Abstract
The cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) of yeasts is involved in an important step in the biosynthesis of ergosterol. Since CYP51A1 is the target of azole antifungal agents, this enzyme is potentially prone to alterations leading to resistance to these agents. Among them, a decrease in the affinity of CYP51A1 for these agents is possible. We showed in a group of Candida albicans isolates from AIDS patients that multidrug efflux transporters were playing an important role in the resistance of C. albicans to azole antifungal agents, but without excluding the involvement of other factors (D. Sanglard, K. Kuchler, F. Ischer, J.-L. Pagani, M. Monod, and J. Bille, Antimicrob. Agents Chemother. 39:2378–2386, 1995). We therefore analyzed in closer detail changes in the affinity of CYP51A1 for azole antifungal agents. A strategy consisting of functional expression in Saccharomyces cerevisiae of the C. albicans CYP51A1 genes of sequential clinical isolates from patients was designed. This selection, which was coupled with a test of susceptibility to the azole derivatives fluconazole, ketoconazole, and itraconazole, enabled the detection of mutations in different cloned CYP51A1 genes, whose products are potentially affected in their affinity for azole derivatives. This selection enabled the detection of five different mutations in the cloned CYP51A1 genes which correlated with the occurrence of azole resistance in clinical C. albicans isolates. These mutations were as follows: replacement of the glycine at position 129 with alanine (G129A), Y132H, S405F, G464S, and R467K. While the S405F mutation was found as a single amino acid substitution in a CYP51A1 gene from an azole-resistant yeast, other mutations were found simultaneously in individual CYP51A1 genes, i.e., R467K with G464S, S405F with Y132H, G129A with G464S, and R467K with G464S and Y132H. Site-directed mutagenesis of a wild-type CYP51A1 gene was performed to estimate the effect of each of these mutations on resistance to azole derivatives. Each single mutation, with the exception of G129A, had a measurable effect on the affinity of the target enzyme for specific azole derivatives. We speculate that these specific mutations could combine with the effect of multidrug efflux transporters in the clinical isolates and contribute to different patterns and stepwise increases in resistance to azole derivatives.
The most common yeast infection in human immunodeficiency virus-infected patients in past years has been oropharyngeal candidiasis caused by Candida albicans (26). These infections are treated with antifungal agents, particularly the triazole derivative fluconazole. However, due to the repeated use of this agent in patients with recurrent oropharyngeal candidiasis, the number of cases of in vitro resistance correlating with clinical failure has risen significantly (23, 25, 32, 39). We have been interested in understanding the molecular mechanisms of resistance to azole antifungal agents in C. albicans, and our work has demonstrated the involvement of efflux multidrug transporters in the development of resistance in C. albicans clinical isolates (28–30). However, other mechanisms of resistance to azole antifungal agents have been described and can exist simultaneously in resistant isolates. One of these mechanisms involves alteration of the ergosterol biosynthetic pathway, particularly a defect in Δ5,6-desaturase, an enzyme responsible for the conversion of ergosta-7,22-dienol into ergosterol (22). The enzyme Δ5,6-desaturase is also thought to be responsible for the formation of the toxic metabolite 14α-methylergosta-8,24(28)-dien-3β,6α-diol when yeast cells are exposed to azole derivatives, and therefore yeasts with a defect in this enzymatic step have a selective advantage when treated with azole antifungal agents (14, 40). Other mechanisms of resistance may involve the target enzyme of azole, which is a cytochrome P-450 catalyzing the 14α demethylation of lanosterol which has been named P-45014dm, Erg11, or CYP51A1. The last designation will be used here as recommended by Nelson et al. (21). The cellular content of CYP51A1 can be increased and therefore can elevate, although modestly, the resistance to azole derivatives, as shown in an azole-resistant Candida glabrata strain (36). The affinity of CYP51A1 for azole derivatives may also be decreased by mutations that contribute to an increase in the MICs of azole derivatives in clinical isolates. Such a phenomenon in a clinical C. albicans isolate has been described by Vanden Bossche et al. (34). In this report, we address the involvement of such a mechanism in sequential clinical isolates of C. albicans from different patients which have acquired resistance to azole derivatives. We reported previously that in the azole-resistant isolates of this collection, multidrug efflux pumps were operating, but without excluding the participation of other resistance mechanisms (30). Here, by using a functional screening strategy enabling the selection of CYP51A1 genes with mutations probably altering the affinity of CYP51A1 for azole derivatives, we show that mutations were indeed present in the CYP51A1 genes of these azole-resistant isolates.
(Part of these results were presented at the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy, New Orleans, La., 15 to 18 September 1996 [27].)
MATERIALS AND METHODS
Strains and media.
C. albicans strains used in this study have been described previously (30). Saccharomyces cerevisiae YKKB-13 (MATa ura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 Δpdr5::TRP1) was used for the expression of CYP51A1 genes in YEp51-derived plasmids. YEp51 is a 2μm-based vector that contains a GAL10 promoter for inducible heterologous gene expression (7, 31). Escherichia coli DH5α was used for propagation of plasmids constructed in this study (10).
C. albicans strains were grown in complex yeast extract-peptone-dextrose (YPD) medium with 1% yeast extract (Difco), 2% peptone (Difco), and 2% glucose. S. cerevisiae was cultured in yeast nitrogen base (YNB) medium (Difco) supplemented with uracil, lysine, adenine, and histidine (each at 50 mg per ml) and containing 2% glucose. For the expression of C. albicans CYP51A1 genes in S. cerevisiae, cells were grown in the same medium but in the presence of 1% galactose and 1% raffinose. YKKB-13 could not grow on galactose, which is required for GAL10 induction, but could grow on raffinose; therefore, both carbon sources were added to the same medium to ensure the simultaneous occurrence of growth of S. cerevisiae and induction of the GAL10 promoter.
Cloning of CYP51A1 genes from C. albicans clinical isolates.
The CYP51A1 genes were cloned from genomic DNA of C. albicans isolates by PCR. DNA was first extracted as previously described (29) and used as a template for amplification of CYP51A1 alleles. PCR was carried out with high-fidelity Pwo DNA polymerase (Boehringer Mannheim), using primers spanning the entire CYP51A1 open reading frame (ORF). These primers, 5′ GCG GAT CCT TAA AAC ATA CAA GTT TCT CTT TT 3′ (CYPCB) and 5′ ACG CGT CGA CAA TAT GGC TAT TGT TGA AAC TGT C 3′ (CYPNS2), were flanked with BamHI and SalI restriction sites to allow the subcloning of amplified CYP51A1 fragments into YEp51 precut by the same enzymes. From each PCR with genomic DNA of a C. albicans isolate, a collection of at least six CYP51A1 expression plasmids was obtained. Individual plasmids were then transformed into S. cerevisiae YKKB-13 by a lithium acetate method (9) in noninducing YNB selective medium with glucose as a carbon source. The expression of CYP51A1 proteins was verified by growth of the Leu+ transformants in inducing YNB selective medium with galactose and raffinose as carbon sources.
Drug susceptibility assays.
Disk diffusion assays with fluconazole were performed with S. cerevisiae Leu+ transformants in raffinose-galactose selective YNB medium. Briefly, S. cerevisiae cells were grown overnight in raffinose-galactose selective YNB medium at 30°C with constant agitation and diluted to a density of 2 × 104 per ml. Plates, each containing 15 ml of raffinose-galactose selective YNB medium with 2% agar, were inoculated with cotton swabs saturated with diluted cell suspension, and a disk containing 50 μg of fluconazole was placed on the center of the agar plate. Plates were incubated for 2 days at 30°C, and the diameter of inhibition, which was visible as a sharp difference in cell growth, was measured with a ruler. MICs of fluconazole for clinical C. albicans isolates were taken from published material (30). MICs of azole antifungal drugs for S. cerevisiae Leu+ transformants were measured by a modification of the National Committee for Clinical Laboratory Standards M27-T microdilution protocol (20). Instead of RPMI 1640 medium, which is usually utilized in this method, a selective YNB medium containing galactose and raffinose was used. Flat-bottomed 96-well microtiter plates containing cell suspension and serial dilutions of the antifungal agents were incubated for 48 h at 30°C and then scanned by a microtiter plate reader (Bio-Rad) at 540 nm. The MIC was defined as the antifungal concentration giving a 50% or less decrease in the optical density at 540 nm (OD540) compared to the OD of the corresponding drug-free incubation medium. Fluconazole was a generous gift of Roerig-Pfizer Inc. (New York, N.Y.). Ketoconazole and itraconazole were provided by Janssen Pharmaceutica (Beerse, Belgium).
Immunoblotting.
