Abstract
Idiopathic pulmonary fibrosis (IPF) is the most common form of interstitial lung disease (ILD). IPF is a complex disease, with environmental and genetic factors variably contributing to disease susceptibility and outcomes. A host of common gene variants with modest effect size impart disease risk in patients with sporadic IPF, while rare variants with large effect size influence disease risk in those with familial interstitial pneumonia. In this review we highlight several common and rare variants underpinning IPF risk and call attention to recently published studies informing our understanding of this risk.
Key Terms: Idiopathic Pulmonary Fibrosis, Genetics, Medical, Familial Interstitial Pneumonia
Introduction
Idiopathic pulmonary fibrosis (IPF), defined as a “chronic, progressive, fibrosing interstitial pneumonia of unknown cause”, is among the most common and deadly of the interstitial lung diseases (ILDs).1 IPF remains relatively rare, with an estimated incidence of roughly 10 cases per 100,000 person years.2 The majority of patients are diagnosed during their 6th and 7th decades of life3 and have median survival of 3 – 5 years, though survival may be improving with the recent approval of anti-fibrotic therapies.4 IPF is a complex disease, with environmental and genetic factors variably contributing to disease susceptibility and outcomes.
The genetic factors influencing IPF susceptibility risk depend largely on whether a patient has sporadic IPF or the familial form, termed familial interstitial pneumonia (FIP). A host of common gene variants with modest effect size impart disease risk in patients with sporadic IPF, while rare variants with large effect size influence disease risk in those with FIP. Patients with FIP were initially thought to represent less than 5% of IPF cases, but recent data suggest that up to one in three cases have a family history of pulmonary fibrosis.5 In this review we highlight several common and rare variants underpinning IPF risk and call attention to recently published studies informing our understanding of this risk.
Common gene variants linked to sporadic IPF
Mucin 5b (MUC5B) Variants
MUC5B encodes mucin-5b, a glycoprotein critical to mucociliary clearance and immune homeostasis.6 The significance of MUC5B in IPF was first identified by linkage analysis of 82 families with FIP and subsequently confirmed in three genome-wide association studies(GWAS).7–9 The linkage analysis conducted by Siebold and colleagues identified a 3.4-mb region of chromosome 11p15 that co-segregated with IPF presence. Further enhancement of this region identified a minor-allele single nucleotide polymorphism (SNP), rs35705950, that was associated with an upstream promoter of MUC5B and predicted to create two new transcription factor binding sites.10
