Abstract
Mutations in the grlA and gyrA genes of 344 clinical strains of Staphylococcus aureus isolated in 1994 in Japan were identified by combinations of single-strand conformation polymorphism analysis, restriction fragment length analysis, and direct sequencing to identify possible relationships to fluoroquinolone resistance. Five types of single-point mutations and four types of double mutations were observed in the grlA genes of 204 strains (59.3%). Four types of single-point mutations and four types of double mutations were found in the gyrA genes of 188 strains (54.7%). Among them, the grlA mutation of TCC→TTC or TAC (Ser-80→Phe or Tyr) and the gyrA mutation of TCA→TTA (Ser-84→Leu) were principal, being detected in 137 (39.8%) and 121 (35.9%) isolates, respectively. The grlA point mutations of CAT→CAC (His-77 [silent]), TCA→CCA (Ser-81→Pro), and ATA→ATT (Ile-100 [silent]) were novel, as was the GAC→GGC (Asp-73→Gly) change in gyrA. A total of 15 types of mutation combinations within both genes were related to ciprofloxacin resistance (MIC ≥ 3.13 μg/ml) and were present in 193 mutants (56.1%). Strains containing mutations in both genes were highly resistant to ciprofloxacin (MIC at which 50% of the isolates are inhibited [MIC50] = 50 μg/ml). Those with the Ser-80→Phe or Tyr alteration in grlA but wild-type gyrA showed a lower level of ciprofloxacin resistance (MIC50 ≤ 12.5 μg/ml). Levofloxacin was active against 68 of 193 isolates (35.2%) with mutations at codon 80 of grlA in the presence or absence of a concomitant mutation at codon 73, 84, or 88 in gyrA (MIC ≤ 6.25 μg/ml). The new fluoroquinolone DU-6859a showed good activity with 186 of 193 isolates (96.4%) for which the MIC was ≤6.25 μg/ml.
Staphylococcus aureus infections, particularly those caused by methicillin-resistant S. aureus, pose serious therapeutic difficulties, because few antimicrobial agents are effective against this pathogen. Fluoroquinolones, broad-spectrum and potent antimicrobial agents, have been effective in the treatment of these infections. With the increasing use of fluoroquinolones, resistance in S. aureus, especially methicillin-resistant S. aureus has become widespread in recent years (1, 12).
Three mechanisms involved in fluoroquinolone resistance have been proposed. One is topoisomerase IV gene mutations (4, 18), a second is DNA gyrase gene mutations (13, 16), and a third is an active efflux pump (8, 19). Recent studies have demonstrated that the primary target of fluoroquinolones in S. aureus is DNA topoisomerase IV, which is composed of the GrlA and GrlB subunits, encoded by the grlA and grlB genes, respectively. DNA gyrase is considered a secondary target (3, 4, 10, 15). In S. aureus clinical strains, mutations in either the grlA or gyrA gene lead to quinolone resistance. grlA mutations are associated with both high- and low-level resistances, while gyrA mutations are responsible for increases in ciprofloxacin (CPFX) resistance in grlA mutants. A combination of mutations in both genes can cause high-level quinolone resistance (4, 5, 13, 15, 16, 18).
Several methods have been used for detection of point mutations of genes. Single-strand conformation polymorphism (SSCP) analysis is a rapid, simple, and effective method in which a mutated sequence is detected by a change in mobility during polyacrylamide gel electrophoresis caused by its altered folded structure (6). It has been applied in detection of DNA gyrase gene mutations in S. aureus (14, 17) and Escherichia coli (11). In SSCP analysis in our study, proper conditions and a new system were used for detection of grlA and gyrA mutations. By using combinations of SSCP analysis, restriction fragment length polymorphism (RFLP) analysis, and direct sequencing, we examined grlA and gyrA mutations in 344 S. aureus strains and studied the relationship between combinations of mutations of both genes and susceptibility of the various mutants to three fluoroquinolones.
MATERIALS AND METHODS
Antimicrobial agents and bacterial strains.
