Abstract
Identification of new potential drug target proteins and their plausible mechanisms for stroke treatment is critically needed. We previously showed that genetic deletion and short-term pharmacological inhibition of P2X4, a purinergic receptor for adenosine triphosphate (ATP), provides acute cerebroprotection. However, potential mechanisms remain unknown. Therefore, we employed RNA-Seq technology to identify the gene expression profiles and pathway analysis followed by qPCR validation of differentially expressed genes (DEGs). This analysis identified roles of DEGs in certain biological processes responsible for P2X4R-dependent cerebroprotection after stroke. We subjected both young and aged male and female global P2X4 receptor knock out (P2X4RKO) and littermate WT (WT) mice to ischemic stroke. After three days, mice were sacrificed, and total RNA was isolated using Trizol and subjected to RNA-Seq and NanoString-mediated qPCR. DESeq2, Gene Ontology (GO), and Ingenuity Pathway Analysis (IPA) were used to identify gene expression profiles and biological pathways. We found 2246 DEGs in P2X4R KO vs. WT tissue after stroke. Out of these DEGs, 1920 genes were downregulated and 325 genes were upregulated in P2X4R KO. GO/IPA analysis of the top 300 DEGs suggests an enrichment of inflammation and extracellular matrix component genes. qPCR validation of the top 30 DEGs revealed downregulation of two common age-independent genes in P2X4R KO mice: Interleukin-6 (Il-6), an inflammatory cytokine, and Cytotoxic T Lymphocyte-Associated Protein 2 alpha (Ctla2a), an immunosuppressive factor. These data suggest that P2X4R-mediated cerebroprotection after stroke is initiated by attenuation of immune modulatory pathways in both young and aged mice of both sexes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11302-023-09956-9.
Keywords: Ischemic stroke, Purinergic receptor P2X4, RNA-Seq analysis, Cerebroprotection, Inflammation
Introduction
Ischemic stroke is the leading cause of disability in the United States. It is a vascular accident that occurs, often without warning, when there is a blockage of cerebral blood flow, and results in damage to the surrounding brain tissue [1]. Despite recent advances, interventions to reduce damage and increase recovery after stroke are limited. Therapies, such as physical, occupational, and speech, are the only currently viable options available for post-stroke recovery [2]. This prompted us to explore pathophysiological events and mechanisms of injury to identify novel drug targets and their downstream pathways. Acute ischemic stroke triggers a series of events: rapid activation of resident microglia, release of chemokines and cytokines in the circulation, mobilization of immune cells from bone marrow, and the infiltration of circulating immune cells in the brain, primarily to phagocytose dead cells [3]. However, excessive infiltration of these immune cells into the ischemic region creates unwanted inflammation during the acute stroke phase by releasing inflammatory cytokines which leads to secondary damage after stroke, thus delaying recovery [4]. Among infiltrating cells, migration of peripheral myeloid cells (i.e., monocytes and neutrophils) to the Central Nervous System (CNS) and their temporal contribution to ischemic injury is a well-known immune response. Peripheral (e.g., monocytes/macrophages) and CNS-resident (microglia) myeloid cells contribute to both acute injury and functional recovery after stroke [5]. Despite the crucial participation of myeloid cells in stroke outcomes, approaches to regulate myeloid cell response remain unsuccessful due to their complex function including both cerebroprotective and damaging roles in stroke.
Myeloid cell function in stroke injury and recovery is further compounded by effects of aging. With age, the immune system has reduced lymphoid cells but increases in myeloid progenitor cells by hematopoietic stem cells [6]. Despite increases in their total systemic number, myeloid cells from aged mice are not as robust as their young counterparts and show diminished phagocytosis, reduced production of cerebroprotective secretory molecules, and increased secretion of inflammatory mediators [7], all of which are likely to influence stroke outcome. We previously showed that aged mice exhibit marked differences in the composition of circulating and infiltrating leukocyte recruitment after stroke [7]. Aged microglia also show increased sensitivity to ATP [8], but whether this contributes to stroke outcomes is unclear. While aging appears to play a role in myeloid cell function in stroke, the mechanism by which it does so remains to be explored.
P2X4, a purinergic receptor for ATP, plays a significant role in modulating synaptic transmission and communication between neurons and neighboring glial cells in the CNS [11]. In the immune system, our previous work revealed that the acute overactivation of P2X4 exacerbated post-ischemic inflammation and hampered recovery [9 Using short-term pharmacological blockade and genetic deletion, we showed that P2X4R can be a potential drug target for the treatment of acute ischemic stroke [10]. We and others have shown that myeloid cells express the highest amount of P2X4R and can modulate neuroinflammation mediated by chemokines, cytokines, and secondary activation of other immune cells such as T cells [10, 12, 13]. To date, however, the molecular mechanisms by which P2X4 impacts many myeloid cell functions, and how this contributes to overall stroke outcomes, has yet to be determined.