When MUC5B expression was evaluated in lung tissue of unaffected individuals carrying the MUC5B promoter variant, this single nucleotide polymorphism (SNP) was associated with a 37.4 fold increase in MUC5B expression compared to those without this risk allele.10 This promoter polymorphism was not associated with increased expression of MUC5B when compared to patients with IPF homozygous for the wild-type allele,10 but immunohistochemical staining of honeycomb cysts showed mucin secretion identical to distal airways, suggesting that these cysts may originate from distal airways.11
Three subsequent genome-wide association studies supported the link between the MUC5B promoter SNP and IPF susceptibility risk. A recent meta-analysis of IPF risk associated with the MU5B promoter SNP showed that this risk allele increased the odds of IPF development by nearly 4-fold (odds-ratio of 3.768 (95% CI 2.935 – 4.863)12. The MUC5B promoter SNP also appears to increase the risk of early ILD after an analysis of the Framingham Heart Study cohort showed an association between interstitial lung abnormalities (ILA) and this risk allele13. While this SNP was not associated with systemic scleroderma associated ILD14 a recent analysis of patients with rheumatoid arthritis showed that this risk allele increases the risk of ILD in this patient population, especially those with a radiographic usual interstitial pneumonia pattern.15
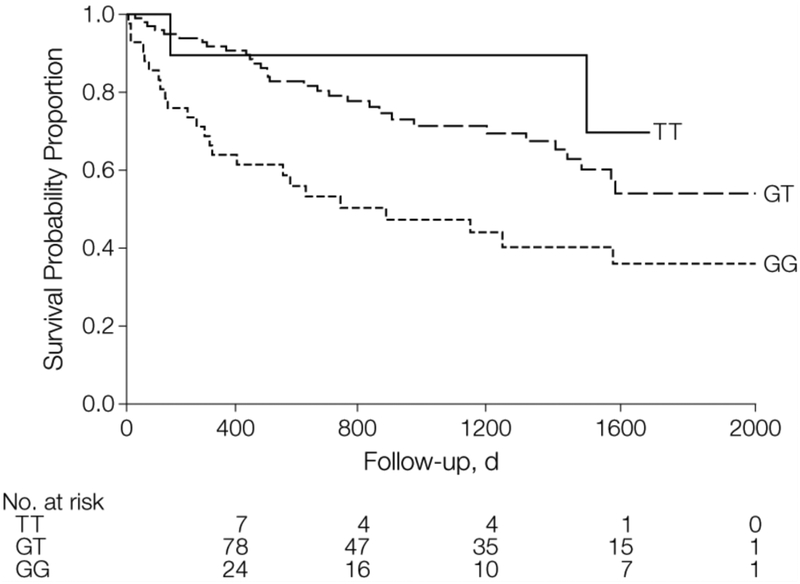
Despite the association with increased susceptibility to IPF, the MUC5B promoter SNP is paradoxically associated with decreased risk of death in IPF. Peljto and colleagues showed that each risk allele was associated with a 2-fold decrease in mortality risk in patients with IPF (HR for TT 0.23 [95% CI 0.05 – 0.49] and TG 0.48 [95% CI 0.31 – 0.72] compared to GG) (Figure 1).16
Figure 1:
Kaplan-Meier Survival Curves by MUC5B Genotypes, Chicago Cohort. Reproduced from (16).
Reprinted with permission from American Medical Association via the Copyright Clearance Center: [JAMA] Peljto AL, Zhang Y, Fingerlin TE, et al. Association between the MUC5B promoter polymorphism and survival in patients with idiopathic pulmonary fibrosis. American Medical Association, 2013.16
These findings may extend to those with ILA as well, as the MUC5B promoter SNP was associated with decreased risk of respiratory exacerbations (OR 0.39, 95% CI 0.22 – 0.69) and longer time to first event (HR 0.56, 95% CI 0.38 – 0.84) in a cohort of smokers with ILA.17 The authors hypothesized that increased MUC5B expression may help normalize the ratio of MUC5B to other mucins, resulting in increased airway clearance in these patients. This risk allele has also been linked to outcomes in patients with chronic hypersensitivity pneumonitis, albeit with opposite effect direction. Ley and colleagues showed that the presence of this SNP predicted worse outcome in two cohorts with chronic hypersensitivity pneumonitis drawn from San Francisco and Dallas (OR 1.91 [95% CI 1.02 – 3.59]).18 While the exact role of MUC5B in fibrosis remains uncertain, the appearance of MUC5B in multiple diseases, some of which with apparently opposing phenotypes, is intriguing.
