Abstract
Substance use disorder (SUD) arises from initiation to subsequent regular, irregular and harmful use of substances such as alcohol, tobacco/nicotine, and cannabis. While thousands of genetic variants have been identified from recent large-scale genome-wide association studies (GWAS), understanding their functions in substance involvement and investigating the mechanisms by which they act in the addiction circuits remains challenging. In this study, we re-analyzed the brain regional transcriptome data from the most comprehensive postmortem transcriptomic study, with a focus on up- or down-regulation of substance-associated protein-coding genes in the addiction circuit-related brain regions (AddictRegions) including six cortical and 11 subcortical regions. We found that substance-associated genes were overrepresented in AddictRegions. Interestingly, we observed a greater degree of genetic overlap between initiation and use, and between use and SUD, than between initiation and SUD. Moreover, substance initiation, use, and SUD-associated genes tend to shift their enriched AddictRegions from the cortical for initiation and to a less extent for substance use to subcortical regions for SUD (e.g., thalamus, substantia nigra, and ventral tegmental area). We also uncovered a pattern of coordinated cortical up-regulation and subcortical down-regulation for the genes associated with tobacco initiation and alcohol use. Moreover, the Gene Ontology terms of glutamate receptor activity and neurotransmitter binding were most significantly overrepresented in AddictRegion-upregulated, substance-associated genes, with the strongest enrichment for those involved in common substance use behaviors. Overall, our analysis provides a resource of AddictRegion-related transcriptomes for studying substance-associated genes and generates intriguing insights into genetic control of substance initiation, use, and SUD.
Keywords: Substance addiction, brain region, genome-wide association study
1. INTRODUCTION
Substance use disorder (SUD) is a mental health condition in which individuals lose the full control of using addictive substances including alcohol, cannabis, and tobacco/nicotine. Understanding the pathogenic process and neurobiological mechanisms of SUD is critical for preventing its occurrence and mitigating its consequences. SUD typically begins with initiation (i.e. the first or experimental use of substances), which triggers increasing use that can progress to harmful levels and ultimately dependence.1–3 Substance initiation, particularly during adolescence, is strongly linked to the subsequent development of problematic substance use or SUD.4–7 While substance use and SUD have been extensively studied, initiation has recently received increasing attention.
Classical genetic studies have shown that substance initiation, use, and SUD are all heritable, with substance use (i.e. use frequency or quantity) and SUD having higher heritability than initiation.3,5,8–10 For example, timing of first alcohol use had heritability of 36%, while heritability of alcohol dependence was 53%.5 Although high genetic correlations were observed between early initiation or age at onset, use frequency or quantity, and SUD (e.g.5,11), genome-wide association studies (GWAS) seem to suggest a limited genetic overlap between initiation, use, and SUD (e.g. cannabis12). As summarized in a recent review on largest GWAS of substance involvement,3 more than 150 GWAS loci reached the genome-wide significance level for alcohol use and/or alcohol use disorder (AUD), approximately 500 GWAS loci for tobacco initiation, use, or tobacco use disorder (TUD), and up to 15 for cannabis initiation, use, or cannabis use disorder (CUD). There seem to be some shared genetic factors between substance use and SUD, but the degree of genetic overlap depends on the substance type. In general, substance use is genetically distinct from dependence, except for nicotine use vs. dependence, possibly due to the highly addictive nature of tobacco.3,13 Therefore, identifying genetic variants contributing to individual substances or specific categories of substance use behaviors, as well as those that are common to multiple substances (i.e., polysubstance) or categories of substance use behaviors will provide insights into molecular mechanisms underlying substance involvement.
To understand the molecular/neurochemical changes that are associated with the addiction circuits during the drug-taking to drug addiction transition, it is essential to parse genes into addiction domains.1,9 Koob and Volkow1 proposed 18 addiction circuits, involving cortical (e.g. the prefrontal cortex/PFC) and subcortical (e.g. nucleus accumbens) regions. Recently, a medial PFC-brain stem circuit was shown to be critical for compulsive alcohol drinking in mouse model,14 and our human brain imaging analysis indicated the importance of a thalamus-PFC-brainstem circuit in alcohol initiation.15 On the other hand, several hypotheses, such as maturational imbalance theory and dual-system model,16,17 were proposed to explain adolescents’ risk-taking behaviors. These models posit that the subcortical region involved in emotion regulation and reward system mature earlier than the PFC, which is critical for executive functions and continues to develop throughout adolescence. Imbalanced development of these two types of brain systems can decrease the ability of adolescents to inhibit impulses. Therefore, identifying the addiction-related brain regions that are affected by substance-associated genes will provide valuable insights into neurobiological mechanisms of drug addiction.
As a first step towards parsing substance-associated genes into the addiction circuit-related brain regions (designated “AddictRegions”), we re-analyzed the human brain transcriptome data, with a particular focus on protein-coding genes that were differentially regulated in at least one of 17 designated AddictRegions. While several brain transcriptomic studies have targeted only one or a few brain regions,18–20 our study aimed to gain a complete profile of AddictRegions transcriptome. To achieve this, we used the most comprehensive postmortem brain transcriptome data,21 which analyzed the expression profiles of approximately 900 anatomically defined brain structures across the whole brain. We also collected the substance-associated single nucleotide polymorphisms (SNPs) obtained from the largest-scale GWAS studies, including 10 publications on alcohol, cannabis, and tobacco12,13,22–29 as summarized by Gelernter and Polimanti,3 as well as five additional publications.30–34 Notably, the most recent publication34 represents the largest-scale GWAS to date, with 3.4 million individuals, leading to identification of nearly 4,000 SNPs associated with alcohol and tobacco use behaviors, including more than 1,900 novel SNPs. Analysis of these substance-associated SNPs will provide a comprehensive view of substance-associated genes. Our goals were to 1) determine patterns of gene expression across AddictRegions, 2) provide an update on the substance-associated genes regarding the overlapping and/or distinct sets of genes based on types of substance and categories of substance use behavior, and 3) assess the relationship between substance-associated genes and up- or down-regulation in AddictRegions.
2. METHODS
2.1. Brain transcriptome data
Normalized expression data from Allen Human Brain Atlas (AHBA) were analyzed.21 The AHBA included DNA microarray data from spatially distinct regions (414 Structure IDs) across cortical, subcortical, and cerebellar regions from six adult postmortem brains.21 Data preprocessing and normalization were performed by the Allen Brain Institute (http://help.brain-map.org/display/humanbrain/Documentation). Additionally, we excluded probes with nondetectable expression levels in at least three out of six brains. Mean expression levels were obtained for each combination of probe and Structure IDs within each brain. Our analyses were restricted to protein-coding genes, using the updated probe-to-gene annotation.35
2.2. Identification of up- or down-regulated genes in AddictRegions
Determination of AddictRegions:
Out of 414 unique Structure IDs, we manually mapped 118 structure IDs into 17 AddictRegions (Table S1), based on the addiction circuits summarized by Koob and colleagues1,9 and the recently reported PFC-brainstem circuit for alcohol.14,15 These AddictRegions include six cortical and 11 subcortical regions: SFG (superior frontal gyrus), MFG (middle frontal gyrus), IFG (inferior frontal gyrus), OFC (orbitofrontal cortex), CgG (cingulate gyrus), Ins (insular gyrus), DS (dorsal striatum), NAc (nucleus accumbens), GP (globus pallidus), Amg (amygdala), Hpc (hippocampus), Tha (Thalamus), Hb (habenula), Hyp (Hypothalamus), SN (substantia nigra), VTA (ventral tegmental area), and PAG (periaqueductal gray, a critical region in brainstem). The control region cohort included 296 Structure IDs not belonging to any AddictRegions. We note that differential analysis between Hb and the control region cohort was not performed, as no expression data was available for the habenula after quality controls.
Determination of up- or down-regulated genes in at least one of 16 AddictRegions:
Gene expression was analyzed at the probe level. For each participant, the probe value from each structure ID within each individual AddictRegion or the control region cohort was regarded as a repeated measure of the gene expression level. Differential expression analyses were performed using the limma package in R (version 4.1.2) to compare the gene expression between each individual AddictRegion and the control region cohort. The duplicateCorrelation approach, a built-in method in limma for analyzing repeated measures data, was used to account for potential correlations in expression level among different structure IDs within the same AddictRegion or the control region cohort. In case two or more probes of a gene were significant, the gene was called differentially expressed only if these probes all showed associations in the same direction. Multiple comparison problems were adjusted by controlling the false discovery rate (FDR) at 0.05 level.
