Abstract
Knowledge of genetic structure at the finest level is essential for the conservation of genetic resources. Despite no visible barriers limiting gene flow, significant genetic structure has been shown in marine species. The common cockle (Cerastoderma edule) is a bivalve of great commercial and ecological value inhabiting the Northeast Atlantic Ocean. Previous population genomics studies demonstrated significant structure both across the Northeast Atlantic, but also within small geographic areas, highlighting the need to investigate fine-scale structuring. Here, we analysed two geographic areas that could represent opposite models of structure for the species: (1) the SW British Isles region, highly fragmented due to biogeographic barriers, and (2) Galicia (NW Spain), a putative homogeneous region. A total of 9250 SNPs genotyped by 2b-RAD on 599 individuals from 22 natural beds were used for the analysis. The entire SNP dataset mostly confirmed previous observations related to genetic diversity and differentiation; however, neutral and divergent SNP outlier datasets enabled disentangling physical barriers from abiotic environmental factors structuring both regions. While Galicia showed a homogeneous structure, the SW British Isles region was split into four reliable genetic regions related to oceanographic features and abiotic factors, such as sea surface salinity and temperature. The information gathered supports specific management policies of cockle resources in SW British and Galician regions also considering their particular socio-economic characteristics; further, these new data will be added to those recently reported in the Northeast Atlantic to define sustainable management actions across the whole distribution range of the species.
Subject terms: Genetic variation, Genetic variation, Evolutionary genetics
Introduction
Knowledge of genetic diversity distribution is crucial for the sustainable management and conservation of natural resources (Leary et al. 2009; Sa-Pinto et al. 2012). This distribution is affected by larval connectivity, demographic parameters and selective processes operating on species populations. Scarcity of physical barriers in marine environments is expected to promote higher connectivity among populations in comparison to terrestrial species (Waples 1998). Moreover, marine species usually show large population sizes, which along with pelagic larval stages, often lasting several weeks, facilitate population genetic homogenisation across wide regions (Sa-Pinto et al. 2012; do Prado et al. 2018). Despite these general features, genetic studies on marine organisms have frequently detected genetic differentiation, even at local scales (i.e., below the geographic scale of effective dispersal of the species studied, known as chaotic genetic patchiness; see Eldon et al. 2016), which can be explained by historical and reproductive/demographic factors (e.g., high fecundity and high mortality in early life stages, sweepstakes reproductive success; see Parrondo et al. 2022), natural selection associated with environmental conditions (Vilas et al. 2015; do Prado et al. 2018; Vera et al. 2019) and oceanic features such as residual currents, bathymetry, coastline shape, upwelling, fronts, gyres and eddies (Vera et al. 2016, 2022; Coscia et al. 2020; Handal et al. 2020; Fisher et al. 2022).
Different types of ocean fronts have been described across the Northeast Atlantic region, encompassing tidal mixing fronts, shelf break fronts, and freshwater fronts separating estuarine freshwater and higher salinity coastal waters (Sharples and Simpson 2019). Examples of these frontal systems on the NW European Shelf include the Celtic Sea Front (NE Celtic Sea), the Irish Sea Front (NW Irish Sea), the Alderney Race (with one of the strongest currents in Europe) and the Ushant Front (W English Channel) (Suberg et al. 2019). These fronts may influence genetic structure acting as barriers to cross-front planktonic dispersal and as conduits through along-front dispersal by frontal jets, with important influences on the pelagic distribution of larvae of marine species (Galarza et al. 2009). Biogeographical barriers can also limit dispersal in marine environments. In the Northeast Atlantic region, Cape Finisterre, the Cornwall Peninsula, the tip of Brittany, the Llyn Peninsula, and the Alderney race along Cotentin Peninsula have been identified as potential barriers to the connectivity of marine organisms due to their oceanographic features, including fish (Abaunza et al. 2008; Larmuseau et al. 2009) and molluscs (Dupont et al. 2007; Piñeira et al. 2008; Martínez et al. 2015; Handal et al. 2020; Vera et al. 2022).
The common cockle, Cerastoderma edule, is a bivalve mollusc naturally distributed throughout the Northeast Atlantic coast, from Senegal, West Africa, to Norway, northern Europe, where it inhabits on intertidal and shallow subtidal soft sediments (Hayward and Ryland 1995). The species is commercially exploited and provides a wealth of services to coastal communities mainly in Ireland, United Kingdom, France, Spain and Portugal, where it is harvested (Flach and de Bruin 1994; Carss et al. 2020; Jackson-Bue et al. 2022). Cockle harvest has been reduced since the 1980s (>100,000 tonnes) to nowadays (~25,000 tonnes in 2019) due to changes in fisheries policies, overfishing, variable recruitment and mass mortalities produced by pollution, climate events and parasites (Villalba et al. 2014; Mahony et al. 2020; Pampín et al. 2023). Furthermore, cockles are considered keystone for ecosystem due to their role as reef engineers, agents of carbon sequestration and their linking between primary producers and higher trophic levels (Norris et al. 1998; Carss et al. 2020). The species is dioecious and can live up to 10 years displaying fast sexual maturation (reached in the first year of life) and high fecundity (Honkoop and van der Meer 1998). The reproductive period occurs from April to August (Malham et al. 2012), but it can be extended to September in more southern European countries such as Portugal (Mahony et al. 2021), and planktonic larvae can remain in the water column for 30 days facilitating widespread dispersal (de Montaudouin et al. 2003; Dare et al. 2004).
Genetic studies throughout the natural cockle’s distribution have identified three main population genetic units: (i) a southern group encompassing the Atlantic coast from Morocco to the Bay of Biscay; (ii) a central group comprising of the Celtic and Irish Seas, the English Channel and the southern North Sea; and (iii) a northern group consisting of the northern North Sea (Beaumont et al. 1980; Hummel et al. 1994; Martínez et al. 2013, 2015). These results have been recently confirmed by Vera et al. (2022) through a wide genome scan (~10,000 single-nucleotide polymorphisms, SNPs), but additionally enabled identifying substructure within the main genetic groups using outlier loci under divergent selection, mostly in accordance with residual current patterns and environmental variables.
However, due to the limited number of markers and/or the scale of sample collection, a comprehensive picture of population connectivity in the common cockle is still incomplete. Information at the microgeographic level, always considering the dispersal capacity of the species (Eldon et al. 2016; Vera et al. 2022), is relevant for the management of fisheries (Bernatchez et al. 2017). Using a wide SNP genomic screening, Coscia et al. (2020) identified three genetic clusters (global FST = 0.021) of cockles in the Celtic and Irish seas and that could be associated with residual ocean currents, salinity and geographical proximity using information on larval dispersal.
This study aimed to analyse the genetic structure of the common cockle at a microgeographic scale using 2b Restriction Associated DNA sequencing (2b-RADseq). Two regions were investigated: (1) the SW British Isles and the English Channel, characterised by putative habitat fragmentation due to tidal mixing fronts and biogeographical barriers; and (2) the Northwest coast of Spain (Galicia), representing a quite homogeneous region according to previous information on other mollusc species (Diz and Presa 2009; Vera et al. 2016). The results confirmed the significant differentiation of cockles’ populations at microgeographic scale, but also the power of larval dispersal to homogenise rather wide coastal areas, thus providing essential information for proper management of this valuable resource.
Material and methods
Sample area and oceanography
Two geographic areas along the Northeast Atlantic coast were investigated (Fig. 1). The first was focused on the British Isles and the English Channel (hereafter called the SW British Isles region), where previous, though incomplete information, supported significant genetic sub-structuring (e.g., Coscia et al. 2020; Vera et al. 2022). The second area was Galicia (Northwest Spain), which may be genetically homogeneous according to information on other mollusc species (Diz and Presa 2009; Vera et al. 2016).
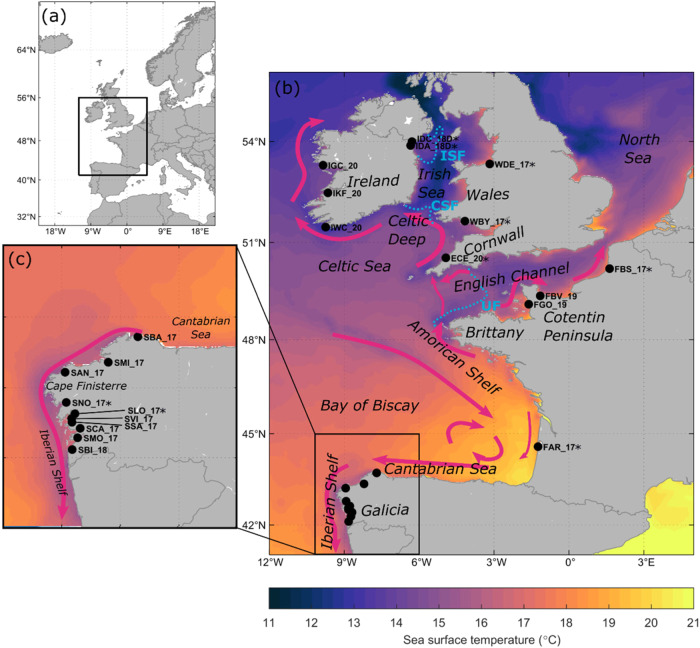
Fig. 1. Geographical distribution of the Cerastoderma edule beds analysed in the present study.
Location of the study area in Europe (a) and focused on SW British Isles (b) and Galicia (c) regions. Summer sea surface temperatures are shaded. Summer sea surface ocean currents are schematically depicted with magenta-coloured arrows. Tidal mixing fronts are indicated with purple dashed lines. UF Ushant Front, CSF Celtic Sea Front, ISF Irish Sea Front. Location codes are shown in Table 1. Beds previously analysed by Vera et al. (2022) are marked with asterisks.
Over the cockle reproductive season (May to September; Mahony et al. 2020), the coastline of Galicia is characterised by wind-driven upwelling of cold waters resulting in sea surface temperatures (SSTs) that are several degrees colder than off-shore SSTs (Supplementary Fig. 1b). Also driven by the predominantly northerly winds in the summer months, the Portugal coastal current transports waters southwards along the coastline of Iberia (Teles-Machado et al. 2016) with residual current strengths along the Galician coastline exceeding 0.15 m/s (Supplementary Fig. 1d). The SW British Isles region is divided into distinct oceanographic regions (the English Channel, the Celtic Deep, the Celtic Sea and the Irish Sea) by diverging current or seasonal frontal systems (Galparsoro et al. 2014). Several tidal mixing fronts separate seasonally stratified and mixed waters (Supplementary Fig. 1a): the Ushant Front (Group “Grepma” 1988), the Celtic Sea Front, and the Irish Sea Front (Simpson and Pingree 1978). The Celtic Sea is characterised by northward flow along the western coast of Cornwall, which merges into the Celtic Sea Front jet and links into the Irish Coastal Current, which transports water clockwise along the south and west coast of Ireland (Supplementary Fig. 1c; Brown et al. 2003; Fernand et al. 2006). Northward currents along the Ushant Front link the American Shelf with the Celtic Sea. The southern English Channel coast is dominated by northeastward flow, with the strongest currents occurring around the Cotentin Peninsula.
