Abstract
Recently, the computational neuroscience community has pushed for more transparent and reproducible methods across the field. In the interest of unifying the domain of auditory neuroscience, naplib-python provides an intuitive and general data structure for handling all neural recordings and stimuli, as well as extensive preprocessing, feature extraction, and analysis tools which operate on that data structure. The package removes many of the complications associated with this domain, such as varying trial durations and multi-modal stimuli, and provides a general-purpose analysis framework that interfaces easily with existing toolboxes used in the field.
Keywords: Python, Auditory neuroscience, iEEG, EcoG, Preprocessing
Code metadata
| Current code version | 1.0.0 |
| Permanent link to code/repository used for this code version | https://github.com/SoftwareImpacts/SIMPAC-2023-268 |
| Permanent link to reproducible capsule | https://codeocean.com/capsule/6656601/tree/v1 |
| Legal code license | MIT License |
| Code versioning system used | git |
| Software code languages, tools and services used | python |
| Compilation requirements, operating environments and dependencies | Linux, macOS, or Windows; matplotlib, numpy, scipy, pandas, statsmodels, hdf5storage, mne, scikit-learn, seaborn |
| If available, link to developer documentation/manual | https://naplib-python.readthedocs.io |
| Support email for questions | nima@ee.columbia.edu |
1. Introduction
With the recent explosion of neural data acquisition and computational power, the field of neuroscience has seen incredible growth in the use of computational methods to analyze neural response patterns and make inferences about the brain. The field of auditory neuroscience is no exception, with the widespread use of computational analyses such as spectro-temporal receptive field (STRF) estimation [1–3] and software such as STRFlab (http://www.strflab.berkeley.edu), mTRF [4], Neural Encoding Model System (NEMS) [5], and others dedicated to these specific techniques. Many papers are now accompanied by small bits of code to reproduce figures or analyses, which greatly aids in the reproducibility of scientific research. However, the explosion of computational methods has also led to a software ecosystem with many highly specialized packages written by, and for, people with very different needs. Even when code is shared openly, it may do little to aid the reproducibility of the experiments because of difficulties running others’ code or omissions of critical details in the original report [6,7]. Therefore, there is a need for a unifying framework which provides comprehensive tools for the auditory neuroscience domain and can easily fit into existing codebases and analysis pipelines used by researchers in the field.
As a Neural Acoustic Processing Library in Python, naplib-python is specifically built for auditory neuroscience. Its library of methods complements those provided by other public toolkits. Python is an ideal language for this purpose, due to its vast open-source ecosystem and the fact that many in the neuroscience community are moving towards Python [8]. In addition to implementing a host of relevant analysis methods that previously had no popular Python implementation, naplib-python expands upon the existing MATLAB toolkit NAPlib [9], which focuses on phoneme response analysis methods, by porting several of these methods to Python and increasing their ease-of-use by ensuring they work with naplib-python data structures. One existing Python package, MNE-Python [10], supports a broad set of functionalities for neural data, including magnetoencephalography (MEG), electroencephalography (EEG), and intracranial EEG (iEEG). However, that package was never meant to be used only for auditory neuroscience, and so it generally offers techniques applicable to many areas of neuroscience while missing functionality that would be useful in the auditory domain, such as linguistic alignment and phonetic feature extraction. In this paper, we first describe the basic data structure and API used in the package, then we provide an overview of the methods available in the package and describe their use in common analysis pipelines in the field.
2. Package description
2.1. Data structure and API
In auditory neuroscience, several problems often arise which make using a general-purpose data structure difficult. Stimuli tend to be non-uniform in duration, especially when using naturalistic stimuli like human speech [11]. Additionally, a large variety of trial-specific metadata may be needed for analyses, such as transcripts of speech stimuli or event labels, which would traditionally need to be loaded and treated as separate variables. The Data object in naplib-python takes care of these problems by seamlessly storing any number of trials containing any number of fields, which may include things like auditory stimulus waveforms and time–frequency representations, neural responses, stimulus transcripts, and trial condition labels. By not enforcing equal durations and allowing various data types through its various fields, the Data class is reminiscent of a pandas DataFrame [12], and in many ways it operates similarly. Fig. 1 shows a visual representation of the most typical information that could be stored in a Data instance. The Data object is designed to enable easy processing by trial, by field, or all together. Many functions in naplib-python can be called by either passing a set of parameters, or simply passing a Data object containing all the necessary fields to fill the parameters. For example, a user can fit a STRF model using the TRF class in naplib-python by passing in a Data instance and the fields for stimulus and response will be automatically extracted and used as the input and output when training the model. Alternatively, certain parameters can be passed in manually without needing them to be stored in a Data instance.