To allow detection of the CYP51A1 protein in yeast extracts, an antibody against this protein was first raised in rabbits. The CYP51A1 protein used for immunization was obtained in E. coli by fusion of a truncated C. albicans CYP51A1 protein (amino acid positions 307 to 518 with respect to the first ATG) with glutathione S-transferase (GST). The CYP51A1-GST fusion protein was prepared by subcloning a PCR fragment obtained with the primers 5′-CGG GAT CCA TGG GTG GTC AAC ATA CTT CT-3′ (CYPN) and 5′-CGG AAT TCC CTG CTG GTT CAG TAG GTA AAA C-3′ (CYPC), using genomic DNA from C. albicans SC5314 as a template. The fragment spanned 653 bp of the C-terminal region of the CYP51A1 protein, starting from nucleotide 916 and ending with nucleotide 1549 with respect to the first ATG of CYP51A1. The obtained fragment was subcloned into the vector pGEX-2T (Amersham) cut with BamHI and EcoRI to allow in-frame fusion with GST. The purification of the CYP51A1-GST protein for preparation of a polyclonal rabbit antibody was achieved by glutathione-agarose affinity chromatography of E. coli cell extracts by standard procedures described by the manufacturer (Amersham). For the detection of yeast CYP51A1 by immunoblotting with the anti-CYP51A1 antibody, total protein extracts were obtained from S. cerevisiae which had been transformed with expression plasmids. Cell extracts were prepared by an alkaline extraction procedure from cells grown to mid-log phase. Briefly, cells (OD540, 5) were resuspended in an Eppendorf microcentrifuge tube with 500 μl of water and 150 μl of a solution containing 1.85 M NaOH and 7.5% β-mercaptoethanol. This mixture was incubated on ice for 10 min. Proteins were then precipitated with 150 μl of a 50% trichloroacetic acid solution, and the suspension was left on ice for another 10 min. Precipitated proteins were sedimented by centrifugation at maximum speed in a microcentrifuge for 15 min. The sediment was then resuspended in 50 μl of loading buffer (40 mM Tris-HCl [pH 6.8], 8 M urea, 5% sodium dodecyl sulfate, 0.1 M EDTA, 1% β-mercaptoethanol, and 0.1 mg of bromphenol blue per ml) and incubated at 37°C for 10 min. Nonsolubilized material was cleared by centrifugation at maximal speed in a microcentrifuge for 10 min. Ten microliters of solubilized yeast protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred by Western blotting onto a nitrocellulose membrane. Immunodetection of CYP51A1 was performed by chemiluminescence with an enhanced chemiluminescence kit (ECL; Amersham) according to the recommendations of the manufacturer.
Site-directed mutagenesis.
Site-directed mutagenesis of the CYP51A1 ORF for the reintroduction of mutations observed in the genes of clinical isolates was performed by a PCR-based approach. Mutations were introduced by amplifying a wild-type CYP51A1 gene with external nonmutagenic primers (CYPCB and CYPNS2) and complementary internal mutagenic primers that overlapped at the site of each mutation. The mutagenic primers were as follows: for G129A, 5′-AGT TTT CGG TAA AGC GGT TAT TTA TGA TT-3′ and 5′-AAT CAT AAA TAA CCG CTT TAC CGA AAA CT-3′; for Y132H, 5′-GTA AAG GGG TTA TTC ATG ATT GTC CAA ATT-3′ and 5′-AAT TTG GAC AAT CAT GAA TAA CCC CTT TAC-3′; for S405F, 5′-ATT ACG TTT TAG TTT TTC CAG GTT ATG CTC-3′ and 5′-GAG CAT AAC CTG GAA AAA CTA AAA CGT AAT-3′; for G464S, 5′-CTT ATT TAC CAT TTA GTG GTG GTA GAC ATA-3′ and 5′-TAT GTC TAC ACA CAC TAA ATG GTA AAT AAG-3′; and for R467K, 5′-ATT TGG TGG TGG TAA ACA TAG ATG TAT TGG-3′ and 5′-CCA ATA CAT CTA TGT TTA CCA CCA CCA AAT-3′. For the introduction of the mutation R467K into CYP51A1-G464S, the following mutagenic primers were used: 5′-ATT TAG TGG TGG TAA ACA TAG ATG TAT TGG-3′ and 5′-CCA ATA CAT CTA TGT TTA CCA CCA CTA AAT-3′. A first round of PCR was carried out with high-fidelity Pwo DNA polymerase, a wild-type CYP51A1 template from clone C27, and each set of external and internal primers to yield two fragments overlapping at the site of the introduced mutation. PCR was performed for 20 cycles, with a 60°C primer annealing temperature. These fragments were purified to remove primers, and a final PCR, using the same conditions as described above, was performed by addition of the two fragments overlapping each mutation and the two external primers only. The final PCR products contained the entire CYP51A1 ORF and the introduced mutations. Sequencing of the final PCR products confirmed the introduction of each mutation. The mutated CYP51A1 genes obtained were reintroduced into YEp51 as described above.
Disruption of the S. cerevisiae CYP51A1 gene.
The procedure used for the disruption of the S. cerevisiae CYP51A1 gene was taken from an article by Kalb et al. (13). Briefly, S. cerevisiae YKKB-13 was first transformed with C. albicans CYP51A1 expression plasmids. Leu+ transformants were grown with raffinose and galactose to induce the different CYP51A1 proteins and transformed by a lithium acetate method (9) with a linear BamHI-HindIII DNA fragment from the p2500H vector described in reference 13, resulting in the disruption of the S. cerevisiae CYP51A1 gene by the URA3 marker. Ura+ colonies were selected, and the disruption of the CYP51A1 gene was verified by PCR and Southern blotting (data not shown).
Sequencing.
The nucleotide sequences of the cloned CYP51A1 genes were determined in both strands by standard protocols, using an AutoRead Kit (Pharmacia, Uppsala, Sweden). The reactions were analyzed on an ALF automated station (Pharmacia). Sequences were obtained by primer elongation using synthesized primers (Microsynth, Balgach, Switzerland).
RESULTS
A strategy enabling the detection of mutations in CYP51A1 genes potentially altering the affinity of CYP51A1 proteins for azole derivatives.
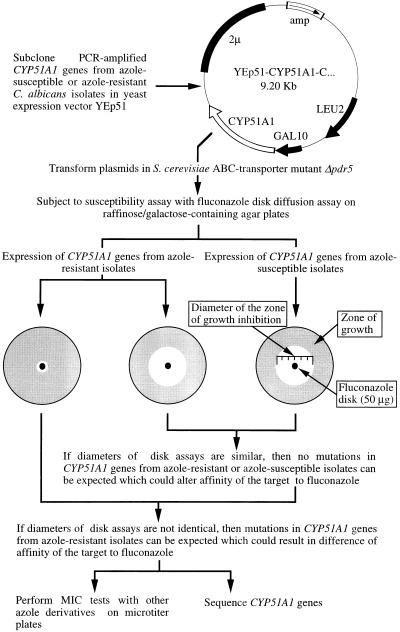
To detect alterations in the affinity of CYP51A1 for azole derivatives, several laboratories have performed biochemical assays on cellular extracts from yeasts (11, 17, 36). These assays consisted of analyzing the accumulation of radiolabeled 14α-methylated sterol metabolites in the presence of different drug concentrations and using cellular extracts from yeast isolates susceptible or resistant to azole antifungal agents. Depending on whether the 50% inhibitory concentrations of ergosterol biosynthesis of a given azole measured by formation of 14α-methylated sterol metabolites were higher in cellular extracts of resistant isolates than those of the azole-susceptible isolates, a change in the affinity of the 14α-demethylase enzyme for azole derivatives could be predicted. Likewise, Vanden Bossche et al. (34) have documented alterations in the affinity of CYP51A1 for azole derivatives by performing binding spectrum studies of CYP51A1-containing microsomal preparations from C. albicans clinical isolates with these agents. However, the evidence that a given mutation was causing these changes was not made available at that time, because no gene encoding CYP51A1 had been cloned from an azole-resistant organism. With the cloning of the C. albicans CYP51A1 gene and the acquisition of its nucleotide sequence (15, 16), the recovery of CYP51A1 genes from azole-resistant isolates became possible. One can isolate and sequence CYP51A1 genes from these organisms and look at differences in the nucleotide sequences which result in substitutions in the deduced amino acid sequence. However, this information does not give evidence that the observed amino acid substitutions are linked to alterations in the affinity of the CYP51A1 protein for azole derivatives. We therefore reasoned that a functional assay enabling the detection of alterations in the affinity of CYP51A1 proteins for azole derivatives would be advantageous. If this assay would be feasible and could reveal such alterations, then the nucleotide sequence should show which amino acid substitution causes the detected alteration. The strategy depicted in Fig. 1 was elaborated. This strategy is based on the PCR cloning, with a high-fidelity polymerase, of CYP51A1 genes from groups of C. albicans isolates with increasing resistance to azole derivatives from given patients. The PCR products are subcloned in the galactose-inducible yeast expression vector YEp51, and the resulting plasmids are transformed in a S. cerevisiae strain for heterologous expression of the C. albicans proteins. We anticipated that the expression in S. cerevisiae of C. albicans CYP51A1 genes with altered affinity for azole derivatives would render S. cerevisiae less susceptible to a panel of different azole derivatives than if a CYP51A1 gene from a C. albicans isolate susceptible to these agents was expressed. Furthermore, by utilizing as the expression host S. cerevisiae YKKB-13, which is defective in the ATP binding cassette transporter Pdr5p (3) and therefore is hypersusceptible to azole derivatives (30), the differences in susceptibility to these agents caused by the expression of the different CYP51A1 genes from C. albicans isolates have the chance to become more apparent. We used this strategy with sequential C. albicans isolates from a human immunodeficiency virus-positive patients, with which we showed recently the implication of multidrug efflux transporters in the appearance of resistance to azole antifungal agents (28, 30).
FIG. 1.
Outline of a strategy for cloning CYP51A1 genes from C. albicans isolates resistant or susceptible to azole derivatives and for predicting alterations of CYP51A1 gene products.
Mutations in CYP51A1 genes from C. albicans isolates resistant to azole antifungal agents.