Toll Interacting Protein (TOLLIP) Variants
In addition to the MUC5B promoter SNP, a recent GWAS identified two independent SNPs in TOLLIP to be associated with IPF susceptibility. TOLLIP is an important regulator of the toll-like receptor (TLR) and variants in this gene have been associated with decreased expression of TLR mRNA and increased susceptibility to pulmonary infection.19 The minor allele of rs5743890 was associated with decreased susceptibility to IPF (OR 0.61, 95% CI 0.52–0.71), while the minor allele of rs5743894 was associated with increased risk (OR 1.49, 95% CI 1.33–1.68). Despite these divergent associations, the presence of each SNP was associated with decreased TOLLIP expression in lung tissue. In outcomes modeling, the minor allele of rs5743890 was also paradoxically associated with increased mortality risk (HR 1.72, 95% CI 1.24 – 2.38), although effect size varied substantially across cohorts.7
Allen et. al. recently completed a GWAS of IPF cases from around the world and confirmed the association between increased IPF risk and the rs5743894 risk allele in TOLLIP via rs111521887, which is in strong linkage disequilibrium with rs5743894. These authors also found a suggestive association between reduced IPF risk and the rs5743890 risk allele, but this association was no longer significant after correction for multiple testing.9
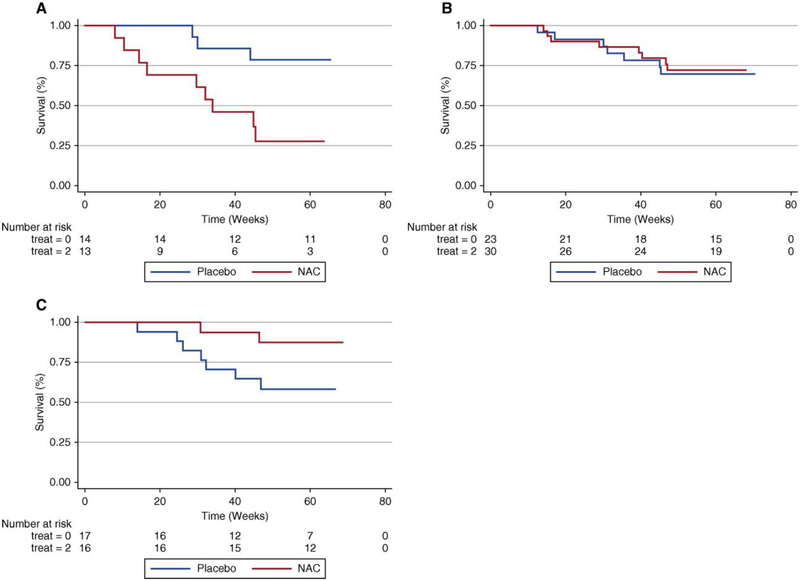
In addition to aforementioned GWAS findings, an additional TOLLIP SNP with suggestive susceptibility association was recently linked to differential treatment response. In a post-hoc analysis of the PANTHER-IPF trial, treatment with N-acetylcysteine (NAC) was associated with benefit in patients with both minor T alleles of rs3750920 (Figure 2). A trend towards harm was observed in those with both common C alleles.20 Although the mechanism for benefit from NAC in the TT genotype is unclear, the authors suggest that the rs3750920 polymorphism may lead to unregulated TLR signaling that is potentially abrogated by NAC therapy.20
Figure 2:
Composite endpoint-free survival between NAC and placebo groups after stratification by rs3750920 (TOLLIP) genotype. In those with a CC genotype (a), NAC therapy is associated with worse survival than placebo (Plogrank ¼ 0.01; hazard ratio, 3.23; 95% confidence interval, 0.79–13.16; P¼0.10). In those with a CT genotype (b), survival is similar between groups (Plogrank ¼ 0.82; HR 0.76; 95% CI 0.27–2.19; P ¼ 0.62). In those with a TT genotype (c), NAC therapy is associated with improved survival compared with placebo (Plogrank ¼ 0.06; HR 0.14; 95% CI 0.02–0.83; P ¼ 0.03). Multivariable Cox regression models adjusted for age, sex, FVC (percentage predicted), and diffusion capacity of the lung for carbon monoxide (percentage predicted) at time of study enrollment. Reprinted with permission of the American Thoracic Society. Copyright (c) 2018 American Thoracic Society. Oldham JM, Ma SF, Martinez FJ, et al. 2015. TOLLIP, MUC5B, and the Response to N-Acetylcysteine among Individuals with Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 192(12):1475.20 The American Journal of Respiratory and Critical Care Medicine is an official journal of the American Thoracic Society. Abbreviations: CI, confidence interval; FVC, forced vital capacity; HR, hazard ratio; NAC, N acetylcysteine.