Fig. 1.
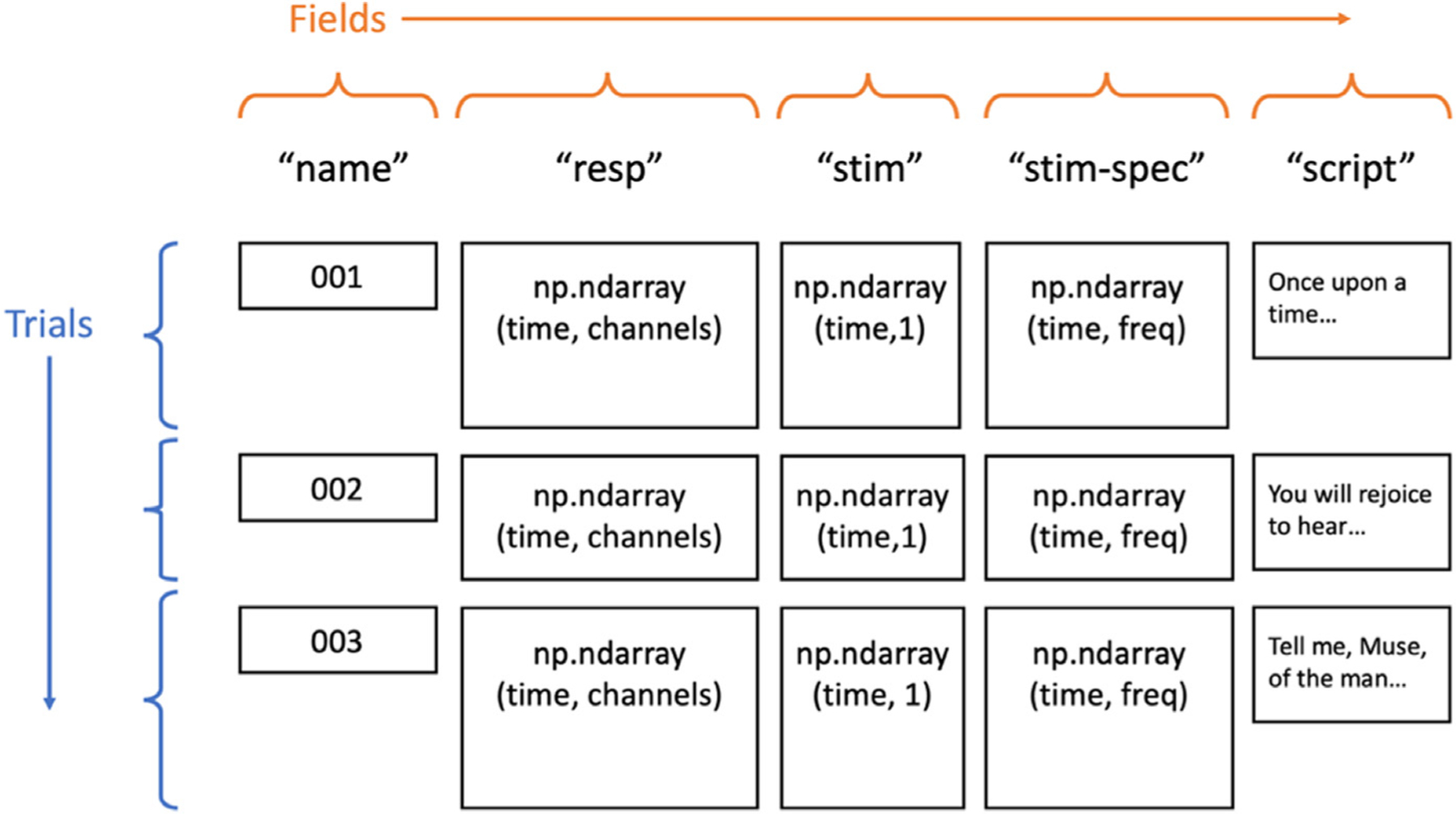
Basic Data object structure for an example task where neural responses to natural language stories were recorded. None of the fields depicted is required for the Data object, but they naturally illustrate what might be included in a typical task.