2.3. Retrieval of substance-associated GWAS data and analysis of genes in relation to substance involvement
Substance-associated SNPs were retrieved from GWAS Catalog (https://www.ebi.ac.uk/gwas/) in which only large-scale alcohol, cannabis and tobacco initiation, use and SUD-related studies were included (Table S2). These consisted of 10 publications12,13,22–29 summarized in the recent comprehensive review3 and five additional publications30–34 retrieved from PubMed or GWAS Catalog. Protein-coding genes were identified by those directly hit by substance-associated SNPs or those nearest to the SNPs and downloaded from GWAS Catalog, except for the most recent study34 in which SNPnexus36 was used for annotation. A total of 1475 protein-coding genes represented by the substance-associated SNPs matched both the HUGO Gene Nomenclature Committee list of protein-coding genes and the protein-coding genes annotated for Probe IDs in the brain transcriptome data.35 For determination of genes overrepresented in each of AddictRegions, Fisher’s Exact test was performed with multiple comparison adjustment controlled at false discovery rate (FDR) of 0.05. Gene Ontology (GO) overrepresentation analysis was performed using the platform PANTHER (http://pantherdb.org)37 with Bonferroni correction.
3. Results
3.1. Identification of differentially regulated genes in AddictRegions
Of the 16,562 protein-coding genes (represented by 37,457 probes) studied, 4,807 were up-regulated (Table S3) and 3,457 down-regulated (Table S4) in at least one of 16 AddictRegions with 2-fold or more expression difference compared to that from the control region cohort. Overall, there were 6,672 differentially expressed genes, indicating that a substantial proportion (40%) of genes were transcriptionally regulated in AddictRegions. Among these genes, 1592 were up-regulated in some AddictRegions but down-regulated in others. Notably, only 1–5% of these up- or down-regulated genes were present in each of 16 AddictRegions. The GP and SN regions had the largest proportions of up-regulated genes (12.7% and 9.5%, respectively), while the Hpc, DS, and GP had the largest proportions of down-regulated genes (7.9%, 6.7% and 5.5%, respectively) (Tables S3 and S4). The majority of these genes (>79%) were up- or down-regulated in one to three AddictRegions, with approximately 50% of them in only one AddictRegion. Only 42 genes were up-regulated in 9–11 AddictRegions and 45 down-regulated in 9–12 AddictRegions, with ANK1, DAO, RELN, and SLC17A6 down-regulated in a total of 12 AddictRegions.
3.2. Genes represented by substance initiation, use, and SUD-associated SNPs
A total of 1475 protein-coding genes were identified mapping in or nearest the SNPs that were associated with substance initiation, use, and/or SUD in the largest-scale GWAS of alcohol, cannabis, and tobacco (Table S5). Of these genes, 461 were associated with alcohol, 9 with cannabis, and 1157 with tobacco (Figure 1A). For different categories of substance use behaviors, 1,060 genes were associated with initiation, 580 with use, and 29 with SUD (Figure 1B). However, there was very little overlap among the genes associated with the three substances or the three categories of substance use behaviors. Only two genes (CADM2 and NCAM1) were common to all three substances. In addition, 145 additional genes were common to alcohol and tobacco, three genes (FOXP2, SDK1, and SMG6) were common to cannabis and tobacco, and no additional gene common to alcohol and cannabis. The same pattern was observed for the different categories of substance use behaviors. Six genes (CHRNA4, FAM163B, FTO, PDE4B, SEMA6D, and SPI1) were common to all three use behaviors. A slightly larger number of genes were common to two categories of substance use behaviors; for instance, additional 166 genes were common to initiation and use, and additional 14 genes were common in use and SUD, and two (CNTLN, and FOXP2) were common to initiation and SUD. Surprisingly, although there were more initiation-associated genes than use-associated genes, less SUD-associated genes overlapped with initiation than with use. The comparison of genes associated with initiation, use, and SUD within a given substance revealed a similar pattern, such as between AUD-associated and alcohol use-associated genes (Figure 1C) and between TUD-associated and tobacco use-associated genes (Figure 1D). Likewise, there were only two genes (CHRNA4 and FAM163B) common to tobacco initiation, use, and TUD (Figure 1D).
FIGURE 1.
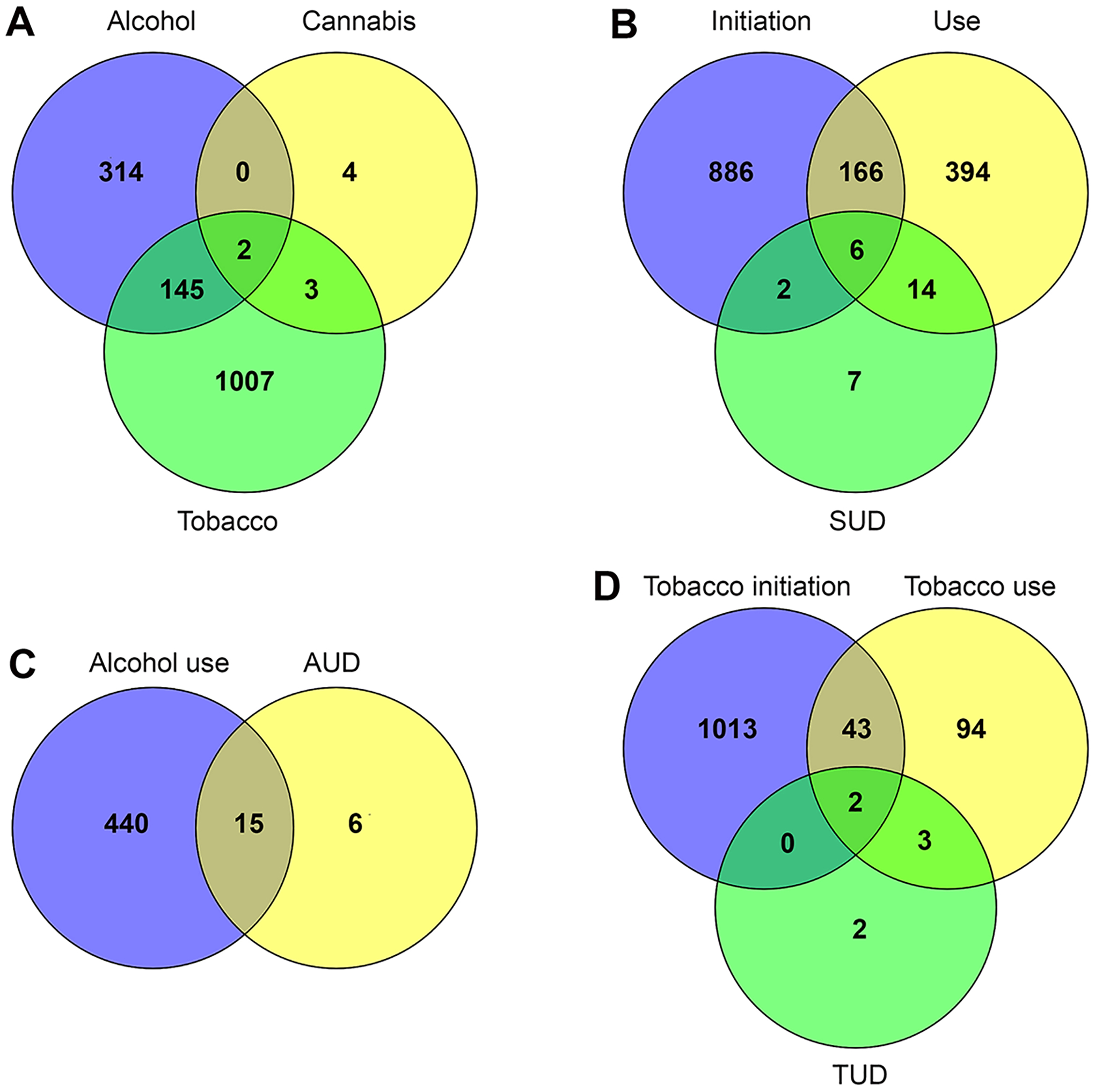
Venn diagrams showing distinct and overlapping number of substance-associated genes. (A) Types of substance. (B) Categories of substance use behaviors. (C) Alcohol use and AUD. (D) Categories of tobacco initiation, use, and TUD.
