Abstract
Elevated impulsivity is a key component of attention-deficit hyperactivity disorder (ADHD), bipolar disorder and juvenile myoclonic epilepsy (JME). We performed a genome-wide association, colocalization, polygenic risk score, and pathway analysis of impulsivity in JME (n = 381). Results were followed up with functional characterisation using a drosophila model. We identified genome-wide associated SNPs at 8q13.3 (P = 7.5 × 10−9) and 10p11.21 (P = 3.6 × 10−8). The 8q13.3 locus colocalizes with SLCO5A1 expression quantitative trait loci in cerebral cortex (P = 9.5 × 10−3). SLCO5A1 codes for an organic anion transporter and upregulates synapse assembly/organisation genes. Pathway analysis demonstrates 12.7-fold enrichment for presynaptic membrane assembly genes (P = 0.0005) and 14.3-fold enrichment for presynaptic organisation genes (P = 0.0005) including NLGN1 and PTPRD. RNAi knockdown of Oatp30B, the Drosophila polypeptide with the highest homology to SLCO5A1, causes over-reactive startling behaviour (P = 8.7 × 10−3) and increased seizure-like events (P = 6.8 × 10−7). Polygenic risk score for ADHD genetically correlates with impulsivity scores in JME (P = 1.60 × 10−3). SLCO5A1 loss-of-function represents an impulsivity and seizure mechanism. Synaptic assembly genes may inform the aetiology of impulsivity in health and disease.
Subject terms: Genetics research, Epilepsy
Introduction
Impulsivity is a heritable behavioural trait leading to actions that are “poorly conceived, prematurely expressed, unduly risky, or inappropriate to the situation and that often result in undesirable consequences”1. Estimates of heritability for impulsivity from a study of twins were between 33% and 56% at ages 11–13 years, and between 19% and 44% at ages 14–162. Raised impulsivity is a key endophenotype of attention-deficit hyperactivity disorder (ADHD)3, bipolar disorder4 and juvenile myoclonic epilepsy (JME)5–7. ADHD is characterised by inattention, hyperactivity and impulsivity. Individuals with ADHD show more signs of impulsivity (attentional, non-planning and motor) compared to controls8. A previous genome-wide association study (GWAS) of impulsive personality traits (UPPS-P Sensation Seeking, Drug Experimentation and UPPS-P Negative Urgency) in 22,861 healthy individuals of European ancestry demonstrated two significant associated loci at 3p12.1 and 22q13.19. Variants at the 3p12.1 locus correlated with predicted Cell Adhesion Molecule–2 (CADM2) expression, in the putamen10, and the 22q13.1 locus near CACNA1I has been previously implicated in schizophrenia11. CADM2 mediates synaptic signalling and is highly expressed in the human cerebral cortex and cerebellum12. Given impulsivity is elevated in neuropsychiatric disorders, there may be shared genetic mechanisms across disorders and/or with impulsivity in the general population, however to our knowledge there has been no GWAS of impulsivity in any neuropsychiatric disorder.
Impulsivity is elevated in different epilepsies, but the evidence across multiple dimensions of impulsivity is strongest in JME5–7. JME is a common adolescent-onset syndrome characterised by awakening myoclonic, generalised tonic-clonic and absence seizures, often triggered by sleep deprivation. Trait impulsivity in JME is associated with the frequency of both myoclonic and absence seizures6, but it is not clear if this indicates a causal relationship or a common mechanism regulating both impulsivity and seizures, though convergent lines of evidence suggest the involvement of overlapping prefrontal-striatal networks in both JME and impulsivity13–20. Finding a shared aetiology would offer new therapeutic approaches for drug-resistant epilepsy.
The overall syndrome of JME has complex inheritance with few replicated susceptibility loci21,22, and other loci with less support22–24. A major challenge for epilepsies of complex inheritance is to explain the wide variation in phenotypic expression and treatment response between individuals. Forty-percent experience antiseizure medication (ASM) resistance or intolerance25. In addition, no current ASM modifies the lifelong disease course of JME and the pharmacological options are sparse, especially for women25. Hence novel treatments based on genetic disease mechanisms, such as those emerging for monogenic channelopathy and mTOR pathway epilepsies, are urgently needed26,27. Our methodological approach is to carry out genome-wide analysis of endophenotypes in JME such as impulsivity and clinically relevant outcomes such as ASM resistance, a strategy with predicted advantages for reducing heterogeneity, increasing statistical power28,29 and improving direct clinical translation for precision medicine.
Results
Genome-wide association analysis with BIS-Brief
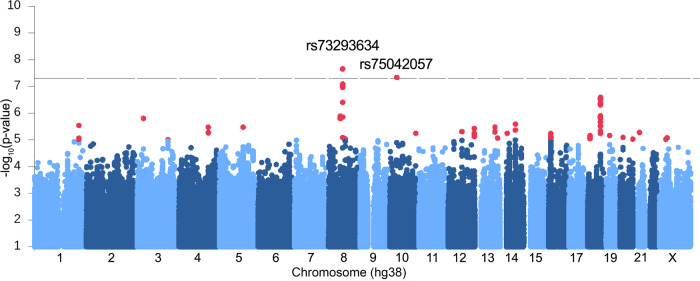
We investigated the influence of 8,950,360 variants on impulsivity in European ancestry patients with JME (n = 324) and a mega-analysis including all ancestries (n = 372), who self-rated their trait impulsivity using the Barratt Impulsivity Scale, eight-item BIS-Brief version30. We conducted a GWAS of BIS-Brief score (Supplementary Fig. 2) in the European subset, adjusted for sex, genotyping batch, age at consent, population stratification, and seizure frequency (Supplementary Table 2). We discovered two genome-wide significant loci, one on chromosome 8 (rs73293634 (G/T)) and one on chromosome 10 (rs75042057 (T/G) (Fig. 1, Table 1, Supplementary Figs. 3 and 4). Given the distribution of BIS-Brief was slightly right skewed, for sensitivity analysis we tested the SNP associations using an inverse rank normal transformed BIS-Brief phenotype as well. Qualitatively similar results were obtained with rs73293634 and rs75042057 demonstrating association with p = 3.1 × 10−8 and p = 1.4 × 10−7, respectively (Supplementary Table 3). The distribution of BIS-Brief by rs73293634 and rs75042057 genotypes are provided in Supplementary Fig. 5. In a mega-analysis comprised of all ancestral groups (Supplementary Fig. 6), these loci were further supported including by a nearby chromosome 8 SNP (rs146866040, r2 = 0.89) with stronger evidence of association in the combined ancestry mega-analysis as measured by the p value (P = 1.57 × 10−9; Table 1), providing cross-ancestral support for the locus.
Fig. 1. Manhattan plot showing GWAS with BIS-Brief score.
Linear regression was used to test association of each SNP with BIS-Brief. Sex, genotyping batch, age at consent, first three PCs, and the frequency of myoclonus or absence seizures were included as covariates in the model. We found two significant genome-wide associations on chromosome 8 (rs73293634 (G/T)) and 10 (rs75042057 (T/G)) in the analysis of 324 European individuals with JME. Variants below −log10P < 1 were omitted in the plot.
Table 1.
Summary of genome-wide associated variants for the GWAS of BIS scores in JME (n = 324).
| European GWAS (n = 324) | Mega-GWAS (n = 372) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variant ID (hg38) | Imputation r2 | MAF | Beta | SE | P value | MAF | Beta | SE | P value |
| chr8:69,884,968* rs73293634 (G/T) | 0.961 | 0.036 (T) | 5.42 | 0.91 | 7.5 × 10−9 | 0.041 | 4.55 | 0.79 | 1.61 × 10−8 |
| chr8:69,876,965 rs146866040 (A/G) | 0.979 | 0.032 (G) | 5.38 | 0.94 | 2.5 × 10−8 | 0.031 | 5.60 | 0.90 | 1.57 × 10−9 |
| chr10:34,202,650 rs75042057 (T/G) | 0.878 | 0.019 (G) | 7.51 | 1.33 | 3.6 × 10−8 | 0.022 | 6.60 | 1.19 | 4.99 × 10−8 |
Linear regression was used to test association of each SNP with BIS-Brief. Sex, genotyping batch, age at consent, first 3 PCs, and the frequency of myoclonus or absence seizures were included as covariates in the model in the European analysis. Sex, genotyping batch, and population stratification were included as covariates in the mega-GWAS.