CPFX, levofloxacin (LVFX), and DU-6859a were synthesized at the New Product Research Laboratories I, Daiichi Pharmaceutical Co., Ltd., Tokyo, Japan. A total of 344 clinical S. aureus strains (one strain per patient) were collected by LVFX surveillance groups from 24 hospitals all over Japan. These were isolated from June to November 1994 and included 215 methicillin-resistant (32 CPFX-susceptible and 183 CPFX-resistant) and 129 methicillin-susceptible (122 CPFX-susceptible and 7 CPFX-resistant) strains. S. aureus FDA 209-P, 891185 (Ser-80→Phe), and 900165 (Glu-84→Lys) were used as controls for detection of grlA mutations, while FDA 209-P, 900165 (Ser-84→Leu), 6859a-r (Glu-88→Lys), 87-53 (Ile-86 [silent]), and 87-20 (Ser-84→Leu, Ile-86 [silent]) were used for detection of gyrA mutations (15, 16).
Determination of MICs.
The MICs were determined by standard agar dilution methods (9) with Mueller-Hinton agar (Difco, Detroit, Mich.). Drug-containing agar plates were incubated with one loopful (5 μl) of an inoculum corresponding to about 104 CFU per spot and were incubated at 37°C for 18 h. The MIC was defined as the lowest drug concentration which prevented visible growth of bacteria.
PCR experiments.
Chromosomal DNA was prepared from 2 μl of an overnight culture heated at 98°C for 5 min. PCR was performed with cycling at 94°C for 30 s, 52°C for 30 s, and 70°C for 1 min for 30 cycles by using 2.5 U of recombinant Taq DNA polymerase (Takara, Shiga, Japan). Oligonucleotides 5′-TTCCGTAAAAGTGCGAAAACAG (nucleotides 178 to 199) and 5′-CGCATTGCCGCTGGCGGATCCTTATCGATAC (complementary to nucleotides 323 to 353) were used for amplification of a 176-bp grlA fragment. For gyrA gene amplification, 5′-CATATAAAAAATCAGCACGTATCGTT (nucleotides 188 to 213) was used as the sense primer, and 5′-TGAGCCATACGTACCATTGC (complementary to nucleotides 265 to 284) (97-bp fragment) and 5′-CGCCATCTCCATCCATTGAACCAAA (complementary to nucleotides 328 to 352) (165-bp fragment) were used as antisense primers for SSCP analysis and direct sequencing, respectively. The amplified products were checked by agarose gel electrophoresis.
SSCP analysis.
PCR products were mixed with denaturant solution (95% formamide, 20 mM EDTA [pH 8.0], 0.05% bromophenol blue, 0.05% xylene cyanol). The mixtures were heated at 80°C for 5 min and then were cooled on ice immediately. Samples were then separated by 12.5% polyacrylamide gel (Daiichi Pure Chemicals, Tokyo, Japan) electrophoresis at 17°C, and DNA bands were visualized by using a silver stain kit (Daiichi Pure Chemicals).
RFLP analysis.
For detection of mutations in the grlA gene, we used restriction endonuclease HinfI (Toyobo, Tokyo, Japan) for the Ser-80→Tyr alteration and BsrGI (New England Biolabs, Beverly, Mass.) for the Glu-84→Lys alteration. The reaction mixtures (1 U of enzyme added to 10 μl of each PCR product) were incubated at 37°C for more than 1 h, and the sizes of the resulting fragments were ascertained by agarose gel electrophoresis.
DNA sequencing.
The PCR-amplified DNA from grlA was cloned into the vector pGEM-T (Promega, Madison, Wis.) and sequenced with an ALFred DNA sequencer (Pharmacia). The amplified fragment from gyrA was sequenced directly with an AutoLoad solid-phase sequencing kit (Pharmacia).
RESULTS
Detection of mutations in the grlA gene.