GO overrepresentation analysis showed that nine GO-Slim terms of biological processes were enriched by 1.5 to 6.8-fold, with a Bonferroni-corrected p-value of <0.05 (Table S6). These include adherens junction organization (6.8-fold), synaptic transmission-glutamatergic (4.9-fold), cell morphogenesis (2.5-fold), neuron projection development (2.3-fold), ion transmembrane transport (2.0-fold), and regulation of transcription by RNA polymerase II (1.5-fold). A total of 154 genes belong to the GO term of regulation of transcription by RNA polymerase II, indicating the potential importance of transcriptional activation for substance involvement. In addition, six GO-Slim terms of molecular function were significantly enriched, including GluR activity (6.5-fold), cell-cell adhesion mediator activity (6.1-fold), voltage-gated calcium channel activity (4.4-fold), cadherin binding (4.3-fold), neurotransmitter binding (4.1-fold), and postsynaptic neurotransmitter receptor activity (4.0-fold).
3.3. Substance-associated genes are differentially up-regulated in AddictRegions
Among 1475 substance-associated genes, 1389 were present in the brain transcriptome data and analyzed further. Of those, 529 were up-regulated in at least one of AddictRegions with a twofold expression difference cutoff (Table S7). This represents a significant overrepresentation (p-value of 2.68E-11) compared to the whole genome, with 38% of the analyzed genes being up-regulated in AddictRegions, versus 29% of the whole genome (4,807 out of 16,562) (Figure 2A). To better understand the functional significance of those AddictRegion-upregulated genes, GO overrepresentation analysis of those 529 genes was performed. We found that 10 GO-Slim terms related to biological processes were overrepresented (Table S8), including regulation of synaptic transmission-glutamatergic (13.3-fold), adherens junction organization (10.2-fold), synaptic transmission-glutamatergic (8.6-fold), axon guidance (5.5-fold), nervous system process (3.8-fold), ion transmembrane transport (2.8-fold), and intracellular signal transduction (2.0-fold). In addition, six GO-Slim terms of molecular function were significantly overrepresented, including glutamate (Glu) receptor (GluR) activity (10.6-fold), neurotransmitter binding (8.5-fold), postsynaptic neurotransmitter receptor activity (7.0-fold), cadherin binding (6.7-fold), and metal ion transmembrane transporter activity (2.6-fold). We also found that several GO-Slim terms in 529 AddictRegion-regulated, substance-associated genes were further enriched compared to that in 1,475 substance-associated genes. These include adherens junction organization, synaptic transmission-glutamatergic, Glu receptor (GluR) activity, and neurotransmitter binding, suggesting the importance of activation of the genes involved in neurotransmission and cell-cell junction in substance involvement. Of note, 22 out of 28 substance-associated transmitter binding genes (i.e., 78.6%) were significantly up-regulated in at least one of 16 AddictRegions (Table 1). This proportion is twice that of the substance-associated genes (38.1%). Most of these genes were associated with alcohol use, tobacco initiation, and tobacco use, indicating a potentially important role of neurotransmitter gene activation in this type of substance use behaviors.
FIGURE 2.
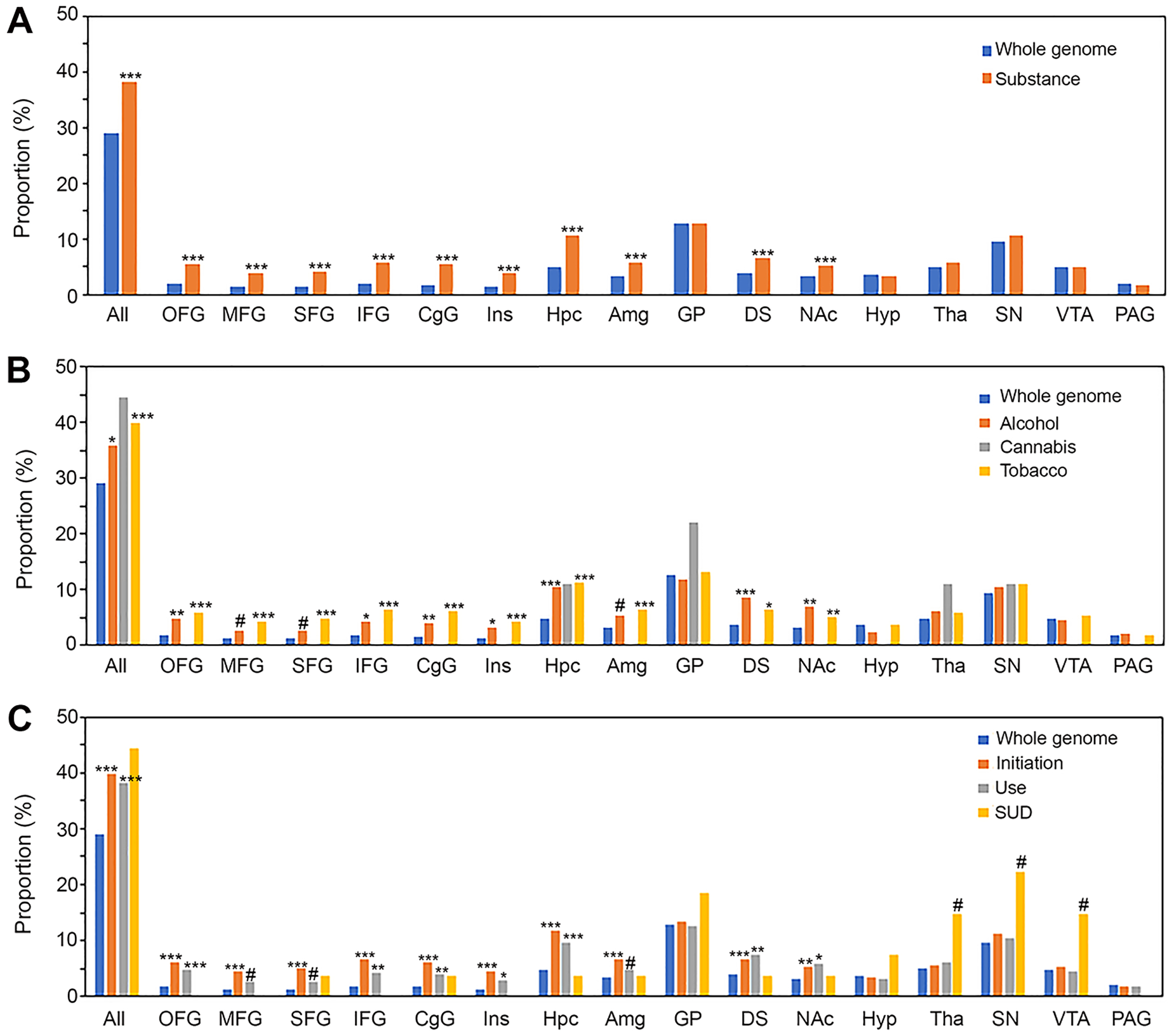
AddictRegion overrepresentation analysis of up-regulated genes. (A) Proportion of all categories of substance-associated genes in each AddictRegion. (B) Proportion of genes in each AddictRegion based on types of substance. (C) Proportion of genes in each AddictRegion based on categories of substance use behavior. All, any of the AddictRegions; *, p<0.05; **, p<0.01; ***, p<0.001 after Bonferroni correction; # indicating p-value <0.05 without Bonferroni correction.
Table 1.