For convenient adoption, naplib-python supports loading data from many sources, such as from the Brain Imaging Data Structure (BIDS) format [13–15] directly into a Data object with a single line of code. This simple integration makes the Data object easy for beginners to adopt and broadly useful for any type of analysis.
2.2. Library overview
In this section we summarize the main modules currently available in the package and the tools offered in each. Full documentation and examples for the functions within these modules, as well as the other utility modules, are available online at https://naplib-python.readthedocs.io.
2.2.1. Features
In auditory neuroscience, a wide variety of features have been proposed to describe both acoustic signals and neural data. An implementation of the auditory spectrogram [16] is provided, a time–frequency representation which models the inner ear and cochlear spectral decomposition of sound waves. Additionally, many linguistic features are available to describe speech signals. A forced aligner is provided based on the Prosodylab-Aligner [17], as well as functions to extract phoneme and word alignment labels from the aligner’s output, which can be used to identify phonetic information and timing in speech stimuli.
2.2.2. Encoding
This module is dedicated to encoding models used by the auditory neuroscience community. For example, a robust temporal receptive field (TRF) class is implemented which interfaces naturally with the Data object. By default, the TRF model uses cross-validated ridge regression, but any class which adheres to the scikit-learn linear model API [18] can be used, meaning that TRFs can be trained from L1-regularized models, elastic net models, or any other linear or non-linear user-defined classes. Users can even use GPU-accelerated linear models from the cuML library [19] if an NVIDIA GPU is available with no additional hassle. This is useful for training both forward STRF models [2,3], which predict neural responses from acoustic stimuli, or backward models for stimulus reconstruction [20,21], which reconstruct acoustic stimuli from neural signals.
2.2.3. Segmentation
Stimulus onset response patterns are often studied in order to understand response properties in auditory neuroscience, as used in studies of evoked potentials [22–24] or stimulus onset-locked encoding [25–27]. The segmentation module contains methods for segmenting multi-trial data based on aligned labels. For example, aligned labels could include phoneme onset labels (where phoneme alignment can be computed using the Features module described above), enabling easy analysis of phoneme onset responses.
2.2.4. Preprocessing
A significant amount of preprocessing is typically involved when analyzing neural signals, so this module includes several functions which are useful for a variety of signal types, especially intracranial recordings. There are functions for extracting the envelope and phase of different frequency bands using a filter bank followed by the Hilbert transform [28], which can be used to extract the well-studied high gamma envelope response for iEEG data, or as an input to further analyses of phase–amplitude coupling [29,30]. There are also generic filtering functions that operate on Data instances which are useful for performing notch filtering to remove line noise or filtering EEG/MEG data into different frequency bands.
2.2.5. Stats
Statistical analysis of data is critical to understand the significance of any findings in neuroscience. This module provides several statistical tools common to auditory neuroscience. For example, one of the first steps in many analysis pipelines is electrode selection, which can be done using a t-test between responses to speech and silence to identify stimulus-responsive electrodes [31]. Another common statistic offered in the package and used in the field is the F-ratio, which is often used to describe the discriminability of neural responses between different stimulus classes [32]. Additionally, a linear mixed effects model is offered to perform linear modeling with the ability to control for effects such as subject identity, which may be needed when studying data across heterogeneous subjects, as is common with iEEG data. Similarly, a generalized t-test method is offered, which can be used to perform t-tests while controlling for additional factors, such as subject identity when testing a distribution with underlying groupings.