Desmoplakin (DSP) Variants
A variant in DSP has been shown to increase the risk in IPF across two GWAS. DSP encodes desmoplakin, which functions in cell-cell adhesion and tethering of the cytoskeleton to the cell membrane.8,9 Expression of DSP in the lung typically decreases with age, but was found to be increased by 2-fold in those with IPF.21 Follow up sequencing of the DSP locus identified one protective variant rs2744371 (OR 0.77, 95% CI 0.66 – 0.91) and one previously described risk variant (rs2076295) (OR 1.36, 95% 1.19 – 1.56) with known functional effects.21
A-Kinase Anchoring Protein 13 (AKAP13) Variants
The most recent GWAS conducted to date identified an additional SNP in AKAP13 (rs62025270[A]) that was associated with IPF.9 This SNP was associated with increased production of AKAP13 regulator factor of rhoA, which is involved in profibrotic signaling pathways.9 The authors additionally observed that AKAP13 was overexpressed in histologic sections of IPF lung compared with controls.9 No association with survival was observed.9
Rare variants linked to FIP
Telomerase complex mutations
Telomeres are the terminal caps on eukaryotic chromosomes.22 The regulation and maintenance of telomeres is accomplished by genes comprising the telomerase complex.22 Telomere shortening or damage serves as a trigger for cell senescence.22 In 2005, Armanios et. al. described a family with autosomal dominant dyskeratosis congenita with anticipation and telomere shortening associated with a mutation in hTERT – an enzyme associated with telomerase.23 Half (3/6) of the family members with confirmed hTERT mutations had been diagnosed with pulmonary fibrosis.23 Subsequently, Stuart et. al. conducted whole exome sequencing in patients with FIP, family members of affected patients, and healthy controls to identify novel mutations associated with disease.24 They identified rare variants in two additional genes in the telomerase complex - PARN and RTEL.24 Several loss of function mutations in PARN, a gene not previously implicated in telomere maintenance, were seen exclusively in the affected 6 probands and no controls.24 Interestingly, five of 13 family members who inherited the PARN mutation had lung disease that did not meet the criteria for IPF, suggesting that familial IPF may present with variable phenotypes.24 Additionally, five heterozygous loss of function variants of RTEL1, a gene involved in telomere maintenance were identified. Biallelic mutations in RTEL1 are associated with the complete dyskeratosis congenita phenotype.24 Concurrent work by Cogan and colleagues found a similar association between rare RETL1 variants and FIP.25
Telomerase complex mutations have also been implicated in sporadic IPF. GWAS studies identified common variants within TERT and TERC to be associated with sporadic IPF,8 although the effects of these variants on telomere length are not well characterized. Petrovski et al. conducted whole exome sequencing and MUC5B promoter SNP genotyping in 262 patients with pulmonary fibrosis and 4,141 controls. In the subgroup with IPF and no family history of fibrotic lung disease, 11.3% of patients had a predicted likely deleterious mutation in TERT, RTEL1, or PARN compared to 0.3% of controls.26 Dressen and colleagues conducted whole genome sequencing and MUC5B promoter SNP genotyping in a cohort of 1510 patients assembled from three large phase III clinical trials a phase II trial, and two observational registries. They identified rare variants in TERT, PARN, TERC, and RTEL1 in 9% of IPF cases and 2% of controls, with those patients with telomerase mutations having shorter telomere lengths and earlier ages of onset.27 Rare TERT variants were found in 3% of patients with MUC5B risk allele compared with 7% of patients with non-risk allele carriers (OR 0.40, 95% CI 0.24 – 0.66), suggesting a potential sub-phenotype within sporadic IPF.