List of 27 substance-associated neurotransmitter binding genes that are differentially regulated in AddictRegions. These include 22 up-regulated genes and 5 genes that were down-regulated only. Fold changes are expressed as log2-transfromed values. AlcUse, alcohol use; AUD; alcohol use disorder; TobIni, tobacco initiation; TobUse, tobacco use; TUD, tobacco use disorder.
| Gene | OFG | MFG | SFG | IFG | CgG | Ins | Hpc | Amg | GP | DS | NAc | Hyp | Tha | SN | VTA | PAG | Substance | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHRM2 | −1.2 | −1.4 | −1.3 | −1.1 | −1.3 | −2.1 | −1.2 | −2.3 | −1.3 | 1.3 | −2.2 | AlcUse | 34 | |||||
| CHRNA2 | −1.1 | 3.6 | TobIni | 25 | ||||||||||||||
| CHRNA3 | −2.3 | −2.5 | −2.3 | −2.4 | −1.9 | −2.1 | −1.5 | 0.0 | −1.6 | −1.1 | 2.0 | TobUse | 25,29,30,32,34 | |||||
| CHRNA4 | −2.0 | −1.5 | 1.1 | 2.0 | 1.8 | 2.1 | TobIni, TobUse, TUD | 13,25,29,31,32,34 | ||||||||||
| CHRNA5 | 1.2 | 1.4 | TobUse | 25,31 | ||||||||||||||
| CHRNA6 | −1.1 | −1.4 | 1.5 | 1.1 | TobUse | 34 | ||||||||||||
| CHRNA7 | −1.2 | −1.2 | 1.0 | AlcUse | 34 | |||||||||||||
| CHRNB2 | 1.3 | 1.3 | TobUse | 25,31,34 | ||||||||||||||
| CHRNB3 | −1.3 | −1.1 | −1.1 | 2.6 | 2.7 | TobUse, TUD | 13,25,31,32,34 | |||||||||||
| DRD2 | 1.2 | 1.3 | 3.1 | 2.7 | 2.1 | 2.3 | 2.1 | 2.5 | AlcUse, AUD, TobUse | 25–28,31,34 | ||||||||
| GABRA4 | 1.2 | 1.3 | 1.4 | 1.2 | 1.1 | 1.4 | 1.0 | 1.1 | −1.7 | 1.6 | 1.7 | −1.3 | 1.1 | −1.3 | AlcUse | 34 | ||
| GABRA6 | −5.4 | −5.9 | −5.7 | −5.2 | −5.0 | TobIni | 25 | |||||||||||
| GABRB1 | 1.4 | 1.4 | 1.5 | 1.1 | TobIni | 34 | ||||||||||||
| GABRB2 | −1.4 | TobIni | 34 | |||||||||||||||
| GABRG1 | 1.5 | 1.3 | 1.2 | 1.7 | 1.6 | AlcUse, TobIni | 34 | |||||||||||
| GLRA3 | 1.4 | −1.4 | 1.0 | 1.4 | 1.2 | 1.3 | AlcUse, TobIni | 34 | ||||||||||
| GRIA4 | −1.7 | −1.3 | −1.1 | −1.8 | −1.4 | −1.0 | TobIni | 34 | ||||||||||
| GRID1 | 1.1 | AlcUse, TobIni | 34 | |||||||||||||||
| GRID2 | −1.2 | −1.1 | −1.1 | −1.1 | −1.1 | −1.7 | −1.5 | −1.1 | TobIni | 25,29,34 | ||||||||
| GRIK2 | −1.1 | −1.1 | −1.1 | −1.3 | −1.3 | TobIni | 34 | |||||||||||
| GRIN2A | 1.2 | 1.3 | 1.2 | 1.3 | 1.1 | 1.1 | 1.5 | −1.2 | −1.1 | −2.1 | −1.4 | TobIni | 25,34 | |||||
| GRIN2B | 1.3 | 1.2 | 1.3 | 1.4 | 1.6 | 1.9 | −1.4 | 1.8 | 1.7 | −1.0 | 1.4 | AlcUse, TobIni | 34 | |||||
| GRM5 | 1.1 | 1.1 | 1.8 | 1.5 | −1.1 | 1.4 | AlcUse, TobIni | 34 | ||||||||||
| HTR2A | 1.7 | 1.9 | 2.0 | 1.6 | 1.4 | −1.4 | −1.1 | −2.6 | −2.5 | −2.0 | −1.2 | −1.8 | −1.2 | −1.1 | TobIni | 34 | ||
| HTR4 | 1.8 | 0.0 | 2.2 | 1.9 | −1.2 | −1.1 | 0.2 | TobIni | 34 | |||||||||
| LY6H | 1.9 | 1.8 | 1.8 | 3.0 | 2.5 | 2.5 | TobIni | 34 | ||||||||||
| OPRM1 | −1.2 | −1 | −1.2 | −1.5 | 1.1 | 1.3 | TobIni | 34 |
When each of 16 AddictRegions was evaluated, all six cortical regions exhibited a 2–3-fold enrichment (3.7–5.8% vs. 1.3–1.9% of the whole genome). However, most subcortical regions did not exhibit any enrichment except for a few regions with relatively mild overrepresentation, such as Hpc (2.2-fold), and Amg, DS and NAc (1.6–1.8 folds) (Figure 2A). A total of 95 genes were up-regulated in at least one of the cortical AddictRegions (Table S9), with 58 of them up-regulated in at least four cortical AddictRegions. Interestingly, we also found that 50 of these genes were up-regulated in Hpc (52 genes), and 28 and 27 genes up-regulated in Amg and DS, respectively. Additionally, we identified 76 genes associated with tobacco initiation and 24 associated with alcohol use, indicating the importance of co-up-regulation of substance-associated genes in cortical regions and Hpc (and to a less extent Amg and DS) for tobacco initiation and alcohol use.
The analysis based on types of substance and categories of substance use behavior revealed interesting findings. Cannabis-associated genes (4 out of 9 genes, i.e., 44.4%) were not significantly overrepresented in any of AddictRegions, while enrichment of alcohol- and tobacco-associated genes was significant (alcohol: 159 out of 443 genes, i.e., 35.9%; tobacco: 433 out of 1087 genes, i.e., 39.8%) (Figure 2B). Tobacco-associated genes were overrepresented in all six cortical regions, with 3.2–3.7-fold enrichment compared to the whole genome, and in four out of 10 subcortical AddictRegions (Hpc: 2.3-fold; Amg: 1.9-fold; DS: 1.7-fold; NAc: 1.6-fold). Alcohol-associated genes were overrepresented in fewer AddictRegions, with 2–2.6-fold enrichment in four cortical regions (OFG, IFG, CgG, Ins) and three subcortical AddictRegions (Hpc, DS, and Nac). Significant enrichment in MFG, SFG, and Amg disappeared after Bonferroni correction for multiple testing (Figure 2B). Regarding substance use behavior, both initiation and use were overrepresented in up-regulated AddictRegions (Figure 2C). With respect to individual AddictRegions, initiation-associated genes were 3.2–3.7-fold enriched in all six cortical regions and four subcortical regions (Hpc, Amg, DS, and Nac, with 1.6–2.4 folds). For use-associated genes, four cortical AddictRegions (OFG, IFG, CgG, and Ins) and three subcortical (Hpc, DS, and Nac) were overrepresented with 1.8–2.5 folds. Although SUD-associated genes (12 out of 27 genes, i.e. 44.4%) were not significantly enriched in AddictRegions, there was 2.3–3.0-fold overrepresentation in three subcortical AddictRegions (Tha, SN, and VTA) which did not pass the significance level after multiple comparison adjustment (Fig. 2C). The lack of significant enrichment after multiple comparison adjustment might be due to small sample size for SUD. Surprisingly, none of eight SUD-associated genes (AUD-associated: DRD2, KLB, PDE4B, SEMA6D, MTTP; CUD-associated: FOXP2; and TUD-associated: CHRNA4 and CHRNB3) that were significantly overrepresented in Tha, SN, and VTA was up-regulated in any of cortical AddictRegions (Table S10).
3.4. AddictRegions-downregulated, substance-associated genes exhibited an opposite pattern.
A total of 438 substance-associated genes were found to be down-regulated in AddictRegion using a twofold cutoff (Table S11). They exhibited similar overrepresentation of GO-Slim terms as those up-regulated genes, such as synaptic transmission-glutamatergic, neurotransmitter binding, GluR activity (Table S8). However, the distribution of overrepresented AddictRegions (Fig. 3) was opposite to that of up-regulated genes (Fig. 2). Specifically, down-regulated genes were only slightly overrepresented in five cortical regions (1.8–2.1 folds) but more strongly in several subcortical regions (e.g., GP, Hyp, Tha, SN, and VTA with 2.2–2.8 folds) for substance involvement (Fig. 3A), two types of substance (Fig. 3B; except cannabis where no gene was down-regulated), or all categories of behavior (Fig. 3C). In addition, while these down-regulated genes had a mild overrepresentation in Hpc and Amg for initiation or tobacco (1.3–1.6 folds), Alc- or use-associated genes had an approximately 2-fold overrepresentation in PAG, although this significance disappeared after multiple comparison adjustment.