All observed sample allele frequencies are comparable to those seen in the European 1000 Genomes (phase 3)60.
*The lead SNP for the mega-GWAS was rs146866040. The LD between them is r2 = 0.89 or D’ = 1.0.
rs73293634 falls in an intergenic region near SLCO5A1. The phenotypic variation explained (PVE) for rs73293634 was 10.1% in the European analysis. Although a second JME cohort with impulsivity measured is not available for replication, Watanabe et al. 31 reported a rs73293634 association with risk taking in the UK Biobank, where they asked the question “Would you describe yourself as someone who takes risks?” (OR (95% CI) = 1.032 (1.001–1.065), p = 0.04, minor allele frequency (MAF) = 0.03, N = 371,049). Association results posted on the same data by the Neale Lab32 with ~23 K fewer participants, provides a similar qualitative conclusion (β (SE) = 0.005 (0.003), p = 0.09, MAF = 0.03, N = 348,549). Two individuals with large structural deletions that include SLCO5A1 are reported in the Decipher Genomics database with seizures and neurodevelopmental disorder (www.deciphergenomics.org/gene/SLCO5A1/patient-overlap/cnvs).
The significant genome-wide association on chromosome 10 (rs75042057) falls in intron 22 of PARD3 (NM_001184785.2). The PVE by the SNP is 9.3%, although there are no variants in linkage disequilibrium with this SNP so further interrogation and confirmation of this locus is required. We note, however, that significant linkage (multipoint max LOD 4.23, alpha 0.34) was previously reported to this locus in French-Canadian families with idiopathic generalised epilepsy (IGE)33, of which JME is a common subtype. As well, rs75042057 was also associated with risk-taking in the UK Biobank (OR (95% CI) = 1.067 (1.029–1.106), p = 4.79E-4, MAF = 0.02, N = 371,049)31.
Colocalization analysis with gene expression
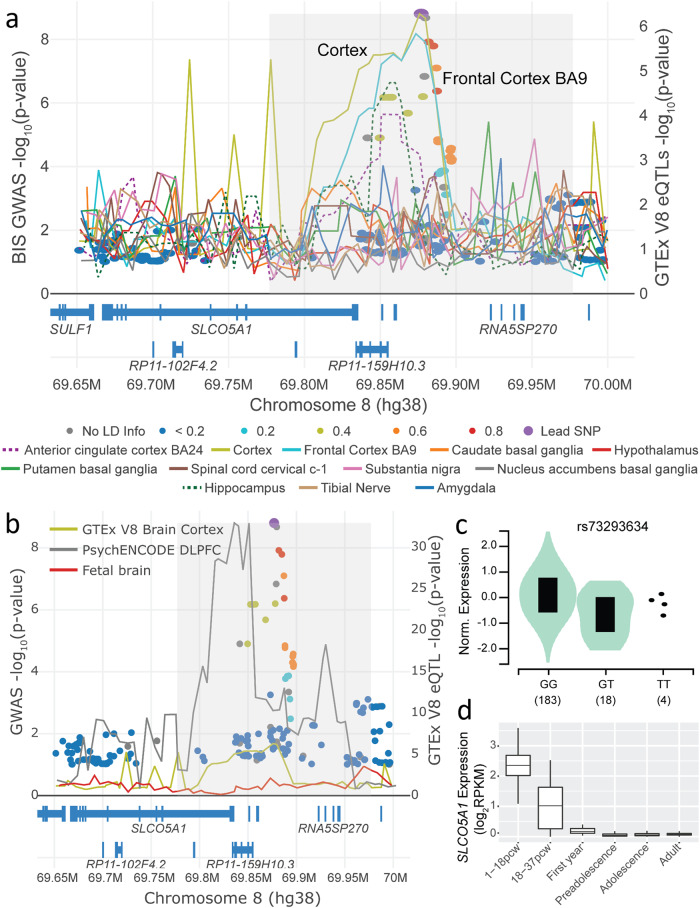
Since the GWAS-associated variants are not exonic, we next assessed whether the variants impact gene expression, and for which gene in which tissue of origin, by assessing colocalization of the genome-wide significant peaks with expression quantitative trait loci (eQTL) in brain tissues. We used eQTLs from the Genotype-Tissue Expression project (GTEx) v812, PsychENCODE34, and human fetal brains35 and combined them with the GWAS summary statistics from the mega-analysis, for colocalization analysis adjusting for multiple hypothesis testing36. Colocalization analysis with eQTLs from GTEx brain and tibial nerve tissues for genes at the locus (chr8:69,650,000–70,000,000, hg38) shows significant colocalization with SLCO5A1 in the cerebral cortex, and no colocalization with other genes in the region (Fig. 2a and Supplementary Fig. 7; Simple Sum 2 colocalization P = 9.5 × 10−3). The minor allele for the lead SNP rs73293634 (T) decreases expression in GTEx cerebral cortex (Fig. 2c). We found no significant colocalization with eQTLs from PsychENCODE34 and fetal brains35, although nearby variants in the locus in adult brains in PsychENCODE have, in general, a clear influence on SLCO5A1 expression (Fig. 2b). According to BrainSpan37,38, SLCO5A1 is highly expressed prenatally, with expression dropping after birth but remains detectable throughout adulthood (Fig. 2d). We did not observe significant colocalization at the chromosome 10 locus with eQTLs from adult brains in GTEx12, PsychENCODE34 or fetal brains35.
Fig. 2. LocusFocus70 plot for the GWAS with BIS-Brief in JME (circles) and eQTLs in GTEx12 brain and tibial nerve tissues for the SLCO5A1 gene (lines).
The Simple Sum 236 and COLOC269 colocalization methods implemented in LocusFocus (v1.4.9)70 were used to test for colocalization of the BIS-Brief genome-wide peaks with eQTL analyses brain tissues from GTEx v812, PsychENCODE34, and fetal brain35. a Colocalization figure from LocusFocus for the SLCO5A1 gene. Lines depict the minimum P value trace in a sliding window for SLCO5A1 eQTLs from GTEx, one line per tissue. Circles depict the GWAS with BIS-Brief, with the lead SNP in purple and pairwise LD with the lead SNP marked as shown in the legend, calculated using the 1000 Genomes Project60 European subset. Significant colocalization is observed for SLCO5A1 eQTLs in GTEx v8 for the cerebral cortex after increasing sample size in a mega-GWAS (n = 367, −log10 Simple Sum 236 P = 9.5 × 10−3). Colocalization analysis with only the Europeans is provided in Supplementary Fig. 7. Colocalization was also tested for all other nearby genes shown in the figure, but no other genes’ eQTLs colocalized with BIS-Brief GWAS (not shown). b Colocalization analysis with PsychENCODE eQTLs in the dorsolateral prefrontal cortex (DLPFC) (n = 1866)34, and eQTLs derived from second trimester fetal brains (n = 120)35, with GTEx’s brain cortex eQTL as in A provided for reference. Colocalization analysis results suggest no colocalization with either PsychENCODE (Simple Sum 2 P = 0.985) or fetal brain eQTLs (does not pass first stage test in Simple Sum 2 for having significant eQTLs in the region). c Violin plot for the eQTL effect of rs73293634 SNP on SLCO5A1 expression in the cerebral cortex from GTEx v8. d Expression change of SLCO5A1 from brains in various developmental stages from BrainSpan37,38. pcw, post conception weeks; preadolescence, 2–12 years old (inclusive); adolescence, 13–19 years old; adult, ≥20 years old (oldest samples are 40 years old). The centre lines represent the 50th percentile (median) and the bounds of the boxes are the 75th and 25th percentiles (interquartile range) with the whiskers being the largest value within 1.5 times the interquartile range above the 75th percentile and smallest values within 1.5 times the interquartile range below the 25th percentile.