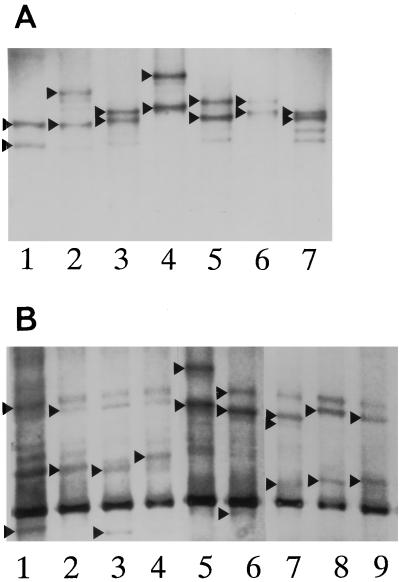
By using a combination of SSCP, RFLP analysis, and sequencing, five single-point mutations and four double mutations were observed in the grlA gene in 204 S. aureus strains (59.3%) (Table 1). Among them, the TCC→TTC or TAC (Ser-80→Phe or Tyr) single-point mutation was principal; it was detected in 137 isolates (39.8%). The double mutation of TCC→TTC or TAC and GAA→AAA (Ser-80→Phe or Tyr and Glu-84→Lys) was found in 49 mutants (14.2%). The CAT→CAC (His-77) and ATA→ATT (Ile-100) single-point mutations were silent mutations, i.e., they led to no amino acid alteration. The TCA→CCA (Ser-81→Pro) was a novel point mutation in the grlA gene. Double-codon alterations of Ser-80→Tyr and Glu-84→Lys, Ser-80→Phe and Ser-81→Pro, and Ser-81→Pro and Glu-84→Lys were also novel grlA mutations and were identified in 41, 1, and 1 isolates, respectively. As shown in Fig. 1A, six of nine types of grlA mutations were distinguishable from the wild type by SSCP analysis.
TABLE 1.
Mutations in the grlA gene in 344 S. aureus strains
| Codon(s) | Mutation(s) | No. of isolates | Method(s) |
|---|---|---|---|
| 77 | His [silent] (CAT→CAC) | 7 | SSCP, sequencing |
| 80 | Ser→Phe (TCC→TTC) | 94 | SSCP |
| Ser→Tyr (TCC→TAC) | 43 | RFLP, sequencing | |
| 84 | Glu→Lys (GAA→AAA) | 5 | RFLP |
| 100 | Ile [silent] (ATA→ATT) | 4 | SSCP, sequencing |
| 80 + 81 | Ser→Phe (TCC→TTC), Ser→Pro (TCA→CCA) | 1 | SSCP, sequencing |
| 80 + 84 | Ser→Phe (TCC→TTC), Glu→Lys (GAA→AAA) | 8 | SSCP, sequencing |
| Ser→Tyr (TCC→TAC), Glu→Lys (GAA→AAA) | 41 | RFLP, sequencing | |
| 81 + 84 | Ser→Pro (TCA→CCA), Glu→Lys (GAA→AAA) | 1 | SSCP, sequencing |
| None | 140 | SSCP |
FIG. 1.
Detection of mutations in the grlA gene (A) and the gyrA gene (B) by SSCP analysis. Mutations in the genes cause amino acid changes as follows. (A) Lane 1, His-77 (CAT→CAC [silent]); lane 2, Ser-80→Phe; lane 3, Ile-100 (ATA→ATT [silent]); lane 4, Ser-80→Phe and Ser-81→Pro; lane 5, Ser-80→Phe and Glu-84→Lys; lane 6, Ser-81→Pro and Glu-84→Lys; lane 7, none (wild type). (B) Lane 1, none (wild type); lane 2, Ser-84→Leu; lane 3, Ser-84→Leu and Ile-86 (ATT→ATC [silent]); lane 4, Ile-86 (silent); lane 5, Ser-84→Leu and Ser-85→Pro; lane 6, Asp-73→Gly; lane 7, Asp-73→Gly and Ser-84→Leu; lane 8, Ser-84→Leu and Glu-88→Lys; lane 9, Glu-88→Lys. Arrowheads indicate the bands used to distinguish differences.
Detection of mutations in the gyrA gene.
As shown in Table 2, 188 S. aureus strains (54.7%) contained mutations in the gyrA gene. By SSCP analysis and direct sequencing, four types of single-point mutations and four types of double mutations were detected at codons 73, 84, 86 (silent), 88, 84 and 73, 84 and 85, 84 and 86, and 84 and 88. The single-point mutations of TCA→TTA (Ser-84→Leu) and GAA→AAA (Glu-88→Lys) were principal; they were found in 121 (35.0%) and 36 (10.5%) isolates, respectively. The GAC→GGC (Asp-73→Gly) change was a novel point mutation in the gyrA gene. The eight types of gyrA mutants and the wild type had distinct SSCP patterns (Fig. 1B); however, the SSCP patterns in lanes 6 (alteration at codon 73) and 7 (alterations at codons 73 and 84) of Fig. 1B were similar, so the mutations of four strains were confirmed by sequencing. There was no strain which possessed amino acid changes in GyrA in the absence of a grlA mutation.