Towards that end, we hypothesized that P2X4R would be more activated in aged mice compared to young mice, and that this increased activation in ischemic stroke would show more pronounced neuroinflammatory cytokine/chemokine gene expression. We also hypothesized that there would be some age independent DEGs contributing to cerebroprotective or cerebrodetrimental effects. In this study, we tested this hypothesis by directly comparing P2X4R response in young and aged mice after stroke. We used bulk RNA-Sequencing (RNA-Seq) analysis using ischemic brain tissue from P2X4R KO and WT mice to identify common pathways and molecular mediators of damage and recovery and NanoString-mediated qPCR validation to confirm those findings. Transcriptomic analysis, which involves measuring the expression levels of thousands of genes simultaneously, provides a comprehensive view of the molecular changes that occur after ischemic stroke and allows for the identification of DEGs that may be involved in the pathophysiology of the disease [14].
The use of transcriptomic analysis in this study can helped us to identify specific genes and pathways that are affected by the knockout of P2X4R and shed light on the potential mechanisms by which P2X4R may be involved in ischemic stroke. This information can be used to develop targeted therapies that specifically modulate the expression or activity of these genes and pathways.
Methods
Animals and diets
We used both young (2–3-month-old) and aged (16–18-month-old) global P2X4 receptor knockout (P2X4R KO) and littermate control (WT) mice of both sexes, generated in-house (C57BL/6 background) at the UConn Health animal facility as published previously [9]. Briefly, P2X4Rfl/fl (RRID:MGI:7,491,919) and HprtCre (RRID:IMSR_JAX:004302) were bred to get global P2X4R KO and littermate WT mice. Mice were fed a standard chow diet and water ad libitum. Standard housing conditions were maintained at a controlled temperature with a 12 h light/dark cycle. All experiments were approved by the Institutional Animal Care and Use Committee of University of Connecticut Health and conducted in accordance with the U.S. National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Experimental design and stroke surgery
A total of 28 young and 26 aged mice (containing both males and females) were used in this study. Of 54 mice, 32 mice were subjected to a transient right middle cerebral artery occlusion (MCAo) for 60 min followed by three days of reperfusion as described previously [9, 10], while 10 mice (4 young and 6 aged) were subjected to sham surgery and the remaining 12 naïve young mice were used for bone marrow cell isolation. For MCAo surgery, a midline ventral neck incision was made to expose the right common carotid artery which was followed by isolation of both the external and internal carotid artery. A unilateral right MCAo was achieved under isoflurane anesthesia by inserting a 6.0 silicone rubber-coated monofilament (size 602,145/602,245/602,345; Doccol Corporation, Sharon, MA) ~ 10–11 mm away from the bifurcation point of the internal carotid artery through an external carotid artery stump. After 60 min of occlusion, monofilament was removed to perfuse the brain tissue for three days before sacrifice. A local injection of bupivacaine (8 mg/kg max) was given at the site of surgery to relieve pain. We have earlier shown that three days post-MCAo represents the peak time point [7, 9, 10] for post stroke inflammation. Further, this time point showed fully matured infarction, which led us to select three days post-MCAo for our experiments. Rectal temperatures were monitored and maintained at 37 ± 0.5 °C with the help of a heating pad. We used laser Doppler flowmetry (DRT 4/Moor Instruments Ltd, Devon, UK) to measure cerebral blood flow and to confirm occlusion (reduction to 15% of baseline cerebral blood flow). Four aged mice (3 WT and 1 P2X4R KO) died after stroke so final data includes 50 mice.
Brain tissue isolation and total RNA extraction
Three days after MCAo, all the mice were sacrificed, and perilesional prefrontal cortex tissue (determined with the help of Triphenyl tetrazolium chloride stain on consecutive brain sections) was isolated from the ischemic hemisphere to extract total RNA using Trizol. We used 500 ng and 200 ng RNA from each sample for RNA-Seq sample preparations (Illumina Hi Seq, Yale Center for Genome Analysis, New Haven, CT) and the NanoString platform (IDDRC molecular genetics core at Boston Children’s Hospital, Boston, MA), respectively.
Bone marrow-derived monocyte (BMDM) isolation
After sacrifice, P2X4R KO and littermate WT mice (8–10 weeks old) were sacrificed and bones (femur and tibia) were harvested after careful dissection in aseptic conditions. The harvested bone was cut from both ends and the internal lumen was flushed with RPMI1640 medium into a collecting tube. Following red blood cell lysis, single cell suspension was poured into a culture dish and fed with 10 ng/ml Granulocyte-Macrophage Colony-Stimulating Factor (GM-CSF) to grow for 7–10 days until they differentiate into macrophages. Mature macrophages can be visualized as more adherent, heterogeneously shaped, and as slightly larger granulated cells, which can also be confirmed by F4/80 staining (not shown here). Mature macrophages were harvested at 10 days after initial seeding to isolate total RNA using Trizol method. A total of 200 ng of RNA from each sample was shipped to IDDRC molecular genetics core at Boston Children’s Hospital in Boston, MA, to complete NanoString qPCR validation.
RNA-seq analysis
Quality control of RNA
Total RNA quality was determined by estimating the A260/A280 and A260/A230 ratios andRNA integrity number (RIN) using a nanodrop and Agilent Bioanalyzer gel respectively. Absorbance ratios provide information about RNA purity while RIN assess the integrity of total RNA samples.