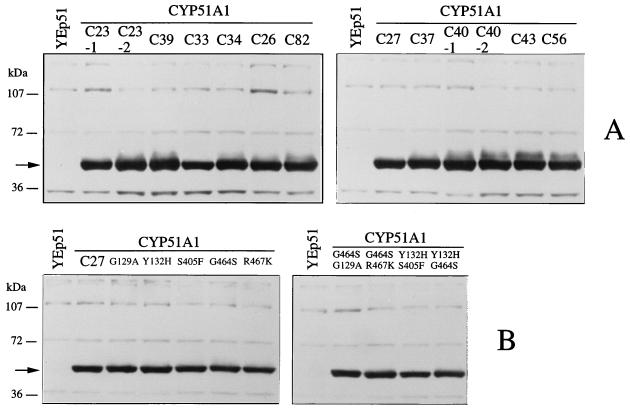
CYP51A1 genes from the C. albicans isolates listed in Table 1 were obtained by PCR with the external primers CYPNS2 and CYPCB and with genomic DNA from these isolates as templates. Fragments of the expected length (1.6 kb) were obtained in each case, and a total of 11 PCR products were subcloned into the yeast expression vector YEp51. From each PCR product subcloning at least six expression plasmids were selected, which were transformed individually into S. cerevisiae YKKB-13. Each yeast Leu+ transformant was subjected to a fluconazole disk diffusion assay on raffinose- and galactose-containing YNB agar. Diameters of inhibition were recorded for each Leu+ transformant, and the results are presented in Table 2; it can be observed that diameters of inhibition for yeasts expressing CYP51A1 genes from isolates that were less susceptible to azoles were decreasing compared to those for the most-susceptible isolates from a given patient. The decrease in the diameter of inhibition probably reflects the fact that alterations in CYP51A1 proteins which translate into a lower susceptibility of YKKB-13 to fluconazole had occurred. In two cases, two distinct diameters of inhibition were measured for YKKB-13 expressing the CYP51A1 genes recovered from the C. albicans isolates C23 and C40. This probably reflects the fact that since C. albicans is a diploid yeast, each allele of the genomic CYP51A1 loci of C23 and C40 was amplified by PCR, and one of these alleles encodes a protein altered in such a way that it decreases the susceptibility of a S. cerevisiae strain expressing the corresponding CYP51A1 allele. One can argue that the decrease in the diameter of inhibition could be caused by different levels of protein produced in S. cerevisiae. To exclude this possibility, the cellular extracts of each S. cerevisiae Leu+ transformant were analyzed by Western blotting with a polyclonal CYP51A1 antibody (Fig. 2). As is evident in Fig. 2, specific bands of equal intensity corresponding to CYP51A1 were detected for each extract. Thus, the decreases in diameter measured in the disk inhibition assays were not caused by differences in the expression of the different CYP51A1 genes but were rather due to the nature of the CYP51A1 proteins produced in S. cerevisiae.
TABLE 1.
MICs of azole derivatives for clinical C. albicans isolates
| Patient no. | C. albicans isolate | MIC (μg/ml) of:
|
||
|---|---|---|---|---|
| Fluconazole | Ketoconazole | Itraconazole | ||
| I | C23 | 1.0 | 0.015 | 0.0625 |
| C39 | 32.0 | 0.125 | 0.125 | |
| III | C33 | 0.25 | 0.015 | 0.0312 |
| C34 | 2.0 | 0.015 | 0.0625 | |
| C26 | >128 | 4.0 | >2.0 | |
| C82 | 32.0 | 0.5 | 1.0 | |
| IV | C27 | 1.0 | 0.015 | 0.0312 |
| C37 | 8.0 | 0.0325 | 0.0625 | |
| C40 | 128.0 | 2.0 | 1.0 | |
| V | C43 | 0.25 | 0.015 | 0.0312 |
| C56 | 128.0 | 4.0 | >2.0 | |
TABLE 2.
Fluconazole susceptibility of S. cerevisiae strains expressing C. albicans CYP51A1 genes as determined by disk diffusion tests
| Patient no. | C. albicans isolate | Expressed CYP51A1 genea | Mean (± SD) zone of inhibition (mm)b | Relative increase in MIC of the following azole for S. cerevisiae Leu+ transformantsc
|
Amino acid mutation(s) | ||
|---|---|---|---|---|---|---|---|
| Fluconazole | Ketoconazole | Itraconazole | |||||
| I | C23 | -C23-1 | 50 ± 2 | 1 | 1 | 1 | |
| C23 | -C23-2 | 38 ± 2 | 4 | 4 | 2 | S405F | |
| C39 | -C39 | 33 ± 3 | 4 | 4 | 2 | S405F | |
| III | C33 | -C33 | 47 ± 3 | 1 | 1 | 1 | |
| C34 | -C34 | 32 ± 2 | 4 | 4 | 2 | S405F | |
| C82 | -C82 | 31 ± 2 | 4 | 4 | 2 | S405F | |
| C26 | -C26 | 0 | >64 | 32 | 8 | S405F, Y132H | |
| IV | C27 | -C27 | 42 ± 3 | 1 | 1 | 1 | |
| C37 | -C37 | 21 ± 2 | 8 | 8 | 2 | G464S, R467K | |
| C40 | -C40-1 | 23 ± 1 | 8 | 8 | 2 | G464S, R467K | |
| C40 | -C40-2 | 0 | >64 | >32 | 8 | G464S, R467K, Y132H | |
| V | C43 | -C43 | 43 ± 3 | 1 | 1 | 1 | |
| C56 | -C56 | 21 ± 2 | 32 | 4 | 1 | G129A, G464S | |
The nomenclature indicates the isolate and, for multiple-protein-encoding genes, the protein; e.g., -C23-1 is an abbreviation for CYP51A1-C23-1, which is a gene encoding one of the two CYP51A1 proteins of isolate C23.
Mean diameters were obtained by measuring the zones of inhibition produced by at least six independent S. cerevisiae Leu+ transformants. The mean diameter (± SD) for YKKB-13 transformed with YEp51 alone was 62 ± 2 mm.
The numbers given are fold increases in MICs of azole derivatives relative to the MICs of the S. cerevisiae strains expressing a C. albicans CYP51A1 gene from the most susceptible isolate of a given patient. Assays of MICs of azole antifungal agents for S. cerevisiae strains expressing the different CYP51A1 genes were repeated three times without changes in obtained values.
FIG. 2.
(A) Western blot analysis of CYP51A1 proteins produced in S. cerevisiae in YNB selective medium containing raffinose and galactose and expressed from CYP51A1 alleles of the C. albicans clinical isolates listed in Table 1. (B) Western blot analysis of CYP51A1 proteins produced in S. cerevisiae and expressed from CYP51A1 genes obtained after site-directed mutagenesis, as outlined in Table 4. Approximately 20 μg of total cellular protein was loaded in each case. The origin of each extract is indicated. An extract (YEp51) from an S. cerevisiae strain transformed with only the parental plasmid YEp51 was loaded as a control to reveal background signals. Molecular mass standards are indicated on the left side. An arrow indicates the position of the CYP51A1-specific signal, with an estimated molecular mass of 58 kDa. No signals appear at this position in the extracts of S. cerevisiae transformed with YEp51, suggesting the absence of cross-reactivity of the antiserum with the S. cerevisiae endogenous CYP51A1 protein.
To quantify more precisely the decrease in susceptibility of each S. cerevisiae Leu+ transformant to fluconazole, MIC assays were performed by a microdilution method in a raffinose- and galactose-containing medium, which is a culture condition necessary, on the one hand, for the induction of the different C. albicans CYP51A1 genes cloned in YEp51 and expressed in S. cerevisiae and, on the other hand, for the growth of the expression host, in this case S. cerevisiae. Not only were the susceptibilities of S. cerevisiae Leu+ transformants to fluconazole tested, but also their susceptibilities to ketoconazole and itraconazole were determined (Table 2). It is apparent from Table 2 that the decrease in susceptibility to fluconazole observed in disk diffusion assays for expression of several CYP51A1 genes could be measured with the same drug in the format of the microdilution assay. The relative increase in the MIC of fluconazole for the expression of several CYP51A1 genes in S. cerevisiae varied from 4-fold to over 64-fold. Very surprisingly, however, the relative increase in the MIC of fluconazole did not always correlate with the same relative increase in the MICs of ketoconazole and itraconazole. For example, the relative increase in the MICs of fluconazole and ketoconazole caused by the expression of CYP51A1-C23-2 and CYP51A1-C39 was elevated by a factor of 4, whereas the relative increase in the MIC of itraconazole changed by a factor of only 2. The same feature was observed for the expression of CYP51A1-C34 and -C82. Likewise, the relative MICs of azole derivatives were increasing stepwise when CYP51A1 genes from sequential isolates with higher degrees of resistance from a given patient were expressed. This is observed, for example, in S. cerevisiae strains expressing CYP51A1-C82 and -C26 or expressing CYP51A1-C40-1 and -C40-2. These observations imply that not only may there be different types of alterations in the expressed CYP51A1 genes, but there may also be additive alterations for the CYP51A1 genes of isolates from a given patient. Most probably, these alterations can be attributed to changes in the affinities of the protein products for the different azole derivatives tested in this study.
These results encouraged us to perform the sequencing of all CYP51A1 genes listed in Table 2 to correlate the differences discussed above with the occurrence of possible mutations. Table 3 gives a comprehensive overview of the differences in the nucleotide sequences of the different CYP51A1 genes compared to a published CYP51A1 sequence (16). Table 3 shows which of the nucleotide changes yield amino acid substitutions. Five amino acid substitutions were found to be linked with a resistance phenotype; these are the substitutions G129A, Y132H, S405F, G464S, and R467K. Other amino acid substitutions, such as D116E, K128T, E266Q, and V437I, were found in CYP51A1 genes from both azole-susceptible and azole-resistant isolates and therefore are not likely to represent mutations linked with an azole antifungal agent resistance phenotype. The S405F mutation was found only in the second CYP51A1 allele of isolate C23 but was in both alleles of isolates C39, C34, C26, and C82. The expression of the genes in S. cerevisiae carrying this mutation increased the relative MICs of fluconazole and ketoconazole by a factor of only 4 and that of itraconazole by a factor of 2. These results suggest that the S405F mutation affected the affinity of the mutant CYP51A1 protein product for fluconazole and ketoconazole more than it affected its affinity for itraconazole. The Y132H mutation appeared in both CYP51A1 alleles of isolate C26 and was added to the already-existing S405F mutation of related isolates C34 and C82. This mutation has a profound effect, as judged by the results of MIC assays with S. cerevisiae expressing CYP51A1-C26. The relative MIC of fluconazole was increased by a factor exceeding 64, and the relative MICs of ketoconazole and itraconazole were increased by factors of 32 and 8, respectively (Table 2). The Y132H mutation was also found in the second CYP51A1 allele of isolate C40, but on the top of two already-existing mutations, namely G464S and R467K, which were present in both CYP51A1 alleles of the related isolates C37 and C40. The effect of both G464S and R467K mutations could be quantified as an eightfold increase in the relative MICs of fluconazole and ketoconazole but only a twofold increase in the relative MIC of itraconazole when the corresponding proteins containing these mutations were produced in S. cerevisiae. As mentioned above, the Y132H mutation was added to these two mutations in CYP51A1-C40-2, with the resulting effect being increases in the relative MICs of S. cerevisiae CYP51A1-expressing strains, by factors exceeding 64 and 32 for fluconazole and ketoconazole, respectively, and by a factor of 8 for itraconazole. Two other mutations, G129A and G464S, were observed in both alleles of CYP51A1-C56, and their combined effect on expression of this gene in S. cerevisiae was a 32-fold relative increase in the MIC of fluconazole, a 4-fold relative increase in the MIC of ketoconazole, but only a 2-fold relative increase in the MIC of itraconazole.