TABLE 2.
Mutations in the gyrA gene in 344 S. aureus strains
| Codon(s) | Mutation(s) | No. of isolates | Method(s) |
|---|---|---|---|
| 73 | Asp→Gly (GAC→GGC) | 1 | SSCP, sequencing |
| 84 | Ser→Leu (TCA→TTA) | 121 | SSCP |
| 86 | Ile [silent] (ATT→ATC) | 8 | SSCP |
| 88 | Glu→Lys (GAA→AAA) | 36 | SSCP |
| 84 + 73 | Ser→Leu (TCA→TTA), Asp→Gly (GAC→GGC) | 3 | SSCP, sequencing |
| 84 + 85 | Ser→Leu (TCA→TTA), Ser→Pro (TCT→CCT) | 6 | SSCP, sequencing |
| 84 + 86 | Ser→Leu (TCA→TTA), Ile [silent] (ATT→ATC) | 9 | SSCP |
| 84 + 88 | Ser→Leu (TCA→TTA), Glu→Lys (GAA→AAA) | 4 | SSCP, sequencing |
| None | 156 | SSCP |
Susceptibilities of mutants to quinolones.
A total of 19 mutation combinations were found in the grlA and gyrA genes of the 344 S. aureus strains (Table 3). Isolates with grlA–gyrA mutation combinations of codon 77–none, codon 100–none, and none–codon 86 were found to have quinolone susceptibilities similar to that of the wild type, because these silent mutations are not responsible for quinolone resistance. The other 15 types observed in 193 mutants (56.1%) were related to CPFX resistance (MIC ≥ 3.13 μg/ml). Overall, those strains mutated in both genes showed higher-level CPFX resistance. The MIC at which 50% of the isolates are inhibited (MIC50) and MIC90 were 50 and 800 μg/ml, respectively. Among such strains, those with combined mutations at codons 84–84, 84–84 plus 86, 80 plus 84–84, 80 plus 84–73 plus 84, 80 plus 84–84 plus 86, 80 plus 84–84 plus 85, and 81 plus 84–84 were highly resistant to CPFX (MIC ≥ 100 μg/ml). The combination of the Ser-80→Phe or Tyr alteration in grlA but wild-type gyrA was present in strains with lower-level resistance to CPFX. The MIC range, MIC50, and MIC90 for these strains were 3.13 to 12.5, 12.5, and 12.5 μg/ml, respectively.
TABLE 3.
Susceptibilities of 344 S. aureus strains with or without mutations in the grlA and gyrA genes
| Codon(s)
|
No. of strains | Drug | MIC (μg/ml)
|
|||
|---|---|---|---|---|---|---|
| grlA | gyrA | Range | 50% | 90% | ||
| 80 | 73 | 1 | CPFX | 6.25 | ||
| LVFX | 1.56 | |||||
| DU-6859a | 0.2 | |||||
| 80 | 84 | 75 | CPFX | 12.5–800 | 25 | 100 |
| LVFX | 3.13–50 | 12.5 | 25 | |||
| DU-6859a | 0.2–6.25 | 0.78 | 1.56 | |||
| 80 | 88 | 35 | CPFX | 12.5–100 | 25 | 100 |
| LVFX | 3.13–12.5 | 6.25 | 12.5 | |||