Telomerase mutations often result in telomere shortening. In an observational cohort, age-adjusted average blood leukocyte telomere length was shorter in patients with IPF when compared to healthy controls.28 The clinical significance of this stems from the observation that shorter telomere length is an independent predictor of decreased survival in IPF.28 These findings were replicated in several additional cohorts from around the world.28,29 Shorter telomere length has also been associated with greater extent of fibrosis, UIP histology, and reduced survival in chronic hypersensitivity pneumonitis.18 Newton and colleagues showed that patients with telomerase mutations have highly variable radiographic and histologic features, but similarly poor survival irrespective of the clinical ILD diagnosis.30 Together, these studies suggest that telomerase mutations in patients with fibrotic ILD result in a biologic endotype more relevant to disease prognostication than the clinically diagnosed ILD phenotype. Others have proposed a similar reordering of ILD diagnostic classification based on an increased understanding of the biologic factors leading to IPF pathogenesis.31
While most telomere studies rely on DNA extracted from peripheral blood leukocytes, Snetselaar and colleagues examined the length of telomeres in lung tissue from surgical biopsy specimens.32 In patients with IPF, the average telomere length was 56% longer in non-fibrotic areas compared to fibrotic areas,32 suggesting that local changes in telomere length may not be reflected in distal sites. Furthermore, shorter than average telomere length in lung tissue when compared to longer than average telomere length was associated with decreased survival.32 More research is needed to determine the optimal compartment and cell type for telomere length determination.
Surfactant producing gene mutations
Surfactant gene mutations were first associated with FIP in 2001 when Nogee et. al. identified surfactant protein C (SFTPC) mutation causing disease in a familial cohort.33 Subsequent research identified mutations in other surfactant producing genes that were also associated with FIP. This included 5 of unrelated 23 patients in a Dutch FIP cohort with mutations in SFTPC.34 The authors used a similar methodology several years later and identified three novel mutations in surfactant protein A2 (SFTPA2) in 39 unrelated patients compared to no control subjects.35 Another unique SFTPC mutation was identified in a large FIP-affected kindred in Japan.36 A type II alveolar(AT2) epithelial cell line with this particular mutation (G100S) was found to have increased endoplasmic reticulum stress and apoptotic cell death.36 Nureki et. al. developed a knock-in murine model of SFTPC mutation that resulted increased markers of AT2 dysfunction and lung fibrosis, further strengthening the association between SFTPC mutations and ILD.37 Exon sequencing of a case-control cohort identified mutations in surfactant genes(SFTPA2 or SFTPC) in 5 of 119 of cases compared with 0 of 178 COPD controls (p = 0.01). Interestingly, only 1 of the 5 surfactant mutations was in a patient with a family history of IPF, suggesting that de novo surfactant mutations may a cause of sporadic IPF.38
Conclusion
We are increasingly identifying genetic risk factors that contribute to sporadic IPF and FIP. Understanding the rare mutations with large effect size provides insight in to the mechanisms of IPF, while the identification of common variants with modest effect size may help with polygenic risk stratification and disease prognostication. Recent discoveries have shown some overlap between the genetic risks for both IPF and FIP, suggesting that these diseases may be two ends of the same continuum. Given the rapid expansion of genomic sequencing technology, the next decade is sure to shed light on the genetic and biologic underpinnings of this devastating disease.
Table 1.
A summary of common SNPs associated with IPF and their effects.
|
Gene |
SNP |
General Population MAFa |
IPF MAF |
IPF Risk |
IPF Mortality Risk |
Reference |
|---|---|---|---|---|---|---|
| MUC5B | rs35705950 | 0.05 | 0.14–0.31 | Increased | Decreased | 7,9,15 |
| TOLLIP | rs5743890 | 0.05 | 0.10 | Decreased | Increased | 7 |
| rs5743894 | 0.08 | 0.20 | Increased | 7 | ||
| DSP | rs2744371 | 0.23 | 0.14 | Decreased | 21 | |
| rs2076295 | 0.43 | 0.46–0.51 | Increased | 8,9,21 | ||
| AKAP13 | rs62025270 | 0.11 | 0.25 | Increased | 9 | |
| TERT | rs2736100 | 0.48 | 0.43 | Increased | 8 |
MAF is the minor allele frequency from the 1000 Genomes Project
Acknowledgments
Conflict of Interest: AB declares no conflicts. JO has received consulting fees from Genentech and BI and is supported by NIH grant K23HL138190. IN has received consulting fees from Genentech and BI is supported by NIH grant R01HL130796.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Andrew Barros, Division of Pulmonary & Critical Care Medicine, Department of Medicine, PO Box 800546, Charlottesville, VA 22908-0546, 434.243.4845.