FIGURE 3.
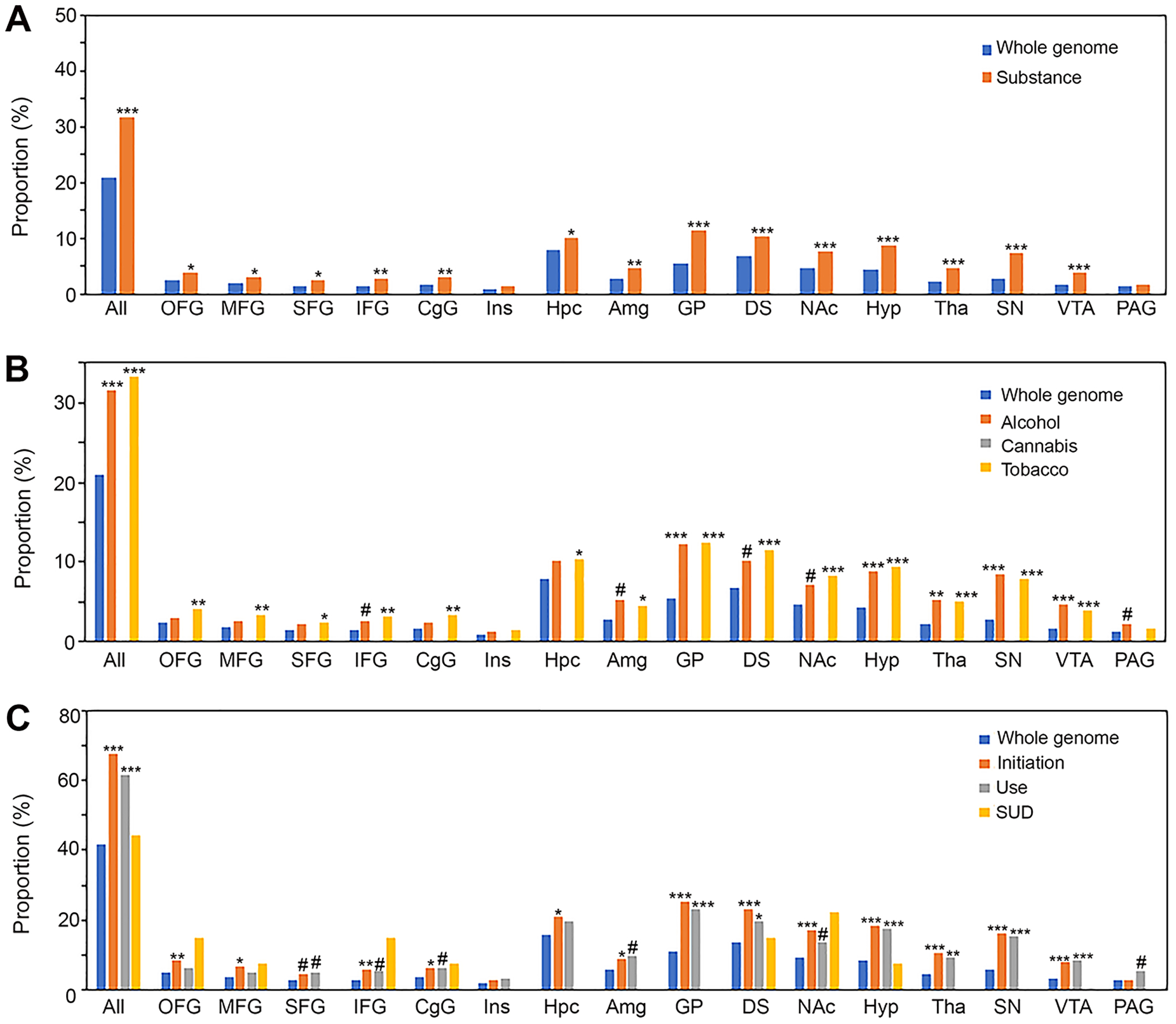
AddictRegion overrepresentation analysis of down-regulated genes. (A) Proportion of all categories of substance-associated genes in each AddictRegion. (B) Proportion of genes in each AddictRegion based on types of substance. (C) Proportion of genes in each AddictRegion based on categories of substance use behavior. All, any of the AddictRegions; *, p<0.05; **, p<0.01; ***, p<0.001 after Bonferroni correction; # indicating p-value <0.05 without Bonferroni correction.
3.5. A subset of substance-associated genes exhibited a pattern of coordinated cortical up-regulation and subcortical down-regulation.
There were 237 substance-associated genes present in both up-regulated and down-regulated gene sets (Table S12), and they were further divided into three clusters (Fig. 4A). Cluster 1 contained 65 genes, indicating that the majority of 95 cortically up-regulated substance-associated genes (Table S9) were up-regulated in cortical AddictRegions (and Hpc or Amg) but down-regulated in other eight subcortical regions. GO-Slim enrichment analysis revealed a significant overrepresentation of transmitter binding and GluR activity compared to the whole genome (13.4 and 30.6-fold, respectively) or all substance-associated genes (3.8 and 5.8-fold, respectively). In addition, this cluster could be split into two groups based on their expression patterns in those eight subcortical regions: one group had 26 genes (with NRGN showing the largest fold change) up-regulated in at least one of eight subcortical regions (Fig. 4B), while the other had 39 genes (with SLC30A3 and NPTX1 exhibiting the largest fold change) not up-regulated in any other subcortical regions (Fig. 4C). The majority of the 39 genes (87%) was associated with tobacco initiation, and eight genes (21%) were associated with alcohol use (Table S12).
FIGURE 4.
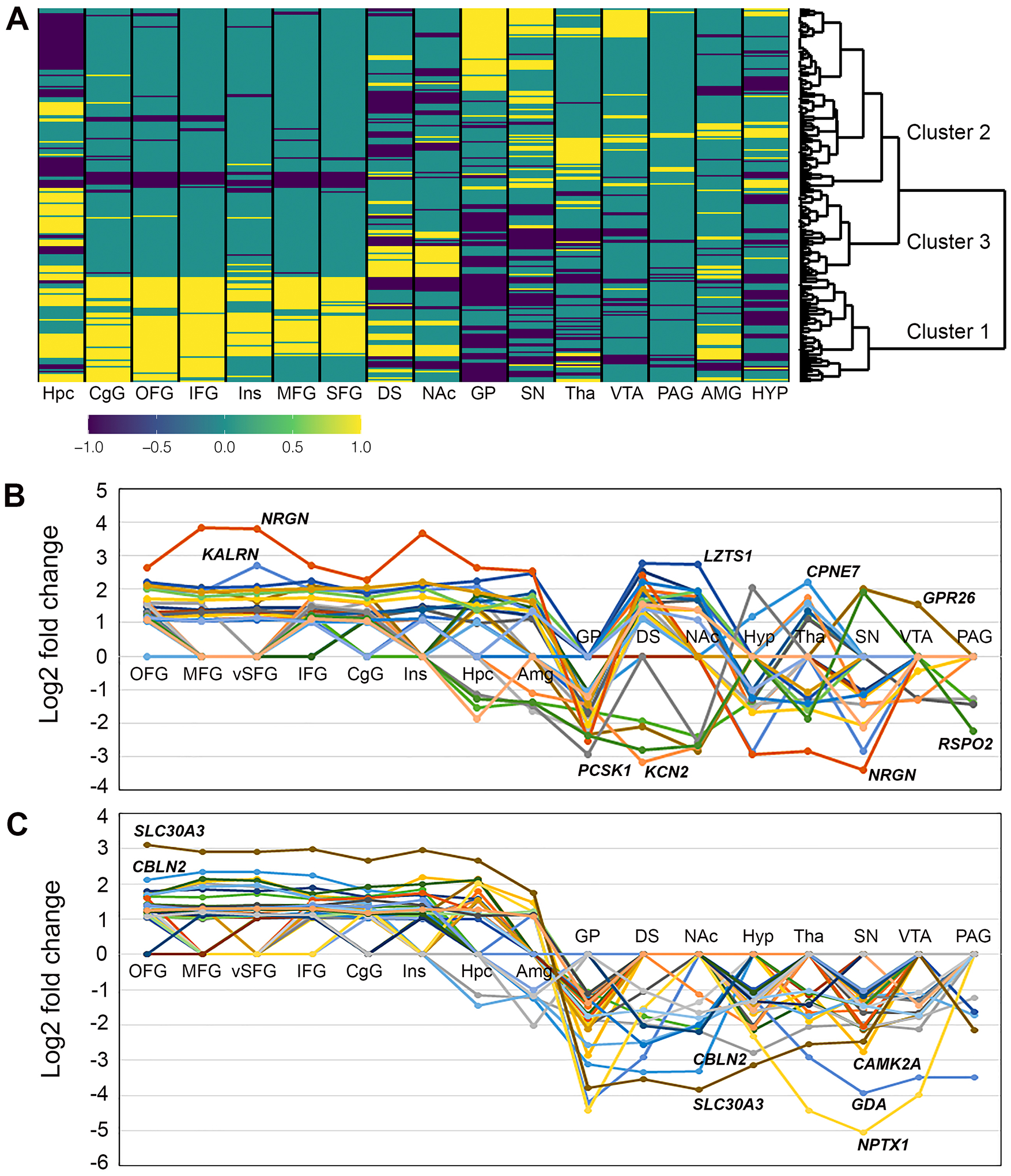
Expression patterns of 231 substance-associated, AddictRegions up- and down-regulated genes. (A) Cluster analysis result. (B and C) Fold changes for genes in two groups of Cluster 1 genes. Genes with the largest fold changes (expressed as log2-transformed values) were indicated.