TABLE 3.
Nucleotides and amino acids substitutions in CYP51A1 genes from C. albicans clinical isolates
| Amino acid substitutiona | Mutation in CYP51A1 alleleb:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C23-1 | C23-2 | C39 | C33 | C34 | C82 | C26 | C27 | C37 | C40-1 | C40-2 | C43 | C56 | |
| V 82 V | A 246 T | A 246 T | A 246 T | A 246 T | |||||||||
| F 105 F | T 315 C | T 315 C | T 315 C | T 315 C | T 315 C | T 315 C | T 315 C | T 315 C | |||||
| D 116 E | T 348 A | T 348 A | T 348 A | T 348 A | T 348 A | T 348 A | T 348 A | T 348 A | |||||
| K 119 K | A 357 G | A 357 G | A 357 G | A 357 G | A 357 G | A 357 G | A 357 G | A 357 G | |||||
| K 128 T | A 383 C | A 383 C | |||||||||||
| G 129 A | G 386 C | ||||||||||||
| Y 132 H | T 394 C | T 394 C | |||||||||||
| S 137 S | C 411 T | C 411 T | C 411 T | C 411 T | C 411 T | C 411 T | C 411 T | C 411 T | |||||
| L 220 L | C 658 T | C 658 T | C 658 T | C 658 T | C 658 T | C 658 T | C 658 T | C 658 T | |||||
| E 266 Q | G 796 C | G 796 C | |||||||||||
| V 332 V | T 996 C | T 996 C | T 996 C | T 996 C | |||||||||
| L 340 L | A 1020 G | A 1020 G | A 1020 G | A 1020 G | A 1020 G | ||||||||
| K 342 K | A 1026 G | ||||||||||||
| S 361 S | A 1083 G | A 1083 G | A 1083 G | A 1083 G | |||||||||
| L 370 L | C 1110 T | C 1110 T | C 1110 T | C 1110 T | C 1110 T | C 1110 T | C 1110 T | C 1110 T | C 1110 T | C 1110 T | |||
| Y 401 Y | T 1203 C | T 1203 C | T 1203 C | T 1203 C | T 1203 C | ||||||||
| S 405 F | C 1214 T | C 1214 T | C 1214 T | C 1214 T | C 1214 T | ||||||||
| P 419 P | T 1257 C | ||||||||||||
| D 428 D | T 1284 C | T 1284 C | T 1284 C | T 1284 C | |||||||||
| V 437 I | G 1309 A | ||||||||||||
| V 456 V | T 1368 A | T 1368 A | T 1368 A | T 1368 A | |||||||||
| G 464 S | G 1390 A | G 1390 A | G 1390 A | G 1390 A | |||||||||
| R 467 K | G 1400 A | G 1400 A | G 1400 A | ||||||||||
| L 480 L | A 1440 G | A 1440 G | A 1440 G | A 1440 G | A 1440 G | A 1440 G | A 1440 G | A 1440 G | A 1440 G | ||||
| N 490 N | T 1470 C | T 1470 C | T 1470 C | T 1470 C | T 1470 C | T 1470 C | T 1470 C | T 1470 C | |||||
Boldface type indicates that the mutation at the corresponding site created a substitution.
The base numbers are with respect to the first ATG codon of CYP51A1.
Other verifications were made to ensure that (i) the expressed CYP51A1 genes yielded functional proteins in S. cerevisiae and (ii) the mutations detected by sequencing PCR-generated products were effectively originating from the genomes of the different C. albicans isolates analyzed in this study.
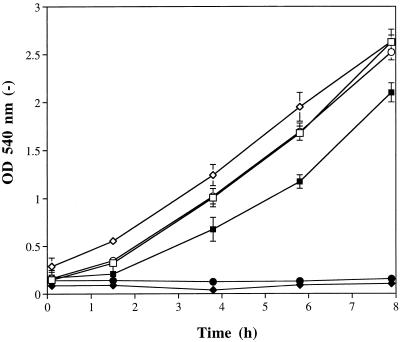
The functionality of C. albicans proteins produced in S. cerevisiae was tested by taking advantage of the fact that the deletion of the S. cerevisiae endogenous CYP51A1 gene is a lethal event when cells are grown under aerobic conditions (2, 13). We reasoned that this lethal event, however, can be rescued only if another functional C. albicans protein replaces the function of the absent endogenous S. cerevisiae CYP51A1 gene product. Since the expression of the C. albicans CYP51A1 genes could be switched on or off, because of the characteristics of the GAL10 promoter on YEp51, by changing the carbon source in the medium, the viability of S. cerevisiae Δcyp51a1 null mutants containing a C. albicans CYP51A1 expression plasmid would be dependent on the presence of an inducible carbon source, e.g., galactose in this case. Therefore, the S. cerevisiae CYP51A1 gene was disrupted in each of the strains expressing the C. albicans CYP51A1 genes listed in Table 2 in the presence of galactose and raffinose, and then the respective Δcyp51a1 null mutants were cultured with glucose as the sole carbon source. Only the results obtained for two such mutants are presented in Fig. 3; however, the results of all experiments indicated that the C. albicans proteins produced were functional in S. cerevisiae. Figure 3 shows that the expression of CYP51A1-C26 and -C82 in a medium containing galactose and raffinose allowed S. cerevisiae Δcyp51a1 mutants to grow at a rate similar to that of cells transformed with the parental plasmid YEp51. However, when glucose replaced raffinose-galactose in the medium, almost no growth of DSY643 and DSY644 was observed.
FIG. 3.
Functional complementation of two C. albicans CYP51A1 genes in S. cerevisiae Δcyp51a1 null mutants. Disruption of the S. cerevisiae CYP51A1 gene was performed as described by Kalb et al. (13) in S. cerevisiae YKKB-13 transformed with plasmids expressing the C. albicans CYP51A1-C82 and CYP51A1-C26 genes. Disruption was performed with cells in which the C. albicans CYP51A1 genes were induced on raffinose-galactose selective YNB medium. The S. cerevisiae Δcyp51a1 disruptants expressing the C. albicans CYP51A1-C82 and CYP51A1-C26 genes (DSY643 and DSY644, respectively) were maintained on this medium and incubated in YNB selective medium containing glucose or raffinose-galactose as the carbon source. As a control for the growth of S. cerevisiae on both glucose and raffinose-galactose, YKKB-13 transformed with the parental expression vector YEp51 was used (DSY610). The OD540s of the cultures were recorded at different time intervals. Closed symbols: DSY610 (▪), DSY643 (⧫), and DSY644 (•) grown with glucose; open symbols: DSY610 (□), DSY643 (◊), and DSY644 (○) grown with raffinose-galactose. Each data point represents the mean value of data from two separate experiments. Standard deviations are indicated (error bars).
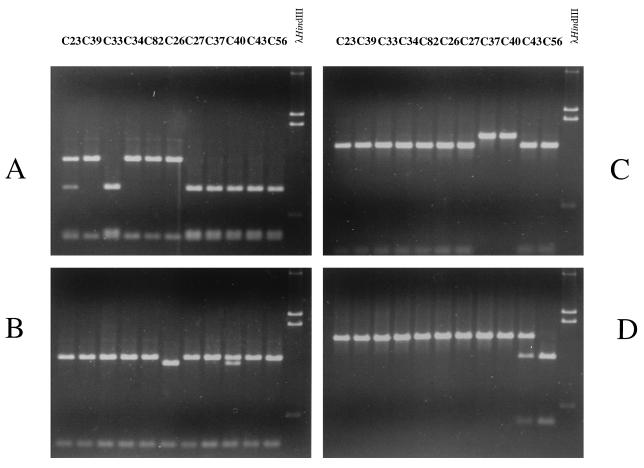
The presence of mutations in the different C. albicans CYP51A1 genes was tested by restriction fragment length polymorphism (RFLP) analysis with PCR fragments directly amplified from the genomes of the C. albicans isolates. This approach is possible only when a given mutation matches with the presence or the absence of a restriction site in CYP51A1. Fortunately, the G129A and Y132H mutations created new AciI and RcaI restriction sites, respectively, whereas the S405F and R467K mutations eliminated BpmI and AccI restriction sites, respectively. No restriction site was found to match with the G464S mutation. The PCR fragments containing the CYP51A1 ORF were digested with each of these enzymes, and the results of these experiments are shown in Fig. 4. As expected, the presence or absence of restriction sites which was predicted by the above-mentioned amino acid substitutions could be observed in the profiles of ethidium bromide-stained bands after agarose gel electrophoresis of the digested PCR products. A single RcaI site in the CYP51A1 ORF was evident in most of the CYP51A1 alleles analyzed (Fig. 4B), but an additional RcaI site, corresponding to the Y132H mutation, was observed on one allele of isolate C40 (i.e., CYP51A1-C40-2), since a mixture of two closely migrating bands of 1,211 and 1,295 bp, resulting from the restriction of DNA at one or two RcaI sites, could be distinguished for this DNA (Fig. 4B, lane C40). An additional RcaI site was observed on two alleles of isolate C26, since only a band of 1,211 bp was detected in this restricted DNA (Fig. 4B, lane C26). The results of this RFLP analysis are in agreement with those of the study of the functional expression of CYP51A1 genes amplified from C40 and C26, in which two different CYP51A1 alleles and only one CYP51A1 allele could be distinguished, respectively (Table 2). The BpmI restriction site is present twice in most of the amplified CYP51A1 alleles but only once in alleles amplified from isolates C39, C34, C82, and C26 (Fig. 4A). This feature can be used to determine the presence of the S405F mutation in these alleles, which was effectively observed by functional expression in S. cerevisiae. Two alleles are distinguished, as expected, for isolate C23, since a mixture of fragments resulting from a single and a double cut of the CYP51A1 ORF with BpmI could be observed (Fig. 4A, lane C23). The AccI restriction site is present in most of the amplified CYP51A1 alleles but is absent from the alleles amplified from C37 and C40 (Fig. 4C), as expected from the presence of the R467K mutations in these alleles. The AciI restriction site is observed only in alleles from isolates C43 and C56 (Fig. 4D). A mixture of cut and uncut DNA is observed in alleles amplified from isolate C43, although these alleles have not been distinguished by the functional screening assay in S. cerevisiae (Table 2). This suggests that the G129A mutation has little effect on the CYP51A1 protein, an observation that will be confirmed below with experimental evidence.