Sample collection
A total of 374 cockles from 14 wild natural beds were collected across the aforementioned two regions in the period 2017–2020 and stored in 100% ethanol for analyses (Table 1). In addition, 231 cockles from eight beds previously analysed (Vera et al. 2022: identified as IDA_18, IDC_18, WDE_17, WBY_17, FBS_17, FAR_17, SNO_17 and SLO_17, where 17 and 18 in the codes represent 2017 and 2018, respectively) were included in the analysis to achieve a comprehensive picture of the areas studied, thus providing an overall total of 605 cockles. To avoid generation overlapping, all samples belonged to the 0+ year age class of their sampling year. No temporal replicates were included considering the temporal genetic stability previously reported by Vera et al. (2022).
Table 1.
Cerastoderma edule beds analysed in the present study.
| All dataset | Polymorphic loci (MAF >0.017) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | Year | Lat (deg N) | Lon (deg E) | Code | Country | N initial | N | Ho | He | FIS | Polymorphic loci | % Polymorphic loci | Ho | He | FIS |
| Galway Connemara | 2020 | 53.306 | −9.846 | IGC_20 | Ireland | 27 | 27 | 0.080 | 0.085 | 0.067 | 4205 | 45.5 | 0.166 | 0.179 | 0.074 |
| Kerry-Feale | 2020 | 52.488 | −9.652 | IKF_20 | Ireland | 30 | 30 | 0.080 | 0.087 | 0.080 | 4362 | 47.2 | 0.161 | 0.176 | 0.088 |
| West Cork-Cockle Beach | 2020 | 51.463 | −9.744 | IWC_20 | Ireland | 30 | 30 | 0.080 | 0.085 | 0.064 | 4294 | 46.4 | 0.163 | 0.175 | 0.070 |
| Dundalk Bay-Annagassan | 2018 | 53.884 | −6.341 | IDA_18 | Ireland | 29 | 29 | 0.074 | 0.080 | 0.082 | 2993 | 32.4 | 0.161 | 0.177 | 0.090 |
| Dundalk Bay-Cooley | 2018 | 53.996 | −6.287 | IDC_18 | Ireland | 22 | 22 | 0.077 | 0.083 | 0.081 | 3493 | 37.8 | 0.173 | 0.190 | 0.087 |
| Dee Estuary | 2017 | 53.343 | −3.174 | WDE_17 | Wales | 30 | 28 | 0.071 | 0.076 | 0.074 | 2561 | 27.7 | 0.167 | 0.182 | 0.084 |
| Burry | 2017 | 51.643 | −4.166 | WBY_17 | Wales | 30 | 30 | 0.073 | 0.080 | 0.091 | 3529 | 38.2 | 0.148 | 0.165 | 0.101 |
| Camel Estuary (Cornwall) | 2020 | 50.531 | −4.930 | ECE_20 | England | 24 | 24 | 0.073 | 0.081 | 0.105 | 3667 | 39.6 | 0.159 | 0.180 | 0.114 |
| Somme Bay | 2017 | 50.201 | 1.627 | FBS_17 | France | 30 | 30 | 0.071 | 0.080 | 0.111 | 3438 | 37.2 | 0.147 | 0.167 | 0.119 |
| Baie des Veys (Brévands) | 2019 | 49.365 | −1.150 | FBV_19 | France | 26 | 26 | 0.079 | 0.081 | 0.022 | 3579 | 38.7 | 0.169 | 0.174 | 0.030 |
| Gouville sur mer | 2019 | 49.105 | −1.612 | FGO_19 | France | 23 | 23 | 0.077 | 0.082 | 0.059 | 4055 | 43.8 | 0.163 | 0.174 | 0.063 |
| Arcachon Bay | 2017 | 44.580 | −1.238 | FAR_17 | France | 30 | 30 | 0.074 | 0.083 | 0.111 | 4335 | 46.9 | 0.140 | 0.159 | 0.120 |
| O Barqueiro | 2017 | 43.722 | −7.701 | SBA_17 | Spain | 30 | 30 | 0.076 | 0.085 | 0.107 | 4595 | 49.7 | 0.140 | 0.158 | 0.115 |
| Miño | 2017 | 43.361 | −8.206 | SMI_17 | Spain | 30 | 30 | 0.073 | 0.081 | 0.102 | 4159 | 45.0 | 0.140 | 0.157 | 0.110 |
| Anllóns | 2017 | 43.220 | −8.943 | SAN_17 | Spain | 30 | 29 | 0.073 | 0.081 | 0.101 | 3526 | 38.1 | 0.143 | 0.161 | 0.110 |
| Ría de Noia | 2017 | 42.790 | −8.923 | SNO_17 | Spain | 30 | 30 | 0.078 | 0.087 | 0.099 | 4885 | 52.8 | 0.139 | 0.156 | 0.107 |
| Lombos do Ulla | 2017 | 42.629 | −8.775 | SLO_17 | Spain | 30 | 30 | 0.075 | 0.085 | 0.113 | 4602 | 49.8 | 0.137 | 0.156 | 0.120 |
| Sarrido | 2017 | 42.507 | −8.826 | SSA_17 | Spain | 30 | 30 | 0.074 | 0.083 | 0.103 | 4317 | 46.7 | 0.140 | 0.158 | 0.112 |
| Vilanova | 2017 | 42.561 | −8.831 | SVI_17 | Spain | 27 | 25 | 0.072 | 0.081 | 0.112 | 3175 | 34.3 | 0.153 | 0.173 | 0.118 |
| Campelo | 2017 | 42.421 | −8.685 | SCA_17 | Spain | 30 | 30 | 0.073 | 0.082 | 0.115 | 4287 | 46.3 | 0.140 | 0.159 | 0.121 |
| Moaña | 2017 | 42.286 | −8.730 | SMO_17 | Spain | 20 | 19 | 0.070 | 0.077 | 0.088 | 2361 | 25.5 | 0.181 | 0.200 | 0.093 |
| Baiona | 2018 | 42.117 | −8.822 | SBI_18 | Spain | 17 | 17 | 0.075 | 0.082 | 0.092 | 3609 | 39.0 | 0.174 | 0.192 | 0.097 |
Location, sampling year, geographical coordinates, code, country, number of individuals collected (N initial) and analysed after quality filtering (N), observed heterozygosity (Ho), expected heterozygosity (He), inbreeding coefficient (FIS) for all datasets and for polymorphic loci are shown. Locations in italics were previously analysed by Vera et al. (2022).
Single-nucleotide polymorphism (SNP) genotyping
Total DNA was extracted from gills using the E.Z.N.A. E-96 mollusc DNA kit (OMEGA Bio-tek), following manufacturer recommendations. 2b-RAD libraries (~90 cockles per run) were constructed using the AlfI IIb restriction enzyme and sequenced in an Illumina NextSeq 500 platform following Maroso et al. (2018; 2019). Bowtie 1.1.2 (Langmead et al. 2009) was used to align reads to the cockle’s genome (Bruzos et al. 2022) allowing a maximum of three mismatches and a unique valid alignment (-v 3 -m 1). The reference-based mode with default parameters in the gstacks module of STACKS 2.0 (Catchen et al. 2013) was used for SNP calling. For genotyping, SNPs were filtered following Vera et al. (2022): (i) SNPs genotyped in >60% individuals; (ii) MAC (minimum allele count) ≥3; (iii) conformance to Hardy–Weinberg expectations (i.e., SNPs with significant FIS values (P < 0.05) in at least 25% of the populations were removed); and (iv) the most polymorphic SNP within each RAD-tag were retained. Individuals with less than 250,000 reads were discarded.
Genetic diversity and population structure
Estimates of genetic diversity (i.e., mean number of alleles per locus (Na), observed (Ho) and expected (He) heterozygosity, proportion of polymorphic loci), departure from Hardy–Weinberg equilibrium and inbreeding coefficients (FIS) were estimated using GENEPOP v4.0 (Rousset 2008) and ARLEQUIN v3.5 (Excoffier and Lischer 2010). Because a minimum allele frequency (MAF) filtering was not applied, ARLEQUIN was also used to estimate Ho, He and FIS exclusively with polymorphic loci (MAF >0.017 according to sample size) for comparison with previous studies.
Global and pairwise coefficients of population differentiation (FST) between cockle beds were calculated with ARLEQUIN v3.5 using 10,000 permutations to test for significance. The variational Bayesian clustering method implemented in the package fastSTRUCTURE v2.3.4 (Raj et al. 2014) was used to estimate the number of genetic population units (K) in the whole studied area and in each region testing from K = 1 to K = number of beds + 1, with an admixture ancestry model, convergence criterion of 1 × 10−8, five cross-validated sets and the simple prior (flat-beta prior). The most likely number of K was estimated using the “chooseK.py” programme included in the fastSTRUCTURE which gives the best K value and the K corresponding with weak population structure in the data using heuristic scores. Summarised outputs were carried out using the software POPHELPER (Francis 2017). Discriminant analyses of principal components (DAPC) were run in ADEGENET package (Jombart et al. 2010; Jombart and Ahmed 2011) for the R platform (R Development Core Team 2014; http://www.r-project.org) with the whole dataset and for each region. Data were transformed using principal component analysis (PCA) and the optimal number of principal components (PC) was calculated using the optim.a.score() command (see Miller et al. 2020). Isolation by distance (IBD) was checked by the correlation between geographical (measured as the shortest oceanic distance between two beds in km) and genetic distance (measured as FST/1-FST; Rousset 1997) matrices using a Mantel test with 10,000 permutations using NTSYS v.2.1 (Rohlf 1993).