3.6. Genes involved in neurotransmitter binding and Glu receptor activity were further enriched for common use behaviors.
Out of 183 genes common to initiation, use, and/or SUD (Table S13) and 148 polysubstance-associated genes (Table S14), 61.3% (n=112) and 63.5% (n=94), respectively, were found to be up- or down-regulated in at least one of 16 AddictRegions. These proportions were significantly higher than that for 1,389 substance-associated genes (p=0.021 and 0.018, respectively). Although, GO-Slim terms of neurotransmitter binding and GluR activity were not significantly enriched for AddictRegions-downregulated genes, they were enriched in the AddictRegion-upregulated genes common to initiation, use, and/or SUD. Compared to 22 neurotransmitter binding genes out of 529 AddictRegions-upregulated, substance-associated genes (Table 1), eight neurotransmitter binding genes (CHRNA4, CHRNB3, DRD2, GABRG1, GLRA3, GRID1, GRIN2B, and GRM5) were present in 75 AddictRegions-upregulated genes that were common to initiation, use, and/or SUD, representing a 2.6-fold enrichment (p=0.046 after multiple comparison adjustment). Five GluR genes (GRID1, GRIN2B, GRM3, GRM5, and GRM7) were AddictRegions-upregulated and common to initiation, use, and/or SUD, although this 3.5-fold enrichment (p=0.0285) was not significant after multiple comparison adjustment (p=0.057).
4. Discussion
Gene expression regulation in the brain plays a critical role in substance addiction. However, studying the living human brain is limited by sample access. To overcome this limitation, postmortem brain tissues have been used as a substitute. Although several studies on postmortem brain transcriptomes of substance-related subjects have been published,33,38–41 they have been limited to only one or a few brain tissues. To provide a broader perspective, we have re-analyzed the most comprehensive brain transcriptome data so far,21 focusing on the 16 AddictRegions, the well-studied brain regions involved in addiction circuits in literature.1,9,14,15 Our results show that 40% of human protein-coding genes are significantly up- or down-regulated in at least one of the 16 AddictRegions, compared to other brain regions. We also observed a slight yet significant enrichment of up- or down-regulation in AddictRegions for the substance-associated genes identified from largest-scale GWAS, suggesting the relevance of altered expression of genes in AddictRegions to substance initiation, use, or SUD. Moreover, we found a regionally preferential transcriptional activation pattern for substance initiation, use, and SUD. It is not clear whether transcriptional regulation in AddictRegions is linked to differential regulation in substance-involved subjects. By comparing genes up- or down-regulated in AddictRegions with those differentially expressed in AUD subjects compared to healthy control,38–43 we found large variations in overlap depending on AddictRegions, with up to 37% of 27 alcohol-associated, thalamus up-regulated genes being also up-regulated in thalamus of AUD subjects and smaller proportions in other Addiction regions (Table S15). Notably, two neurotransmitter binding genes, GRIN2B and DRD2, which were 2.6- and 4.9-fold up-regulated in thalamus (Table 1), had higher expression (21.8- and 5.5-fold, respectively) in thalamus of AUD subjects.41 Similarly, CPNE7, which was associated with tobacco use31 and up-regulated in hippocampus, amygdala, and thalamus, had higher expression in these three regions of AUD subjects.39,41,42 Taken together, our work provides a useful resource for addiction research by revealing the AddictRegion-related transcriptome features. Integrating these transcriptome features into future studies on substance GWAS or functional validation of those candidate genes will help elucidate the neurobiological mechanisms of addiction.
In the search for genetic variants involved in substance involvement, understanding the degree of genetic overlap among different substances or different use behaviors is crucial. Our findings suggest limited genetic overlap across all three substances and behaviors, with only eight out of 1,475 substance-associated genes commonly associated with all three substances or behaviors. For tobacco, which has been studied most intensively with the largest number (1,157) of tobacco-associated genes identified, only two genes were common to tobacco initiation, use, and TUD. While our findings suggest that genetic control of substance involvement differs significantly, overlap increases when only two use behavior were examined. However, this kind of genetic overlap seems to change with the shift from initiation to use and to SUD. It is noteworthy that although tobacco use and nicotine dependence have been reported to share more genetic factors than that in alcohol or cannabis,3,13 our comparison based on the proportion of overlapping genes between alcohol use and AUD, or between tobacco use and TUD, does not seem to support such substance-type differences. More importantly, our result indicates that when substance involvement shifts from initiation to use and to SUD, the common genetic components also shift during the substance addiction process, resulting in fewer genes overlapped between initiation and SUD than between initiation and use or between use and SUD. This implies that genetic factors underlying each stage of the disease progression are both distinct and partially overlapped. Interestingly, although there are genes associated with initiation, use, and SUD that are up-regulated in both cortical and subcortical AddictRegions, our results from overrepresentation analysis of substance-associated genes up-regulated in each AddictRegions suggest that the stronger enrichment in cortical up-regulation of initiation-associated genes shifts to weaker enrichment for use-associated genes and that the enrichment tends to shift further to subcortical up-regulation of SUD-associated genes.
The analysis of substance-associated genes up-regulated in AddictRegions has provided additional insights. First, our results highlight the importance of co-activation of genes in cortical AddictRegions and hippocampus and down-regulation in subcortical AddictRegions for initiation. While only one large-scale GWAS on cannabis initiation and none for alcohol initiation have been published, the analysis of tobacco initiation may provide clues to genetic control of substance initiation. We found that 47 tobacco initiation-associated genes were up-regulated in at least four of six cortical AddictRegions and that half of those genes were frequently up-regulated in hippocampus (Table S9). We also identified a subset of 39 genes that exhibited a pattern of coordinated cortical up-regulation and subcortical down-regulation, particularly for genes associated with tobacco initiation (Table S12; Fig. 4C). One possible explanation for the stronger transcriptional activation in the cortical AddictRegions (and to a less extent hippocampus) and inhibition in other subcortical regions could be related to the alteration in the PFC-controlled executive functions and subcortical AddcitRegions-controlled novelty seeking and/or emotion processing. According to the dual systems or maturational imbalance models,16,17 the PFC during early- and mid-adolescence is not developed fully, compared to early maturing subcortical regions, and adolescents lack normal inhibitory control, leading to a risky decision-making including the tendency to experiment or initiate substance use. It would be interesting to investigate whether the substance initiation risk alleles are linked to up- or down-regulation of those genes mapped in or nearest to these SNPs and whether such mis-regulated expression is causally linked to initiation.