FIG. 4.
Restriction site analysis of CYP51A1 alleles amplified from C. albicans isolates and digested with BpmI (A), RcaI (B), AccI (C), and AciI (D). The origin of amplified DNA is indicated for each lane. Approximately 5% of the total PCR product was digested in a total volume of 20 μl and analyzed by 1% agarose gel electrophoresis. For each analysis, DNA molecular size standards (λHindIII) were loaded, with the following lengths: 4.3, 2.3, 2.0, and 0.56 kb. To allow the visualization of small fragments, only the lower part of the agarose gel is shown. Expected sizes of products obtained by the digestion of CYP51A1 alleles amplified by primers CYPNS2 and CYPCB are as follows: for BpmI, with the S405F mutation, 1,241 and 362 bp; for BpmI, without the S405F mutation, 947, 394, and 362 bp; for AccI, with the R467K mutation, 1,603 bp; for AccI, without the R467K mutation, 197 and 1,406 bp; for AciI, with the A129G mutation, 1,603 bp; for AciI, without the A129G mutation, 394 and 1,309 bp; for RcaI, with the Y132H mutation, 1,211, 308, and 84 bp; and for RcaI, without the Y132H mutation, 1,295 and 308 bp.
Taken together, the results of the RFLP analyses showed that the mutations detected by nucleotide sequencing were not due to artifacts of PCRs performed with genomic DNA from the C. albicans isolates. Since RFLP analysis could detect either homozygotic mutations or allelic variations among these mutations which were predicted by the functional assay, the introduction of errors by the PCRs is unlikely, emphasizing that the mutations detected in CYP51A1 genes were effectively present in the different genomes of the yeasts investigated here.
Reintroduction of mutations in CYP51A1 by site-directed mutagenesis.
Most of the mutations detected in the CYP51A1 genes from the C. albicans isolates investigated here were found in combination, with the exception of the S405F mutation, which was the only amino acid substitution found in CYP51A1-C23-1 and in CYP51A1-C32. To address the effect of each mutation on the S. cerevisiae susceptibility test system, each mutation was reintroduced by site-directed mutagenesis into a CYP51A1 gene from a susceptible C. albicans isolate, namely C27. In some CYP51A1 genes, two mutations were reintroduced sequentially to mimic the situation found in the CYP51A1 genes from clinical isolates. The expression of each CYP51A1 mutant form was verified by immunoblotting, and no differences in the expression levels of these proteins were detected (Fig. 2B). Each of the S. cerevisiae strains expressing the mutated CYP51A1 genes was subjected to the microbroth dilution MIC assay with fluconazole, ketoconazole, and itraconazole. The results are presented in Table 4. It was observed that all single mutations increased the relative MICs of azole derivatives when the corresponding genes were expressed in S. cerevisiae, with the exception of the G129A mutation, which in CYP51A1-G129A had no effect in this experimental setting. The relative increases in the MICs of fluconazole, ketoconazole, and itraconazole resulting from the expression of CYP51A1-S405F in S. cerevisiae were identical to those measured for CYP51A1 genes carrying the S405F mutation (for example, CYP51A1-C23-2). The relative increases in the MICs of azole derivatives resulting from the expression of CYP51A1-Y132H, -G464S, and -R467K were slightly lower than those measured when some of the mutations introduced into these genes were combined (i.e., G464S with R467K and Y132H with S405F), as was observed for the CYP51A1 genes from clinical isolates. When, however, two mutations were introduced sequentially by site-directed mutagenesis, the relative MICs of azole derivatives when the corresponding genes were expressed in S. cerevisiae increased to levels similar to those observed when these mutations were combined in the genes from clinical isolates. For example, the relative MICs of fluconazole, ketoconazole, and itraconazole measured by the expression of CYP51A1-G464S/R476K were increased by factors of eight-, four-, and twofold, respectively (Table 4), while they were increased by factors of eight- and twofold, respectively, for CYP51A1-C37 (Table 2). Surprisingly, when the G129A mutation, which is without effect in our test system, was added to the G464S mutation, the relative increases in the MICs of azole derivatives rose compared to the values obtained when only the G464S mutation alone was introduced. This suggests that some mutations can exert an effect only when they are combined with other mutations. The site-directed mutagenesis experiments therefore confirmed the role of distinct mutations in the capacity to cause alterations in CYP51A1, most probably resulting in a decreased affinity of the target enzyme for azole derivatives.
TABLE 4.
Effect of mutations introduced by site-directed mutagenesis on MICs of azoles for yeasts expressing the mutated CYP51A1 forms
| Expressed CYP51A1 gene(s)a | Relative increase in MIC of the following azole for S. cerevisiae Leu+ transformantsb
|
Introduced amino acid substitution(s) | ||
|---|---|---|---|---|
| Fluconazole | Ketoconazole | Itraconazole | ||
| -C27c | 1 | 1 | 1 | |
| -G129A | 1 | 1 | 1 | G129A |
| -S405F | 4 | 4 | 2 | S405F |
| -Y132H | 4 | 16 | 2 | Y132H |
| -G464S | 4 | 4 | 2 | G464S |
| -R467K | 4 | 4 | 2 | R467K |
| -S405F/Y132H | >64 | 32 | 8 | S405F, Y132H |
| -467S/Y132H | 32 | 32 | 4 | G464S, Y132H |
| -G464S/R467K | 8 | 4 | 2 | G464S, R467K |
| -G464S/G129A | 16 | 4 | 1 | G464S, G129A |
Unless otherwise noted, the nomenclature indicates the amino acid substitution(s) resulting; e.g., -G129A is an abbreviation for CYP51A1-G129A, a gene encoding a CYP51A1 protein with a substitution of Ala for Gly-129.
See footnote c of Table 2.
The gene from isolate C27 that encodes CYP51A1.
DISCUSSION
Resistance to azole antifungal agents has been shown to be attributable to a variety of mechanisms (12, 37, 38). Only recently have approaches aimed to reveal these mechanisms at the molecular level been undertaken. In a group of C. albicans isolates from AIDS patients that we investigated, we observed that azole antifungal agents failed to accumulate to the levels measured in azole-susceptible isolates (30). The cause of this effect was linked to the enhanced expression of efflux multidrug transporters genes, namely CDR1, CDR2, and BENr (28, 30). The overexpression of the ATP binding cassette transporter genes CDR1 and CDR2 is capable of mediating cross-resistance to known azole derivatives, including fluconazole, ketoconazole, and itraconazole (28, 30). The overexpression of the major facilitator gene BENr is linked only with the acquisition of resistance to fluconazole (1, 28, 30). Of the azole-resistant isolates investigated in this study, only one (C40) overexpressed the BENr gene without a corresponding overexpression of CDR1 and CDR2. One would have expected this isolate to have had a fluconazole-specific increase of resistance compared to a parental susceptible isolate. When the MICs of the three azole derivatives for this isolate were examined, however, all were increased by a substantial factor. This contradicted our prediction, and therefore we searched for other possible mechanisms of resistance in this isolate. The only unexamined possibility was an alteration of the affinity of the target of these antifungal agents, CYP51A1, for azole derivatives. Since our focus is on the molecular aspects of resistance to antifungal agents, we looked for a strategy enabling a direct diagnosis of the presence of such an alteration. We were encouraged to follow this approach since ergosterol biosynthesis inhibition experiments on cellular extracts from C40 and C27 (the parent azole-susceptible isolate of C40) with 14C-labeled mevalonic acid as the precursor indicated that the 50% inhibitory concentrations of azole derivatives for ergosterol biosynthesis were much higher in extracts from C40 than in those from C27 (data not shown). A favorable approach to uncovering the molecular mechanisms which was not yet used by others was the functional expression of PCR-amplified CYP51A1 alleles in a heterologous host with known and controlled genetics, such as S. cerevisiae. A first batch of experiments was undertaken with the CYP51A1 alleles amplified from the isolates from patient IV (C27 and C40). They were expressed in the azole-hypersusceptible Δpdr5 mutant of S. cerevisiae (YKKB-13). The functional screening assay in S. cerevisiae, involving disk diffusion assays and comparison of diameters of inhibition measured when CYP51A1 alleles from these isolates were expressed (Table 2), revealed that mutations in the CYP51A1 alleles could be present. This was confirmed by nucleotide sequencing of these alleles (Table 3). These results confirmed the soundness of our approach, and we therefore decided to screen the other C. albicans isolates of our study. To our surprise, other CYP51A1 alleles from all other azole-resistant isolates contained amino acid substitutions which could contribute to the resistance of these strains to azole derivatives.