RNA-seq library preparation
mRNA was purified from approximately 500 ng of total RNA with oligo-dT beads and sheared by incubation at 94oC. Following first-strand synthesis with random primers, second strand synthesis was performed with dUTP for generating strand-specific sequencing libraries. The cDNA library was then end-repaired, A-tailed, adapters were ligated, and second-strand digestion was performed by Uracil-DNA-Glycosylase. Indexed libraries that met appropriate cut-offs for both were quantified by qRT-PCR using a commercially available kit (KAPA Biosystems) and insert size distribution determined with the LabChip GX or Agilent Bioanalyzer. Samples with a yield of ≥ 0.5 ng/µl were used for sequencing.
Flow cell preparation and sequencing
Sample concentrations were normalized to 10 nM and loaded onto Illumina HiSeq4000 flow cells at a concentration that yields 300 million passing filter clusters per lane. Samples were sequenced using 100 bp paired-end sequencing according to Illumina protocols. The 8 bp index was read during an additional sequencing read that automatically follows the completion of read 1. Data generated during sequencing runs were simultaneously transferred to the YCGA high-performance computing cluster (Yale). A positive control (prepared bacteriophage Phi X library) provided by Illumina was spiked into every lane at a concentration of 0.3% to monitor sequencing quality in real time.
Primary RNA-seq analysis and storage
Signal intensities were converted to individual base calls during a run using the system’s Real Time Analysis (RTA) software. Base calls were transferred from the machine’s dedicated personal computer to the High-Performance Computing cluster at Yale Center for Research Computing (New Haven, CT) via a 1 Gigabit network mount for downstream analysis.
Secondary RNA-seq data analysis
Our analysis was carried out as follows. The reads were trimmed for quality using custom scripts. The minimum length accepted was 45 bases. The trimmed reads were then aligned to the mm10 reference genome using gencode annotation (Frankish et al., 2019) using HISAT2 for alignment, and StringTie for transcript abundance estimation (Kim et al., 2015). The generated counts were processed with DESeq2 (Love et al., 2014) in R to determine significantly expressed genes. Internally, the DESEq2 uses Wald’s test for each gene to determine if the log fold change is statistically significant. Wald’s test itself is a variation of χ2 test (this is one reason why one needs to use the multiple hypothesis testing correction to determine truly statistically significant genes). For the current analysis, we used adjusted p-value < 0.05 (effectively accepting 5% false discovery rate). The adjustment applied to the p-value is one of the mildest corrections.
Differentially expressed gene (DEG) analysis
Fragments per kilobase of exon model per million mapped reads (FPKM) is the most commonly used method for measuring gene expression levels (Mortazavi et al., 2008). The gene expression level of each sample was analyzed by HTSeq software (Princeton University, USA), using union as the counting model. We used a cutoff value of FPKM > 1 to define the gene expression. DEG analysis was performed using DESeq (Anders and Huber, 2010) (Bioconductor, USA). Genes with differential expression were screened and hierarchical clustering analysis was performed.
Gene ontology (GO) analysis
Gene ontology analysis was performed on the sets of top 10% up and down regulated genes in P2X4R KO mice from RNA-Seq data after stroke. Using the GO database (http://www.geneontology.org/), biological processes with an FDR threshold of ≤ 0.05 were filtered and biological process-related categories were selected and grouped by hierarchy [15].
Pathway and network analysis by ingenuity pathway analysis (IPA)
Similar to GO analysis, a list of DEGs (top 300) containing gene identifiers and corresponding expression values were uploaded into the IPA software (Qiagen) [16]. The “core analysis” function included in the software was used to interpret the differentially expressed data, which included biological processes, canonical pathways, upstream transcriptional regulators, and gene networks. We used two different metrics to identify the most important downstream effects of DEGs: activation z-score and p-value. A negative z-score (Blue bars) indicates pathway inhibition in P2X4R KO vs. WT. and a positive z-score (orange bar) indicates pathway activation. The p-value, calculated with the Fischer’s exact test, indicates the likelihood that the association between a set of genes in our dataset and a biological function is significant. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathway Knowledge Base (IPKB).
QPCR validation of top DEGs by NanoString technology
A total of 200 ng of RNA per sample was sent to IDDRC Molecular Genetics Core Facility at Boston Children’s Hospital in Boston, MA, for gene expression profiling using the nCounter platform and custom Codeset of the 30 top DEGs and housekeeping genes (Table 1). Prior to qPCR, quality control of each sample was performed. Data was normalized as per manufacturer’s instructions by using the nSolver Software (NanoString Technologies) and used for differential expression as in [10].
Table 1.