Second, we found that GO terms of neurotransmitter binding and GluR activity are overrepresented in substance-associated and/or AddictRegion-upregulated genes. This enrichment is even stronger for common substance use behavior and to a less extent for polysubstance. These findings are consistent with the known importance of synaptic transmission in addiction.44–48 There are at least four types of neurotransmitters, each of which binds to distinct receptors, that exert excitatory, inhibitory or modulatory effect on the target cells responsible for various addictive behaviors: Glu, acetylcholine (ACh), gamma-aminobutyric acid (GABA), and dopamine. GluR activity is critical for mediating fast excitation and synaptic plasticity modulation in mesocorticolimbic pathways including VTA and NAc.48 Emerging evidence suggests a role for GluRs in Amg, a key brain region for emotional and appetitive memories; however, their role in other key addiction behavior-related brain regions remains largely unknown.48 Our result indicates that certain GluR genes, which are associated with substance use behaviors (especially polysubstance or common use), are also up-regulated in cortical regions (e.g. GRIN2B and GRM5) and Hpc (GRIN2B, GRM5, and GRM7). Future studies are needed to determine how these GluR genes contribute to substance involvement. It is encouraging that a GRM5/mGluR5-specific negative allosteric modulator HTL0014242 (now called TMP-301) has been approved to enter Phase I clinical trial for treatment of alcohol and other substance disorders.49 Ach receptors, which may excite or inhibit target cells, can be either nicotinic or muscarinic receptors. Subunits (alpha, beta, gamma, delta, and epsilon) of the nicotinic ACh receptors (nAChRs) are encoded by 17 CHOLINERGIC RECEPTOR NICOTINIC (CHRN) genes CHRNA1 to CHRNA10, CHRNB1 to CHRNB4, CHRND, CHRNE, and CHRNG. The importance of alpha and beta subunits of nAChRs in substance can be reflected by the identification of nine substance-associated CHRN genes (CHRNA2-A7, and CHRNB2-B4). The CHRNA3-A5-B4 gene cluster located on chromosome 15 (15q25.1) represents the best-studied nAChRs in nicotine addiction.46,47 They were all associated with tobacco use only, as reported by at least two large-scale GWAS studies (Table S4). In addition, the most recent and largest GWAS34 not only replicated several CHRN-substance associations, but also reported novel associations (CHRNA6-tobacco use, and CHRNA7-alcohol use). Except CHRNB4, which was excluded in the brain transcriptome analysis due to an undetectable expression level in postmortem tissues, eight substance-associated CHRNA and CHRNB genes analyzed here all exhibited significant up-regulation in AddictRegions. Surprisingly, they were only up-regulated in subcortical AddctRegions, with a strong preference in Tha, SN, and VTA, supporting a critical role for reward cognition and information relay impacted by activation of these nAChRs genes in these regions. The third type of neurotransmitter binding proteins is GABA type A and type B receptors which exert fast inhibitory effects in many parts of the brain and play important role in addiction.44 Among six substance-associated, GABA receptor subunit-encoding genes, three (GABRA4, GABRB1, and GABRG1) were up-regulated in AddictRegions. Their association with alcohol use or tobacco initiation was only reported in the most recent and largest GWAS.34 GABRA4 has the largest number of up-regulated AddictRegions among all 4823 genes, including all six cortical and six subcortical AddictRegions, indicating its global involvement in the brain for alcohol use. In contrast, GABRB1 and GABRG1 are up-regulated in subcortical AddictRegions in particular Amg, NAc, and Hyp, suggesting the possible involvement of emotion regulation and reward cognition mediated by these GABA receptors in alcohol use or tobacco initiation. The last type of neurotransmitter binding protein discussed here is dopamine receptor. Out of five dopamine receptor genes, only DRD2 was repeatedly reported in largest-scale GWAS to associate with AUD,26,28 alcohol use,27,28,31,34 and tobacco use.25,34 However, it’s important to note that the DRD2-AUD association may need to be re-assessed due to spurious low allele frequencies for a SNP in controls, according to a meta-analysis.50 Despite this, the association of DRD2 with alcohol use and tobacco use have been replicated in the largest GWAS.34 Interestingly, DRD2 was up-regulated in almost all 10 subcortical AddictRegions (except Hpc and PAG) but not in any of six cortical AddictRegions. This up-regulation in NAc, Amg, VTA and other subcortical AddictRegions is consistent with the important role of dopamine in the reward system, emotional arousal, and regulation of motor behavioral and pleasures related to motivation.45 It’s possible that DRD2, in mediating these brain functions, is critical for substance use only rather than initiation and SUD.
Limitations
Although our analyses have yielded intriguing insights, there are limitations to this approach. First, the definition of AddictRegions and the control region cohort is based on current literature, which may not fully capture the complexity of addiction-related circuitry in the brain. Second, discrete subregions within individual AddictRegions may have distinct functions in addiction and other psychiatric disorders. Thus, while summary measures of gene expression changes in those AddictRegions allow to align with the findings of the prior addiction imaging studies that target AddictRegions, future research is needed to closely examine gene expression in discrete subregions or even individual cells within each AddictRegion. Third, we performed a replication analysis using data from a smaller-scale brain transcriptomic study20 that sampled only 16 brain regions, with eight of them treated as the control region cohort. We found that a considerable proportion of the genes identified in our study were replicated in subcortical regions (see Table S16). However, replication in cortical regions was poor, possibly due to various factors, including the use of a different control region cohort and different platforms for gene analysis (DNA microarray vs. RNA-sequencing). Fourth, given the prevalence of smoking in the AUD subjects, AUD-associated genes from existing literature might need to be cautiously interpreted if the smoking status of AUD subjects was not included as a confounder in prior studies. Finally, while our analysis provides valuable insights into the overall gene expression differences between AddictRegions and the control region cohort, future research could consider incorporating intra-AddictRegion comparison, as it has the potential to unveil region-preferential expression profiles that may contribute to a unique or predominant role of a specific AddictRegion.
CONCLUSIONS
Our work provides a valuable resource of transcriptome features related to addiction, which can be useful in future investigations into substance-associated genes. Moreover, our findings provide intriguing insights into the genetic mechanisms underlying substance involvement, such as the genetic overlap between categories of use behaviors, shift of cortical to subcortical up-regulation from initiation to SUD, and a critical role for synaptic transmission in substance initiation, use, and SUD.
Supplementary Material
ACKNOWLEDGEMENTS
The study is partly supported by a grant from National Institute of Alcohol Abuse and Alcoholism (5R01AA029611 to YZ) and in part by a grant from National Institute of Mental Health (1RF1MH128614 to YZ).
Footnotes
CONFLICT OF INTEREST
None of the authors has any financial or intellectual conflict of interest in this research.
SUPPORTING INFORMATION
Table S1. Structure IDs in each of 17 AddictRegions.
Table S2. Large-scale GWAS on substance involvement.
Table S3. List of 4807 genes up-regulated in AddictRegions.
Table S4. List of 3457 genes down-regulated in AddictRegions.
Table S5. List of 1475 substance-associated genes.
Table S6. List of significantly enriched GO terms for 1475 substance-associated genes.
Table S7. List of 529 substance-associated genes that are significantly up-regulated in AddictRegions.
Table S8. List of significantly enriched GO terms for genes associated with substance and differentially regulated in AddictRegions.
Table S9. List of 95 substance-associated genes that are significantly up-regulated in cortical AddictRegions.
Table S10. List of eight SUD-associated genes that have a trend showing twofold enrichment in up-regulation in subcortical AddictRegions, Tha, SN, and VTA.
Table S11. List of 438 substance-associated genes that are significantly down-regulated in AddictRegions.
Table S12. List of 231 substance-associated genes that are both up- and down-regulated in AddictRegions.
Table S13. List of 112 AddictRegions-differentially regulated genes that are commonly associated with substance initiation, use, and/or SUD
Table S14. List of 94 AddictRegions-differentially regulated genes that are associated with polysubstance.
Table S15. List of substance-associated, AddictRegions-reguated genes that are found in prior studies to be up- or down-regulated in postmortem brain regions from subjects with alcohol use disorder.
Table S16. List of AddictRegion-differentially expressed genes that can be replicated in an independent study.