The analysis of the nucleotide sequences of CYP51A1 alleles from sequential isolates from a given patient could reveal the extent to which these strains are related. Assuming that the nucleotide sequences of the CYP51A1 alleles from an azole-susceptible isolate and an azole-resistant isolate are identical except for the nucleotide change responsible for the resistance, this supports a high degree of relatedness between these strains and strengthens the idea that no strain replacement occurred during the appearance of resistance in the treated patient. In a previous study, strains listed in Table 1 were analyzed by different typing methods, and it was concluded that a high degree of relatedness between strains isolated from the same patient existed (5). In the CYP51A1 alleles sequenced in this study, such a high degree of relatedness could be seen between isolates C27, C37, and C40 from patient IV, since no nucleotide changes other than those resulting in amino acid substitutions important for resistance were detected (Table 3). This high-level sequence conservation is, however, not as evident in the CYP51A1 genes of other isolates from a given patient. A comparison of the CYP51A1 alleles of isolates C43 and C56 from patient V revealed only one nucleotide change besides those implicated in azole resistance (G796C; Table 3). When the CYP51A1 alleles of isolates C33, C34, C82, and C26 from patient III were compared, only for the CYP51A1 allele of isolate C33 were several nucleotide changes evident, namely, G796C, A1026G, T1203C, T1257C, and G1309A (Table 3). The CYP51A1 nucleotide sequences from isolates C34, C82, and C26 are identical except for the changes responsible for resistance. This suggests that strain C33 is not as closely related to the group consisting of strains C34, C82, and C26, which were isolated at different time intervals from patient III. It is therefore possible that a strain replacement in patient III had occurred after the isolation of strain C33. A comparison of the CYP51A1 alleles of isolates C23 and C39 from patient I revealed several nucleotide changes in the latter (T315C, T348A, A357G, A383C, C411T, and C658T), suggesting a lower degree of relatedness between these strains. Taken together, the analysis of nucleotide sequences of CYP51A1 alleles from yeasts isolated at different time intervals from a given patient is helpful for revealing strain differences which cannot be observed with conventional typing techniques. Moreover, CYP51A1 nucleotide sequences exhibited differences, supporting the idea of allelic microheterogeneity in the C. albicans population.
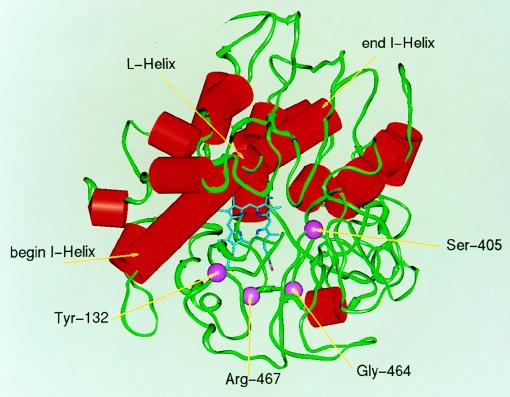
A total of five different nucleotide changes in CYP51A1 alleles investigated here yielded amino acid substitutions, namely G129A, Y132H, S405F, G464S, and R467K. These substitutions were linked to increases in the MICs of azole derivatives for C. albicans isolates and to increases in the MICs of these agents when the alleles carrying these mutations were expressed in S. cerevisiae. The decreases in diameter in disk diffusion assays and the relative increases in MICs of some azole derivatives measured when the CYP51A1 alleles carrying these mutations were expressed in S. cerevisiae were considered to be a strong indication of the existence of alterations affecting the affinity of azoles for the CYP51A1 protein products. As a validation of this hypothesis, Lamb et al. (18) recently expressed a C. albicans CYP51A1 gene with the mutation T315A in an S. cerevisiae host by a technique similar to that described here. The resulting mutant protein was purified, and it exhibited, as deduced from biochemical experiments, a twofold-reduced enzyme activity and approximately a fourfold-reduced affinity for fluconazole and ketoconazole. The same mutant protein produced in S. cerevisiae increased the MICs of fluconazole and ketoconazole by factors of four- and fivefold, respectively. Therefore, a direct correlation between relative MIC increase and relative decrease in affinity of a mutant protein for azole derivatives could be made, thus enabling us to make similar correlations between our MIC assay in S. cerevisiae and a decrease in affinity for the protein product. We added, however, the advantage of using an S. cerevisiae Δpdr5 multidrug transporter mutant in the MIC assay, a mutant which is hypersusceptible to azole derivatives because it cannot efflux these drugs and thus is more likely to detect small MIC differences when several C. albicans CYP51A1 alleles are compared. The CYP51A1 expression system used in S. cerevisiae coupled with an MIC assay for azole derivatives has been employed here to estimate alterations in affinity of the protein products for fluconazole, ketoconazole, and itraconazole. Another feature of this method is that it not only represents a simple assay but also will allow the testing of other or new azole derivatives. The properties of these agents with regard to their affinity for the wild-type and mutated targets enzymes could be thus compared with each other, which is an advantage for the design and development of new drugs not affected by mutations. However, future experimental work is needed to confirm by biochemical analyses the correlation between the above-described in vivo assay and the alterations in the affinity of the mutated enzymes for azole derivatives or for the substrate, i.e., lanosterol. The heterologous expression of the different C. albicans CYP51A1 genes in S. cerevisiae is an adequate tool for the production of the large amounts of CYP51A1 proteins needed for purification and further biochemical assays.
All single mutations reintroduced into CYP51A1 resulted in a change in the affinity of the target enzyme for azoles, with the exception of G129A. The alteration is, however, dependent on the type of mutation and on the type of azole used in the assay; while a fourfold decrease in affinity is measured for CYP51A1-S405F, -G464S, and -R467K with fluconazole and ketoconazole, only a twofold decrease is measured with itraconazole. For CYP51A1-Y132H, 4-fold, 16-fold, and 2-fold decreases in affinity are measured when fluconazole, ketoconazole, and itraconazole, respectively, are used in the assay. The same statement is valid when the mutations are combined, with the difference being that the decrease in affinity is more pronounced than in the case in which only a single mutation is introduced. The most remarkable effects were observed when the Y132H mutation was combined with the S405F or the R467K mutation (Table 4).
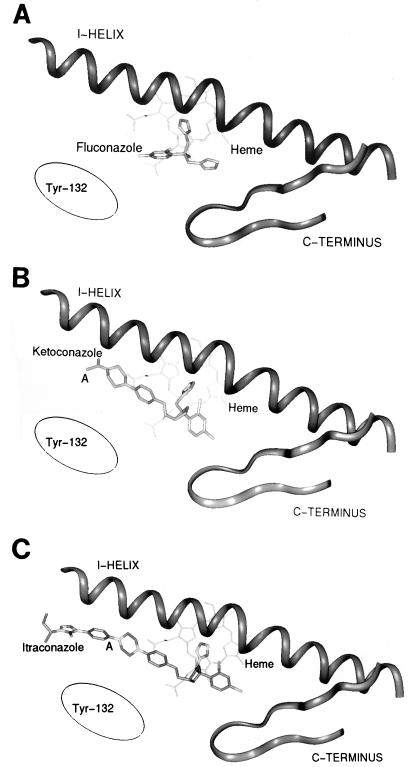
Azole derivatives inhibit CYP51A1 by interacting with the heme molecule. It is believed that the unhindered nitrogen atom of the azole ring (N3 in imidazole or N4 in derivatives) binds to the heme iron at its sixth coordinate position (35, 43, 44). The blocking of this position, which is required by the activated oxygen for the hydroxylation of the 14α methyl group of lanosterol, prevents initiation of the hydroxylation reaction. The structure, lipophilicity, and stereochemical orientation of the N-1 side chain of azole derivatives play roles not only in the affinity of these antifungal agents for their targets but also in their selectivity (45). Azole derivatives also fit in the CYP51A1 substrate pocket, which normally accepts lanosterol as a natural substrate. How do the mutations described here affect CYP51A1 and produce what might be alterations in its affinity for azoles? Two possibilities can be envisaged: (i) the amino acid affected by the mutation is in direct contact with some part of the azole molecule and thus the substitution results in a less efficient binding of the azole to the mutated protein; or (ii) the amino acid substitution displaces the three-dimensional arrangement of structures (α-helices or β-sheets) important for the optimal binding of the azole molecule and thus also results in a reduction in the affinity of the mutated protein for azoles. Using a model predicting the position of α-helices in the C. albicans CYP51A1 protein as proposed by Boscott and Grant (6), it is possible to place the mutations responsible for azole affinity alterations in this proposed structure (Fig. 5 and 6). As shown in the putative model of CYP51A1 in Fig. 6, it is remarkable that the mutations described here are found near the heme molecule and are not located at the surface of the protein. The G464S and R467K mutations are very near the cysteine residue (Cys-470) which generally serves in all cytochrome P-450 molecules as the fifth heme thiolate ligand (4, 24). These positions are also very conserved among lanosterol demethylases from other organisms (Fig. 7C). These positions, shown in Fig. 5 and 6, are very close to the L helix, which is a helix situated below the planar shape of the heme molecule. The heme molecule itself (shown in blue in Fig. 6) is embedded between the L and I helices (6). Thus, Gly-464 and Arg-467 are not likely to be in direct contact with the azole molecule, since azole derivatives fit between the I helix and the heme molecule. The S405F mutation is also situated in a highly conserved position in the lanosterol demethylases from other organisms, with the exception of Ustilago maydis, in which Ser is replaced by Ala (Fig. 7B). It is also difficult to speculate on the effect of such a mutation within the proposed model of Boscott and Grant (6); however, since it is close to the K helix and, thus, near the substrate binding pocket, this position could be in contact with azole molecules. The Y132H and A129G mutations are situated in a region which is also highly conserved among lanosterol 14α-demethylases (Fig. 7A). However, we observed that the A129G mutation had little effect on the affinity for azole derivatives, and therefore we will only concentrate on the Y132H mutation. The region matching with this mutation is situated in the B-B′ helix cluster, a region believed to play a role in the entry of the substrate in the substrate pocket, as documented for other cytochrome P-450 molecules whose three-dimensional structures have been resolved. One could speculate that a mutation in this region might also affect the entry of an azole derivative in the substrate pocket so that the imidazole or triazole ring could reach the sixth coordinate of the heme iron. In an attempt to construct a model of the C. albicans CYP51A1 protein superimposed on the known crystal structures of three bacterial cytochrome P-450 molecules (CYP101, CYP102, and CYP108), as done for other cytochromes P-450 (8), the position of the Tyr-132 residue was estimated relative to a model of the I helix and the C terminus of CYP51A1, which was proposed recently (33). Interestingly, the location of Tyr-132 was expected to be close to the point where the chemical structures of ketoconazole and itraconazole begin to diverge (apart from the imidazole and triazole rings). Attached to the piperazine ring of ketoconazole is an aldehyde group, whereas in itraconazole this is a phenyltriazone moiety (point A in Fig. 8B and C). If Tyr-132 is interacting with these parts of the molecules, it can be envisioned that changing Tyr-132 to His will have different effects on ketoconazole and itraconazole. In the case of fluconazole, Tyr-132 might interact with the fluorophenyl moiety (Fig. 8A).