Functional characterisation of SLCO5A1
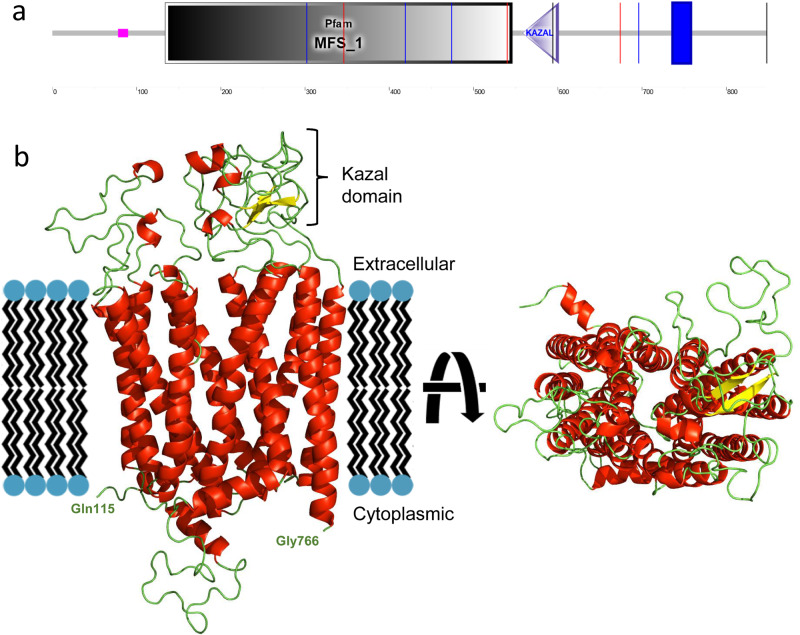
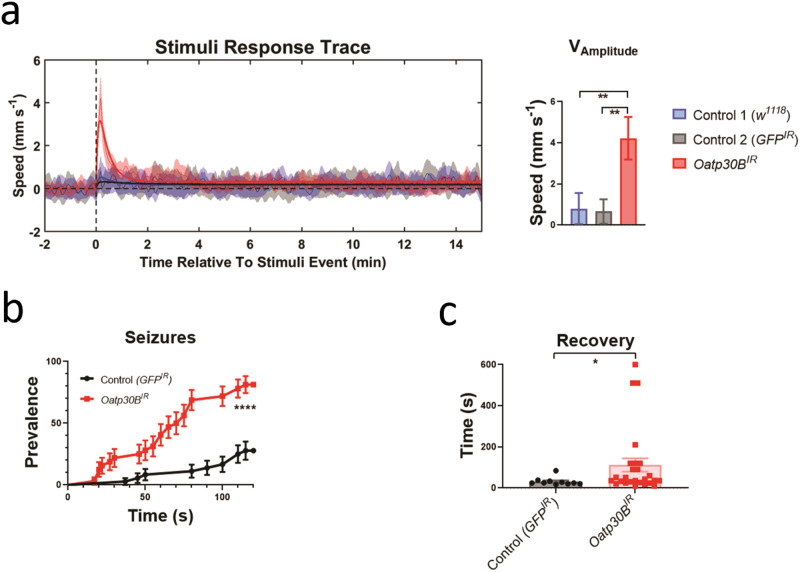
SLCO5A1 is a membrane-bound organic anion transporter with no known substrate39 (Fig. 3). We performed a full protein BLAST (BLASTp) search of the SLCO5A1 polypeptide sequence (NP_112220.2) on Drosophila melanogaster to identify the closest matching sequence alignment. While several members of the Oatp family were found to have significant homology, Oatp30B was the family member with the highest homology and a 37.66% identity and E-value of 2 × 10−150 (NP_995667.1). SLCO5A1 was the closest human analogue of Oatp30B also in a reverse BLASTp. Indeed, BLASTp of Oatp30B polypeptide sequence (Q9VLB3) across all species for conserved domains reveals this gene has conserved major facilitator superfamily (MFS), OATP, and Kazal domains (Fig. 3 and Supplementary Fig. 8). We therefore used an effective RNAi transgenic line (Supplementary Fig. 9A) to assess the effect of pan-neuronal adult knockdown of Oatp30B/SLCO5A1. Flies with reduced Oatp30B levels displayed a small but significant shortening of their lifespan (Supplementary Fig. 9B) and a striking over-reaction to vibration stimuli applied through the automated Drosophila Arousal Tracking (DART) system40, which elicit an otherwise modest activity response in two separate control fly genotypes (Fig. 4a). Additional analysis of locomotor behaviour clarifies that Oatp30B knockdown did not alter the speed of flies or the duration of each activity bout or the interval in between bouts of action (Supplementary Fig. 9C–E), indicating a specific defect in excessive response to stimuli. Furthermore, Oatp30B knockdown led to a dramatic increase in the frequency of seizure-like events (Fig. 4b) when exposed to hyperthermia, a trigger for seizures in Drosophila41. Recovery to full motility after seizure-like events was also significantly slower in flies with Oatp30B knockdown (Fig. 4c). These data establish a common causal link between Oatp30B/SLCO5A1 downregulation, startling behaviour, and susceptibility to seizure-like events.
Fig. 3. Domain architecture of human SLCO5A1.
a Schematic representation of the protein with the indication of recognised domains. A SMART analysis to identify structural domains confirmed the presence of two modules, Major Facilitator Superfamily (MFS) and a Kazal domain, interspaced with potentially unstructured sequences. The MFS transporters are membrane proteins capable of transporting small solutes in response to chemiosmotic ion gradients72,73. They are represented in many organisms from Archaea to Homo sapiens. MFS proteins target a wide range of substrates, including ions, carbohydrates, lipids, amino acids and peptides, nucleosides and other small molecules and transport them in both directions across the membrane74. The Kazal domain is an evolutionary conserved module usually acting as a serine-protease inhibitor. b Predicted model of the monomeric form of SLCO5A1 from amino acids 115–766, built using the SwissModel homology server (https://swissmodel.expasy.org) and utilising the template structure pdb:7eeb. Red: alpha helices; Yellow: Beta strands; Green: Loops.
Fig. 4. Startling reaction to trains of vibrations, increased seizure prevalence and increased post-seizure recovery time in flies with Oatp30B knockdown.
a Startling reaction to trains of vibrations. The UAS-Oatp30BIR (GD12775) transgenic or the control UAS-GFPIR were driven with nSyb-Gal4 and Ubi-Gal80ts. The w1118 strain is a control for the genetic background in absence of transgenes. Mean ±SEM **P < 0.01, One Way ANOVA, Tukey’s post-hoc test. Units are the vibration events experienced 6 times for each fly, n = 174–210. b Increased seizure prevalence. The UAS-Oatp30BIR (GD12775) transgenic or the control UAS-GFPIR were driven with nSyb-Gal4 and Ubi-Gal80ts. Percent ±SE ****P < 0.0001, Log-rank (Mantel-Cox) test, X2 24.68 for 1 df, n = 34–36. c Increased post-seizure recovery time. The UAS-Oatp30BIR (GD12775) transgenic or the control UAS-GFPIR were driven with nSyb-Gal4 and Ubi-Gal80ts. Mean ±SEM *P < 0.05, Mann Whitney non-parametric test, two tails, n = 10–26. Only flies that displayed a seizure within 120 s as in b have been included in the analysis.
Gene enrichment analyses
We next sought to assess whether there was additional signal in the GWAS where sub-GWAS significant variants could inform additional contributing genes or pathways and whether there were shared genetic contributions with other psychiatric or epilepsy phenotypes. We selected all variants displaying P ≤ 5 × 10−4 and annotated these variants to the transcription start site of the nearest gene resulting in 810 unique genes. Gene enrichment analysis using one-sided hypergeometric tests42 identified a 12.7-fold enrichment of associated genes from the presynaptic membrane organisation gene set (five out of nine genes; gene ontology (GO):0097090) and a 14.3-fold enrichment of associated genes from the presynaptic membrane assembly gene set (five out of eight genes; GO:0097105). These genes were PTEN, NLGN1, PTPRD, IL1RAPL1 and NLGN4X (Table 2). The combined PVE for the lead variants annotated to these five genes was 15.6% (25.8% with the addition of rs73293634 from the SLCO5A1 locus and rs75042057 from the PARD3 locus).
