacromolecules 19 (3) (2018) 761–772. 10.1021/acs.biomac.7b01539. [DOI] [PubMed] [Google Scholar]
- [29].Craig AF, Clark EE, Sahu ID, Zhang R, Frantz ND, Al-Abdul-Wahid MS, Dabney-Smith C, Konkolewicz D, Lorigan GA, Tuning the size of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) using RAFT polymerization for biophysical studies, Biochim. Biophys. Acta, Biomembr 1858 (11) (2016) 2931–2939. 10.1016/j.bbamem.2016.08.004. [DOI] [PubMed] [Google Scholar]
- [30].Ravula T, Hardin NZ, Ramadugu SK, Cox SJ, Ramamoorthy A, Formation of pH-Resistant Monodispersed Polymer–Lipid Nanodiscs, Angew. Chem. Int. Ed 57 (5) (2018) 1342–1345. 10.1002/anie.201712017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [31].Fiori MC, Jiang Y, Altenberg GA, Liang H, Polymer-encased nanodiscs with improved buffer compatibility, Sci. Rep 7 (1) (2017) 7432. 10.1038/s41598-017-07110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ravula T, Hardin NZ, Ramadugu SK, Ramamoorthy A, pH Tunable and Divalent Metal Ion Tolerant Polymer Lipid Nanodiscs, Langmuir 33 (40) (2017) 10655–10662. 10.1021/acs.langmuir.7b02887. [DOI] [PubMed] [Google Scholar]
- [33].Ravula T, Ramadugu SK, Di Mauro G, Ramamoorthy A, Bioinspired, Size-Tunable Self-Assembly of Polymer–Lipid Bilayer Nanodiscs, Angew. Chem. Int. Ed 56 (38) (2017) 11466–11470. 10.1002/anie.201705569. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [34].Lindhoud S, Carvalho V, Pronk JW, Aubin-Tam M-E, SMA-SH: Modified Styrene–Maleic Acid Copolymer for Functionalization of Lipid Nanodiscs, Biomacromolecules 17 (4) (2016) 1516–1522. 10.1021/acs.biomac.6b00140. [DOI] [PubMed] [Google Scholar]
- [35].Hall SCL, Tognoloni C, Charlton J, Bragginton ÉC, Rothnie AJ, Sridhar P, Wheatley M, Knowles TJ, Arnold T, Edler KJ, Dafforn TR, An acid-compatible co-polymer for the solubilization of membranes and proteins into lipid bilayer-containing nanoparticles, Nanoscale 10 (22) (2018) 10609–10619. 10.1039/C8NR01322E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Neville GM, Edler KJ, Price GJ, Fluorescent styrene maleic acid copolymers to facilitate membrane protein studies in lipid nanodiscs, Nanoscale 14 (15) (2022) 5689–5693. 10.1039/D1NR07230G. [DOI] [PubMed] [Google Scholar]
- [37].Krishnarjuna B, Ravula T, Ramamoorthy A, Detergent-free extraction, reconstitution and characterization of membrane-anchored cytochrome-b5 in native lipids, Chem. Commun 56 (48) (2020) 6511–6514. 10.1039/D0CC01737J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kopf AH, Lijding O, Elenbaas BOW, Koorengevel MC, Dobruchowska JM, van Walree CA, Killian JA, Synthesis and Evaluation of a Library of Alternating Amphipathic Copolymers to Solubilize and Study Membrane Proteins, Biomacromolecules 23 (3) (2022) 743–759. 10.1021/acs.biomac.1c01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Glueck D, Grethen A, Das M, Mmeka OP, Patallo EP, Meister A, Rajender R, Kins S, Raschle M, Victor J, Chu C, Etzkorn M, Köck Z, Bernhard F, Babalola JO, Vargas C, Keller S, Electroneutral Polymer Nanodiscs Enable Interference-Free Probing of Membrane Proteins in a Lipid-Bilayer Environment, Small 18 (47) (2022) 2202492. 10.1002/smll.202202492. [DOI] [PubMed] [Google Scholar]
- [40].Workman CE, Cawthon B, Brady NG, Bruce BD, Long BK, Effects of Esterified Styrene–Maleic Acid Copolymer Degradation on Integral Membrane Protein Extraction, Biomacromolecules 23 (11) (2022) 4749–4755. 10.1021/acs.biomac.2c00928. [DOI] [PubMed] [Google Scholar]