Outlier tests
The Bayesian FST-based method implemented in BAYESCAN v2.1 (Foll and Gaggiotti 2008) was used to identify outlier loci subjected to selection. BAYESCAN was run using default parameters (i.e., 20 pilot runs; prior odds value of 10; 100,000 iterations; burn-in of 50,000 iterations and a sample size of 5000, hereafter “BY10”), but we also explored increasing prior odds value to 1000 (hereafter “BY1000”). Despite high prior odds tend to remove false positives, they also reduce the power for detection loci under selection (Foll 2012). Loci with a false discovery rate (FDR, q value) <0.05 were considered as outliers. Moreover, the PC-based method implemented in R package PCADAPT v4.0 (Luu et al. 2017; Prive et al. 2020) was also applied. This method renders low false-positive rates and uses individual information, not requiring a priori population assignment. For the analysis, the number of PC retained was performed with the “chooseK” option. The outlier identification was carried out with an FDR <0.05. We considered as outliers those loci identified by any of the two approaches, but additionally, those shared between all approaches as the most confident ones.
Seascape analyses
Effects of spatial (latitude and longitude) and relevant abiotic factors in coastal and marine environments (SST, °C); sea bottom temperature (SBT, °C); sea surface salinity (SSS, psu); sea bottom salinity (SBS, psu); bottom shear stress (BSS, N·m−2); net primary productivity (NPP, mg·m−3·day−1); see Coscia et al. 2020; Vera et al. 2022) shaping genetic differentiation across beds in the studied areas were assessed using a canonical redundancy analysis (RDA) implemented in the VEGAN software (Oksanen 2015) in R. This abiotic information was retrieved as monthly averages from the IBI_REANALYSIS_PHYS_005_002 ocean reanalysis model (https://resources.marine.copernicus.eu/?option=com_csw&task=results?option=com_csw&view=details&product_id=IBI_REANALYSIS_PHYS_005_002) and IBI_REANALYSIS_BIO_005_003 model (https://resources.marine.copernicus.eu/?option=com_csw&task=results?option=com_csw&view=details&product_id=IBI_REANALYSIS_BIO_005_003) for the period 2014–2018 (Supplementary Table 1), respectively. The nearest model cell classified as the ocean was selected to extract the data (average distance between the sampling location and centre of the nearest model grid cell edge = 11.6 km). Then, averages for the spawning season (i.e., from April to September, see Malham et al. 2012; Mahony et al. 2020), winter (i.e., from January to March) and summer (i.e., from July to September) were calculated for each bed. Allele frequencies were calculated for each bed with ADEGENET package using the “makefreq” option. Loci with missing values were removed from the analysis. The significance of the variance associated with the different variables was tested with 1000 random permutations. Variance inflation factor (VIF) was estimated to explore collinearity (correlation) between seascape variables in the dataset, with VIF values >10 suggesting important collinearity problems (Marquardt 1970). The selection model was performed using an automatic stepwise model-building algorithm based on permutation p values tests. This procedure was performed with the ordistep function included in VEGAN. The reduced panel of explanatory variables was used to recalculate the total proportion of genetic variation in the variance partitioning. The weight of the different loci on the significant environmental vectors was calculated using VEGAN. All these analyses were performed separately for the whole, neutral and divergent outlier SNP datasets in the regions studied.
Potential correlations between allele frequencies and seascape variables were investigated with BAYENV2 (Coop et al. 2010; Gunther and Coop 2013) and results were compared with the mentioned RDA analyses. The method implemented in this software allows controlling the neutral genetic structure, because the fit improvement for a given genetic variant between a model including the environmental factor and a model including only neutral genetic structure is tested (Rellstab et al. 2015). BAYENV2 was carried out using the whole SNP datasets from SW British Isles and Galicia, respectively. First, analyses were performed with 100,000 iterations across five independent runs to obtain the average covariance matrix for each subset. Secondly, the correlation between each SNP and the different variables was calculated using 100,000 iterations to obtain Bayes factors (BFs). As in the previous step, five independent runs were used. Only SNPs with a BF >10 and Spearman’s coefficient (rho, ρ) thresholds >1% for any variable in all runs were considered well-supported environment-associated SNPs. Finally, significantly correlated SNPs were compared with the outliers identified in the BAYESCAN and PCADAPT analyses.
Gene mining and functional enrichment
RAD-tags including divergent outlier SNPs were mapped in the C. edule genome (Bruzos et al. 2022) and their position compared with the consistent genomic windows under divergent selection previously reported by Vera et al. (2022) in the Northeast Atlantic Ocean. The very low genetic differentiation with neutral markers in the studied areas precluded the detection of consistent genomic regions under stabilising selection. Thus, we could verify in more restricted geographical scenarios (SW British Isles and Galicia) the consistency of the genomic regions under divergent selection previously detected. In addition, we looked for new regions under selection considering the singularity of the new sample collections of this study following a similar methodology to that proposed by Vera et al. (2022). Briefly, we defined a consistent window when ≥2 consecutive outliers were detected; then, we expanded the region ±250 kb from the external outliers of the seed to define a genomic window for mining. Genes included in those genomic windows were identified using the cockle’s transcriptome assembled and annotated by Pardo et al. (2022), which was used as a reference to detect Gene Ontology (GO) functional enrichment of the genomic regions under selection (FDR 5%) using GOfuncR (Grote 2022). Furthermore, we also analysed genomic windows around the SNPs correlated with environmental variables for mining; since we could not identify consecutive SNPs as with outliers, we were more conservative and defined smaller windows around each SNP (±100 kb).
Results
Genetic diversity and differentiation: whole sample and SNP dataset
A total of 599 cockles were analysed since six specimens that exhibited a low number of reads (<250,000 reads) from WDE_17 (two individuals), SAN_17 (one individual), SVI_17 (two individuals) and SMO_17 (one individual), were removed. After quality filtering, the number of SNPs retained in the whole dataset was 9250. This number was slightly lower than the number used in the macrogeographical study carried out by Vera et al. (2022) (9309 markers), because 59 of these markers were monomorphic in the studied regions. All the 9250 markers were included in the “9309 markers” dataset and their genomic information is available at https://onlinelibrary.wiley.com/doi/10.1111/eva.13340, where the SNP code from Vera et al. (2022) has been maintained for comparison between studies.
Observed (Ho) and expected (He) heterozygosities ranged respectively from 0.070 (SMO_17, Spain) to 0.080 (IWC_20, IGC_20 and IKF_20, Ireland; mean ± SD = 0.075 ± 0.003) and from 0.076 (WDE_17, Wales) to 0.087 (SNO_17, Spain and IKF_20, Ireland; mean ± SD = 0.082 ± 0.003) (Table 1). All FIS values per locus and bed were positive, suggesting heterozygote deficit, but low (always <0.115) and not significant, and all beds met HW expectations (P < 0.0022; 0.05/22 populations), an expected outcome considering the HW filtering applied to retain SNPs. The percentage of polymorphic loci ranged from 25.5% in SMO_17 (Spain) to 52.8% in SNO_17 (Spain) (mean ± SD = 41.3 ± 7.2%). When only polymorphic loci within each bed were considered, Ho ranged from 0.137 in SLO_17 (Spain) to 0.181 in SMO_17 (Spain) (mean ± SD = 0.155 ± 0.014), showing these two beds also the lowest (0.156) and highest (0.200) He (mean ± SD = 0.171 ± 0.012). No differences in genetic diversity were found between the SW British Isles and Galician regions (Mann–Whitney U tests P > 0.250 for Ho, He with all loci and with polymorphic loci). Genetic diversity was in the range of previous values reported by Vera et al. (2022) for the whole Atlantic area using the same methodology.
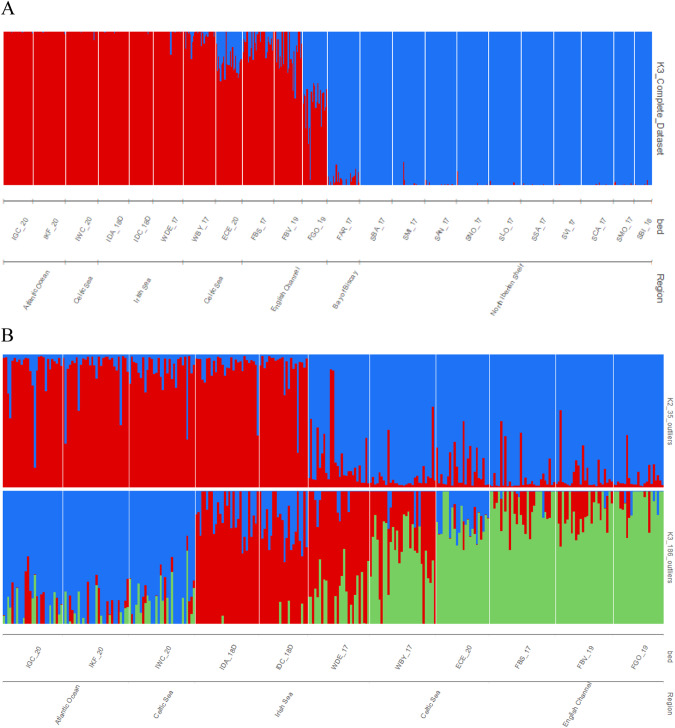
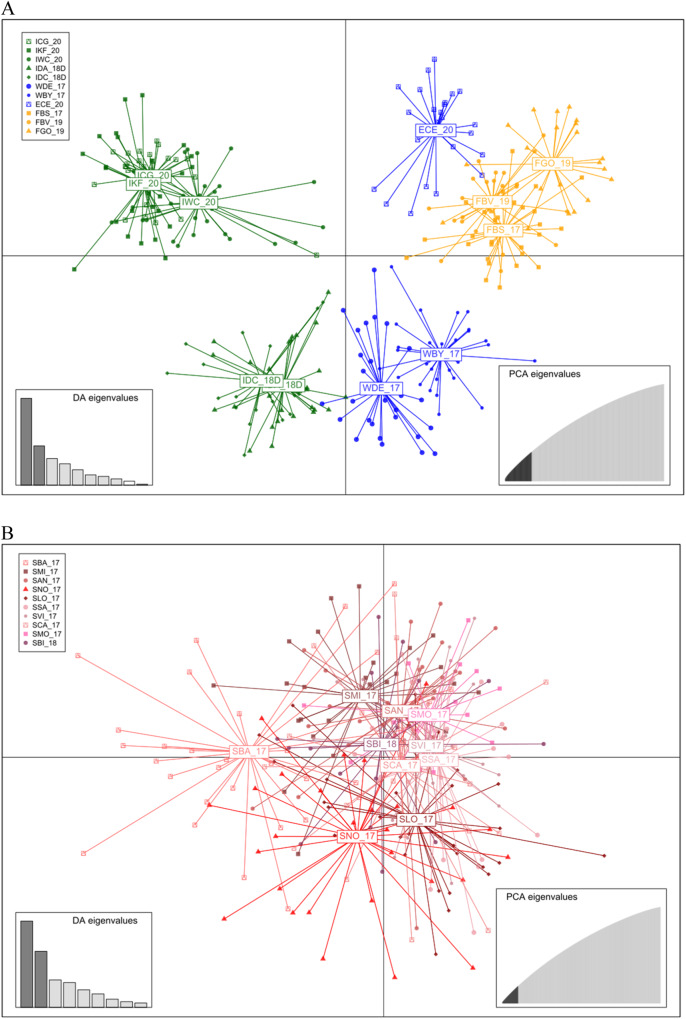
Global FST for all beds was 0.02118 (P < 0.001), pairwise FST ranged from 0 (non-significant ≠ 0) for many bed pairs up to a maximum of 0.05040 (P < 0.001) between IDC_18 and SVI_17 (Supplementary Table 2). Most pairwise comparisons were significant excluding those from Galicia. Average pairwise FST between the SW British Isles and Galicia was 0.03400 (P < 0.001), while 0.01374 (P < 0.001) within the SW British Isles and −0.00529 (P = 1.000) within Galicia. The two beds from the Cotentin Peninsula (FBV_19 and FGO_19, SW British Isles region), separated by 190 km, showed significant genetic differentiation (FST = 0.01207, P < 0.001). The most likely K values inferred by fastSTRUCTURE were 1 and 3. When K = 3 was plotted, two main groups were identified differentiating the SW British Isles (IGC_20, IKF_20, IWC_20, IDA_18, IDC_18, WDE_17, WBY_17, ECE_20, FBS_17, FBV_19 and FGO_19) from Galicia (plus Arcachon) (FAR_17, SBA_17, SMI_17, SAN_17, SNO_17, SLO_17, SSA_17, SVI_17, SCA_17, SMO_17, SBI_18) (Fig. 2A). FGO_19 (France, Cotentin Peninsula) showed a high component of the southern group, also detectable in all samples from the English Channel (ECE_20, FBS_17 and FBV_18), suggesting some introgression between the two groups. The DAPC representation on the SW British Isles also suggested differentiation of the English Channel samples from the northernmost populations across the second component, while the first one, indicated a remarkable divergence of the IKF_20 sample from the remaining ones (Fig. 3A). The DAPC from Galicia showed most of the samples grouped excluding SNO_17, in the middle of the distribution, below Cape Finisterre, and SBI_18, the southernmost one (Fig. 3B).