2.2.6. Input-output
Input from and output to files is supported in the IO module, including functions to save and load files, as well as read from or write to third-party file structures commonly used in the field of computational neuroscience like MATLAB, BIDS format, and the Continuous-event Neural Data (CND) format. Convenient data loading functions are available for common raw data recording structures including Neurodata Without Borders format (NWB) [33], European Data Format (EDF) [34], and raw data stored by Tucker Davis Technologies (TDT) machines. These functions make naplib-python easy to use no matter how a researcher’s data is currently stored or what stage of the analysis pipeline a researcher wants to use naplib-python.
3. Software impact
With its large suite of implemented methods, naplib-python enables researchers in the field of auditory neuroscience to run common analysis pipelines in only a few lines of code, all while using a general data framework that applies to nearly any type of neural recording data. This allows for collaborations and easier code sharing across disciplines, even when data or recording methods differ significantly between researchers. Furthermore, the ability to interface between naplib-python and other commonly used packages greatly extends the utility of all related toolkits, since researchers can rely on the naplib-python framework and data structure for basic analyses but still utilize state-of-the-art methods available elsewhere without needing to write new code. There are multiple tutorial notebooks available on the online documentation illustrating how to integrate naplib-python with other toolkits, such as fitting encoding models on naplib-python data using NEMS and plotting EEG analysis results using MNE visualization tools.
In addition to offering an analysis framework which can interface with various third-party packages, naplib-python offers a wide array of support for methods which are commonly used in the field but lack standard open-source implementations. This means naplib-python is well-positioned to become the standard toolbox for researchers in the field. Introducing this package will help reduce the overhead of writing new preprocessing and analysis code, especially for researchers with minimal computational experience, and improve the reproducibility of research by making code-sharing easier. The package was critical in a recent study of noise adaptation mechanisms [35], where it was used for phoneme- and word-alignment, segmentation, statistical testing, and visualization, as well as a study of multitalker speech encoding [36], where it was used to extract a wide variety of stimulus features which were used for fitting models to predict neural responses. In both these studies, the package aided in the data analysis stage by providing the computational framework for various feature extraction and testing methods.
4. Limitations and future improvements
The main limitation of naplib-python is because all operations are performed in-memory, processing of large (hours-long) datasets becomes highly inefficient. This could be enhanced in the future with dynamic data loading and saving for individual trial data. Additionally, while data can be imported from various sources, there is currently limited ability to save data to a wide variety of file structures, since naplib-python is primarily useful for the later stages of data analysis, beginning with preprocessing after raw data have been collected and stored.
Acknowledgments
We thank the members of the Neural Acoustic Processing Lab who provided feedback on the package and its applications. This work was supported by National Institutes of Health, United States grant R01DC016234 and National Institute on Deafness and Other Communication Disorders, United States grant R01DC014279, as well as a grant from the Marie-Josée and Henry R. Kravis Foundation. GM was supported in part by the National Science Foundation Graduate Research Fellowship Program, United States under grant DGE2036197.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The code (and data) in this article has been certified as Reproducible by Code Ocean: (https://codeocean.com/). More information on the Reproducibility Badge Initiative is available at https://www.elsevier.com/physical-sciences-and-engineering/computer-science/journals.