FIG. 5.
Secondary structure of the C. albicans CYP51A1 protein as proposed by Boscott and Grant (6). The names of the predicted α-helices (from A to L) are given to the right of the corresponding underlined amino acid segments. The positions of mutations found in the CYP51A1 genes of C. albicans clinical isolates are given below the CYP51A1 amino acid sequence and are circled. The position of the fifth heme ligand (Cys-470), found in all P-450 proteins, is boxed.
FIG. 6.
Three-dimensional model of the C. albicans CYP51A1 structure adapted from reference 6. The structure was drawn with available coordinates and is presented from the top view, looking into the substrate binding pocket. The α-helices are shown with red cylinders, while the rest of the structure is shown with green contours. The I and L helices, corresponding to the distal and proximal heme binding sites, respectively, are labelled. The heme molecule is in blue, and the positions of the mutations described in this study are marked by purple spheres.
FIG. 7.
Alignments of lanosterol 14α-demethylase amino acid sequences from yeasts, fungi, and mammals. The alignments were generated by the program CLUSTAL implemented in the GCG software package of the University of Wisconsin with the entire amino acid sequences. Only the regions around the mutations detected in C. albicans CYP51A1 are shown for mutations G129A Y132H (A), S405F (B), and G464S R467K (C). Stars show conserved amino acids; periods show conserved replacements. The sequence sources and their GenBank accession numbers are as follows: rat, D55681; Homo sapiens, D55653; C. albicans, X13296; C. tropicalis, M23673; C. glabrata, S75389; S. cerevisiae, M18109; U. maydis, Z48164; and P. italicum, Z49750.
FIG. 8.
Top view of the estimated position of Tyr-132 relative to the I helix and C terminus of CYP51A1 from C. albicans. The position of Tyr-132 is indicated by an ellipse, which represents the area where this amino acid residue might be situated. The three models show the positions of the azole molecules fluconazole (A), ketoconazole (B), and itraconazole (C). The imidazole and triazole rings of azole derivatives lie between the I helix and the heme molecule. Point A in panels B and C marks the position of divergence between ketoconazole and itraconazole.
Some of the mutations in the C. albicans CYP51A1 genes described here have been reported recently by two different laboratories. White (42) reported the occurrence of the R467K mutation in a group of azole-resistant isolates in which efflux multidrug-dependent resistance mechanisms were also operating (41). This author could not, however, estimate the contribution of this mutation to an alteration of the target enzyme but could correlate the occurrence of this mutation with increases in the MICs of azole derivatives for the clinical isolate. Löffler et al. (19) reported sequence variations among CYP51A1 genes cloned from azole-resistant isolates. Among these mutations was G464S, but there was no experimental evidence of an effect of this mutation on the target enzyme. It is, however, interesting that some of the mutations described here were also found independently by others in the CYP51A1 genes from other isolates obtained in different locations. This illustrates that there may be preferential amino acid positions able to confer a phenotype of resistance to azole derivatives. It would be necessary to look at other CYP51A1 nucleotide sequences from other C. albicans isolates before considering them as possible azole-resistance “hot spots”. By using RFLP with the restriction enzymes employed in this study, screening for the occurrence of mutations can be accomplished without the cumbersome sequencing of many different C. albicans CYP51A1 genes. RFLP analysis will still cover only a limited number of mutations, but their number is likely to be increased in the future.
The effect of the mutations in the different CYP51A1 alleles has to be combined now with the other mechanisms of resistance already described in the yeasts employed in the present study. In a previous study, we showed that enhanced expression of multidrug transporters was one of the factors responsible for increases in MICs of azole derivatives. Table 5 gives an overview of the resistance mechanisms characterized so far and the steps at which they occur in the yeast isolates for which there is evidence of increasing MICs of azole derivatives. One can observe that a stepwise relative increase in MIC can be related to the appearance of distinct resistance mechanisms. For example, the overexpression of the multidrug transporter gene BENr in isolate C40 was identified previously as one of the determinants of resistance to azole derivatives. However, since we know that the overexpression of BENr can render yeasts resistant to fluconazole only, it was still not clear why in C40 the MICs of all azole derivatives were increasing. Now that it has been established here that C40 carries the R476K and G464S mutations in both CYP51A1 alleles and that the Y132H mutation is on only one CYP51A1 allele, an increase in the MICs of all azole derivatives can be expected, since these mutations affect the affinity for azole derivatives, although to different extents (Table 2). In other isolates, it might be difficult to distinguish the relative contributions of distinct mechanisms of resistance to increases in MICs measured for isolates from a given patient. For example, both the overexpression of efflux multidrug transporter genes (CDR1 and CDR2) and the mutations G464S and G129A in CYP51A1 are observed simultaneously in isolate C56. The relative increases in MICs of fluconazole, ketoconazole, and itraconazole for C56 versus C43 are 512-, 256-, and 64-fold, respectively (Table 5). While the affinity of CYP51A1 from C56 for fluconazole and ketoconazole is reduced by 32- and 4-fold, respectively, one may speculate that the differences between these values and the relative increases in MICs for clinical isolates are due to the effect of efflux multidrug transporters. The construction of C. albicans mutants carrying individual or combined resistance mechanisms is possible and might lead to the precise determination of the contribution of each resistance mechanism to the increase in MICs. It is also very clear from Table 5 that each isolate seems to have developed its own combination of mechanisms of resistance to azole derivatives. This apparent randomization of mechanisms of resistance to azole derivatives can yield different degrees of resistance, which are reflected by the measurement of MICs dependent on the type of azole used. By including a larger number of yeast isolates in a study of the molecular epidemiology of mechanisms of resistance to azole derivatives, it might be possible not only to reveal the emergence of major resistance mechanisms, or a preferred manner of their combination, but also to discover new resistance mechanisms. Such an analysis is under way in our laboratory.
TABLE 5.
Summary of resistance mechanisms observed in sequential yeast isolates from AIDS patients
| Patient no. | C. albicans isolate | Relative (fold) increase in MIC of:
|
Resistance mechanism(s) linked to MIC increase | ||
|---|---|---|---|---|---|
| Fluconazole | Ketoconazole | Itraconazole | |||
| I | C23 | 1 | 1 | 1 | S405F mutation on one allele |
| C39 | 32 | 8 | 2 | S405F mutation on two alleles; increase in CDR1 and CDR2 mRNAs | |
| III | C33 | 1 | 1 | 1 | |
| C34 | 8 | 1 | 2 | S405F mutation | |
| C82 | 128 | 32 | 32 | Increase in CDR1 and CDR2 mRNAs | |
| C26 | 512 | 256 | 64 | Y132H mutation | |
| IV | C27 | 1 | 1 | 1 | |
| C37 | 8 | 2 | 2 | G464S and R467K mutations | |
| C40 | 128 | 128 | 32 | Y132H mutation on one allele; increase in BENr mRNA | |
| V | C43 | 1 | 1 | 1 | |
| C56 | 512 | 256 | 64 | G129A and G464S mutations | |
| Increase in CDR1 and CDR2 mRNAs | |||||
ACKNOWLEDGMENTS
This work was supported by a grant from the Swiss Research National Foundation (no. 3100-045716) to D.S. and partially by a grant from the Office Fédéral de la Santé Publique (no. 93.7125) to J.B. and D.S.
We thank K. Kuchler (University of Vienna Biocenter) for providing strain YKKB-13.