Fig. 2. Population structure of Cerastoderma edule at different geographical scales using fastSTRUCTURE.
Each vertical bar represents one individual, and the colour proportion of each bar represents the posterior probability of assignment of each individual to the different clusters (K) inferred by the programme. The most likely K = 3 using the whole dataset (A), and K = 2 using the 35 shared divergent outliers between methodologies and K = 3 using the total 186 divergent outliers (B) for the SW British Isles are represented. Codes are shown in Table 1. Plots for all the K values tested for the different datasets are shown in Supplementary Figures.
Fig. 3. Representation of discriminant analysis of principal components (DAPC) results using complete SNP datasets.
DAPC plots of Cerastoderma edule beds belonging to the SW British Isles (A) and Galicia (B) are shown. The weight of retained discriminant analysis (DA) and principal components selected are shown on left bottom box and right bottom box, respectively. Codes are shown in Table 1.
Genetic structure within regions: demographic and selective factors
To understand the factors underlying genetic differentiation within the SW British Isles and Galician regions, we first identified those loci under selection using three different statistical approaches. BY10, BY1000 and PCADAPT detected 159, 47 and 84 outliers in the SW British Isles, respectively, all of them under divergent selection and representing a total of 186 different outliers (Supplementary Table 3). Thirty-five markers were shared between the three methods. The number of outliers in Galicia was much lower (BY10 = 15, BY1000 = 2, PCADAPT = 39), two of them shared between the three methods and representing a total of 51 outliers, all of them putatively under divergent selection. Among the whole outlier dataset, 15 were shared between SW British Isles and Galicia. Then, by discounting the total number of outliers to the whole dataset in each region, a total of 9064 neutral markers were identified in the SW British Isles and 9199 in Galicia, representing the neutral datasets for each region.
Small but significant genetic differentiation was detected among the SW British Isles beds using neutral markers (FST = 0.00778, P < 0.001), suggesting limitations to larval dispersion in this area by biogeographical barriers. As expected, the 186 total outliers rendered a much higher global FST (0.10959, P < 0.001), being more accentuated when using the shared set of outliers between methods (FST = 0.17411, P < 0.001), which suggests selective factors increasing structuring. Pairwise FST ranged from −0.03095 (IKF_20 – WDE_17) to 0.02366 (IGC_20 – FGO_19 pair) for neutral markers; from 0.00061 (IDA_18 – IDC_18 pair) to 0.17142 (IDA_18 – FGO_19 pair) for the 186 total outliers; and from −0.00345 (IDA_18 – IDC_18 pair) to 0.27313 (IKF_20 – FBS_17 pair) for the 35 shared outliers (Supplementary Table 4). IBD was significant with the shared and total outlier datasets (r = 0.63169 and 0.55136, respectively; P < 0.001), but not with the neutral dataset (r = 0.08578, P = 0.330). These results suggest that correlations could be a by-product of the unequal spatial distribution of the environmental factors responsible for selective forces shaping the cockle’s genome, since IBD patterns should be reflected by the balance between drift and migration on neutral markers. The fastSTRUCTURE analyses identified K = 1, K = 2 and K = 3 as the most likely values for the neutral, 35 shared outlier and 186 total outlier datasets, respectively (Fig. 2B), which consistently differentiated the Celtic Sea and the Northwest Irish cluster (IGC_20, IKF_20 and IWC_20), not studied to date, and the English Channel cluster (ECE_20, FBS_17, FBV_19, FGO_19). In contrast, the Irish Sea appeared as a rather differentiated group with 186 outliers, which was split into two clusters, the Irish side (IDA_18 and IDC_18) most closely associated with the Celtic and Atlantic Ocean cluster, and the Welsh side (WDE_17 and WBY_17), most closely linked with the English Channel cluster, when using the 35 outlier loci. The differentiation of the Irish Sea from the other samples, and the contrast between the Welsh and Irish (east and west, respectively) samples of the Irish Sea, was shown when exploring a scenario with a larger K value, with both datasets displaying a very similar structure with K = 4 (Supplementary Figs. 2 and 3). The DAPC analysis with neutral markers showed a very similar picture to that described with the whole SNP dataset (Supplementary Fig. 4A); however, the 186 and 35 outlier datasets displayed a very distinct picture, both separating the English Channel (ECE_20, FGO_19, FBV_19 and FBS_17) from the Welsh populations, but also from the Irish populations, which were further divided into two groups, the westernmost Northeast Atlantic Ocean group (IWC_20, ICG_20 and IKF_20) and the Irish/Celtic Seas group (IDC_18 and IDA_18) (Supplementary Fig. 4B, C).
In contrast to the SW British Isles, no population differentiation was found in Galicia with the neutral dataset (FST = 0.00552, P = 1.000), also supported by the fastSTRUCTURE (K = 1) and DAPC, as previously outlined with the whole dataset (Fig. 3B and Supplementary Fig. 4D). However, low but significant differentiation was detected with the 51 outliers (FST = 0.00870, P < 0.001), the pairwise FST supporting a significant differentiation of the two northernmost samples (SMI_17 and especially SBA_17; Supplementary Table 5) from the rest. This differentiation was not disclosed with fastSTRUCTURE (K = 1; see Supplementary Fig. 5) and only suggested with DAPC (Supplementary Fig. 4E).
Seascape analysis
RDA analyses in SW British Isles region suggested longitude as the main driver for the observed differentiation with all datasets and seasons (Table 2). Latitude was also supported as a driver for many models, especially for those related to the 186 total outliers. SBS was suggested for all seasons with the 186 outlier dataset, while BSS was for reproductive and summer seasons using the whole and neutral datasets (Table 2). When longitude and latitude were removed, SST was suggested for all the datasets in the summer season, and in the reproductive and winter seasons only with the whole and 186 outlier datasets, respectively. SBT was suggested for the reproductive and summer season with the 186 outlier dataset. SBS and SSS were suggested with the 186 outlier dataset for the summer and winter seasons, respectively. NPP was suggested for all datasets in the winter season and for the 186 outlier dataset for the reproductive season. Finally, BSS was suggested in all seasons for the neutral dataset and in the reproductive and summer seasons for the complete dataset. In Galician region, no associations were found, except for latitude in all periods analysed using the 51 outliers, and for BSS during winter when latitude and longitude were removed (Table 2). However, VIF values were usually high (>10), suggesting that results should be taken with caution due to the high collinearity among the variables in many cases.
Table 2.
Results of the redundancy analysis (RDA) on the SW British Isles region of Cerastoderma edule.
| SW British Isles | Complete dataset | Neutral dataset | 186 total outlier dataset | |||||
|---|---|---|---|---|---|---|---|---|
| Model | Season | Variable | P value | Adjusted R2 | P value | Adjusted R2 | P value | Adjusted R2 |
| All seascape variables | Reproductive period | Latitude | – | 0.102 | – | 0.098 | 0.005 | 0.414 |
| Longitude | 0.001 | 0.001 | 0.001 | |||||
| SBS | – | – | 0.051 | |||||
| BSS | 0.004 | 0.001 | – | |||||
| Winter | Latitude | 0.001 | 0.087 | 0.001 | 0.080 | 0.009 | 0.423 | |
| Longitude | 0.016 | 0.015 | 0.001 | |||||
| SBS | – | – | 0.030 | |||||
| Summer | Latitude | – | 0.102 | – | 0.098 | 0.012 | 0.410 | |
| Longitude | 0.002 | 0.003 | 0.001 | |||||
| SBS | – | – | 0.055 | |||||
| BSS | 0.001 | 0.001 | – | |||||
| Only abiotic variables | Reproductive period | SST | – | 0.053 | – | 0.053 | 0.013 | 0.319 |
| SBT | – | – | 0.001 | |||||
| BSS | 0.002 | 0.002 | – | |||||
| NPP | – | – | 0.027 | |||||
| Winter | SST | 0.003 | 0.067 | – | 0.069 | – | 0.202 | |
| SSS | – | – | 0.041 | |||||
| BSS | – | 0.010 | – | |||||
| NPP | 0.017 | 0.008 | 0.019 | |||||
| Summer | SST | 0.011 | 0.086 | 0.014 | 0.083 | 0.001 | 0.355 | |
| SBT | – | – | 0.009 | |||||
| SBS | – | – | 0.083 | |||||
| BSS | 0.001 | 0.001 | – | |||||
| Galicia | Complete dataset | Neutral dataset | 51 total outlier dataset | |||||
|---|---|---|---|---|---|---|---|---|
| Model | Season | Variable | P value | Adjusted R2 | P value | Adjusted R2 | P value | Adjusted R2 |
| All seascape variables | Reproductive period | Latitude | – | – | – | – | 0.012 | 0.124 |
| Winter | Latitude | – | – | – | – | 0.011 | 0.124 | |
| Summer | Latitude | – | – | – | – | 0.008 | 0.124 | |
| Only abiotic variables | Reproductive period | – | – | – | – | – | – | – |
| Winter | BSS | – | – | – | – | 0.045 | 0.083 | |
| Summer | – | – | – | – | – | – | – | |
Only variables included by the forward selection model are shown. Adjusted R2 and P value associated with each variable of its selection stage.