RNA-Seq Metrics for quality control
| Samples | nReads | p.Unique | p.Aln | n.Unique | p.Coding. UTR | p.Intronic | p.Ribo | p.Intergenic |
|---|---|---|---|---|---|---|---|---|
| WT1 | 37,281,768 | 67.65% | 86.72% | 25,218,490 | 76.93% | 11.89% | 2.22% | 8.99% |
| WT2 | 52,533,142 | 66.41% | 86.83% | 34,881,254 | 67.07% | 16.46% | 2.38% | 14.11% |
| WT3 | 41,843,688 | 73.13% | 90.13% | 30,595,240 | 76.50% | 11.71% | 1.38% | 10.43% |
| KO1 | 49,133,748 | 65.67% | 85.76% | 32,259,060 | 68.85% | 15.61% | 3.37% | 12.21% |
| KO2 | 48,974,522 | 70.05% | 87.96% | 34,301,356 | 71.52% | 16.01% | 2.54% | 9.98% |
| KO3 | 50,459,042 | 67.94% | 87.70% | 34,277,640 | 72.12% | 13.33% | 1.83% | 12.74% |
Results
We performed RNA-Seq using total RNA isolated from perilesional ipsilateral cortex of young P2X4R KO (2 M + 1 F) and WT mice (2 M + 1 F). After quality filtering and normalizing the raw sequencing data, we identified DEGs based on the following criteria: fold change > 2 or < 0.5, and false discovery rate (FDR) < 0.05. To understand the molecular and cellular impacts of these DEGs, we analyzed the top 300 genes (consisting of both up- and down-regulated genes) among P2X4R KO vs. WT mice after stroke using GO and IPA analysis. Among the top gene candidates, we validated about 30 biologically relevant genes using NanoString technology.
Quality control of RNA-seq data
To ensure the quality, reliability, and accuracy of downstream analyses, the following common parameters were used by our data analyst and therefore reported with data: nReads, p.Unique, p.Aln, n.Unique, p.Coding.UTR, p.Intronic, p.Ribo, p.Intergenic, etc. (Table 1). These parameters are derived from different aspects of the sequencing process. Parameter definitions are provided in Supplementary File 1.
DEGs identification and enrichment
Compared with WT control, 2245 genes were differentially expressed in the P2X4R KO group, with FDR (p-adjusted) value < 0.05, including 325 up-regulated genes and 1920 down-regulated genes (Supplementary File 1). The top 300 DEGs were then subjected to GO and IPA analysis for pathway enrichment and network analysis. These analyses helped us to obtain a comprehensive understanding of the functional significance, underlying pathways, and regulatory networks associated with these DEGs, enabling further exploration of the biological mechanisms involved in the studied condition. Other comparisons between Sham P2X4R KO and WT, and sham and stroke are given in Supplementary File 1.
GO analysis
GO analysis is a widely used approach to annotate DEG function. It helps to understand biological Interpretation and biological Insights such as key pathways involved to identify potential underlying mechanisms or regulatory networks. It also helps us to generate hypotheses. Using the top 300 DEGs, we identified 17 significantly enriched GO terms (Table 2) with 11 related to biological processes and 6 related to molecular functions. The top hit in the biological process and molecular function category were related to extracellular matrix, cellular origination, angiogenesis, and inflammatory response function (Fig. 1). These data suggest that P2X4R may be involved in these processes after stroke, which highlighted its important role in other biological process apart from inflammation.
Table 2.
Results of the GO enrichment analysis
| Name of the process | Description and GO accession | No of DEG | Fold enrichment | FDR | Total read |
|---|---|---|---|---|---|
| Biological process | extracellular matrix organization (GO:0030198) | 24 | 7.72 | 7.97e-10 | 291 |
| Biological process | regulation of cellular component movement (GO:0051270) | 40 | 3.16 | 5.29e-07 | 291 |
| Biological process | blood vessel morphogenesis (GO:0048514) | 24 | 4.83 | 9.09e-07 | 291 |
| Biological process | positive regulation of angiogenesis (GO:0045766) | 15 | 7.58 | 3.97e-06 | 291 |
| Biological process | inflammatory response (GO:0006954) | 23 | 4.1 | 1.48e-05 | 291 |
| Biological process | cell adhesion (GO:0007155) | 31 | 3.17 | 1.51e-05 | 291 |
| Biological process | RNA processing (GO:0006396) | 0 | < 0.01 | 2.20e-02 | 291 |
| Biological process | RNA metabolic process (GO:0016070) | 3 | 0.21 | 4.38e-02 | 291 |
| Biological process | sensory perception of smell (GO:0007608) | 1 | 0.08 | 1.10e-02 | 291 |
| Biological process | sensory perception of chemical stimulus (GO:0007606) | 1 | 0.07 | 4.10e-03 | 291 |
| Biological process | sensory perception (GO:0007600) | 5 | 0.27 | 3.08e-02 | 291 |
| Molecular function | extracellular matrix structural constituent (GO:0005201) | 16 | 9.54 | 2.57e-07 | 291 |
| Molecular function | calcium ion binding (GO:0005509) | 27 | 3.81 | 1.37e-05 | 291 |
| Molecular function | integrin binding (GO:0005178) | 12 | 6.97 | 5.61e-04 | 291 |
| Molecular function | glycosaminoglycan binding (GO:0005539) | 14 | 5.35 | 9.12e-04 | 291 |
| Molecular function | collagen binding (GO:0005518) | 8 | 8.94 | 4.85e-03 | 291 |
| Molecular function | immune receptor activity (GO:0140375) | 10 | 6.62 | 4.94e-03 | 291 |
Fig. 1.