REFERENCES
- 1.Koob GF, Volkow ND. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry. 2016;3(8):760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez-Roige S, Palmer AA, Clarke TK. Recent Efforts to Dissect the Genetic Basis of Alcohol Use and Abuse. Biol Psychiatry. 2020;87(7):609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gelernter J, Polimanti R. Genetics of substance use disorders in the era of big data. Nat Rev Genet. 2021;22(11):712–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crews F, He J, Hodge C. Adolescent cortical development: a critical period of vulnerability for addiction. Pharmacol Biochem Behav. 2007;86(2):189–199. [DOI] [PubMed] [Google Scholar]
- 5.Sartor CE, Lynskey MT, Bucholz KK, Madden PA, Martin NG, Heath AC. Timing of first alcohol use and alcohol dependence: evidence of common genetic influences. Addiction. 2009;104(9):1512–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordan CJ, Andersen SL. Sensitive periods of substance abuse: Early risk for the transition to dependence. Dev Cogn Neurosci. 2017;25:29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray KM, Squeglia LM. Research Review: What have we learned about adolescent substance use? J Child Psychol Psychiatry. 2018;59(6):618–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verweij KJ, Vinkhuyzen AA, Benyamin B, et al. The genetic aetiology of cannabis use initiation: a meta-analysis of genome-wide association studies and a SNP-based heritability estimation. Addict Biol. 2013;18(5):846–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reilly MT, Noronha A, Goldman D, Koob GF. Genetic studies of alcohol dependence in the context of the addiction cycle. Neuropharmacology. 2017;122:3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deak JD, Johnson EC. Genetics of substance use disorders: a review. Psychol Med. 2021;51(13):2189–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillespie NA, Neale MC, Jacobson K, Kendler KS. Modeling the genetic and environmental association between peer group deviance and cannabis use in male twins. Addiction. 2009;104(3):420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson EC, Demontis D, Thorgeirsson TE, et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry. 2020;7(12):1032–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quach BC, Bray MJ, Gaddis NC, et al. Expanding the genetic architecture of nicotine dependence and its shared genetics with multiple traits. Nat Commun. 2020;11(1):5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siciliano CA, Noamany H, Chang CJ, et al. A cortical-brainstem circuit predicts and governs compulsive alcohol drinking. Science. 2019;366(6468):1008–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Constable RT, Hien D, Chung T, Potenza MN. Brain anatomical covariation patterns linked to binge drinking and age at first full drink. Neuroimage Clin. 2021;29:102529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casey B, Jones RM, Somerville LH. Braking and Accelerating of the Adolescent Brain. J Res Adolesc. 2011;21(1):21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shulman EP, Smith AR, Silva K, et al. The dual systems model: Review, reappraisal, and reaffirmation. Dev Cogn Neurosci. 2016;17:103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang HJ, Kawasawa YI, Cheng F, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu C, Li Q, Efimova O, et al. Human-specific features of spatial gene expression and regulation in eight brain regions. Genome Res. 2018;28(8):1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, Santpere G, Imamura Kawasawa Y, et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362(6420). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minica CC, Verweij KJH, van der Most PJ, et al. Genome-wide association meta-analysis of age at first cannabis use. Addiction. 2018;113(11):2073–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasman JA, Verweij KJH, Gerring Z, et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci. 2018;21(9):1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walters RK, Polimanti R, Johnson EC, et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 2018;21(12):1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu M, Jiang Y, Wedow R, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51(2):237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kranzler HR, Zhou H, Kember RL, et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun. 2019;10(1):1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evangelou E, Gao H, Chu C, et al. New alcohol-related genes suggest shared genetic mechanisms with neuropsychiatric disorders. Nat Hum Behav. 2019;3(9):950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou H, Sealock JM, Sanchez-Roige S, et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci. 2020;23(7):809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu K, Li B, McGinnis KA, et al. Genome-wide association study of smoking trajectory and meta-analysis of smoking status in 842,000 individuals. Nat Commun. 2020;11(1):5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matoba N, Akiyama M, Ishigaki K, et al. GWAS of smoking behaviour in 165,436 Japanese people reveals seven new loci and shared genetic architecture. Nat Hum Behav. 2019;3(5):471–477. [DOI] [PubMed] [Google Scholar]
- 31.Brazel DM, Jiang Y, Hughey JM, et al. Exome Chip Meta-analysis Fine Maps Causal Variants and Elucidates the Genetic Architecture of Rare Coding Variants in Smoking and Alcohol Use. Biol Psychiatry. 2019;85(11):946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erzurumluoglu AM, Liu M, Jackson VE, et al. Meta-analysis of up to 622,409 individuals identifies 40 novel smoking behaviour associated genetic loci. Mol Psychiatry. 2020;25(10):2392–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapoor M, Chao MJ, Johnson EC, et al. Multi-omics integration analysis identifies novel genes for alcoholism with potential overlap with neurodegenerative diseases. Nat Commun. 2021;12(1):5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saunders GRB, Wang X, Chen F, et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature. 2022;612(7941):720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnatkeviciute A, Fulcher BD, Fornito A. A practical guide to linking brain-wide gene expression and neuroimaging data. Neuroimage. 2019;189:353–367. [DOI] [PubMed] [Google Scholar]
- 36.Oscanoa J, Sivapalan L, Gadaleta E, Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of human genome sequence variation (2020 update). Nucleic Acids Res. 2020;48(W1):W185–W192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas PD, Ebert D, Muruganujan A, Mushayahama T, Albou LP, Mi H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022;31(1):8–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Z, Yuan Q, Mash DC, Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc Natl Acad Sci U S A. 2011;108(16):6626–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farris SP, Arasappan D, Hunicke-Smith S, Harris RA, Mayfield RD. Transcriptome organization for chronic alcohol abuse in human brain. Mol Psychiatry. 2015;20(11):1438–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapoor M, Wang JC, Farris SP, et al. Analysis of whole genome-transcriptomic organization in brain to identify genes associated with alcoholism. Transl Psychiatry. 2019;9(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hade AC, Philips MA, Reimann E, et al. Chronic Alcohol Use Induces Molecular Genetic Changes in the Dorsomedial Thalamus of People with Alcohol-Related Disorders. Brain Sci. 2021;11(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32(5):1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mamdani M, Williamson V, McMichael GO, et al. Integrating mRNA and miRNA Weighted Gene Co-Expression Networks with eQTLs in the Nucleus Accumbens of Subjects with Alcohol Dependence. PLoS One. 2015;10(9):e0137671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nudmamud-Thanoi S, Veerasakul S, Thanoi S. Pharmacogenetics of drug dependence: Polymorphisms of genes involved in GABA neurotransmission. Neurosci Lett. 2020;726:134463. [DOI] [PubMed] [Google Scholar]
- 45.Wise RA, Robble MA. Dopamine and Addiction. Annu Rev Psychol. 2020;71:79–106. [DOI] [PubMed] [Google Scholar]
- 46.Wittenberg RE, Wolfman SL, De Biasi M, Dani JA. Nicotinic acetylcholine receptors and nicotine addiction: A brief introduction. Neuropharmacology. 2020;177:108256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Icick R, Forget B, Cloez-Tayarani I, Pons S, Maskos U, Besson M. Genetic susceptibility to nicotine addiction: Advances and shortcomings in our understanding of the CHRNA5/A3/B4 gene cluster contribution. Neuropharmacology. 2020;177:108234. [DOI] [PubMed] [Google Scholar]
- 48.Chiamulera C, Piva A, Abraham WC. Glutamate receptors and metaplasticity in addiction. Curr Opin Pharmacol. 2021;56:39–45. [DOI] [PubMed] [Google Scholar]
- 49.Bennett KA, Sergeev E, MacSweeney CP, Bakker G, Cooper AE. Understanding Exposure-Receptor Occupancy Relationships for Metabotropic Glutamate Receptor 5 Negative Allosteric Modulators across a Range of Preclinical and Clinical Studies. J Pharmacol Exp Ther. 2021;377(1):157–168. [DOI] [PubMed] [Google Scholar]
- 50.Jung Y, Montel RA, Shen PH, Mash DC, Goldman D. Assessment of the Association of D2 Dopamine Receptor Gene and Reported Allele Frequencies With Alcohol Use Disorders: A Systematic Review and Meta-analysis. JAMA Netw Open. 2019;2(11):e1914940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