- [41].Oluwole AO, Danielczak B, Meister A, Babalola JO, Vargas C, Keller S, Solubilization of Membrane Proteins into Functional Lipid-Bilayer Nanodiscs Using a Diisobutylene/Maleic Acid Copolymer, Angew. Chem. Int. Ed 56 (7) (2017) 1919–1924. 10.1002/anie.201610778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Oluwole AO, Klingler J, Danielczak B, Babalola JO, Vargas C, Pabst G, Keller S, Formation of Lipid-Bilayer Nanodiscs by Diisobutylene/Maleic Acid (DIBMA) Copolymer, Langmuir 33 (50) (2017) 14378–14388. 10.1021/acs.langmuir.7b03742. [DOI] [PubMed] [Google Scholar]
- [43].Ball LE, Riley LJ, Hadasha W, Pfukwa R, Smith CJI, Dafforn TR, Klumperman B, Influence of DIBMA Polymer Length on Lipid Nanodisc Formation and Membrane Protein Extraction, Biomacromolecules 22 (2) (2021) 763–772. 10.1021/acs.biomac.0c01538. [DOI] [PubMed] [Google Scholar]
- [44].Danielczak B, Rasche M, Lenz J, Pérez Patallo E, Weyrauch S, Mahler F, Agbadaola MT, Meister A, Babalola JO, Vargas C, Kolar C, Keller S, A bioinspired glycopolymer for capturing membrane proteins in native-like lipid-bilayer nanodiscs, Nanoscale 14 (5) (2022) 1855–1867. 10.1039/DlNR03811G. [DOI] [PubMed] [Google Scholar]
- [45].Janson K, Zierath J, Kyrilis FL, Semchonok DA, Hamdi F, Skalidis I, Kopf AH, Das M, Kolar C, Rasche M, Vargas C, Keller S, Kastritis PL, Meister A, Solubilization of artificial mitochondrial membranes by amphiphilic copolymers of different charge, Biochim. Biophys. Acta, Biomembr 1863 (12) (2021) 183725. 10.1016/j.bbamem.2021.183725. [DOI] [PubMed] [Google Scholar]
- [46].Yasuhara K, Arakida J, Ravula T, Ramadugu SK, Sahoo B, Kikuchi J.-i., Ramamoorthy A, Spontaneous Lipid Nanodisc Fomation by Amphiphilic Polymethacrylate Copolymers, J. Am. Chem. Soc 139 (51) (2017) 18657–18663. 10.1021/jacs.7b10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Janata M, Čadová E, Angelisová P, Charnavets T, Hořejší V, Raus V, Tailoring Butyl Methacrylate/Methacrylic Acid Copolymers for the Solubilization of Membrane Proteins: The Influence of Composition and Molecular Weight, Macromol. Biosci 22 (10) (2022) 2200284. 10.1002/mabi.202200284. [DOI] [PubMed] [Google Scholar]
- [48].Hardin NZ, Ravula T, Mauro GD, Ramamoorthy A, Hydrophobic Functionalization of Polyacrylic Acid as a Versatile Platform for the Development of Polymer Lipid Nanodisks, Small 15 (9) (2019) 1804813. 10.1002/smll.201804813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Timcenko M, Autzen AAA, Autzen HE, Characterization of Divalent Cation Interactions with AASTY Nanodiscs, ACS Appl. Polym. Mater 4 (2) (2022) 1071–1083. 10.1021/acsapm.1c01507. [DOI] [Google Scholar]
- [50].Ravula T, Ramamoorthy A, Synthesis, Characterization, and Nanodisc Formation of Non-ionic Polymers, Angew. Chem. Int. Ed 60 (31) (2021) 16885–16888. 10.1002/anie.202101950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Krishnarjuna B, Marte J, Ravula T, Ramamoorthy A, Enhancing the stability and homogeneity of non-ionic polymer nanodiscs by tuning electrostatic interactions, J. Colloid Interface Sci 634 (2023) 887–896. 10.1016/j.jcis.2022.12.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Esmaili M, Brown CJ, Shaykhutdinov R, Acevedo-Morantes C, Wang YL, Wille H, Gandour RD, Turner SR, Overduin M, Homogeneous nanodiscs of native membranes formed by stilbene–maleic-acid copolymers, Nanoscale 12 (32) (2020) 16705–16709. 10.1039/D0NR03435E. [DOI] [PubMed] [Google Scholar]