Gene ontology (GO) enrichment analysis for top 300 DEGs (A) Biological Process and (B) Molecular Function. Each pie displays the distribution of DEGs according to biological process and molecular function. The top 6 significantly enriched biological processes and molecular functions are shown along with the p-value after FDR and number inside the Pie suggest number of DEGs contributing those pathways. The enriched process and functions related to immunomodulation (inflammatory response, calcium binding, immune receptor activity and angiogenesis) are marked in bold in each case. The GO enrichment analysis was performed using the PANTHER Classification System
Networks analysis of candidate genes by IPA
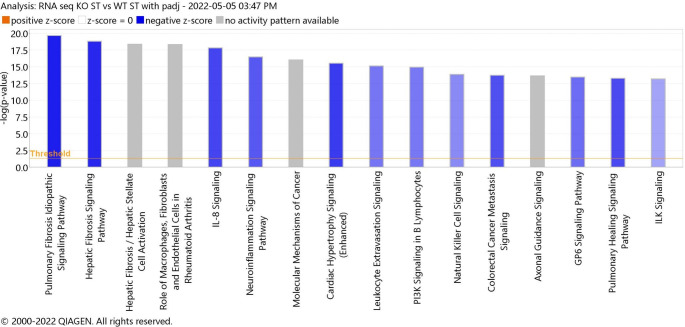
We subjected the top 300 DEGs to IPA gene network analysis. IPA explores the set of input genes to generate networks by using Ingenuity Pathway Knowledge Base for interactions between genes (Fig. 2). Analysis of the top 16 pathways identified that P2X4R regulates several molecular functions and biological processes, a result similar to the GO analysis outcome (Fig. 1). Among these, neuroinflammatory signaling pathways and immune cell activation were prominent, but already well-known pathways associated with P2X4R activity. Biological processes such as angiogenesis, however, were identified, but have not been previously characterized in relation to P2X4R. This data warrants further study into the role of P2X4R in angiogenesis.
Fig. 2.
Histogram of the top sixteen canonical pathways (p < 0.05) for young P2X4R KO vs. WT stroke group. Ingenuity Pathway Analysis (IPA) of top 16 canonical pathways for each genotype. The most statistically significant canonical pathways identified in the brain tissue are listed according to their p value (− log; orange line). Blue bars: negative z-score; orange bars: positive z-score; gray bars: no activity pattern available. The y–axis represent the -log (p-value)
qPCR validation of the 30 biologically relevant genes using NanoString
qPCR validation of 30 DEGs (obtained from RNA-Seq data) was carried out in the perilesional cortical tissue from both young and aged mice after stroke. In addition, we also used BMDMs for cell specific changes in these DEGs. Hierarchical clustering analysis of NanoString data of the 30 biologically relevant DEGs (Fig. 3) showed that most differentially regulated genes were related to extracellular matrix, cell-cell adhesion, and immunomodulatory pathways in young P2X4R KO and WT mice. Along with most significant DEGs, we also added other members of P2X receptor family in our analysis to observe the effect of loss of P2X4R on the expression of other P2X receptors. As suspected, we found that P2x7r expression was reduced in P2X4R KO mice. This data helped us to delineate the effect of passenger mutations in other family member genes [17, 18]. We also identified several significantly modulated inflammatory cytokines or chemokines in young brain tissue (Fig. 4a) and BMDMs (Fig. 4b) and aged brain tissue (Fig. 4c). These data found both common genes that are reduced after stroke in both young and aged mice (Ctla2a and Il-6), but also diverse inflammation related genes (transforming growth factor beta 1 Tgfb1, Mannose receptor 1 Mrc1 and purinergic receptor P2x7 in young, and Interleukin 1 beta Il-1b and Tissue necrosis factor Tnf in aged mice) that were reduced at different ages in P2X4R KO mice after stroke. Tissue RNAseq data represents mixed brain cells, and macrophages show highest P2X4R expression among various brain and immune cells [10]. Thus, we further explored the macrophage-specific differential expression on the top 30 DEGS using GM-CSF activated BMDMs from naïve P2X4R KO and WT mice. Among several validated DEGs, Mrc1 was a common gene between tissue and BMDMs data of young mice.
Fig. 3.
Heat map of top 30 DEGs hierarchical clustering analysis. This heat map represents expression data of top 30 immune related genes validated by NanoString panel using mouse brain tissue of young P2X4R KO (n = 2 M + 1 F) and WT mice (n = 2 M + 1 F) after stroke. The right 3 columns represent WT controls, and the left 3 columns represent P2X4R KO mice. Each row represents a single gene. The color change from yellow to black represents log (TPM) value ranging from high to low
Fig. 4.