Justin Oldham, Division of Pulmonary, Critical Care and Sleep Medicine, Department of Internal Medicine, The University of California at Davis, 4150 V Street Suite 3400, Sacramento, CA 95817, 916-734-1551.
Imre Noth, Division of Pulmonary & Critical Care Medicine, Department of Medicine, PO Box 800546, Charlottesville, VA 22908-0546, 434.243.4845.
References
- 1.Raghu G, Remy-Jardin M, Myers JL, et al. Diagnosis of Idiopathic Pulmonary Fibrosis. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. AmJRespirCritCare Med. 2018;198(5):e44. [DOI] [PubMed] [Google Scholar]
- 2.Ley B, Collard HR. Epidemiology of idiopathic pulmonary fibrosis. ClinEpidemiol. 2013;5:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raghu G, Chen SY, Yeh WS, et al. Idiopathic pulmonary fibrosis in US Medicare beneficiaries aged 65 years and older: incidence, prevalence, and survival, 2001–11. Lancet RespirMed. 2014;2(7):566. [DOI] [PubMed] [Google Scholar]
- 4.Fisher M, Nathan SD, Hill C, et al. Predicting Life Expectancy for Pirfenidone in Idiopathic Pulmonary Fibrosis. J Manag Care Spec Pharm. 2017;23(3-b Suppl):S17–S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez BA, Fox G, Bhatia R, et al. A Newfoundland cohort of familial and sporadic idiopathic pulmonary fibrosis patients: clinical and genetic features. RespirRes. 2012;13:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy MG, Livraghi-Butrico A, Fletcher AA, et al. Muc5b is required for airway defence. Nature. 2014;505(7483):412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noth I, Zhang Y, Ma SF, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet RespirMed. 2013;1(4):309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fingerlin TE, Murphy E, Zhang W, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. NatGenet. 2013;45(6):613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen RJ, Porte J, Braybrooke R, et al. Genetic variants associated with susceptibility to idiopathic pulmonary fibrosis in people of European ancestry: a genome-wide association study. Lancet Respir Med. 2017;5(11):869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seibold MA, Wise AL, Speer MC, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. NEnglJMed. 2011;364(16):1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seibold MA, Smith RW, Urbanek C, et al. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PLoS One. 2013;8(3):e58658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee MG, Lee YH. A meta-analysis examining the association between the MUC5B rs35705950 T/G polymorphism and susceptibility to idiopathic pulmonary fibrosis. InflammRes. 2015;64(6):463. [DOI] [PubMed] [Google Scholar]
- 13.Hunninghake GM, Hatabu H, Okajima Y, et al. MUC5B promoter polymorphism and interstitial lung abnormalities. NEnglJMed. 2013;368(23):2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borie R, Crestani B, Dieude P, et al. The MUC5B variant is associated with idiopathic pulmonary fibrosis but not with systemic sclerosis interstitial lung disease in the European Caucasian population. PLoS One. 2013;8(8):e70621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juge PA, Lee JS, Ebstein E, et al. MUC5B Promoter Variant and Rheumatoid Arthritis with Interstitial Lung Disease. N Engl J Med. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peljto AL, Zhang Y, Fingerlin TE, et al. Association between the MUC5B promoter polymorphism and survival in patients with idiopathic pulmonary fibrosis. Jama. 2013;309(21):2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ash SY, Harmouche R, Putman RK, et al. Association between acute respiratory disease events and the MUC5B promoter polymorphism in smokers. Thorax. 2018;73(11):1071–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ley B, Newton CA, Arnould I, et al. The MUC5B promoter polymorphism and telomere length in patients with chronic hypersensitivity pneumonitis: an observational cohort-control study. Lancet Respir Med. 2017;5(8):639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah JA, Vary JC, Chau TT, et al. Human TOLLIP regulates TLR2 and TLR4 signaling and its polymorphisms are associated with susceptibility to tuberculosis. JImmunol. 