Table 2.
List of top variants annotated to the five presynaptic assembly genes enriched in the European GWAS of BIS in JME (n = 324).
| Gene | Location | Size | rsid | Beta | P value | PVE |
|---|---|---|---|---|---|---|
| PTPRD | chr9:8,314,246–10,613,002 | 2,298,757 | rs1781264 | 1.827 | 1.19E-04 | 0.042 |
| NLGN1 | chr3:173,398,448–174,286,644 | 888,197 | rs73177088 | 6.191 | 9.95E-04 | 0.044 |
| NLGN4X | chrX:5,890,042–6,228,867 | 338,826 | rs146813567 | −2.898 | 3.06E-04 | 0.039 |
| IL1RAPL1 | chrX:28,587,446–29,956,718 | 1,369,273 | rs5943492 | 1.039 | 8.73E-04 | 0.043 |
| PTEN | chr10:87,862,563–87,971,930 | 109,368 | rs112050451 | 5.158 | 1.27E-03 | 0.041 |
Variants with P ≤ 5 × 10−4 were annotated to the gene with the nearest transcription start site using the Ensembl Variant Effect Predictor (v94)62. This gene set was used as input in a GO enrichment analysis63,64, to test for enrichment in annotated pathways. One-sided hypergeometric tests were completed to identify over-representation of pathways42. To reduce the risk of false positive results, a permutation procedure65 was employed by randomly shuffling GWAS p values 2000 times, each time re-applying the P ≤ 5 × 10−4 threshold and calculating the hypergeometric test statistics. For each pathway, the final permutation-based p value was calculated as the percentage of the 2000 permutations that produced a p value less than or equal to the p value calculated from the non-permuted data. A pseudo count was added during this calculation to prevent calculating p values equal to 0.
PTPRD, Protein Tyrosine Phosphatase Receptor Type D; NLGN1, Neuroligin 1; NLGN4X, Neuroligin 4 X-Linked; IL1RAPL1, Interleukin 1 Receptor Accessory Protein Like 1; PTEN, Phosphatase and Tensin Homologue.
The permutation tests of presynaptic membrane organisation (GO:0097090) over-enrichment and of presynaptic membrane assembly (GO:0097104) over-enrichment both produced permutation-based p values of 0.0005.
Investigation of these 810 genes revealed further43–45 that there was a significant overlap with genes reported in the GWAS catalogue that contribute to phenotypes relevant to the predominance of JME seizures on awakening, impulsivity and metabolism: chronotype (66 out of 522 genes overlap, P = 2.92 × 10−12), obesity-related traits (77 out of 662 overlap, P = 2.69 × 10−12), general risk tolerance (30 out of 238 overlap, P = 2.30 × 10−5), and adventurousness (21/134, P = 3.70 × 10−5).
Polygenic risk score analysis
Given impulsivity is a major component of ADHD, risk taking, bipolar disorder and epilepsy, we tested and found that a higher ADHD polygenic risk score (PRS) was significantly associated with a higher BIS-Brief score (p = 1.60 × 10−3) (Supplementary Fig. 10). It should be noted that the lead SLCO5A1 SNP, rs73293634, was not present in the ADHD GWAS from which the PRS was calculated, but rs146866040 which is in high LD did not show evidence of association itself with ADHD (OR (SE) = 0.9481(0.0562), p = 0.34)46. The rs75042057 SNP on Chr10 was also not present in the ADHD dataset nor was there a proxy with R2 > 0.6 available. The risk-taking PRS was also nominally associated with a higher BIS-Brief score (p = 0.018). PRSs for bipolar disorder, generalised and focal epilepsy did not reach statistical significance for association with BIS-Brief score at the 5% or Bonferroni-corrected level of 1% (P = 0.08, 0.33 and 0.96, respectively) (Supplementary Table 4). Altogether this suggests that the impulsive trait seen in JME is an endophenotype that shares genetic architecture with impulsivity in the general population as well as with individuals diagnosed with ADHD.
Discussion
This is a GWAS of trait impulsivity in a neuropsychiatric disorder and we present convergent evidence for the role of SLCO5A1 in impulsivity and seizure susceptibility through triangulation47 with GWAS, independent replication, colocalization with gene expression and functional evaluation in Drosophila48. While several Oatp family members display significant homology to SLCO5A1, the identified Oatp30B was the closest polypeptide in a BLASTp search and SLCO5A1 was the human polypeptide with the highest homology in a reverse BLASTp search. Therefore, whereas our analysis does not rule out some contribution by other closely related Oatp genes, for instance Oatp26F, it has identified a major role of Oatp30B in regulating startling and seizure-like behaviour in Drosophila. In contrast to human SLCO5A1, Oatp30B is expressed in the nervous system at constant low to moderate levels throughout fly stages, from development to adulthood. This enables investigation of gene function in vivo, in adult flies, although it limits generalisation as an SLCO5A1-linked disease model.
One GWAS of impulsive traits in the general population identified genome-wide significant association with variants in the CADM2 gene. CADM2 encodes a cell adhesion protein from the SynCam Immunoglobulin superfamily of recognition molecules, important for synaptic organisation and specificity; association of variants at the CACNA1I locus has been observed in previous studies with schizophrenia9. Our GWAS did not show significant association with these previously reported general population-associated variants at the CADM2 and CACNA1I loci9 (P = 0.152, beta = −0.52 for rs139528938; and P = 0.32, beta = −0.35 for rs4522708; the latter a SNP with r2 = 0.87 with the reported SNP, rs199694726, in our BIS-Brief dataset). Genome-wide summary statistics were not available to make additional comparisons. Genome-wide summary statistics were available for the risk-taking phenotype in the UK Biobank31, in which we observed replication of our lead genome-wide significant SLCO5A1 variant, rs73293634.
Previous expression studies show that SLCO5A1 upregulates gene sets implicated in cell adhesion, synapse assembly and organisation, principally belonging to the cadherin superfamily39; and the enrichment for presynaptic membrane assembly and organisation pathways in our dataset includes genes encoding trans-synaptically interacting proteins that are implicated in a wide range of neuropsychiatric disorders49,50. Genetic correlation between ADHD and the BIS-Brief score suggests converging genetic influences across ADHD and epilepsy. Taken together, these results support an important role for specific cell recognition molecules in the organisation of synaptic connections as a mechanism for variation in impulsivity across health and disease51.
While prefrontal-striatal inhibitory control networks are implicated in impulse control, specifically between mPFC and nucleus accumbens18,20, a role for these limbic networks has only been hinted at in epilepsy. Striato-nigral circuits, preferentially involving the ventral striatum, have long ago been implicated in the regulation of generalised seizures in rodent models of generalised epilepsy19. Recently, an initiating role for cortico-striatal networks in absence seizures with generalised spike-and-wave discharges has been shown in the mouse model of genetic epilepsy caused by haploinsufficiency of STXBP152, specifically by reduced cortical excitatory transmission onto striatal fast-spiking interneurons. The startling and the seizure-like phenotype of the SLCO5A1/Oatp30B knockdown in Drosophila suggests the genetic co-causality of startling and seizures. While it is not possible to define startling as the Drosophila equivalent of impulsivity, the two traits share some commonality in the lack of moderation in behaviour. This offers some additional support to the idea that excitatory-inhibitory imbalance in the prefrontal-striatal network may predispose simultaneously to epilepsy and impulsivity substrates and invites new approaches to neuromodulation of generalised seizures.