qPCR validation of top 30 DEGs in various group. (A) show data of significantly different expressed genes from brain tissue of young P2X4 KO (2 M + 1 F) & WT mice (2 M + 1 F). (B) show data of significantly different expressed genes from GM-CSF stimulated BMDMs from P2X4R KO (3 M + 3 F) and WT (3 M + 3 F) mice. (C) show data of significantly different expressed gene from Aged P2X4 KO (n = 4 M + 4 F) & WT mice (n = 3 M + 5 F).*p < 0.05 vs. WT [ unpaired t-test, multiple comparison test with FDR (5% probability) followed by two-stage step-up (Benjamini, Krieger, and Yekutieli) method]. Data are shown mean ± S.D and analyzed by graph pad prism version 9.5
Discussion
Our previous study demonstrated that Global P2X4R KO protects mice from further damage after ischemic stroke, yet the underlying molecular networks associated with P2X4R in the neuro-inflammatory response to cortical injury due to ischemic stroke remain unexplored [9]. We used next-generation RNA-Seq to analyze gene expression profiles generated from P2X4R KO and littermate WT mice to identify potential biological and molecular function pathways and related signaling networks in the absence of P2X4R during acute neuro-inflammation after ischemic stroke injury using transient MCAo mouse model. Here, we have identified many up- and down-regulated genes, revealing that the absence of P2X4R has widespread effects after ischemic stroke. Gene ontology (GO) enrichment analysis of DEGs suggest many biological processes such as cellular integrity, organization, positive regulation of angiogenesis, and inflammatory response modulation are among the top biological processes which are affected by the absence of P2X4R. The dependence of these biological processes on P2X4R was further supported by molecular function pathways by GO analysis and canonical pathways by IPA. These two independent analysis platforms suggest that immune cell activation/neuroinflammation and leukocyte extravasation pathways/function are dominantly altered by the absence of P2X4R. We and others have previously shown that P2X4R activation exacerbates inflammation and acute ischemic injury [3, 5, 10]. We previously showed that short-term blockade of P2X4R reduces acute ischemic injury by reducing neuroinflammation and by reducing leukocyte infiltration into the brain after stroke [10]. These data also suggest that P2X4R plays a significant role in blood-brain barrier (BBB) integrity, either by activation of P2X4R on myeloid cells or its interaction with endothelial cells. However, it is not yet clear if these effects on BBB permeability are mediated by myeloid or endothelial P2X4R activation [10, 13]. Our qPCR data with BMDM cells suggest that the absence of P2X4R increases genes like Ct1a2a, Lox, and Loxl2. These genes are involved in rebuilding of ECM by cross-linking collagen and elastin fibers, which can improve the brain’s structural integrity and support its recovery following a stroke. It is well established that ECM integrity is compromised after stroke [19]. These findings support the notion that myeloid P2X4R activation might play a major role in ECM degradation and cell to cell adhesion, thus may affect BBB integrity.
Our qPCR data validated data from young, stroked mice show that the absence of P2X4R reduced several inflammatory genes like P2x7r, Tgfb1, and Mrc1. Among them, MRC1, which is a member of the C-type lectin (CLEC) family of mannose receptor, has diverse roles and can bind and internalize a variety of endogenous and pathogen-associated ligands [20]. Because of these properties, its role in endocytosis and antigen processing and presentation has been studied intensively and is consistent with the role of P2X4R in endocytosis [21]. Our data suggest that the endocytosis roles of P2X4R might be mediated by MRC1. Besides endocytosis, it can also directly influence the activation of various immune cells by its expression during ischemic stroke [21]. Both ischemic brain tissue in young mice and BMDM data show significant decrease in Mrc1 genes in P2X4R KO group, indicating that cerebroprotective effects of P2X4R blockade might be mediated by MRC1-mediated immune cell activation. MRC1 also has a regulatory effect on the induction of immune responses by regulating T cell activation, a function distinct from antigen uptake and presentation [23]. This regulatory effect of the MRC1 was mediated by a direct interaction with CD45 on the T cell, inhibiting its phosphatase activity, which resulted in up-regulation of cytotoxic T-lymphocyte-associated protein CTLA-2a. Given that the mannose receptor plays an important role in phagocytosis and clearance of cellular debris [24], it will be worthwhile to study how P2X4R blockade affects phagocytic uptake and T-cell activation after ischemic stroke. Our data suggest that MRC1-P2X4R may play important diverse roles during acute ischemic injury. Interestingly, our data found an age-independent decrease in expression of genes Ctla2a in P2X4R KO mice after stroke. This finding suggests that the effects Mrc1 on T cells might be mediated by Ctla2a in a P2X4R-dependent manner persisting in aging.
CTLA-2a is a cysteine proteinase inhibitor protein which was originally discovered in mouse-activated T cells and mast cells. Previously it has been shown that T cell activation is detrimental during acute ischemia, and lymphocyte-deficient mice are protected in models of focal ischemia as discussed in detail in [3]. The cytotoxic activity of T cells may be related to innate functions of T cells. Further, P2X4R activation increases T cell activation and their migration to injured tissue [13]. This evidence suggests that loss/blockade of P2X4R in the brain reduces Ctla-2a expression on T cells to inhibit their activation and migration to ischemic tissue, which is consistent with IP analysis data. Ctla2a was reduced both in young and aged mice after stroke and stroke mainly occurs in aging population. This observation suggests that CTLA-2a may be a downstream target of P2X4R and can be a potential therapeutic target to treat ischemic stroke. Similar to Ctla-2a, we also found reduced expression of Il-6 after stroke in both young and aged P2X4R KO mice. Although the pro-inflammatory role of Il-6 is well-defined after acute stroke injury, reduced Il-6 levels may indicate cerebroprotective effects of P2X4R KO during early stroke injury timepoints. Although the exact mechanism of how P2X4R activates Il-6 is not clear, we and others have shown that pro-inflammatory effects of P2X4R activation might indirectly contribute to stroke injury [25]. We also want to highlight a potential limitation of this work due to small animal number (n = 3 per genotype) in our initial RNA-seq work, which may lead to type 1 or 2 statistical error. Nevertheless, qPCR validation works in BMDMs and aged mice were done in optimum number of animals to rule out such possibility of errors.