SST sea surface temperature, SBT sea bottom temperature, SSS sea surface salinity, SBS sea bottom salinity, BSS bottom shear stress, NPP net primary production.
While no correlations were identified in Galicia with BAYENV2, a total of 54 markers were correlated with different environmental variables in the SW British Isles (Supplementary Table 6). Thirty of these markers (55.6%) were previously identified as outliers by the different methodologies applied. Markers were mainly correlated with latitude, longitude, temperature and salinity. The main variable correlated with genetic markers in the reproductive season and summer scenarios was SBT, while SSS and NPP were in the winter scenario.
Gene mining around outliers and environmentally correlated markers
Genetic markers associated with divergent selection or correlated with environmental variables were mapped in the common cockle genome to look for functional interpretation (Supplementary Tables 3 and 6). Outliers identified in the SW British Isles area were scattered across all chromosomes, between one in C18 and 22 in C3, while five chromosomes (C8, C11, C13, C14, C17) did not bear any outlier in Galicia, the maximum being detected in C1 (11 outliers) (Table 3). The 51 outliers detected in Galicia only identified a single consistent genomic region (window) under selection according to our criteria and other five outliers were distributed across four confident genomic windows previously reported by Vera et al. (2022) (Supplementary Tables 7 and 8). However, among the 186 outliers detected in the SW British Isles, 14 defined five new consistent genomic windows under divergent selection and other 45 mapped on genomic windows previously reported by Vera et al. (2022) (Supplementary Tables 7 and 8). Most outliers detected in Galicia were specific to this region, while an important number of outliers from the SW British Isles were shared with the Northern region previously analysed by Vera et al. (2022) (Supplementary Fig. 6). Still, a notable proportion of outliers in the North were specific of each study (North-Vera et al. (2022): 137 vs SW British Isles: 101) suggesting specific evolutionary factors related to each scenario. Among the genes annotated in the five new windows, several related to oxidative stress, hypoxia and immunity were identified in a 200 kb region in C2 and in a 340 kb region in C3 (Supplementary Table 9) (Gerdol and Venier 2015; Grandi et al. 2016; Sokolov et al. 2019). Also, in a 480 kb region in C5, some genes involved in signalling and detoxification (Wang et al. 2018; Kron 2022; Thoma et al. 2022) were identified. Finally, a gene associated with ocean acidification (Lim et al. 2021) was identified in C19. Despite the low number of genes handled, a significantly enriched GO Molecular Function was detected (protein serine/threonine phosphatase activity; GO:0004722) taking as background the common cockle transcriptome reported by Pardo et al. (2022).
Table 3.
Distribution of divergent outliers and markers correlated with environmental variables in the SW British Isles and Galicia across the Cerastoderma edule genome (version 4.0).
| Outlier loci (divergent selection) | |||||
|---|---|---|---|---|---|
| Mega-scaffold (chromosome) | Chromosome length (bp) | British Isles | Galicia | Shared | Markers correlated environmental variablesa |
| C1 | 64,609,245 | 21 | 11 | 3 | 5 (3) |
| C2 | 56,319,168 | 14 | 2 | 7 (3) | |
| C3 | 55,987,847 | 22 | 3 | 1 | 6 (5) |
| C4 | 52,087,795 | 18 | 5 | 2 | 7 (6) |
| C5 | 50,828,891 | 11 | 1 | 1 | 3 (2) |
| C6 | 40,237,005 | 13 | 3 | 2 | 4 (1) |
| C7 | 39,934,596 | 2 | 1 | ||
| C8 | 39,684,391 | 9 | 2 (1) | ||
| C9 | 39,070,162 | 11 | 3 | 2 (1) | |
| C10 | 38,264,924 | 14 | 8 | 2 | 1 |
| C11 | 38,197,540 | 2 | 1 | ||
| C12 | 36,327,582 | 6 | 1 | 1 (1) | |
| C13 | 35,955,507 | 10 | 5 (2) | ||
| C14 | 33,816,358 | 5 | |||
| C15 | 31,726,440 | 3 | 1 | 1 | 3 (1) |
| C16 | 31,510,408 | 10 | 8 | 1 | 2 (2) |
| C17 | 26,587,828 | 4 | 2 (1) | ||
| C18 | 22,603,465 | 1 | 1 | 1 | |
| C19 | 21,711,631 | 4 | 1 | 1 | 1 |
| Other scaffolds | 6 | 2 | 2 (1) | ||
| Total | 186 | 51 | 15 | 54 (30) | |
aOnly detected in SW British Isles; in parentheses, those markers also identified as outliers for divergent selection.
Markers correlated with environmental variables were scattered across most chromosomes, excluding C7, C14 and C18, and a higher number (seven markers) were detected in two big chromosomes (C2 and C4) (Table 3). An important number of correlated markers were also identified as outliers for divergent selection (55.6%), some of them associated with consistent genomic windows (Supplementary Table 8). Of note, the three markers were detected in one of the most consistent genomic windows in C4. We also mined the cockle genome around the correlated marker dataset (Supplementary Table 10) and detected several genes related to nervous system development and physiology. These genes were mostly clustered at C1 around 142462_31 (correlated with SBT) and C2 around 210318_7 (correlated with SST), respectively. Furthermore, some of these genes were previously associated with temperature stress and oxygen depletion stress or differentially expressed under specific experimental conditions in other mollusc species (Woo et al. 2011; Chen et al. 2022). Another important group of genes scattered around different markers in the cockle genome were related to immunity and defence and had been previously reported in other mollusc species in response to viruses and bacteria (Barbosa et al. 2022; Saco et al. 2023) (Supplementary Table 9).
Discussion
Assessment of the distribution of genetic variability across populations, incorporating historical processes and local adaptation framed within the dispersal range of the focal organism (Richardson et al. 2014), is essential to develop management actions to preserve exploited species (Bernatchez et al. 2017). In the present study, two different patterns of genetic structure at microgeographic scale were identified in two regions within the natural distribution of C. edule, highlighting the need to perform analyses at multiple spatial scales (Hoffman et al. 2012), to provide information supporting the management of this valuable resource.
Heterogeneous pattern of microgeographic structure in the common cockle
The two geographic areas studied, the SW British Isles region and Galicia, were selected by their different habitat fragmentation patterns. Both areas were slightly differentiated (FST = 0.03400), in accordance with their location in the major northern and southern regions of the species’ range separated around French Brittany (Vera et al. 2022), but did not show differences in genetic diversity, unlike Vera et al. (2022), who reported a slight, but significant higher diversity in the southern region.
The extensive analysis performed in Galicia (10 natural beds) suggested the presence of a single panmictic unit in this area, as previously reported for other molluscs, with similar pelagic larval periods (Donax trunculus, Nanton et al. 2017; Ensis siliqua, Arias-Perez et al. 2012; Mitilus galloprovincialis, Diz and Presa 2009; Ostrea. edulis, Vera et al. 2016; Polititapes rhomboides, Chacón et al. 2021), and for other marine species (Hippocampus guttulatus, Lopez et al. 2015; Pollicipes pollicipes, Parrondo et al. 2022). Our data does not support Cape Finisterre as a biogeographical barrier for the species as previously suggested (Lopez-jamar et al. 1992; Piñeira et al. 2008; Martínez et al. 2013; Cruz et al. 2020) since no differentiation was detected between beds on both sides of the Cape with the whole and neutral datasets. However, when using outlier loci, the two northernmost Galician beds showed significant differentiation from the remaining ones, especially the bed closest to the Cantabrian Sea (SBA_17) (average FST = 0.03145), which could be related to the higher temperature regime in the Cantabrian Sea (Marquina et al. 2015), but a more detailed study in the Cantabrian Sea would be necessary to confirm this observation. Oceanographic dynamics on the Galician coast indicate that the cold-upwelled water usually penetrates estuaries on the west, while it only occurs during very intense events on the north (Alvarez et al. 2010). Thus, water temperature decreases from north to west, with an SST average value of 19.5 °C on the Cantabrian coast compared with the 18.5 °C measured on the west coast for the 1985–2005 period (Gomez-Gesteira et al. 2008). Larval dispersal modelling carried out by Vera et al. (2022) (see their Fig. 7) confirmed that cockle beds are well connected with each other by larval transport in Galicia, but the connection between the Rias and the sites to the northeast of Cape Finisterre was weaker, though present. Furthermore, whilst the beds along the northwest coast of the Iberian Peninsula are affected by very similar oceanographic conditions, during the late spring and late summer, temperatures at the most north-easterly site can differ markedly from those at the other beds due to its location at the edge of the upwelling system and at the inception point of the Portugal Coastal Current (STT two degrees higher in the northern beds (mean = 18.51 °C) than in the southern ones (mean = 16.47 °C) during the summer; see Supplementary Table 1). Despite the genotype-environment association methods did not identify sea temperature as driver, latitude, which is highly correlated with temperature, was suggested by the RDA analysis as a potential driver in the region.