Methods
Human participants
We collected cross-sectional clinical and genetic data from the Biology of Juvenile Myoclonic Epilepsy (BIOJUME) consortium study, which focuses on gathering cases with JME (n = 864)25. Inclusion criteria have been discussed previously6. BIOJUME is a study across 50 sites in 10 countries (Appendix). Furthermore, all participants’ medical history was reviewed by a phenotyping committee to validate the diagnosis of JME. Written informed consent was obtained from all participants prior to inclusion in the study and ethical approval from the UK Health Research Authority, South Central Oxford C Research Ethics Committee (16/SC/0266) and all other collaborating sites was obtained. The SickKids Research Ethics Board of The Hospital for Sick Children (1000033784) also gave ethical approval for this work.
Barratt impulsivity scale-brief (BIS-Brief)
We collected self-rating of trait impulsivity through the BIS-brief6,30. The BIS-Brief is a short version of the BIS, one of the most commonly used measures of impulsiveness. The current version of BIS (BIS-11) includes 30-items measuring 3 theoretical subtraits: attentional, motor, and non-planning impulsiveness. BIS-Brief is a unidimensional scale including 8 of the original BIS-11 items generating a total score ranging from 8 to 32. BIS-Brief demonstrated similar indices of construct validity observed for the BIS-11 total score. Using BIS-Brief in large epidemiological studies of psychiatric disorders reduces the burden on respondents without loss of information29.
Genotyping quality control
DNA was extracted from blood by each consortium site and sent to The Centre for Applied Genomics at The Hospital for Sick Children in Toronto for genotyping. We genotyped participants’ DNA in four batches (n = 702) using the Illumina Omni 2.5 array. SNPs were called using the self-clustering method in Genome Studio. We performed quality control (QC) for each genotyped batch using PLINK v1.90b6.1853 and custom in-house scripts. Briefly, we removed individuals and variants with call rates below 90%; samples with sex mismatches and/or high heterozygosity; males with heterozygous calls for X chromosome markers (non-pseudoautosomal region); and females with non-missing calls for markers on the Y chromosome. We retained heterozygous calls for mitochondrial markers in both sexes (i.e., due to heteroplasmy). We obtained an unrelated sample by using KING v.2.2.4 software’s54 --unrelated option (that is, those with estimated kinship coefficient <0.088). We corrected and updated the ped file with all found relationships, and identified markers with Mendelian errors using PEDSTATS 0.6.1255. We flagged 399 markers but did not remove those out of Hardy–Weinberg Equilibrium (P < 10−4). We conducted principal component analysis adjusted using the kinship matrix output by KING using PC-AiR in the GENESIS v2.16.0 package56.
We performed quality control on each genotyping batch separately, followed by removal of ambiguous A/T, G/C SNPs, chr0 SNPs, indels, monomorphic variants, and duplicate variants; and performed strand alignment using Will Rayner’s alignment files (www.well.ox.ac.uk/~wrayner/strand/), then merged all batches. We re-analysed and removed cryptic relationships across batches. The final merged set contained 1,489,917 variants, 695 individuals (241 males, 454 females) including 23 related pairs (for association analyses however, an unrelated set was selected).
Genotype imputation
We used the McCarthy Tools v4.3.0 to prepare the genotype data for imputation (www.well.ox.ac.uk/~wrayner/tools/HRC-1000G-check-bim-v4.3.0.zip) using TOPMed as the reference panel (r2@1.0.0) on the TOPMed imputation server57–59. We converted coordinates from hg37 to hg38 coordinates using strand files (www.well.ox.ac.uk/~wrayner/strand/InfiniumOmni2-5-8v1-4_A1-b38-strand.zip). We merged the pseudoautosomal region (PAR) using PLINK’s --merge-x option and checked variants using the HRC checking tool. We removed a total of 282,660 variants due to no matches in the reference (but still analysed for association with BIS-Brief afterwards), and 1,739,329 variants remained for imputation on the server. We used Eagle v2.4 for phasing, and minimac v4 v1.0.2 for imputation. We kept variants with imputation quality score r2 > 0.4 and MAF > 1% for analysis. A total of 8,950,360 variants remained for association analysis.
Genome-wide association analysis
We included for analysis 381 individuals who passed phenotype QC with complete BIS-Brief rating. From these, four failed genotyping QC, and one individual was removed due to cryptic relatedness (n = 376). The mega-GWAS analysis consisted of a total of 372 unrelated individuals adjusted for sex, genotyping batch, and population stratification (Supplementary Fig. 1). The mega-GWAS was used for colocalization analysis of the genome-wide association peak on chromosome 8. We identified 329 patients as European ancestry (defined as within 6 standard deviations from the 1000 Genomes60 European cluster in a principal component analysis). Among these, five patients had missing information on seizure frequency, so we used 324 individuals for the genome-wide association analysis. The current sample size is sufficient to detect genetic variants that explain 12% of the variance in the BIS-Brief score with 80% power after adjusting for multiple hypothesis testing at the genome-wide significance level. We adjusted for sex, genotyping batch, age at consent, population stratification, and the frequency of myoclonus or absence seizures. The relationship of the frequency of myoclonus or absence seizures, and its relationship with ASM and sex with trait impulsivity in JME, has been described previously and was thus adjusted for in current regression analyses6,25. All analyses were conducted in the European subset unless noted otherwise. Chromosome X (non-pseudoautosomal region) was analysed with males coded as zero for the reference allele and two for the alternate allele, under the assumption of X-inactivation61. Genome-wide significant loci were further investigated for replication of association with risk-taking phenotypes in the general population using publicly available summary statistics31,32.
Gene enrichment analysis
Variants with P ≤ 5 × 10−4 were annotated to the gene with the nearest transcription start site using the Ensembl Variant Effect Predictor (v94)62. This gene set was used as input in a GO enrichment analysis63,64, to test for enrichment in annotated pathways. One-sided hypergeometric tests were completed to identify over-representation of pathways42. To reduce the risk of false positive results, a permutation procedure65 was employed by randomly shuffling GWAS p values 2000 times, each time re-applying the P ≤ 5 × 10−4 threshold and calculating the hypergeometric test statistics. For one pathway, the final permutation-based p value was calculated as the percentage of the 2000 permutations that produced a p value less than or equal to the p value calculated from the non-permuted data. A pseudo count was added during this calculation to prevent calculating p values equal to 0.
Phenome-wide association study (PheWAS) analysis
We queried the top associated genome-wide variant and the top associated variant for each of the nine presynaptic assembly enriched genes across PheWAS databases: GWAS Atlas (https://atlas.ctglab.nl/), Global Biobank Engine66, PheWeb67, and Gene Atlas68.
We used PheWeb portals:
UK Biobank: https://pheweb.org/MGI-freeze2/
Oxford Brain Imaging Genetics (BIG) Project: http://big.stats.ox.ac.uk/
fastGWA: https://yanglab.westlake.edu.cn/resources/ukb_fastgwa/imp/
PRS analysis
Clumping and thresholding were used to calculate ADHD, risk taking, bipolar disorder, generalised epilepsy, and focal epilepsy PRS in individuals of European ancestry using PLINK v1.953. Five PRS were calculated. A Bonferroni-corrected critical value for significance would therefore be p < 0.05/5 = 0.01. The source of summary statistics used, variant filtering, clumping and thresholding details are summarised in Supplementary Table 1. PRS values were generated by weighting selected SNPs after clumping and thresholding by the additive scale effect (log10(OR)/Beta), and then summing over the variants. The PRS values were then centred to the mean. Association of PRSs with BIS-Brief was tested using linear regression with age, sex, and frequency of absence/myoclonic seizure as covariates in the model.
Colocalization analysis
We used the Simple Sum 236 and COLOC269 colocalization methods as implemented in LocusFocus70 (v1.4.9) to test for colocalization of the genome-wide peaks with eQTL analyses in brain tissues in GTEx v812, PsychENCODE34, and fetal brain35. For the genome-wide associated locus on chromosome 8, we performed colocalization analysis using both the mega-GWAS and Europeans-only GWAS. The required significance threshold, after multiple testing of all colocalization datasets analysed was 0.01.