Taken together, our RNA-Seq data support the hypothesis that P2X4R modulates BBB integrity, inflammatory response, and immune cell activation and infiltration. This data also identifies two novel potential downstream targets of P2X4R during ischemic stroke: MRC1 and CTLA-2a. Data from this work support not only a key role for P2X4R in modulating the complex networks of cell death and the immune response in myeloid cells after ischemic stroke but also suggest a new role in T cell activation and a potential mechanism for this T cell activation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Authors’ contributions
RV and BL conceptualized the manuscript, DGT and CC performed the experiment and analyzed data. RV wrote the manuscript.
Funding
This work was supported American Heart Association grant 18CDA34110011 and NINDS grant 1R01NS125405 (to R.V.) and NHLBI grant 1R41HL156322 to B Liang.
Data Availability
Some of the raw data of RNA-Seq is provided in supplementary file and other data will be available on demand from corresponding author due process mandated by UConn Health from an authorized entity.
Declarations
Ethics approval and consent to participate
All the animal protocol was approved by Institutional Animal Care and Use Committee (IACUC) of UConn Health with Public Health Service (PHS) assurance number - A3471-01 (D16-00295).
Consent to participate
All the authors consented to participate.
Consent to publish
All the authors gave consent to publish.
Competing interests
No competing interests of any author
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Centers for Disease Control and Prevention. Underlying Cause of Death, 1999–2018. CDC WONDER Online Database. Centers for Disease Control and Prevention (2018) ; Accessed March 12, 2020
- 2.Dobkin BH, Dorsch A (2013 Jun) New evidence for therapies in stroke rehabilitation. Curr Atheroscler Rep 15(6):331. 10.1007/s11883-013-0331-y [DOI] [PMC free article] [PubMed]
- 3.Iadecola C, Buckwalter MS, Anrather J Immune responses to stroke: mechanisms, modulation, and therapeutic potential. J Clin Invest. 2020 Jun 1;130(6):2777–2788. doi: 10.1172/JCI135530 [DOI] [PMC free article] [PubMed]
- 4.Kim JY, Kawabori M, Yenari MA. Innate inflammatory responses in stroke: mechanisms and potential therapeutic targets. Curr Med Chem. 2014;21(18):2076–2097. doi: 10.2174/0929867321666131228205146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spiteri AG, Wishart CL, Pamphlett R, Locatelli G, King NJC (2022 Feb) Microglia and monocytes in inflammatory CNS disease: integrating phenotype and function. Acta Neuropathol 143(2):179–224. 10.1007/s00401-021-02384-2 [DOI] [PMC free article] [PubMed]
- 6.Montecino-Rodriguez E, Berent-Maoz B, Dorshkind K (2013 Mar) Causes, consequences, and reversal of immune system aging. J Clin Invest 123(3):958–965. 10.1172/JCI64096 [DOI] [PMC free article] [PubMed]
- 7.Ritzel RM, Lai YJ, Crapser JD, Patel AR, Schrecengost A, Grenier JM, Mancini NS, Patrizz A, Jellison ER, Morales-Scheihing D, Venna VR, Kofler JK, Liu F, Verma R, McCullough LD Aging alters the immunological response to ischemic stroke. Acta Neuropathol 2018 Jul;136(1):89–110. doi: 10.1007/s00401-018-1859-2 [DOI] [PMC free article] [PubMed]
- 8.Spittau B (2017 Jun) Aging Microglia-Phenotypes, functions and implications for age-related neurodegenerative Diseases. Front Aging Neurosci 14:9:194. 10.3389/fnagi.2017.00194 [DOI] [PMC free article] [PubMed]
- 9.Verma R, Cronin CG, Hudobenko J, Venna VR, McCullough LD, Liang BT (2017 Nov) Deletion of the P2X4 receptor is neuroprotective acutely, but induces a depressive phenotype during recovery from ischemic stroke. Brain Behav Immun 66:302–312. 10.1016/j.bbi.2017.07.155 [DOI] [PMC free article] [PubMed]
- 10.Srivastava P, Cronin CG, Scranton VL, Jacobson KA, Liang BT, Verma R (2020 Jul) Neuroprotective and neuro-rehabilitative effects of acute purinergic receptor P2X4 (P2X4R) blockade after ischemic stroke. Exp Neurol 329:113308. 10.1016/j.expneurol.2020.113308 [DOI] [PMC free article] [PubMed]
- 11.Stokes L, Layhadi JA, Bibic L, Dhuna K, Fountain SJ P2X4 receptor function in the nervous system and current breakthroughs in Pharmacology. Front Pharmacol 2017 May 23;8:291. doi: 10.3389/fphar.2017.00291 [DOI] [PMC free article] [PubMed]