References
- [1].Aertsen AMHJ, Olders JHJ, Johannesma PIM, Spectro-temporal receptive fields of auditory neurons in the grassfrog - III. Analysis of the stimulus-event relation for natural stimuli, Biol. Cybern 39 (3) (1981) 10.1007/BF00342772. [DOI] [PubMed] [Google Scholar]
- [2].Theunissen FE, Sen K, Doupe AJ, Spectral-temporal receptive fields of nonlinear auditory neurons obtained using natural sounds, J. Neurosci 20 (6) (2000) 10.1523/jneurosci.20-06-02315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Theunissen FE, David S.v., Singh NC, Hsu A, Vinje WE, Gallant JL, Estimating spatio-temporal receptive fields of auditory and visual neurons from their responses to natural stimuli, Network: Comput. Neural Syst 12 (3) (2001) 10.1088/0954-898X/12/3/304. [DOI] [PubMed] [Google Scholar]
- [4].Crosse MJ, di Liberto GM, Bednar A, Lalor EC, The multivariate temporal response function (mTRF) toolbox: A MATLAB toolbox for relating neural signals to continuous stimuli, Front. Hum. Neurosci 10 (NOV2016) (2016) 10.3389/fnhum.2016.00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].David S.v., Incorporating behavioral and sensory context into spectro-temporal models of auditory encoding, in: Hearing Research, vol. 360, 2018, 10.1016/j.heares.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Easterbrook SM, Open code for open science?, Nat. Geosci 7 (11) (2014) 10.1038/ngeo2283. [DOI] [Google Scholar]
- [7].Miłkowski M, Hensel WM, Hohol M, Replicability or reproducibility? On the replication crisis in computational neuroscience and sharing only relevant detail, J. Comput. Neurosci 45 (3) (2018) 10.1007/s10827-018-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Muller E, Bednar JA, Diesmann M, Gewaltig MO, Hines M, Davison AP, Python in neuroscience, Front. Neuroinform 9 (APR) (2015) 10.3389/fninf.2015.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Khalighinejad B, Nagamine T, Mehta A, Mesgarani N, NAPLib: An open source toolbox for real-time and offline neural acoustic processing, in: ICASSP, IEEE International Conference on Acoustics, Speech and Signal Processing - Proceedings, 2017, 10.1109/ICASSP.2017.7952275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gramfort A, Luessi M, Larson E, Engemann DA, Strohmeier D, Brodbeck C, Goj R, Jas M, Brooks T, Parkkonen L, Hämäläinen M, MEG and EEG data analysis with MNE-Python, Front. Neurosci 7 DEC (2013) 10.3389/fnins.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hamilton LS, Huth AG, The revolution will not be controlled: Natural stimuli in speech neuroscience, Lang., Cogn. Neurosci 35 (5) (2020) 10.1080/23273798.2018.1499946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].McKinney W, Pandas: A foundational Python library for data analysis and statistics, Python High Perform. Sci. Comput (2011). [Google Scholar]
- [13].Gorgolewski KJ, Auer T, Calhoun VD, Craddock RC, Das S, Duff EP, Flandin G, Ghosh SS, Glatard T, Halchenko YO, Handwerker DA, Hanke M, Keator D, Li X, Michael Z, Maumet C, Nichols BN, Nichols TE, Pellman J, et al. , The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments, Sci. Data 3 (2016) 10.1038/sdata.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Niso G, Gorgolewski KJ, Bock E, Brooks TL, Flandin G, Gramfort A, Henson RN, Jas M, Litvak V, Moreau JT, Oostenveld R, Schoffelen JM, Tadel F, Wexler J, Baillet S, MEG-BIDS, the brain imaging data structure extended to magnetoencephalography, Sci. Data 5 (2018) 10.1038/sdata.2018.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pernet CR, Appelhoff S, Gorgolewski KJ, Flandin G, Phillips C, Delorme A, Oostenveld R, EEG-BIDS, an extension to the brain imaging data structure for electroencephalography, Sci. Data 6 (1) (2019) 10.1038/s41597-019-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yang X, Wang K, Shamma SA, Auditory representations of acoustic signals, IEEE Trans. Inform. Theory 38 (2) (1992) 10.1109/18.119739. [DOI] [Google Scholar]
- [17].Gorman K, Howell J, Wagner M, Prosodylab-aligner: A tool for forced alignment of laboratory speech, Can. Acoust. - Acoustique Canadienne 39 (3) (2011). [Google Scholar]
- [18].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É, Scikit-learn: Machine learning in Python, J. Mach. Learn. Res 12 (2011). [Google Scholar]