Domain architecture of SLCO5A1
A BLAST search against the entire Protein Data Base (PDB) identified only one hit with a convincingly high E-value (1e-55) that pointed to the Chain L of the Kazal-like domain containing mice protein (7EEB). The search had a 26% identity and a coverage of 74%. After this hit, the other four identified sequences had E values > 0.002, clearly distinguishing between significant and non-significant hits. 7EEB is a large complex containing several subunits, among which is SLCO6C1, which is the region scoring for SLCO5A1.
Phenotypic variance explained
To assess the PVE by a SNP or a group of SNPs, we calculated the partial r2 as the proportion of the residual sum of squares (RSS) reduced when adding the SNP (or group of SNPs) to the base regression model with all covariates.
siRNA probe design and knockdown of Oatp30B in Drosophila melanogaster
Drosophila
Flies were maintained and crossed at 18 °C. All ageing was done in a controlled environment of 29 °C and 60% humidity.
Stocks
ubiGal80ts // UAS-Oatp30BIR (GD12775) obtained from the VDRC // w1118, nSyb-Gal4, TubGal4 and UAS-GFPIR obtained from the BDSC.
Lifespan
Lifespan analysis was performed as previously reported41. All crosses were maintained at 18 °C during the developmental stages of the progeny. Newly eclosed adult flies were collected within 5 days at 18 °C. Females and males were pooled together and equally distributed within vials.
Motor behaviour assay
Single-fly tracking was carried out as previously described41. In each of three experiments, up to 12 flies per genotype, aged 15 days (adult stage) at 29 °C to allow RNAi expression and knockdown, were placed into individual round six-wells arenas. The protocol used consisted of 6 stimuli events equally split during a period of 2 h and 15 min, the first one starting after 30 min of recording, and the last one 30 min before the end of the protocol. Each stimuli event was composed of five vibrations of 200 ms spaced by 500 ms. The x/y position of each single fly was tracked and analysed using the DART software in order to evaluate the relative speed and activity before, during and after the stimuli event. The speed analysis was used for the “Stimuli Response Trace” and the general activity used to deduce “Active Speed”, “Mean Bout Length” and “Inter-Bout Interval”, using a custom-made modification of the DART software40.
Heat-induced seizure assay
Flies aged 15 days at 29 °C to allow RNAi expression and knockdown were isolated into new plastic vials without food for 10–20 min before immersion in a 42 °C water bath for 120 seconds. Each tube was video recorded during and post immersion and seizures were defined as a period of brief leg twitches, convulsions, and failure to maintain standing posture. Flies were, thereafter, allowed to recover at room temperature and the time to recover from seizure was calculated only for flies that had undergone seizures. All experiments were randomised and double-blinded.
RNA extraction and qPCR
RNA was extracted as previously reported71 from 15 adult flies of both sexes, aged 15 days at 29 °C to allow RNAi expression and knockdown, using TriZol (Thermo-Fischer). cDNA was generated using SuperScript III Reverse Transcriptase (Thermo-Fischer). Quantitative PCR was performed in combination with qPCRBIO SyGreen Blue mix (PCR Biosystems) on Quantstudio 7 from real-time PCR system (Thermo-Fischer). eIF4a was used as housekeeping control. The following oligos were used: Oatp30B Fw (GAATCCGACCAACCGCCTGA), Oatp30B Rv (ATGGATTCCTGCCGCCTGTG), eIF4a Fw (CGTGAAGCAGGAGAACTGG), eIF4a Rv (CATCTCCTGGGTCAGTTG).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work was supported by the Canadian Institutes of Health Research (CIHR) Operating Grant, FRN: 142405 (D.K.P., L.J.S.) and CIHR Foundation Grant, FRN: 167282 (L.J.S.); UK Medical Research Council, Centre for Neurodevelopmental Disorders MR/N026063/1 (D.K.P., M.P.R.); UK Medical Research Council, Programme Grant MR/K013998/1, (M.P.R.); PhD stipend from UK Medical Research Council and the Sackler Institute for Translational Neurodevelopment (A.S.); NIHR Specialist Biomedical Research Centre for Mental Health of South London and Maudsley National Health Service Foundation Trust (D.K.P., M.P.R.); UK Engineering and Physical Sciences Research Council, Centre for Predictive Modelling in Healthcare (EP/N014391/1 (M.P.R.)); DINOGMI Department of Excellence of MIUR 2018–2022 (legge 232 del 2016 (P.S.)); Wales BRAIN Unit and Research Delivery Staff funded by Welsh Government through Health and Care Research Wales (K.H.); Biomarin srl, ENECTA srl, GW Pharmaceuticals, Kolfarma srl. and Eisai (P.S.); South-Eastern Regional Health Authority, Norway (Project Number 2016129 (K.K.S.)); The Research Council of Norway (Project Number 299266 (M.S.)); Epilepsy Research UK (RHT, M.P.R.); Health & Care Research Wales (M.P.R.), Wales Gene Park (M.P.R.), Abertawe Bro Morgannwg University NHS R&D (M.P.R.); UCB (G.R.); Nationwide Children’s Hospital (D.A.G.); Odense University Hospital (J.G.); University of Southern Denmark (17/18517 (C.P.B.)); Grants NC/V001051/1 from the NC3Rs (M.F.), European Union’s Horizon 2020 Research and Innovation Programme (765912 - DRIVE - H2020-MSCA-ITN-2017 (H.J.)) and Action Medical Research (GN2446 (H.J., M.F.)). L.J.S. is a Canada Research Chair and this research was undertaken, in part, thanks to funding from the Canada Research Chairs Programme. Work supported by #NEXTGENERATIONEU (NGEU) and funded by the Ministry of University and Research (MUR), National Recovery and Resilience Plan (NRRP), project MNESYS (PE0000006)–A Multiscale integrated approach to the study of the nervous system in health and disease (DN. 1553 11.10.2022)- IRCCS ‘G. Gaslini’ is a meber of ERN-Epicare.
Author contributions
L.J.S. and D.K.P. contributed to the conception and study design. D.R., N.P., A.S., A.C., F.L., A.H., K.K., D.K.P. and L.J.S. contributed to data management and project administration. D.M.A., C.P.B., C.Y.F., E.G., J.G., D.A.G., C.D., F.M., K.H., K.S.L., R.S.M., C.C.N., A.O., K.K.S., G.R., P.S., M.S., I.T., R.H.T., J.Z., M.P.R., D.K.P. and L.J.S. contributed to acquisition of study data. D.R., E.J.S., N.P., A.S., C.D., F.M., S.T., H.J., M.P.R., A.P., M.F., D.K.P. and L.J.S. contributed to analysis of data. D.R., E.J.S., N.P., A.S., M.F., L.J.S., and D.K.P. contributed to drafting the manuscript. Members of the BIOJUME consortium are listed in the appendix.
Data availability
eQTL data are available for download from GTEx (https://gtexportal.org/home), PsychENCODE (http://resource.psychencode.org/), and fetal brains (10.6084/m9.figshare.6881825). GWAS summary statistics for this study are available for download from our website (https://lab.research.sickkids.ca/strug/softwareandresources/).