- [53].Marconnet A, Michon B, Le Bon C, Giusti F, Tribet C, Zoonens M, Solubilization and Stabilization of Membrane Proteins by Cycloalkane-Modified Amphiphilic Polymers, Biomacromolecules 21 (8) (2020) 3459–3467. 10.1021/acs.biomac.0c00929. [DOI] [PubMed] [Google Scholar]
- [54].Marconnet A, Michon B, Prost B, Solgadi A, Le Bon C, Giusti F, Tribet C, Zoonens M, Influence of Hydrophobic Groups Attached to Amphipathic Polymers on the Solubilization of Membrane Proteins along with Their Lipids, Anal. Chem 94 (41) (2022) 14151–14158. 10.1021/acs.analchem.2c01746. [DOI] [PubMed] [Google Scholar]
- [55].Yu T, Omarova M, Zhang M, Hossain I, Chen J, Darvish O, John VT, Zhang D, Uncovering the Optimal Molecular Characteristics of Hydrophobe-Containing Polypeptoids to Induce Liposome or Cell Membrane Fragmentation, Biomacromolecules 24 (3) (2023) 1511–1521. 10.1021/acs.biomac.3c00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mahler F, Meister A, Vargas C, Durand G, Keller S, Self-Assembly of Protein-Containing Lipid-Bilayer Nanodiscs from Small-Molecule Amphiphiles, Small 17 (49) (2021) 2103603. 10.1002/smll.202103603. [DOI] [PubMed] [Google Scholar]
- [57].McCalpin SD, Ravula T, Ramamoorthy A, Saponins Form Nonionic Lipid Nanodiscs for Protein Structural Studies by Nuclear Magnetic Resonance Spectroscopy, J. Phys. Chem. Lett 13 (7) (2022) 1705–1712. 10.1021/acs.jpclett.1c04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Grime RL, Logan RT, Nestorow SA, Sridhar P, Edwards PC, Tate CG, Klumperman B, Dafforn TR, Poyner DR, Reeves PJ, Wheatley M, Differences in SMA-like polymer architecture dictate the conformational changes exhibited by the membrane protein rhodopsin encapsulated in lipid nanoparticles, Nanoscale 13 (31) (2021) 13519–13528. 10.1039/D1NR02419A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Real Hernandez LM, Levental I, Lipid packing is disrupted in copolymeric nanodiscs compared with intact membranes, Biophys. J (2023). 10.1016/j.bpj.2023.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Morrison KA, Wood L, Edler KJ, Doutch J, Price GJ, Koumanov F, Whitley P, Membrane extraction with styrene-maleic acid copolymer results in insulin receptor autophosphorylation in the absence of ligand, Sci. Rep 12 (1) (2022) 3532. 10.1038/s41598-022-07606-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kamilar E, Bariwal J, Zheng W, Ma H, Liang H, SMALPs Are Not Simply Nanodiscs: The Polymer-to-Lipid Ratios of Fractionated SMALPs Underline Their Heterogeneous Nature, Biomacromolecules 24 (4) (2023) 1819–1838. 10.1021/acs.biomac.3c00034. [DOI] [PubMed] [Google Scholar]
- [62].Ravula T, Hardin NZ, Bai J, Im S-C, Waskell L, Ramamoorthy A, Effect of polymer charge on functional reconstitution of membrane proteins in polymer nanodiscs, Chem. Commun 54 (69) (2018) 9615–9618. 10.1039/C8CC04184A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Overduin M, Klumperman B, Advancing membrane biology with poly(styrene-co-maleic acid)-based native nanodiscs, Eur. Polym. J 110 (2019) 63–68. 10.1016/j.eurpolymj.2018.11.015. [DOI] [Google Scholar]