Previous data from the SW British Isles suggested significant structure in C. edule related both to current dynamics as well as to abiotic factors, such as salinity and temperature (Coscia et al. 2020; Vera et al. 2022), as reported in other shellfish species such as the horse mussel Modiolus modiolus (Gormley et al. 2015) and the great scallop Pecten maximus (Vendrami et al. 2019; Hold et al. 2021). However, some regions in this area are still poorly sampled in the common cockle (English Channel) or without information (West Irish coast, Northeast Atlantic). Outlier markers showed a moderate pairwise genetic differentiation between beds (FST = 0.10959 and 0.17411 with the 186 and 35 outlier datasets, respectively), higher than that observed with neutral markers (FST = 0.00778), as expected, suggesting selective factors shaping specific genomic regions in a small geographic area. An important proportion of divergent outliers (68 markers) were shared with those reported by Vera et al. (2022) for the northern group (210 outliers), which gives robustness to our observations; however, data also suggest specific selective factors shaping the cockle’s genome associated with the new sampling in the SW British Isles (117 new outlier loci; 31 within consistent genomic windows). In fact, five new confident genomic windows were identified, including relevant genes related to oxidative stress and immunity that would deserve further studies as candidates to explain the association observed with environmental factors. Despite biotic factors, such as pathogens, could not be contemplated in our study, their diversity and distribution (influenced by abiotic factors) are important drivers shaping the genome and distribution of species (Theodosopoulos et al. 2019) and specifically in cockles (Vera et al. 2022; Pampín et al. 2023). Furthermore, we also deepened into the correlation of specific SNPs with environmental factors and could identify, by mining in the cockle genome several genes related to nervous transmission and immunity, arranged in clusters or scattered in different chromosomes, that had been previously reported in other mollusc associated with temperature or oxidative stress (Woo et al. 2011; Barbosa et al. 2022; Chen et al. 2022).
The population structure observed in the SW British Isles region may be in part explained by the residual ocean currents and ocean fronts that characterise this area, but also by selective factors such as salinity gradients, variable BSS (due to large tidal variability) and sea temperature gradients (driven by ocean currents, stratification and mixing, and latitudinal gradients); however, spatial seascape results should be taken with caution due to the collinearity detected among variables. Both outlier datasets could identify four genetic clusters following two main west-east and north-south axes, which could explain the correlation observed between genetic and geographic distances for outlier loci, but also the identification of longitude and latitude as two main drivers in the seascape analysis. According to the outlier information, the new sampled beds from Western Ireland (IGC_20, IKF_20 and IWC_20) (Northeast Atlantic) would constitute a new cluster. These sites are connected by the Irish coastal current (Brown et al. 2003; Fernand et al. 2006) and larval dispersal modelling (see Fig. 7 in Vera et al. 2022) showed that the beds along the southwest coast of Ireland are well interconnected. The Irish Sea can be split into two different clusters associated with the Irish and Welsh sides, as previously suggested by Coscia et al. (2020). Sites along the southeast coast of the Irish Sea are generally connected by northward currents and sites along the north coast of Wales by eastward currents. In contrast, the two sites on the west coast of the Irish Sea (IDC_18 and IDA_18) appear genetically separated from the remainder of the Irish Sea. This may be driven by the Irish Sea Front acting as a barrier which also drives warmer temperatures in the northwest Irish Sea than in the well-mixed northeast Irish Sea. Finally, the English Channel forms a fourth cluster including the ECE_20 bed from Cornwall with the southern beds limited by the Ushant front. Interestingly, the Cotentin Peninsula, previously identified as a physical barrier to dispersal in other molluscs, such as the slipper limpet Crepidula fomicata (Dupont et al. 2007) and P. maximus (Nicolle et al. 2016; Handal et al. 2020), showed significant differentiation between samples on its west and east sides (FGO_19 and FBV_19) with neutral markers (FST = 0.01045, P < 0.001) and higher with outlier loci (FST 35 outliers = 0.06654, P < 0.001; FST 186 outliers = 0.05876, P < 0.001), suggesting additional selective factors differentiating both sides. Oceanic distance between the two Cotentin beds (~190 km) is shorter than the longest distance between Galician beds (~300 km), where no genetic differentiation was detected with neutral markers. Of note, FGO_19 showed an important genomic component of the South group, suggesting introgression from the south especially in the west coast of the Cotentin Peninsula.
Management implications
The present study represents a refined analysis of the population structure of C. edule in two geographic areas of small-medium size representing differentiated models that could aid in obtaining a more comprehensive picture for improving the management and conservation of this valuable commercial and ecological resource. Galician beds were suggested to constitute a panmictic population and this region could be managed as a single genetic unit. The fisheries in this region are exclusively commercial and their exploitation management can be through territorial concessions leased by shellfisher guilds or directly by Galician regional government (i.e., free access shellfish areas). This genetic information should be included in the Galician legislation, thus allowing translocations from high-production areas (Ría de Noia) to depleted ones by different factors, such as the parasite M. cochillia (Ría de Arousa; Villalba et al. 2014). However, caution should be taken considering biotic factors not evaluated in our study, such as emergent pathologies (e.g., marteiliosis), which will require specific recommendations within the general framework depicted in our study. A sharp fragmentation was displayed by the SW British Isles region, especially with divergent outliers, mostly representing adaptive management units (Bernatchez et al. 2017). Thus, Western (Northeast Atlantic) Irish beds would represent a differentiated group from those previously described, while subtle genetic sub-structuring was identified along the English Channel, with a significant effect at the Cotentin Peninsula representing as a biogeographic barrier. Furthermore, the Irish Sea, a narrow water body mass between Wales and Ireland, appears to represent differentiated units on its both sides, according to our information. All these population units should be individually managed, avoiding translocations between them. Finally, our results could help to improve cockles’ production by founding appropriate broodstock to enhance depleted populations and by tracing samples to check undesirable transferences among regions.
Supplementary information
Acknowledgements
The authors wish to thank L. Insua, S. Sánchez-Darriba and S. Gómez from the ACUIGEN group (USC) for their technical support. Supercomputing Center of Galicia (http://www.cesga.es) provided computing facilities for genotyping. A. Casanova (ACC) was funded by a Xunta de Galicia-Campus Terra postdoctoral fellow. The authors are also indebted to COCKLES Interreg European project partners who provided samples included in this study. Finally, the authors are grateful to Prof. S. Goodacre, Dr X. Zhan and three anonymous reviewers for their helpful comments on the earlier version of this manuscript.
Author contributions
MV, AV and PM designed and supervised the study. DI, AC, KM, FO, SKM and SL performed field collections. PM, SCC, SL, PER, SKM and FO provided funding. FM, MH and AB analysed bioinformatically genomic sequences and created genotyping files. SBW and PER provided information about oceanography, environmental variables and developed geographic maps included in the figures. MV, CB, ACC, AB and PM performed the genetic analyses. MV and PM wrote the manuscript with contributions from all authors. All of them read the manuscript and gave their approval.
Funding
This study has been supported by the COCKLES project (grant number: EAPA_458/2016) of the INTERREG EUROPEAN PROGRAMME and the NERC-SHEAR project (NE/W001217/1).
Data availability
Data for this study are available at Dryad Digital Repository (10.5061/dryad.xpnvx0kmr) and Supplementary material.
Competing interests
The authors declare no competing interests.
Footnotes
Associate editor Xiangjiang Zhan.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Manuel Vera, Email: manuel.vera@usc.es.
Paulino Martínez, Email: paulino.martinez@usc.es.
Supplementary information
The online version contains supplementary material available at 10.1038/s41437-023-00646-1.
References
- Abaunza P, Murta AG, Campbell N, Cimmaruta R, Comesana AS, Dahle G, et al. Stock identity of horse mackerel (Trachurus trachurus) in the Northeast Atlantic and Mediterranean Sea: integrating the results from different stock identification approaches. Fish Res. 2008;89:196–209. doi: 10.1016/j.fishres.2007.09.022. [DOI] [Google Scholar]
- Alvarez I, Gomez-Gesteira M, DeCastro M, Gomez-Gesteira JL, Dias JM. Summer upwelling frequency along the western Cantabrian coast from 1967 to 2007. J Mar Syst. 2010;79:218–226. doi: 10.1016/j.jmarsys.2009.09.004. [DOI] [Google Scholar]
- Arias-Perez A, Fernandez-Tajes J, Gaspar MB, Mendez J. Isolation of microsatellite markers and analysis of genetic diversity among East Atlantic populations of the sword razor shell Ensis siliqua: a tool for population management. Biochem Genet. 2012;50:397–415. doi: 10.1007/s10528-011-9484-y. [DOI] [PubMed] [Google Scholar]
- Barbosa M, Schwaner C, Pales Espinosa E, Allam B. A Transcriptomic analysis of phenotypic plasticity in Crassostrea virginica larvae under experimental acidification. Genes. 2022;13:1529. doi: 10.3390/genes13091529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont AR, Day TR, Gade G. Genetic variation at the octopine dehydrogenase locus in the adductor muscle of Cerastoderma edule (L) and 6 other bivalve species. Mar Biol Lett. 1980;1:137–148. [Google Scholar]