- [19].Raschka S, Patterson J, Nolet C, Machine learning in python: Main developments and technology trends in data science, machine learning, and artificial intelligence, Information (Switzerland) 11 (4) (2020) 10.3390/info11040193. [DOI] [Google Scholar]
- [20].Bialek W, Rieke F, de Ruyter Van Steveninck RR, Warland D, Reading a neural code, Science 252 (5014) (1991) 10.1126/science.2063199. [DOI] [PubMed] [Google Scholar]
- [21].Mesgarani N, David S.v., Fritz JB, Shamma SA, Influence of context and behavior on stimulus reconstruction from neural activity in primary auditory cortex, J. Neurophysiol 102 (6) (2009) 10.1152/jn.91128.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gage N, Poeppel D, Roberts TPL, Hickok G, Auditory evoked M100 reflects onset acoustics of speech sounds, Brain Res. 814 (1–2) (1998) 10.1016/S0006-8993(98)01058-0. [DOI] [PubMed] [Google Scholar]
- [23].Näätänen R, Gaillard AWK, Mäntysalo S, Early selective-attention effect on evoked potential reinterpreted, Acta Psychol. 42 (4) (1978) 10.1016/0001-6918(78)90006-9. [DOI] [PubMed] [Google Scholar]
- [24].Picton T, Hearing in time: Evoked potential studies of temporal processing, Ear Hear. 34 (4) (2013) 10.1097/AUD.0b013e31827ada02. [DOI] [PubMed] [Google Scholar]
- [25].Gwilliams L, Linzen T, Poeppel D, Marantz A, In spoken word recognition, the future predicts the past, J. Neurosci 38 (35) (2018) 10.1523/JNEUROSCI.0065-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Phillips DP, Hall SE, Boehnke SE, Central auditory onset responses, and temporal asymmetries in auditory perception, Hear. Res 167 (1–2) (2002) 10.1016/S0378-5955(02)00393-3. [DOI] [PubMed] [Google Scholar]
- [27].Hamilton LS, Edwards E, Chang EF, A spatial map of onset and sustained responses to speech in the human superior temporal gyrus, Curr. Biol 28 (12) (2018). [DOI] [PubMed] [Google Scholar]
- [28].Edwards E, Soltani M, Kim W, Dalal SS, Nagarajan SS, Berger MS, Knight RT, Comparison of time-frequency responses and the event-related potential to auditory speech stimuli in human cortex, J. Neurophysiol 102 (1) (2009) 10.1152/jn.90954.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Canolty RT, Edwards E, Dalal SS, Soltani M, Nagarajan SS, Kirsch HE, Berger MS, Barbare NM, Knight RT, High gamma power is phase-locked to theta oscillations in human neocortex, Science 313 (5793) (2006) 10.1126/science.1128115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tort ABL, Komorowski R, Eichenbaum H, Kopell N, Measuring phase–amplitude coupling between neuronal oscillations of different frequencies, J. Neurophysiol 104 (2) (2010) 10.1152/jn.00106.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mesgarani N, Chang EF, Selective cortical representation of attended speaker in multi-talker speech perception, Nature 485 (7397) (2012) 10.1038/nature11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Khalighinejad B, Patel P, Herrero JL, Bickel S, Mehta AD, Mesgarani N, Functional characterization of human Heschl’s gyrus in response to natural speech, NeuroImage 235 (2021) 10.1016/j.neuroimage.2021.118003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Teeters JL, Godfrey K, Young R, Dang C, Friedsam C, Wark B, Asari H, Peron S, Li N, Peyrache A, Denisov G, Siegle JH, Olsen SR, Martin C, Chun M, Tripathy S, Blanche TJ, Harris K, Buzsáki G, et al. , Neurodata without borders: Creating a common data format for neurophysiology, Neuron 88 (4) (2015) 10.1016/j.neuron.2015.10.025. [DOI] [PubMed] [Google Scholar]
- [34].Kemp B, Värri A, Rosa AC, Nielsen KD, Gade J, A simple format for exchange of digitized polygraphic recordings, Electroencephalogr. Clin. Neurophysiol 82 (5) (1992) 10.1016/0013-4694(92)90009-7. [DOI] [PubMed] [Google Scholar]
- [35].Mischler G, Keshishian M, Bickel S, Mehta AD, Mesgarani N, Deep neural networks effectively model neural adaptation to changing background noise and suggest nonlinear noise filtering methods in auditory cortex, NeuroImage 119819 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Raghavan VS, O’Sullivan J, Bickel S, Mehta AD, Mesgarani N, Distinct neural encoding of glimpsed and masked speech in multitalker situations, PLOS Biol. 21 (6) (2023) e3002128, 10.1371/journal.pbio.3002128. [DOI] [PMC free article] [PubMed] [Google Scholar]