- [64].Krishnarjuna B, Sharma G, Ravula T, Ramamoorthy A, Factors influencing the detergent-free membrane protein isolation using synthetic nanodisc-forming polymers, bioRxiv (2023) 2023.05.12.540572. 10.1101/2023.05.12.540572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Dobrynin AV, Rubinstein M, Theory of polyelectrolytes in solutions and at surfaces, Prog. Polym. Sci 30 (11) (2005) 1049–1118. 10.1016/j.progpolymsci.2005.07.006. [DOI] [Google Scholar]
- [66].Janata M, Kůdela V, Gromadzki D, štěpánek P, Nallet F, Diat O, Vlček P, Toman L, Synthesis of highly sulfonated polystyrene-based block copolymers soluble in tetrahydrofuran, e-Polymers 6 (1) (2006). 10.1515/epoly.2006.6.1.702. [DOI] [Google Scholar]
- [67].Baigl D, Seery TAP, Williams CE, Preparation and Characterization of Hydrosoluble, Partially Charged Poly(styrenesulfonate)s of Various Controlled Charge Fractions and Chain Lengths, Macromolecules 35 (6) (2002) 2318–2326. 10.1021/ma011707o. [DOI] [Google Scholar]
- [68].Elabd YA, Napadensky E, Sulfonation and characterization of poly(styrene-isobutylene-styrene) triblock copolymers at high ion-exchange capacities, Polymer 45 (9) (2004) 3037–3043. 10.1016/j.polymer.2004.02.061. [DOI] [Google Scholar]
- [69].Jamshad M, Grimard V, Idini I, Knowles TJ, Dowle MR, Schofield N, Sridhar P, Lin Y, Finka R, Wheatley M, Thomas ORT, Palmer RE, Overduin M, Govaerts C, Ruysschaert J-M, Edler KJ, Dafforn TR, Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins, Nano Res. 8 (3) (2015) 774–789. 10.1007/s12274-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Morrison KA, Akram A, Mathews A, Khan ZA, Patel JH, Zhou C, Hardy DJ, Moore-Kelly C, Patel R, Odiba V, Knowles TJ, Javed M.-u.-H., Chmel NP, Dafforn TR, Rothnie AJ, Membrane protein extraction and purification using styrene–maleic acid (SMA) copolymer: effect of variations in polymer structure, Biochem. J 473 (23) (2016) 4349–4360. 10.1042/BCJ20160723. [DOI] [PubMed] [Google Scholar]
- [71].Swainsbury DJK, Scheidelaar S, Foster N, van Grondelle R, Killian JA, Jones MR, The effectiveness of styrene-maleic acid (SMA) copolymers for solubilisation of integral membrane proteins from SMA-accessible and SMA-resistant membranes, Biochim. Biophys. Acta, Biomembr 1859 (10) (2017) 2133–2143. 10.1016/j.bbamem.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sezgin E, Levental I, Mayor S, Eggeling C, The mystery of membrane organization: composition, regulation and roles of lipid rafts, Nat. Rev. Mol. Cell Biol 18 (6) (2017) 361–374. 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Angelisová P, Ballek O, Sýkora J, Benada O, Čajka T, Pokorná J, Pinkas D, Hořejší V, The use of styrene-maleic acid copolymer (SMA) for studies on T cell membrane rafts, Biochim. Biophys. Acta, Biomembr 1861 (1) (2019) 130–141. 10.1016/j.bbamem.2018.08.006. [DOI] [PubMed] [Google Scholar]
- [74].Sahoo BR, Genjo T, Moharana KC, Ramamoorthy A, Self-Assembly of Polymer-Encased Lipid Nanodiscs and Membrane Protein Reconstitution, J. Phys. Chem. B 123 (21) (2019) 4562–4570. 10.1021/acs.jpcb.9b03681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bjørnestad VA, Orwick-Rydmark M, Lund R, Understanding the Structural Pathways for Lipid Nanodisc Formation: How Styrene Maleic Acid Copolymers Induce Membrane Fracture and Disc Formation, Langmuir 37 (20) (2021) 6178–6188. 10.1021/acs.langmuir.1c00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in Supporting Information or on request from the authors.