REFERENCES
- 1.Albertson G D, Niimi M, Cannon R D, Jenkinson H F. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob Agents Chemother. 1996;40:2835–2841. doi: 10.1128/aac.40.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bard M, Lees N D, Turi T, Craft D, Cofrin L, Barbuch R, Koegel C, Loper J C. Sterol synthesis and viability of erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids. 1993;28:963–967. doi: 10.1007/BF02537115. [DOI] [PubMed] [Google Scholar]
- 3.Bissinger P H, Kuchler K. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. A yeast ABC transporter conferring mycotoxin resistance. J Biol Chem. 1994;269:4180–4186. [PubMed] [Google Scholar]
- 4.Black S D. Cytochrome P450 structure and function. In: Schenkman J B, Greim H, editors. Cytochrome P450. Berlin, Germany: Springer-Verlag; 1993. pp. 155–168. [Google Scholar]
- 5.Boerlin P, Boerlin-Petzold F, Goudet J, Durussel C, Pagani J-L, Chave J-P, Bille J. Typing Candida albicans oral isolates from human immunodeficiency virus-infected patients by multilocus enzyme electrophoresis and DNA fingerprinting. J Clin Microbiol. 1996;34:1235–1248. doi: 10.1128/jcm.34.5.1235-1248.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boscott P E, Grant G H. Modeling cytochrome P450 14 alpha-demethylase (Candida albicans) from P450cam. J Mol Graph. 1994;12:185–192. doi: 10.1016/0263-7855(94)80086-3. [DOI] [PubMed] [Google Scholar]
- 7.Broach J R, Li Y Y, Wu L-C C, Jayaram M. Vectors for high-level, inducible expression of cloned genes in yeast. In: Inoye M, editor. Experimental manipulation of gene expression. New York, N.Y: Academic Press; 1983. pp. 83–117. [Google Scholar]
- 8.De Groot M J, Vermeulen N P, Kramer J D, van Acker F A, Donne-Op den Kelder G M. A three-dimensional protein model for human cytochrome P450 2D6 based on the crystal structures of P450 101, P450 102, and P450 108. Chem Res Toxicol. 1996;9:1079–1091. doi: 10.1021/tx960003i. [DOI] [PubMed] [Google Scholar]
- 9.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning. A practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 109–135. [Google Scholar]
- 11.Joseph-Horne T, Hollomon D, Löffler J, Kelly S L. Altered P450 activity associated with direct selection for fungal azole resistance. FEBS Lett. 1995;374:174–178. doi: 10.1016/0014-5793(95)01102-k. [DOI] [PubMed] [Google Scholar]
- 12.Joseph-Horne T, Hollomon D W. Molecular mechanisms of azole resistance in fungi. FEMS Microbiol Lett. 1997;149:141–149. doi: 10.1111/j.1574-6968.1997.tb10321.x. [DOI] [PubMed] [Google Scholar]
- 13.Kalb V F, Woods C W, Turi T G, Dey C R, Sutter T R, Loper J C. Primary structure of the P450 lanosterol demethylase gene from Saccharomyces cerevisiae. DNA Cell Biol. 1987;6:529–537. doi: 10.1089/dna.1987.6.529. [DOI] [PubMed] [Google Scholar]
- 14.Kelly S L, Lamb D C, Corran A J, Baldwin B C, Kelly D E. Mode of action and resistance to azole antifungals associated with the formation of 14-α-methylergosta-8,24(28)-dien-3-β,6-α-diol. Biochem Biophys Res Commun. 1995;207:910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 15.Kirsch D R, Lai M H, O’Sullivan J. Isolation of the gene for cytochrome P450 L1A1 (lanosterol 14 alpha-demethylase) from Candida albicans. Gene. 1988;68:229–237. doi: 10.1016/0378-1119(88)90025-x. [DOI] [PubMed] [Google Scholar]
- 16.Lai M H, Kirsch D R. Nucleotide sequence of cytochrome P450 L1A1 (lanosterol 14 alpha-demethylase) from Candida albicans. Nucleic Acids Res. 1989;17:804. doi: 10.1093/nar/17.2.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb D C, Corran A, Baldwin B C, Kwon-Chung J, Kelly S L. Resistant P450 51A1 activity in azole antifungal tolerant Cryptococcus neoformans from AIDS patients. FEBS Lett. 1995;368:326–330. doi: 10.1016/0014-5793(95)00684-2. [DOI] [PubMed] [Google Scholar]
- 18.Lamb D C, Kelly D E, Schunck W H, Shyadehi A Z, Akhtar M, Lowe D J, Baldwin B C, Kelly S L. The mutation T315A in Candida albicans sterol 14-alpha-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 19.Löffler J, Kelly S L, Hebart H, Schumacher U, Lass-Flörl C, Einsele H. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol Lett. 1997;151:263–268. doi: 10.1111/j.1574-6968.1997.tb12580.x. [DOI] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeast. Tentative standard M27-T. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1995. [Google Scholar]
- 21.Nelson D R, Koymans L, Kamataki T, Stegeman J J, Feyereisen R, Waxman D J, Waterman M R, Gotoh O, Coon M J, Estabrook R W, Gunsalus I C, Nebert D W. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Nolte F S, Parkinson T, Falconer D J, Dix S, Williams J, Gilmore C, Geller R, Wingard J R. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob Agents Chemother. 1997;41:196–199. doi: 10.1128/aac.41.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Odds F C. Resistance of clinically important yeasts to antifungal agents. Int J Antimicrob Agents. 1996;6:145–147. doi: 10.1016/0924-8579(95)00048-8. [DOI] [PubMed] [Google Scholar]
- 24.Ortiz de Montellano P R, Graham-Lorence S E. Structure of cytochrome P450: heme-binding site and heme reactivity. In: Schenkman J B, Greim H, editors. Cytochrome P450. Berlin, Germany: Springer-Verlag; 1993. pp. 169–181. [Google Scholar]
- 25.Redding S, Smith J, Farinacci G, Rinaldi M, Fothergill A, Rhinechalberg J, Pfaller M. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS—documentation by in vitro susceptibility testing and DNA subtype analysis. Clin Infect Dis. 1994;18:240–242. doi: 10.1093/clinids/18.2.240. [DOI] [PubMed] [Google Scholar]
- 26.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanglard D, Ischer F, Bille J. Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1996. Mutations in the Candida albicans cytochrome P450 14α-demethylase gene (CYP51A1) conferring increased resistance to azole antifungal agents, abstr. C69; p. 46. [Google Scholar]
- 28.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents—characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 29.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanglard D, Kuchler K, Ischer F, Pagani J-L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seghezzi W, Meili C, Ruffiner R, Kuenzi R, Sanglard D, Fiechter A. Identification and characterization of additional members of the cytochrome P450 multigene family CYP52 of Candida tropicalis. DNA Cell Biol. 1992;11:767–780. doi: 10.1089/dna.1992.11.767. [DOI] [PubMed] [Google Scholar]
- 32.Troillet N, Durussel C, Bille J, Glauser M P, Chave J P. Correlation between in vitro susceptibility of Candida albicans and fluconazole-resistant oropharyngeal candidiasis in HIV-infected patients. Eur J Clin Microbiol Infect Dis. 1993;12:911–915. doi: 10.1007/BF01992164. [DOI] [PubMed] [Google Scholar]
- 33.Vanden Bossche H, Koymans L, Moereels H. P450 inhibitors of use in medical treatments: focus on mechanisms of action. Pharmacol Ther. 1995;67:79–100. doi: 10.1016/0163-7258(95)00011-5. [DOI] [PubMed] [Google Scholar]
- 34.Vanden Bossche H, Marichal P, Gorrens J, Bellens D, Moereels H, Janssen P A J. Mutation in cytochrome P450-dependent 14α-demethylase results in decreased affinity for azole antifungals. Biochem Soc Trans. 1990;18:56–59. doi: 10.1042/bst0180056. [DOI] [PubMed] [Google Scholar]
- 35.Vanden Bossche H, Marichal P, Gorrens J, Coene M-C, Willemsens G, Bellens D, Roels I, Moereels H, Janssen P A J. Biochemical approaches to selective antifungal activity. Mycoses. 1989;32:35–52. doi: 10.1111/j.1439-0507.1989.tb02293.x. [DOI] [PubMed] [Google Scholar]
- 36.Vanden Bossche H, Marichal P, Odds F C, Le Jeune L, Coene M-C. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1992;36:2602–2610. doi: 10.1128/aac.36.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanden Bossche H, Marichal P, Odds F C. Molecular mechanisms of drug resistance in fungi. Trends Microbiol. 1994;2:393–400. doi: 10.1016/0966-842x(94)90618-1. [DOI] [PubMed] [Google Scholar]
- 38.Vanden Bossche H V, Warnock D W, Dupont B, Kerridge D, Sengupta S, Improvisi L, Marichal P, Odds F C, Provost F, Ronin O. Mechanisms and clinical impact of antifungal drug resistance. J Med Vet Mycol. 1994;32:189–202. doi: 10.1080/02681219480000821. [DOI] [PubMed] [Google Scholar]
- 39.Vuffray A, Durussel C, Boerlin P, Boerlin-Petzold F, Bille J, Glauser M P, Chave J P. Oropharyngeal candidiasis resistant to single-dose therapy with fluconazole in HIV-infected patients. AIDS. 1994;8:708–709. doi: 10.1097/00002030-199405000-00023. [DOI] [PubMed] [Google Scholar]
- 40.Watson P F, Rose M E, Kelly S L. Isolation and analysis of ketoconazole resistant mutants of Saccharomyces cerevisiae. J Med Vet Mycol. 1988;26:153–162. doi: 10.1080/02681218880000231. [DOI] [PubMed] [Google Scholar]
- 41.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White T C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;41:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkinson C F, Hetnarski H, Hicks L J. Substituted imidazoles as inhibitor of microsomal oxidation and insecticide synergists. Pestic Biochem Physiol. 1974;4:299–312. [Google Scholar]
- 44.Wilkinson C F, Hetnarski H, Yellin Y O. Imidazole derivatives. A new class of microsomal enzyme inhibitors. Biochem Pharmacol. 1972;21:3187–3192. doi: 10.1016/0006-2952(72)90147-5. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida Y, Aoyama Y. Interaction of azole fungicides with yeast cytochrome P-450 which catalyzes lanosterol 14α-demethylation. In: Iwata K, Vanden Bossche H, editors. In vitro and in vivo evaluation of antifungal agents. Amsterdam, The Netherlands: Elsevier Science Publishers; 1986. pp. 123–134. [Google Scholar]