Competing interests
D.A., K.K.S., R.H.T. and J.Z. report honoraria from UCB Pharma (manufacturer of levetiracetam) and R.H.T. reports honoraria from Sanofi (manufacturer of sodium valproate). K.H. reports honoraria from UCB Pharma, Eisai and GW Pharma. M.S. reports honoraria from UCB Pharma and Eisai. G.R. reports honoraria from UCB Pharma (manufacturer of levetiracetam), from EISAI (manufacturer of perampanel), from Angelini Pharma (manufacturer of cenobamate). R.H.T. reports honorarium from Arvelle/Angelini, Bial, Eisai, GW Pharma/Jazz, Zogenix. All other authors report no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Deb K. Pal, Email: deb.pal@kcl.ac.uk
Lisa J. Strug, Email: Lisa.Strug@utoronto.ca
the BIOJUME Consortium:
Zuzana Šobíšková, Cechovaz Pracoviste, Michaela Kajsova, Rikke S. Møller, Elena Gardella, Maria Miranda, Pasquale Striano, Alessandro Orsini, Pronab Bala, Amy Kitching, Kate Irwin, Lorna Walding, Lynsey Adams, Uma Jegathasan, Rachel Swingler, Rachel Wane, Julia Aram, Nikil Sudarsan, Dee Mullan, Rebecca Ramsay, Vivien Richmond, Mark Sargent, Paul Frattaroli, Matthew Taylor, Marie Home, Sal Uka, Susan Kilroy, Tonicha Nortcliffe, Halima Salim, Kelly Holroyd, Alison McQueen, Dympna Mcaleer, Dina Jayachandran, Dawn Egginton, Bridget MacDonald, Michael Chang, David Deekollu, Alok Gaurav, Caroline Hamilton, Jaya Natarajan, Inyan Takon, Janet Cotta, Nick Moran, Jeremy Bland, Rosemary Belderbos, Heather Collier, Joanne Henry, Matthew Milner, Sam White, Michalis Koutroumanidis, William Stern, Jennifer Quirk, Javier Peña Ceballos, Anastasia Papathanasiou, Ioannis Stavropoulos, Dora Lozsadi, Andrew Swain, Charlotte Quamina, Jennifer Crooks, Tahir Majeed, Sonia Raj, Shakeelah Patel, Michael Young, Melissa Maguire, Munni Ray, Caroline Peacey, Linetty Makawa, Asyah Chhibda, Eve Sacre, Shanaz Begum, Martin O’ Malley, Lap Yeung, Claire Holliday, Louise Woodhead, Karen Rhodes, Shan Ellawela, Joanne Glenton, Verity Calder, John Davis, Paul McAlinden, Sarah Francis, Lisa Robson, Karen Lanyon, Graham Mackay, Elma Stephen, Coleen Thow, Margaret Connon, Martin Kirkpatrick, Susan MacFarlane, Anne Macleod, Debbie Rice, Siva Kumar, Carolyn Campbell, Vicky Collins, William Whitehouse, Christina Giavasi, Boyanka Petrova, Thomas Brown, Catie Picton, Michael O’Donoghue, Charlotte West, Helen Navarra, Seán J. Slaght, Catherine Edwards, Andrew Gribbin, Liz Nelson, Stephen Warriner, Heather Angus-Leppan, Loveth Ehiorobo, Bintou Camara, Tinashe Samakomva, Rajiv Mohanraj, Vicky Parker, Rajesh Pandey, Lisa Charles, Catherine Cotter, Archana Desurkar, Alison Hyde, Rachel Harrison, Markus Reuber, Rosie Clegg, Jo Sidebottom, Mayeth Recto, Patrick Easton, Charlotte Waite, Alice Howell, Jacqueline Smith, Shyam Mariguddi, Zena Haslam, Elizabeth Galizia, Hannah Cock, Mark Mencias, Samantha Truscott, Deirdre Daly, Hilda Mhandu, Nooria Said, Mark Rees, Seo-Kyung Chung, Owen Pickrell, Beata Fonferko-Shadrach, Mark Baker, Fraser Scott, Naveed Ghaus, Gail Castle, Jacqui Bartholomew, Ann Needle, Julie Ball, Andrea Clough, Shashikiran Sastry, Charlotte Busby, Amit Agrawal, Debbie Dickerson, Almu Duran, Muhammad Khan, Laura Thrasyvoulou, Eve Irvine, Sarah Tittensor, Jacqueline Daglish, Sumant Kumar, Claire Backhouse, Claire Mewies, Julia Aram, Nikil Sudarsan, Dee Mullan, Rebecca Ramsay, Vivien Richmond, Denise Skinner, Mark Sargent, Rahul Bharat, Sarah-Jane Sharman, Arun Saraswatula, and Helen Cockerill
Supplementary information
The online version contains supplementary material available at 10.1038/s41525-023-00370-z.
References
- 1.Daruna, J. H. & Barnes, P. A. In: The impulsive client: theory, research and treatment (eds W. G. McCown, J. L. Johnson, & M. B. Shure) 23–37 (American Psychological Association, 1993).
- 2.Niv S, Tuvblad C, Raine A, Wang P, Baker LA. Heritability and longitudinal stability of impulsivity in adolescence. Behav. Genet. 2012;42:378–392. doi: 10.1007/s10519-011-9518-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalley JW, Robbins TW. Fractionating impulsivity: neuropsychiatric implications. Nat. Rev. Neurosci. 2017;18:158–171. doi: 10.1038/nrn.2017.8. [DOI] [PubMed] [Google Scholar]
- 4.Ramirez-Martin A, Ramos-Martin J, Mayoral-Cleries F, Moreno-Kustner B, Guzman-Parra J. Impulsivity, decision-making and risk-taking behaviour in bipolar disorder: a systematic review and meta-analysis. Psychol. Med. 2020;50:2141–2153. doi: 10.1017/S0033291720003086. [DOI] [PubMed] [Google Scholar]
- 5.Smith A, Syvertsen M, Pal DK. Meta-analysis of response inhibition in juvenile myoclonic epilepsy. Epilepsy Behav. 2020;106:107038. doi: 10.1016/j.yebeh.2020.107038. [DOI] [PubMed] [Google Scholar]
- 6.Shakeshaft, A. et al. Trait impulsivity in juvenile myoclonic epilepsy. Ann Clin Transl Neurol, 10.1002/acn3.51255 (2020). [DOI] [PMC free article] [PubMed]
- 7.Wandschneider B, et al. Risk-taking behavior in juvenile myoclonic epilepsy. Epilepsia. 2013;54:2158–2165. doi: 10.1111/epi.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malloy-Diniz L, Fuentes D, Leite WB, Correa H, Bechara A. Impulsive behavior in adults with attention deficit/ hyperactivity disorder: characterization of attentional, motor and cognitive impulsiveness. J. Int. Neuropsychol. Soc. 2007;13:693–698. doi: 10.1017/S1355617707070889. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez-Roige S, et al. Genome-wide association studies of impulsive personality traits (BIS-11 and UPPS-P) and drug experimentation in up to 22,861 adult research participants identify loci in the CACNA1I and CADM2 genes. J. Neurosci. 2019;39:2562–2572. doi: 10.1523/JNEUROSCI.2662-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbeira AN, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GTEx Consortium. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Muircheartaigh J, et al. Focal structural changes and cognitive dysfunction in juvenile myoclonic epilepsy. Neurology. 2011;76:34–40. doi: 10.1212/WNL.0b013e318203e93d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller SS, et al. Microstructural and volumetric abnormalities of the putamen in juvenile myoclonic epilepsy. Epilepsia. 2011;52:1715–1724. doi: 10.1111/j.1528-1167.2011.03117.x. [DOI] [PubMed] [Google Scholar]
- 15.Landvogt C, Buchholz H-G, Bernedo V, Schreckenberger M, Werhahn KJ. Alteration of dopamine D2/D3 receptor binding in patients with juvenile myoclonic epilepsy: alteration of dopamine D2/D3 receptor binding in JME. Epilepsia. 2010;51:1699–1706. doi: 10.1111/j.1528-1167.2010.02569.x. [DOI] [PubMed] [Google Scholar]
- 16.Ciumas C, et al. Reduced dopamine transporter binding in patients with juvenile myoclonic epilepsy. Neurology. 2008;71:788–794. doi: 10.1212/01.wnl.0000316120.70504.d5. [DOI] [PubMed] [Google Scholar]
- 17.Dalley JW, Everitt BJ, Robbins TW. Impulsivity, compulsivity, and top-down cognitive control. Neuron. 2011;69:680–694. doi: 10.1016/j.neuron.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Dalley JW, Roiser JP. Dopamine, serotonin and impulsivity. Neuroscience. 2012;215:42–58. doi: 10.1016/j.neuroscience.2012.03.065. [DOI] [PubMed] [Google Scholar]
- 19.Deransart C, Vercueil L, Marescaux C, Depaulis A. The role of basal ganglia in the control of generalized absence seizures. Epilepsy Res. 1998;32:213–223. doi: 10.1016/s0920-1211(98)00053-9. [DOI] [PubMed] [Google Scholar]
- 20.Cho SS, et al. Morphometric correlation of impulsivity in medial prefrontal cortex. Brain Topogr. 2013;26:479–487. doi: 10.1007/s10548-012-0270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos BPD, et al. Genetic susceptibility in juvenile myoclonic epilepsy: systematic review of genetic association studies. PLoS One. 2017;12:e0179629. doi: 10.1371/journal.pone.0179629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International League Against Epilepsy Consortium on Complex Epilepsies. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat. Commun. 2018;9:5269. doi: 10.1038/s41467-018-07524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai D, et al. DNA variants in coding region of EFHC1: SNPs do not associate with juvenile myoclonic epilepsy. Epilepsia. 2008;50:1184–1190. doi: 10.1111/j.1528-1167.2008.01762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey JN, et al. Variant intestinal-cell kinase in juvenile myoclonic epilepsy. N. Engl. J. Med. 2018;378:1018–1028. doi: 10.1056/NEJMoa1700175. [DOI] [PubMed] [Google Scholar]