- Bernatchez L, Wellenreuther M, Araneda C, Ashton DT, Barth JMI, Beacham TD, et al. Harnessing the power of genomics to secure the future of seafood. Trends Ecol Evol. 2017;32:665–680. doi: 10.1016/j.tree.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Brown J, Carrillo L, Fernand L, Horsburgh KJ, Hill AE, Young EF, et al. Observations of the physical structure and seasonal jet-like circulation of the Celtic Sea and St. George’s Channel of the Irish Sea. Cont Shelf Res. 2003;23:533–561. doi: 10.1016/S0278-4343(03)00008-6. [DOI] [Google Scholar]
- Bruzos AL, Santamarina M, Garcia-Souto D, Diaz S, Rocha S, Zamora J et al. (2022) The evolution of two transmissible leukaemias colonizing the coasts of Europe. Preprint at Biorxiv 10.1101/2022.08.06.503021
- Carss DN, Brito AC, Chainho P, Ciutat A, de Montaudouin X, Fernandez Otero RM, et al. Ecosystem services provided by a non-cultured shellfish species: the common cockle Cerastoderma edule. Mar Environ Res. 2020;158:104931. doi: 10.1016/j.marenvres.2020.104931. [DOI] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Mol Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón GM, Arias-Perez A, Freire R, Martinez L, Ojea J, Insua A. Genetic characterization of wild, broodstock and seed samples of Polititapes rhomboides (Bivalvia: Veneridae): Implications for hatchery seed production. Aquac Rep. 2021;20:100658. doi: 10.1016/j.aqrep.2021.100658. [DOI] [Google Scholar]
- Chen J, Leng T, Jiang YM, Chen XB, Liu ZM. RNA-seq analysis of the differential response to low-temperature stress in two morphs of mud crabs (Scylla paramamosain) Comp Biochem Physiol D Genom Proteom. 2022;43:101010. doi: 10.1016/j.cbd.2022.101010. [DOI] [PubMed] [Google Scholar]
- Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscia I, Wilmes SB, Ironside JE, Goward-Brown A, O’Dea E, Malham SK, et al. Fine-scale seascape genomics of an exploited marine species, the common cockle Cerastoderma edule, using a multimodelling approach. Evol Appl. 2020;13:1854–1867. doi: 10.1111/eva.12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz A, da Costa F, Fernandez-Perez J, Nanton A, Fernandez-Boo S, Insua A, et al. Genetic variability in Ruditapes decussatus clam combined with Perkinsus infection level to support founder population selection for a breeding program. PeerJ. 2020;8:e9728. doi: 10.7717/peerj.9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dare PJ, Bell MC, Walker P, Bannister RCA (2004) Historical and current status of cockle and mussel stocks in The Wash. CEFAS
- de Montaudouin X, Bachelet G, Sauriau PG. Secondary settlement of cockles Cerastoderma edule as a function of current velocity and substratum: a flume study with benthic juveniles. Hydrobiologia. 2003;503:103–116. doi: 10.1023/B:HYDR.0000008493.83270.2d. [DOI] [Google Scholar]
- Diz AP, Presa P. The genetic diversity pattern of Mytilus galloprovincialis in Galician Rias (NW Iberian estuaries) Aquaculture. 2009;287:278–285. doi: 10.1016/j.aquaculture.2008.10.029. [DOI] [Google Scholar]
- do Prado FD, Vera M, Hermida M, Bouza C, Pardo BG, Vilas R, et al. Parallel evolution and adaptation to environmental factors in a marine flatfish: implications for fisheries and aquaculture management of the turbot (Scophthalmus maximus) Evol Appl. 2018;11:1322–1341. doi: 10.1111/eva.12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont L, Ellien C, Viard F. Limits to gene flow in the slipper limpet Crepidula fornicata as revealed by microsatellite data and a larval dispersal model. Mar Ecol Prog Ser. 2007;349:125–138. doi: 10.3354/meps07098. [DOI] [Google Scholar]
- Eldon B, Riquet F, Yearsley J, Jollivet D, Broquet T. Current hypotheses to explain genetic chaos under the sea. Curr Zool. 2016;62:551–566. doi: 10.1093/cz/zow094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fernand L, Nolan GD, Raine R, Chambers CE, Dye SR, White M, et al. The Irish coastal current: a seasonal jet-like circulation. Cont Shelf Res. 2006;26:1775–1793. doi: 10.1016/j.csr.2006.05.010. [DOI] [Google Scholar]
- Fisher MC, Helser TE, Kang S, Gwak W, Canino MF, Hauser L. Genetic structure and dispersal in peripheral populations of a marine fish (Pacific cod, Gadus macrocephalus) and their importance for adaptation to climate change. Ecol Evol. 2022;12:e8474. doi: 10.1002/ece3.8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach EC, de Bruin W. Does the activity of cockles, Cerastoderma edule (L.) and lugworms, Arenicola marina L., make corophium-volutator pallas more vulnerable to epibenthic predators: a case of interaction modification. J Exp Mar Biol Ecol. 1994;182:265–285. doi: 10.1016/0022-0981(94)90056-6. [DOI] [Google Scholar]
- Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M (2012) BayeScan v2.1 User Manual. http://cmpg.unibe.ch/software/BayeScan/files/BayeScan2.1_manual.pdf
- Francis RM. POPHELPER: an R package and web app to analyse and visualize population structure. Mol Ecol Resour. 2017;17:27–32. doi: 10.1111/1755-0998.12509. [DOI] [PubMed] [Google Scholar]
- Galarza JA, Carreras-Carbonell J, Macpherson E, Pascual M, Roques S, Turner GF, et al. The influence of oceanographic fronts and early-life-history traits on connectivity among littoral fish species. Proc Natl Acad Sci USA. 2009;106:1473–1478. doi: 10.1073/pnas.0806804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galparsoro I, Borja A, Uyarra MC. Mapping ecosystem services provided by benthic habitats in the European North Atlantic Ocean. Front Mar Sci. 2014;1:23. doi: 10.3389/fmars.2014.00023. [DOI] [Google Scholar]
- Gerdol M, Venier P. An updated molecular basis for mussel immunity. Fish Shellfish Immunol. 2015;46:17–38. doi: 10.1016/j.fsi.2015.02.013. [DOI] [PubMed] [Google Scholar]
- Gomez-Gesteira M, de Castro M, Alvaez I, Gomez-Gesteira JL. Coastal sea surface temperature warming trend along the continental part of the Atlantic Arc (1985–2005) J Geophys Res Oceans. 2008;113:C04010. doi: 10.1029/2007JC004315. [DOI] [Google Scholar]
- Gormley K, Mackenzie C, Robins P, Coscia I, Cassidy A, James J, et al. Connectivity and dispersal patterns of protected biogenic reefs: implications for the conservation of Modiolus modiolus (L.) in the Irish sea. PLOS One. 2015;10:e0143337. doi: 10.1371/journal.pone.0143337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi A, Santi A, Campagnoli S, Parri M, De Camilli E, Song C, et al. ERMP1, a novel potential oncogene involved in UPR and oxidative stress defense, is highly expressed in human cancer. Oncotarget. 2016;7:63596–63610. doi: 10.18632/oncotarget.11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote S (2022) GOfuncR: Gene ontology enrichment using FUNC. R package version 1.18.0
- Group “Grepma” A physical, chemical and biological characterization of the Ushant tidal front. Int Rev Ges Hydrobiol Hydrogr. 1988;73:511–536. doi: 10.1002/iroh.19880730503. [DOI] [Google Scholar]
- Gunther T, Coop G. Robust identification of local adaptation from allele frequencies. Genetics. 2013;195:205–220. doi: 10.1534/genetics.113.152462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handal W, Szostek C, Hold N, Andrello M, Thiebaut E, Harney E, et al. New insights on the population genetic structure of the great scallop (Pecten maximus) in the English Channel, coupling microsatellite data and demogenetic simulations. Aquat Conserv. 2020;30:1841–1853. doi: 10.1002/aqc.3316. [DOI] [Google Scholar]
- Hayward PJ, Ryland JS (1995) Handbook of the marine fauna of north-west Europe. Oxford University Press
- Hoffman JI, Clarke A, Clark MS, Fretwell P, Peck LS. Unexpected fine-scale population structure in a broadcast-spawning Antarctic marine mollusc. PLOS One. 2012;7:e32415. doi: 10.1371/journal.pone.0032415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hold N, Robins P, Szostek CL, Lambert G, Lincoln H, Le Vay L, et al. Using biophysical modelling and population genetics for conservation and management of an exploited species, Pecten maximus L. Fish Oceanogr. 2021;30:740–756. doi: 10.1111/fog.12556. [DOI] [Google Scholar]
- Honkoop PJC, van der Meer J. Experimentally induced effects of water temperature and immersion time on reproductive output of bivalves in the Wadden Sea. J Exp Mar Biol Ecol. 1998;220:227–246. doi: 10.1016/S0022-0981(97)00107-X. [DOI] [Google Scholar]
- Hummel H, Wolowicz M, Bogaards RH. Genetic variability and relationships for populations of Cerastoderma edule and of the C. glaucum complex. Neth J Sea Res. 1994;33:81–89. doi: 10.1016/0077-7579(94)90053-1. [DOI] [Google Scholar]
- Jackson-Bue M, Brito AC, Cabral S, Carss DN, Carvalho F, Chainho P, et al. Inter-country differences in the cultural ecosystem services provided by cockles. People Nat. 2022;4:71–87. doi: 10.1002/pan3.10252. [DOI] [Google Scholar]
- Jombart T, Ahmed I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron NS. In search of the Aplysia immunome: an in silico study. BMC Genom. 2022;23:543. doi: 10.1186/s12864-022-08780-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larmuseau MHD, Van Houdt JKJ, Guelinckx J, Hellemans B, Volckaert FAM. Distributional and demographic consequences of Pleistocene climate fluctuations for a marine demersal fish in the north-eastern Atlantic. J Biogeogr. 2009;36:1138–1151. doi: 10.1111/j.1365-2699.2008.02072.x. [DOI] [Google Scholar]
- Leary D, Vierros M, Hamon G, Arico S, Monagle C. Marine genetic resources: a review of scientific and commercial interest. Mar Policy. 2009;33:183–194. doi: 10.1016/j.marpol.2008.05.010. [DOI] [Google Scholar]
- Lim Y-K, Cheung K, Dang X, Roberts SB, Wang X, Thiyagarajan V. DNA methylation changes in response to ocean acidification at the time of larval metamorphosis in the edible oyster, Crassostrea hongkongensis. Mar Environ Res. 2021;163:105214. doi: 10.1016/j.marenvres.2020.105214. [DOI] [PubMed] [Google Scholar]
- Lopez A, Vera M, Planas M, Bouza C. Conservation genetics of threatened Hippocampus guttulatus in vulnerable habitats in NW Spain: temporal and spatial stability of wild populations with flexible polygamous mating system in captivity. PLOS One. 2015;10:0117538. doi: 10.1371/journal.pone.0117538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Jamar E, Cal RM, Gonzalez G, Hanson RB, Rey J, Santiago G, et al. Upwelling and outwelling effects on the benthic regime of the continental-shelf off Galicia, NW Spain. J Mar Res. 1992;50:465–488. doi: 10.1357/002224092784797584. [DOI] [Google Scholar]
- Luu K, Bazin E, Blum MGB. Pcadapt: an R package to perform genome scans for selection based on principal component analysis. Mol Ecol Resour. 2017;17:67–77. doi: 10.1111/1755-0998.12592. [DOI] [PubMed] [Google Scholar]