| DU-6859a | 0.2–0.78 | 0.39 | 0.78 | |||
| 80 | 13 | CPFX | 3.13–12.5 | 12.5 | 12.5 | |
| LVFX | 0.39–6.25 | 3.13 | 3.13 | |||
| DU-6859a | 0.05–0.39 | 0.2 | 0.39 | |||
| 80 | 73 + 84 | 2 | CPFX | 50 | ||
| LVFX | 12.5 | |||||
| DU-6859a | 0.78 | |||||
| 80 | 84 + 86 | 8 | CPFX | 50–100 | ||
| LVFX | 12.5–25 | |||||
| DU-6859a | 0.78–1.56 | |||||
| 80 | 84 + 88 | 3 | CPFX | 50–800 | ||
| LVFX | 12.5–50 | |||||
| DU-6859a | 0.78–3.13 | |||||
| 84 | 84 | 4 | CPFX | 100–800 | ||
| LVFX | 25–100 | |||||
| DU-6859a | 1.56–6.25 | |||||
| 84 | 84 + 88 | 1 | CPFX | 800 | ||
| LVFX | 400 | |||||
| DU-6859a | 50 | |||||
| 80 + 84 | 84 | 41 | CPFX | 100–>800 | 800 | 800 |
| LVFX | 25–100 | 50 | 100 | |||
| DU-6859a | 0.78–6.25 | 6.25 | 6.25 | |||
| 80 + 84 | 73 + 84 | 1 | CPFX | 200 | ||
| LVFX | 50 | |||||
| DU-6859a | 1.56 | |||||
| 80 + 84 | 84 + 85 | 6 | CPFX | ≧800 | ||
| LVFX | ≧800 | |||||
| DU-6859a | 25 | |||||
| 80 + 84 | 84 + 86 | 1 | CPFX | 200 | ||
| LVFX | 50 | |||||
| DU-6859a | 3.13 | |||||
| 80 + 81 | 88 | 1 | CPFX | 100 | ||
| LVFX | 12.5 | |||||
| DU-6859a | 0.78 | |||||
| 81 + 84 | 84 | 1 | CPFX | 200 | ||
| LVFX | 50 | |||||
| DU-6859a | 3.13 | |||||
| 77 (silent) | 7 | CPFX | 0.2–1.56 | |||
| LVFX | 0.2–0.39 | |||||
| DU-6859a | 0.025–0.05 | |||||
| 100 (silent) | 4 | CPFX | 0.39 | |||
| LVFX | 0.2 | |||||
| DU-6859a | 0.025–0.05 | |||||
| 86 (silent) | 8 | CPFX | 0.39–3.13 | |||
| LVFX | 0.2–0.78 | |||||
| DU-6859a | 0.025–0.1 | |||||
| 132 | CPFX | 0.2–3.13 | 0.78 | 1.56 | ||
| LVFX | 0.1–0.78 | 0.39 | 0.39 | |||
| DU-6859a | 0.013–0.1 | 0.05 | 0.05 | |||
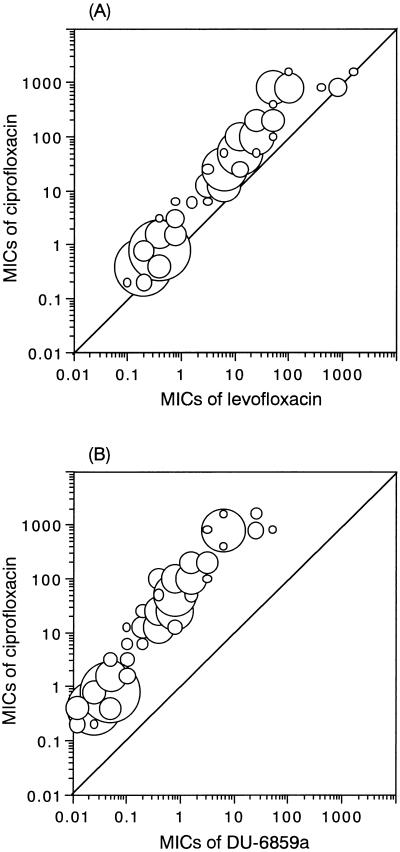
With respect to LVFX and DU-6859a, the distribution of susceptibilities of mutants had a tendency similar to that of susceptibility to CPFX (Fig. 2). LVFX was active against 68 of 193 mutants (35.2%) which contained alterations codon as codon 80–73, 80–84, 80–88, or 80–none (MIC of LVFX ≤ 6.25 μg/ml). The other mutants were moderately or highly resistant to LVFX. DU-6859a had good activity against 120 of the mutants (62.2%; MIC of DU-6859a ≤ 0.78 μg/ml). The MICs of DU-6859a for 66 of the mutants (34.2%) were from 1.56 to 6.25 μg/ml. Six mutants with the codon 80 plus 84–84 plus 85 alterations and one with the codon 84–84 plus 88 alterations were highly resistant to DU-6859a (MIC ≥ 25 μg/ml).