- 25.Shakeshaft A, et al. Sex-specific disease modifiers in juvenile myoclonic epilepsy. Sci. Rep. 2022;12:2785. doi: 10.1038/s41598-022-06324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M, et al. Antisense oligonucleotide therapy reduces seizures and extends life span in an SCN2A gain-of-function epilepsy model. J. Clin. Invest. 2021;131:e152079. doi: 10.1172/JCI152079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karalis V, Bateup HS. Current approaches and future directions for the treatment of mTORopathies. Dev. Neurosci. 2021;43:143–158. doi: 10.1159/000515672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall M-H, Smoller JW. A new role for endophenotypes in the GWAS era: functional characterization of risk variants. Harv. Rev. Psychiatry. 2010;18:67–74. doi: 10.3109/10673220903523532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manchia M, et al. The impact of phenotypic and genetic heterogeneity on results of genome wide association studies of complex diseases. PLoS One. 2013;8:e76295. doi: 10.1371/journal.pone.0076295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinberg L, Sharp C, Stanford MS, Tharp AT. New tricks for an old measure: the development of the Barratt Impulsiveness Scale-Brief (BIS-Brief) Psychol. Assess. 2013;25:216–226. doi: 10.1037/a0030550. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe K, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019;51:1339–1348. doi: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 32.Neale’s Lab UK Biobank GWAS Results Round 2 (Imputed v3 - File Manifest Release 20180731), http://www.nealelab.is/uk-biobank.
- 33.Kinirons P, et al. A novel locus for idiopathic generalized epilepsy in French-Canadian families maps to 10p11. Am. J. Med. Genet. A. 2008;146A:578–584. doi: 10.1002/ajmg.a.32139. [DOI] [PubMed] [Google Scholar]
- 34.Wang, D. et al. Comprehensive functional genomic resource and integrative model for the human brain. Science362, 10.1126/science.aat8464 (2018). [DOI] [PMC free article] [PubMed]
- 35.O’Brien HE, et al. Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol. 2018;19:194. doi: 10.1186/s13059-018-1567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F, Panjwani N, Wang C, Sun L, Strug LJ. A flexible summary statistics-based colocalization method with application to the mucin cystic fibrosis lung disease modifier locus. Am. J. Hum. Genet. 2022;109:253–269. doi: 10.1016/j.ajhg.2021.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.BrainSpan Atlas of the Developing Human Brain [Internet], http://brainspan.org.
- 38.Sunkin SM, et al. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013;41:D996–D1008. doi: 10.1093/nar/gks1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sebastian K, et al. Characterization of SLCO5A1/OATP5A1, a solute carrier transport protein with non-classical function. PLoS One. 2013;8:e83257. doi: 10.1371/journal.pone.0083257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faville R, Kottler B, Goodhill GJ, Shaw PJ, van Swinderen B. How deeply does your mutant sleep? Probing arousal to better understand sleep defects in Drosophila. Sci. Rep. 2015;5:8454. doi: 10.1038/srep08454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazaud D, et al. Transcriptional regulation of the glutamate/GABA/glutamine cycle in adult glia controls motor activity and seizures in drosophila. J. Neurosci. 2019;39:5269–5283. doi: 10.1523/JNEUROSCI.1833-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 43.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demontis D, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawlor DA, Tilling K, Davey Smith G. Triangulation in aetiological epidemiology. Int. J. Epidemiol. 2016;45:1866–1886. doi: 10.1093/ije/dyw314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parker L, Howlett IC, Rusan ZM, Tanouye MA. Seizure and epilepsy: studies of seizure disorders in Drosophila. Int. Rev. Neurobiol. 2011;99:1–21. doi: 10.1016/B978-0-12-387003-2.00001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uhl GR, Martinez MJ. PTPRD: neurobiology, genetics, and initial pharmacology of a pleiotropic contributor to brain phenotypes. Ann. N Y Acad. Sci. 2019;1451:112–129. doi: 10.1111/nyas.14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Z, Xiao X, Zhang Z, Li M. Genetic insights and neurobiological implications from NRXN1 in neuropsychiatric disorders. Mol. Psychiatry. 2019;24:1400–1414. doi: 10.1038/s41380-019-0438-9. [DOI] [PubMed] [Google Scholar]
- 51.Sanes JR, Zipursky SL. Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell. 2020;181:536–556. doi: 10.1016/j.cell.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Miyamoto H, et al. Impaired cortico-striatal excitatory transmission triggers epilepsy. Nat. Commun. 2019;10:1917. doi: 10.1038/s41467-019-09954-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 56.Conomos MP, Miller MB, Thornton TA. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Das S, et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taliun D, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Genomes Project Consortium, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen B, Craiu RV, Strug LJ, Sun L. The X factor: a robust and powerful approach to X-chromosome-inclusive whole-genome association studies. Genet. Epidemiol. 2021;45:694–709. doi: 10.1002/gepi.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McLaren W, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gene Ontology, C. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Backes C, et al. Systematic permutation testing in GWAS pathway analyses: identification of genetic networks in dilated cardiomyopathy and ulcerative colitis. BMC Genomics. 2014;15:622. doi: 10.1186/1471-2164-15-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McInnes G, et al. Global Biobank Engine: enabling genotype-phenotype browsing for biobank summary statistics. Bioinformatics. 2019;35:2495–2497. doi: 10.1093/bioinformatics/bty999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gagliano Taliun SA, et al. Exploring and visualizing large-scale genetic associations by using PheWeb. Nat. Genet. 2020;52:550–552. doi: 10.1038/s41588-020-0622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat. Genet. 2018;50:1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dobbyn A, et al. Landscape of conditional eQTL in dorsolateral prefrontal cortex and co-localization with schizophrenia GWAS. Am. J. Hum. Genet. 2018;102:1169–1184. doi: 10.1016/j.ajhg.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Panjwani N, et al. LocusFocus: web-based colocalization for the annotation and functional follow-up of GWAS. PLoS Comput. Biol. 2020;16:e1008336. doi: 10.1371/journal.pcbi.1008336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Napoletano F, et al. Polyglutamine Atrophin provokes neurodegeneration in Drosophila by repressing fat. EMBO J. 2011;30:945–958. doi: 10.1038/emboj.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walmsley AR, Barrett MP, Bringaud F, Gould GW. Sugar transporters from bacteria, parasites and mammals: structure-activity relationships. Trends Biochem. Sci. 1998;23:476–481. doi: 10.1016/s0968-0004(98)01326-7. [DOI] [PubMed] [Google Scholar]
- 74.Madej MG, Dang S, Yan N, Kaback HR. Evolutionary mix-and-match with MFS transporters. Proc. Natl Acad. Sci. USA. 2013;110:5870–5874. doi: 10.1073/pnas.1303538110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
eQTL data are available for download from GTEx (https://gtexportal.org/home), PsychENCODE (http://resource.psychencode.org/), and fetal brains (10.6084/m9.figshare.6881825). GWAS summary statistics for this study are available for download from our website (https://lab.research.sickkids.ca/strug/softwareandresources/).