2012;189(4):1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oldham JM, Ma SF, Martinez FJ, et al. TOLLIP, MUC5B, and the Response to N-Acetylcysteine among Individuals with Idiopathic Pulmonary Fibrosis. AmJRespirCritCare Med. 2015;192(12):1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathai SK, Pedersen BS, Smith K, et al. Desmoplakin Variants Are Associated with Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2016;193(10):1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackburn EH, Epel ES, Lin J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science. 2015;350(6265):1193. [DOI] [PubMed] [Google Scholar]
- 23.Armanios M, Chen JL, Chang YP, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. ProcNatlAcadSciUSA. 2005;102(44):15960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuart BD, Choi J, Zaidi S, et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet. 2015;47(5):512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cogan JD, Kropski JA, Zhao M, et al. Rare variants in RTEL1 are associated with familial interstitial pneumonia. Am J Respir Crit Care Med. 2015;191(6):646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrovski S, Todd JL, Durheim MT, et al. An Exome Sequencing Study to Assess the Role of Rare Genetic Variation in Pulmonary Fibrosis. Am J Respir Crit Care Med. 2017;196(1):82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dressen A, Abbas AR, Cabanski C, et al. Analysis of protein-altering variants in telomerase genes and their association with MUC5B common variant status in patients with idiopathic pulmonary fibrosis: a candidate gene sequencing study. Lancet Respir Med. 2018;6(8):603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuart BD, Lee JS, Kozlitina J, et al. Effect of telomere length on survival in patients with idiopathic pulmonary fibrosis: an observational cohort study with independent validation. Lancet RespirMed. 2014;2(7):557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai J, Cai H, Li H, et al. Association between telomere length and survival in patients with idiopathic pulmonary fibrosis. Respirology. 2015;20(6):947. [DOI] [PubMed] [Google Scholar]
- 30.Newton CA, Batra K, Torrealba J, et al. Telomere-related lung fibrosis is diagnostically heterogeneous but uniformly progressive. Eur Respir J. 2016;48(6):1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolters PJ, Blackwell TS, Eickelberg O, et al. Time for a change: is idiopathic pulmonary fibrosis still idiopathic and only fibrotic? Lancet Respir Med. 2018;6(2):154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snetselaar R, van Batenburg AA, van Oosterhout MFM, et al. Short telomere length in IPF lung associates with fibrotic lesions and predicts survival. PLoS One. 2017;12(12):e0189467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nogee LM, Dunbar AE 3rd, Wert SE, Askin F, Hamvas A, Whitsett JA. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N Engl J Med. 2001;344(8):573–579. [DOI] [PubMed] [Google Scholar]
- 34.van Moorsel CH, van Oosterhout MF, Barlo NP, et al. Surfactant protein C mutations are the basis of a significant portion of adult familial pulmonary fibrosis in a dutch cohort. Am J Respir Crit Care Med. 2010;182(11):1419–1425. [DOI] [PubMed] [Google Scholar]
- 35.van Moorsel CH, Ten Klooster L, van Oosterhout MF, et al. SFTPA2 Mutations in Familial and Sporadic Idiopathic Interstitial Pneumonia. Am J Respir Crit Care Med. 2015;192(10):1249–1252. [DOI] [PubMed] [Google Scholar]
- 36.Ono S, Tanaka T, Ishida M, et al. Surfactant protein C G100S mutation causes familial pulmonary fibrosis in Japanese kindred. Eur Respir J. 2011;38(4):861–869. [DOI] [PubMed] [Google Scholar]
- 37.Nureki SI, Tomer Y, Venosa A, et al. Expression of mutant Sftpc in murine alveolar epithelia drives spontaneous lung fibrosis. J Clin Invest. 2018;128(9):4008–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coghlan MA, Shifren A, Huang HJ, et al. Sequencing of idiopathic pulmonary fibrosis-related genes reveals independent single gene associations. BMJ Open Respir Res. 2014;1(1):e000057. [DOI] [PMC free article] [PubMed] [Google Scholar]