FIG. 2.
Distribution of the MICs of LVFX and DU-6859a versus the MICs of ciprofloxacin. The sizes of the circles reflect the numbers of strains.
DISCUSSION
In clinical S. aureus strains, five types of grlA mutations (causing Ser-80→Phe, Ser-80→Tyr, Glu-84→Lys, Glu-84→Gln, and Ser-80→Phe plus Glu-84→Lys changes) were demonstrated to be responsible for quinolone resistance (2–4, 10, 15, 18). In gyrA, 10 types of mutations (causing Ser-84→Leu, Ser-84→Ala, Ser-84→Val, Ser-85→Pro, Glu-88→Lys, Glu-88→Gly, Ser-84→Leu plus Ser-85→Pro, Ser-84→Leu plus Ile-86 [silent], Ser-84→Leu plus Glu-88→Lys, and Ser-84→Leu plus Glu-88→Gly changes) were found in strains with high-level quinolone resistance (5, 7, 13–17).
In this study, we examined 344 clinical isolates of S. aureus by a combination of SSCP, RFLP analysis, and direct sequencing and found nine types of mutations in grlA and eight types in gyrA. Among them, five types in grlA and two types in gyrA were novel. The grlA mutation at codon 80 and the gyrA mutation at codon 84 were principal, being found in 137 of 204 GrlA mutants (67.2%) and 121 of 188 GyrA mutants (64.4%), respectively. This result is consistent with findings reported for clinical isolates (4, 14). grlA mutations at codons 80 and 84 and gyrA mutations at codons 84, 85, and 88 were detected in the CPFX-resistant strains (MIC ≥ 3.13 μg/ml), which is in agreement with other reports that these point mutations are responsible for quinolone resistance (5, 18). Further study is necessary to determine whether the Ser-81→Pro alteration encoded by grlA and the Asp-73→Gly alteration encoded by gyrA directly contributed to quinolone resistance.
Fifteen types of mutation combinations of both genes observed in this investigation were related to CPFX resistance. Among them, 13 mutants with lower-level CPFX resistance contained a grlA mutation at codon 80 but no mutation in gyrA. This supports the notion that in S. aureus topoisomerase IV is a primary target of fluoroquinolones. The finding that grlA-gyrA double mutants exhibited higher-level CPFX resistance (MIC50 = 50 μg/ml) confirmed previous genetic studies (10, 18). The combinations of mutations including the Glu-84→Lys alteration encoded by grlA and the Ser-84→Leu alteration encoded by gyrA conferred high-level quinolone resistance (MIC of CPFX ≥ 100 μg/ml).
Diversity in the MICs of fluoroquinolones for strains with same mutation combination suggests that the strains may possess other mechanisms of resistance. Some strains with no mutations in both genes or with a silent mutation in either gene showed low-level CPFX resistance (MIC, 1.56 to 3.13 μg/ml), which suggests that other resistance mechanisms are present. In this study, the alteration at codon 116 in the sequence encoding GrlA, which considered a DNA binding site (10), was not checked because the length of the DNA fragment was limited for detection by SSCP analysis. The amino acid change in the remaining region of GrlA and GyrA, alteration in GrlB and GyrB, and the association of the quinolone efflux system were possibly the cause of the diversity in the MICs. The numerous and complicated mutations seen may explain the rapid and widespread development of quinolone resistance described for S. aureus. DU-6859a showed good activity against CPFX- or LVFX-resistant mutants because of its high inhibitory activity against both topoisomerase IV and DNA gyrase as demonstrated by Tanaka et al. (15).
In this study, SSCP analysis was found to be a rapid, simple, and effective method for detection of point mutations in both the grlA gene and the gyrA gene of S. aureus strains. The phenomenon that one strand could be separated into two bands due to different stable conformations (6) was observed as well under our SSCP conditions.
ACKNOWLEDGMENTS
This work was supported by grants from the Japan Health Sciences Foundation.
We thank Yoshikuni Onodera and Takaaki Akasaka for their technical assistance and Yuki Nagano for synthesizing the primers used in this study.