- Mahony KE, Lynch SA, Egerton S, Cabral S, de Montaudouin X, Fitch A, et al. Mobilisation of data to stakeholder communities. Bridging the research-practice gap using a commercial shellfish species model. PLOS One. 2020;15:0238446. doi: 10.1371/journal.pone.0238446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony KE, Lynch SA, Egerton S, Laffan RE, Correia S, de Montaudouin X, et al. Latitudinal influence on gametogenesis and host-parasite ecology in a marine bivalve model. Ecol Evol. 2021;11:7029–7041. doi: 10.1002/ece3.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malham SK, Hutchinson TH, Longshaw M. A review of the biology of European cockles (Cerastoderma spp.) J Mar Biol Assoc U K. 2012;92:1563–1577. doi: 10.1017/S0025315412000355. [DOI] [Google Scholar]
- Maroso F, Casanova A, do Prado FD, Bouza C, Pardo BG, Blanco A, et al. Species identification of two closely exploited flatfish, turbot (Scophthalmus maximus) and brill (Scophthalmus rhombus), using a ddRADseq genomic approach. Aquat Conserv. 2018;28:1253–1260. doi: 10.1002/aqc.2932. [DOI] [Google Scholar]
- Maroso F, Perez de Gracia C, Iglesias D, Cao A, Diaz S, Villalba A, et al. A useful SNP panel to distinguish two cockle species, Cerastoderma edule and C. glaucum, co-occurring in some European beds, and their putative hybrids. Genes. 2019;10:760. doi: 10.3390/genes10100760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt DW. Generalized inverses, ridge regression, biased linear estimation and nonlinear estimation. Technometrics. 1970;12:59. doi: 10.2307/1267205. [DOI] [Google Scholar]
- Marquina D, Fernandez-Alvarez FA, Norena C. Five new records and one new species of Polycladida (Platyhelminthes) for the Cantabrian coast (North Atlantic) of the Iberian Peninsula. J Mar Biol Assoc U K. 2015;95:311–322. doi: 10.1017/S0025315414001106. [DOI] [Google Scholar]
- Martínez L, Mendez J, Insua A, Arias-Perez A, Freire R. Genetic diversity and population differentiation in the cockle Cerastoderma edule estimated by microsatellite markers. Helgol Mar Res. 2013;67:179–189. doi: 10.1007/s10152-012-0314-3. [DOI] [Google Scholar]
- Martínez L, Freire R, Arias-Perez A, Mendez J, Insua A. Patterns of genetic variation across the distribution range of the cockle Cerastoderma edule inferred from microsatellites and mitochondrial DNA. Mar Biol. 2015;162:1393–1406. doi: 10.1007/s00227-015-2676-y. [DOI] [Google Scholar]
- Miller JM, Cullingham CI, Peery RM. The influence of a priori grouping on inference of genetic clusters: simulation study and literature review of the DAPC method. Heredity. 2020;125:269–280. doi: 10.1038/s41437-020-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanton A, Arias-Perez A, Freire R, Fernandez-Perez J, Novoa S, Mendez J. Microsatellite variation in Donax trunculus from the Iberian Peninsula, with particular attention to Galician estuaries (NW Spain) Estuar Coast Shelf Sci. 2017;197:27–34. doi: 10.1016/j.ecss.2017.08.011. [DOI] [Google Scholar]
- Nicolle A, Moitie R, Ogor J, Dumas F, Foveau A, Foucher E, et al. Modelling larval dispersal of Pecten maximus in the English Channel: a tool for the spatial management of the stocks. ICES J Mar Sci. 2016;74:1812–1825. doi: 10.1093/icesjms/fsw207. [DOI] [Google Scholar]
- Norris K, Bannister RCA, Walker PW. Changes in the number of oystercatchers Haematopus ostralegus wintering in the Burry Inlet in relation to the biomass of cockles Cerastoderma edule and its commercial exploitation. J Appl Ecol. 1998;35:75–85. doi: 10.1046/j.1365-2664.1998.00279.x. [DOI] [Google Scholar]
- Oksanen J (2015) Multivariate analysis of ecological communities in R: vegan tutorial. http://cc.oulu.fi/~jarioksa/opetus/metodi/vegantutor.pdf
- Pampín M, Casanova A, Fernandez C, Blanco A, Hermida M, Vera M, et al. Genetic markers associated with divergent selection against the parasite Marteilia cochillia in common cockle (Cerastoderma edule) using transcriptomics and population genomics data. Front Mar Sci. 2023;10:1057206. doi: 10.3389/fmars.2023.1057206. [DOI] [Google Scholar]
- Pardo BG, Fernandez C, Pampin M, Blanco A, Iglesias D, Cao A et al. (2022) Transcriptome characterization of the common cockle (Cerastoderma edule) after exposure to a Marteilia cochillia outbreak. Preprint at Biorxiv 10.1101/2022.10.18.512677
- Parrondo M, Moran P, Ballenghien M, Acuna JL, Aguion A, Arrontes J, et al. Chaotic genetic patchiness in the highly valued atlantic stalked barnacle Pollicipes pollicipes from the iberian peninsula: implications for fisheries management. Front Mar Sci. 2022;9:801780. doi: 10.3389/fmars.2022.801780. [DOI] [Google Scholar]
- Piñeira J, Quesada H, Rolan-Alvarez E, Caballero A. Genetic discontinuity associated with an environmentally induced barrier to gene exchange in the marine snail Littorina saxatilis. Mar Ecol Prog Ser. 2008;357:175–184. doi: 10.3354/meps07278. [DOI] [Google Scholar]
- Prive F, Luu K, Vilhjalmsson BJ, Blum MGB. Performing highly efficient genome scans for local adaptation with R Package pcadapt Version 4. Mol Biol Evol. 2020;37:2153–2154. doi: 10.1093/molbev/msaa053. [DOI] [PubMed] [Google Scholar]
- Raj A, Stephens M, Pritchard JK. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics. 2014;197:573–U207. doi: 10.1534/genetics.114.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellstab C, Gugerli F, Eckert AJ, Hancock AM, Holderegger R. A practical guide to environmental association analysis in landscape genomics. Mol Ecol. 2015;24:4348–4370. doi: 10.1111/mec.13322. [DOI] [PubMed] [Google Scholar]
- Richardson JL, Urban MC, Bolnick DI, Skelly DK. Microgeographic adaptation and the spatial scale of evolution. Trends Ecol Evol. 2014;29:165–176. doi: 10.1016/j.tree.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Rohlf F (1993) NTSYS-pc. Numerical taxonomy and multivariate analysis system, Version 2.1. Setauket, New York
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. GENEPOP ‘ 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Saco A, Suárez H, Novoa B, Figueras A. A genomic and transcriptomic analysis of the C-type lectin gene family reveals highly expanded and diversified repertoires in bivalves. Mar Drugs. 2023;21:254. doi: 10.3390/md21040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa-Pinto A, Branco MS, Alexandrino PB, Fontaine MC, Baird SJE. Barriers to gene flow in the marine environment: insights from two common intertidal limpet species of the Atlantic and Mediterranean. PLOS One. 2012;7:0050330. doi: 10.1371/journal.pone.0050330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples J, Simpson JH (2019) Shelf sea and shelf slope fronts. In: Elías S (ed) Reference module in Earth systems and environmental sciences, vol. 1. Elsevier, pp 24–34
- Simpson JH, Pingree RD (1978). Shallow sea fronts produced by tidal stirring. In: Bowman MJ, Esaias WE (eds) Oceanic fronts in coastal processes. Springer, Berlin, Heidelberg
- Sokolov EP, Markert S, Hinzke T, Hirschfeld C, Becher D, Ponsuksili S, et al. Effects of hypoxia-reoxygenation stress on mitochondrial proteome and bioenergetics of the hypoxia-tolerant marine bivalve Crassostrea gigas. J Proteom. 2019;194:99–111. doi: 10.1016/j.jprot.2018.12.009. [DOI] [PubMed] [Google Scholar]
- Suberg LA, Miller PI, Wynn RB. On the use of satellite-derived frontal metrics in time series analyses of shelf-sea fronts, a study of the Celtic Sea. Deep Sea Res. Part I Oceanogr. Res Pap. 2019;149:103033. [Google Scholar]
- Teles-Machado A, Peliz Á, McWilliams JC, Couvelard X, Ambar I. Circulation on the Northwestern Iberian Margin: Vertical structure and seasonality of the alongshore flows. Prog. Oceanogr. 2016;140:134–153. doi: 10.1016/j.pocean.2015.05.021. [DOI] [Google Scholar]
- Theodosopoulos AN, Hund AK, Taylor SA. Parasites and host species barriers in animal hybrid zones. Trends Ecol Evol. 2019;34:19–30. doi: 10.1016/j.tree.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Thoma J, Stenitzer D, Grabherr R, Staudacher E. Identification, characterization, and expression of a beta-Galactosidase from Arion species (Mollusca) Biomolecules. 2022;12:1578. doi: 10.3390/biom12111578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrami DLJ, De Noia M, Telesca L, Handal W, Charrier G, Boudry P, et al. RAD sequencing sheds new light on the genetic structure and local adaptation of European scallops and resolves their demographic histories. Sci Rep. 2019;9:7455. doi: 10.1038/s41598-019-43939-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera M, Carlsson J, El Carlsson J, Cross T, Lynch S, Kamermans P, et al. Current genetic status, temporal stability and structure of the remnant wild European flat oyster populations: conservation and restoring implications. Mar Biol. 2016;163:239. doi: 10.1007/s00227-016-3012-x. [DOI] [Google Scholar]
- Vera M, Maroso F, Wilmes SB, Hermida M, Blanco A, Fernandez C, et al. Genomic survey of edible cockle (Cerastoderma edule) in the Northeast Atlantic: a baseline for sustainable management of its wild resources. Evol Appl. 2022;15:262–285. doi: 10.1111/eva.13340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera M, Pardo BG, Cao A, Vilas R, Fernandez C, Blanco A, et al. Signatures of selection for bonamiosis resistance in European flat oyster (Ostrea edulis): new genomic tools for breeding programs and management of natural resources. Evol Appl. 2019;12:1781–1796. doi: 10.1111/eva.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilas R, Vandamme SG, Vera M, Souza C, Maes GE, Volckaert FAM, et al. A genome scan for candidate genes involved in the adaptation of turbot (Scophthalmus maximus) Mar Genom. 2015;23:77–86. doi: 10.1016/j.margen.2015.04.011. [DOI] [PubMed] [Google Scholar]
- Villalba A, Iglesias D, Ramilo A, Darriba S, Parada JM, No E, et al. Cockle Cerastoderma edule fishery collapse in the Ria de Arousa (Galicia, NW Spain) associated with the protistan parasite Marteilia cochillia. Dis Aquat Org. 2014;109:55–80. doi: 10.3354/dao02723. [DOI] [PubMed] [Google Scholar]
- Wang M, Wang L, Ni D, Yi Q, Wang X, Jia Z, et al. The mRNA expression profiles demonstrating versatile roles of glutathione S-transferase genes in the mollusk Chlamys farreri. Invertebr Surviv J. 2018;15:302–315. [Google Scholar]
- Waples RS. Separating the wheat from the chaff: Patterns of genetic differentiation in high gene flow species. J Hered. 1998;89:438–450. doi: 10.1093/jhered/89.5.438. [DOI] [Google Scholar]
- Woo S, Jeon HY, Kim SR, Yum S. Differentially displayed genes with oxygen depletion stress and transcriptional responses in the marine mussel, Mytilus galloprovincialis. Comp Biochem Physiol D Genom Proteom. 2011;6:348–356. doi: 10.1016/j.cbd.2011.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for this study are available at Dryad Digital Repository (10.5061/dryad.xpnvx0kmr) and Supplementary material.