- 12.Ulmann L, Hirbec H, Rassendren F P2X4 receptors mediate PGE2 release by tissue-resident macrophages and initiate inflammatory pain. EMBO J. 2010 Jul 21;29(14):2290 – 300. doi: 10.1038/emboj.2010.126 [DOI] [PMC free article] [PubMed]
- 13.Ledderose C, Liu K, Kondo Y, Slubowski CJ, Dertnig T, Denicoló S, Arbab M, Hubner J, Konrad K, Fakhari M, Lederer JA, Robson SC, Visner GA, Junger WG Purinergic P2X4 receptors and mitochondrial ATP production regulate T cell migration. J Clin Invest. 2018 Aug 1;128(8):3583–3594. doi: 10.1172/JCI120972 [DOI] [PMC free article] [PubMed]
- 14.Kim J, Kang SW, Mallilankaraman K, Baik SH, Lim JC, Balaganapathy P, She DT, Lok KZ, Fann DY, Thambiayah U, Tang SC, Stranahan AM, Dheen ST, Gelderblom M, Seet RC, Karamyan VT, Vemuganti R, Sobey CG, Mattson MP, Jo DG, Arumugam TV (2018) Transcriptome analysis reveals intermittent fasting-induced genetic changes in ischemic stroke. Hum Mol Genet. 2018 May 1;27(9):1497–1513. doi: 10.1093/hmg/ddy057. Erratum in: Hum Mol Genet. Jul 1;27(13):2405
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000 May) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25(1):25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed]
- 16.Krämer A, Green J, Pollard J Jr, Tugendreich S Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014 Feb 15;30(4):523 – 30. doi: 10.1093/bioinformatics/btt703. Epub 2013 Dec 13 [DOI] [PMC free article] [PubMed]
- 17.Er-Lukowiak M, Duan Y, Rassendren F, Ulmann L, Nicke A, Ufer F, Friese MA, Koch-Nolte F, Magnus T, Rissiek B A P2rx7 passenger mutation affects the vitality and function of T cells in congenic mice. iScience. 2020 Nov 27;23(12):101870. doi: 10.1016/j.isci.2020.101870 [DOI] [PMC free article] [PubMed]
- 18.Ellegaard M, Hegner T, Ding M, Ulmann L, Jørgensen NR (2021 Jun) Bone phenotype of P2X4 receptor knockout mice: implication of a P2X7 receptor mutation? Purinergic Signal. 17(2):241–246. 10.1007/s11302-021-09784-9. Epub 2021 Apr 15 [DOI] [PMC free article] [PubMed]
- 19.Romanic AM, White RF, Arleth AJ, Ohlstein EH, Barone FC (1998 May) Matrix metalloproteinase expression increases after cerebral focal ischemia in rats: inhibition of matrix metalloproteinase-9 reduces infarct size. Stroke 29(5):1020–1030. 10.1161/01.str.29.5.1020 [DOI] [PubMed]
- 20.Staines K, Hunt LG, Young JR, Butter C (2014) Evolution of an expanded mannose receptor gene family. PLoS One. Nov 12;9(11):e110330. doi: 10.1371/journal.pone.0110330 [DOI] [PMC free article] [PubMed]
- 21.Gaudet P, Livstone MS, Lewis SE, Thomas PD (2011 Sep) Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform 12(5):449–462. 10.1093/bib/bbr042Epub 2011 Aug 27. PMID: 21873635; PMCID: PMC3178059 [DOI] [PMC free article] [PubMed]
- 22.McKenzie EJ, Taylor PR, Stillion RJ, Lucas AD, Harris J, Gordon S, Martinez-Pomares L Mannose receptor expression and function define a new population of murine dendritic cells. J Immunol 2007 Apr 15;178(8):4975–83. doi: 10.4049/jimmunol.178.8.4975 [DOI] [PubMed]
- 23.Schuette V, Embgenbroich M, Ulas T, Welz M, Schulte-Schrepping J, Draffehn AM, Quast T, Koch K, Nehring M, König J, Zweynert A, Harms FL, Steiner N, Limmer A, Förster I, Berberich-Siebelt F, Knolle PA, Wohlleber D, Kolanus W, Beyer M, Schultze JL, Burgdorf S Mannose receptor induces T-cell tolerance via inhibition of CD45 and up-regulation of CTLA-4. Proc Natl Acad Sci U S A 2016 Sep 20;113(38):10649–54. doi: 10.1073/pnas.1605885113 [DOI] [PMC free article] [PubMed]
- 24.Gazi U, Martinez-Pomares L. Influence of the mannose receptor in host immune responses. Immunobiology. 2009;214(7):554–561. doi: 10.1016/j.imbio.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Ma J, Gao J, Niu M, Zhang X, Wang J, Xie A P2X4R overexpression Upregulates Interleukin-6 and exacerbates 6-OHDA-Induced Dopaminergic Degeneration in a rat model of PD. Front Aging Neurosci 2020 Nov 4;12:580068. doi: 10.3389/fnagi.2020.580068 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Some of the raw data of RNA-Seq is provided in supplementary file and other data will be available on demand from corresponding author due process mandated by UConn Health from an authorized entity.