REFERENCES
- 1.Ball, P. 1990. Emergent resistance to ciprofloxacin amongst Pseudomonas aeruginosa and Staphylococcus aureus: clinical significance and therapeutic approaches. J. Antimicrob. Chemother. 26(Suppl. F):165–179. [DOI] [PubMed]
- 2.Blanche F, Cameron B, Bernard F-X, Maton L, Manse B, Ferrero L, Ratet N, Lecoq C, Goniot A, Bisch D, Crouzet J. Differential behaviors of Staphylococcus aureus and Escherichia coli type II DNA topoisomerases. Antimicrob Agents Chemother. 1996;40:2714–2720. doi: 10.1128/aac.40.12.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrero L, Cameron B, Manse B, Lagneaux D, Crouzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 5.Goswitz J J, Willard K E, Fasching C E, Peterson L R. Detection of gyrA gene mutations associated with ciprofloxacin resistance in methicillin-resistant Staphylococcus aureus: analysis by polymerase chain reaction and automated direct DNA sequencing. Antimicrob Agents Chemother. 1992;36:1166–1169. doi: 10.1128/aac.36.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi K. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Applications. 1991;1:34–38. doi: 10.1101/gr.1.1.34. [DOI] [PubMed] [Google Scholar]
- 7.Ito H, Yoshida H, Bogaki-Shonai M, Niga T, Hattori H, Nakamura S. Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2014–2023. doi: 10.1128/aac.38.9.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaatz G W, Seo S M, Ruble C A. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1086–1094. doi: 10.1128/aac.37.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A2. 2nd ed. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1990. [Google Scholar]
- 10.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouabdesselam S, Hooper D C, Tankovic J, Soussy C J. Detection of gyrA and gyrB mutations in quinolone-resistant clinical isolates of Escherichia coli by single-strand conformational polymorphism analysis and determination of levels of resistance conferred by two different single gyrA mutations. Antimicrob Agents Chemother. 1995;39:1667–1670. doi: 10.1128/aac.39.8.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shalit I, Berger S A, Gorea A, Frimerman H. Widespread quinolone resistance among methicillin-resistant Staphylococcus aureus isolates in a general hospital. Antimicrob Agents Chemother. 1989;33:593–594. doi: 10.1128/aac.33.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sreedharan S, Oram M, Jensen B, Peterson L R, Fisher L M. DNA gyrase gyrA mutations in ciprofloxacin-resistant strains of Staphylococcus aureus: close similarity with quinolone resistance mutations in Escherichia coli. J Bacteriol. 1990;172:7260–7262. doi: 10.1128/jb.172.12.7260-7262.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takenouchi T, Ishii C, Sugawara M, Tokue Y, Ohya S. Incidence of various gyrA mutants in 451 Staphylococcus aureus strains isolated in Japan and their susceptibilities to 10 fluoroquinolones. Antimicrob Agents Chemother. 1995;39:1414–1418. doi: 10.1128/aac.39.7.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka M, Onodera Y, Uchida Y, Sato K, Hayakawa I. Inhibitory activity of quinolones against DNA gyrase and topoisomerase IV purified from Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:2362–2366. doi: 10.1128/aac.41.11.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka M, Zhang Y X, Ishida H, Akasaka T, Sato K, Hayakawa I. Mechanisms of 4-quinolone resistance in quinolone-resistant Staphylococcus aureus isolates from Japan and China. J Med Microbiol. 1995;42:214–219. doi: 10.1099/00222615-42-3-214. [DOI] [PubMed] [Google Scholar]
- 17.Tokue Y, Sugano K, Saito D, Noda T, Ohkura H, Shimosato Y, Sekiya T. Detection of novel mutations in the gyrA gene of Staphylococcus aureus by nonradioisotopic single-strand conformation polymorphism analysis and direct DNA sequencing. Antimicrob Agents Chemother. 1994;38:428–431. doi: 10.1128/aac.38.3.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamagishi J-I, Kojima T, Oyamada Y, Fujimoto K, Hattori H, Nakamura S, Inoue M. Alterations in the DNA topoisomerase IV grlA gene responsible for quinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1157–1163. doi: 10.1128/aac.40.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida H, Bogaki M, Nakamura S, Ubukata K, Konno M. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol. 1990;172:6942–6949. doi: 10.1128/jb.172.12.6942-6949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]