Abstract
Here, we describe the isolation and draft genome sequence of Paenibacillus sp. CCS19. Paenibacillus sp. CCS19 was isolated from leaf soil collected in Japan and identified based on similarity of the 16S rRNA sequence with related Paenibacillus type strains. The draft genome sequence of Paenibacillus sp. CCS19 consisted of a total of 107 contigs containing 6,816,589 bp, with a GC content of 51.5% and comprising 5,935 predicted coding sequences.
Keywords: Paenibacillus, Oligotroph, Enzyme, 16S rRNA, Draft genome sequence
Introduction
The isolation and identification of previously unknown bacteria could lead to the development of new industrial technologies. For example, in a previous study by Akita et al., thermostable NADP+-dependent meso-diaminopimelate dehydrogenase (meso-DAPDH) was newly discovered from Ureibacillus thermosphaericus, a bacterium isolated from compost 1. Moreover, NADP+-dependent D-amino acid dehydrogenase (D-AADH) was created from meso-DAPDH by protein engineering 2. D-Amino acids are important chiral intermediates for agrochemicals, semisynthetic antibiotics, and pharmaceutical drugs, but their utilization is limited by high production costs. D-AADH can be used in one-step production of several D-amino acids by reductive amination of the corresponding 2-oxo acids with ammonia and NADPH in a process involving fewer steps than current industrial-scale production methods 2. Thus, production methods using D-AADH have potential for industrial use because manufacturing costs can be reduced.
Several types of oligotrophic microorganisms exhibit high growth rates under low-nutrient conditions; thus, the metabolic enzymes from oligotrophs may have high catalytic activity. Moreover, as mentioned above, such enzymes are also potentially useful in industrial applications. Recently, using our newly developed method for the isolation and identification of novel oligotrophs, we identified Deinococcus kurensis KR-1T 3, Enterobacter oligotrophicus CCA6T 4, Pseudomonas humi CCA1T 5, and Paenibacillus glycanilyticus subsp. hiroshimensis CCI5T 6. Here, we report the isolation and identification of another oligotroph, strain CCS19.
Materials and Methods
Soil samples such as compost, leaf soil, mud, and peat moss were collected at Narashino City in Chiba Prefecture, Japan. Oligotrophs were isolated from the soil samples on 1.5% agar plates (pH 7.2) containing sulfate (<0.4%), calcium (<0.1%), iron (<0.01%), and a few fatty acids and/or other minerals at concentrations of less than 0.01%. Soil samples were suspended separately at 10% (w/v) in sterilized water and then filtered. The filtrates were inoculated separately onto 1.5% agar plates and incubated for 2 days at 37°C. Individual colonies were successively re-streaked onto fresh 1.5% agar plates at least three times to obtain pure colonies, and the pure colonies were used for phylogenetic characterization.
After incubation in R2A broth at 37°C for 2 days, the cultures were washed with sterile water. Subsequently, genomic DNA was extracted using an IllustraTM bacteria genomicPrep Mini Spin Kit (GE Healthcare, Chicago, IL, USA) according to the manufacturer's instructions. The concentration and purity of the extracted genomic DNA were measured using a Quant-iT dsDNA Assay Kit (Invitrogen) and a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific), respectively.
Using the extracted genomic DNA as a template, the 16S rRNA gene was amplified using KOD-plus DNA Polymerase (TOYOBO, Osaka, Japan) with the bacterial universal primers 27f 7 and 1391r 8. After the amplified PCR product was purified using a Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA), the purified product was cloned into the pTA2 vector (TOYOBO), yielding pTA2/16S, which was then sequenced. The sequence of the 16S rRNA gene (accession number: LC763767; 1512 bp) was compared with reference sequences available in the GenBank/EMBL/DDBJ databases using BLAST. Multiple alignment and construction of a maximum-likelihood tree were performed using MEGA-X 9 with the Tamura-Nei model 10.
Sequence libraries for genome sequencing were prepared using a Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer's instructions. The resulting libraries were sequenced using a MiSeq sequencer (Illumina) with a MiSeq Reagent Kit v3 (Illumina). Default parameters were used for all software unless otherwise specified. Quality control and de novo assembly were carried out using Trimmomatic ver.0.39 11 and Shovill ver.1.1.0 12, respectively. Genome annotation was carried out using DFAST ver.1.2.0. 13.
Results and Discussion
To isolate the objective oligotrophs, we screened samples on 1.5% agar plates without a carbon source and other medium components, and a few colonies were formed when filtrate prepared from leaf soil was plated. After standard dilution plating was carried out for the purification of colonies, a colony exhibiting rapid growth was obtained and designated strain CCS19.
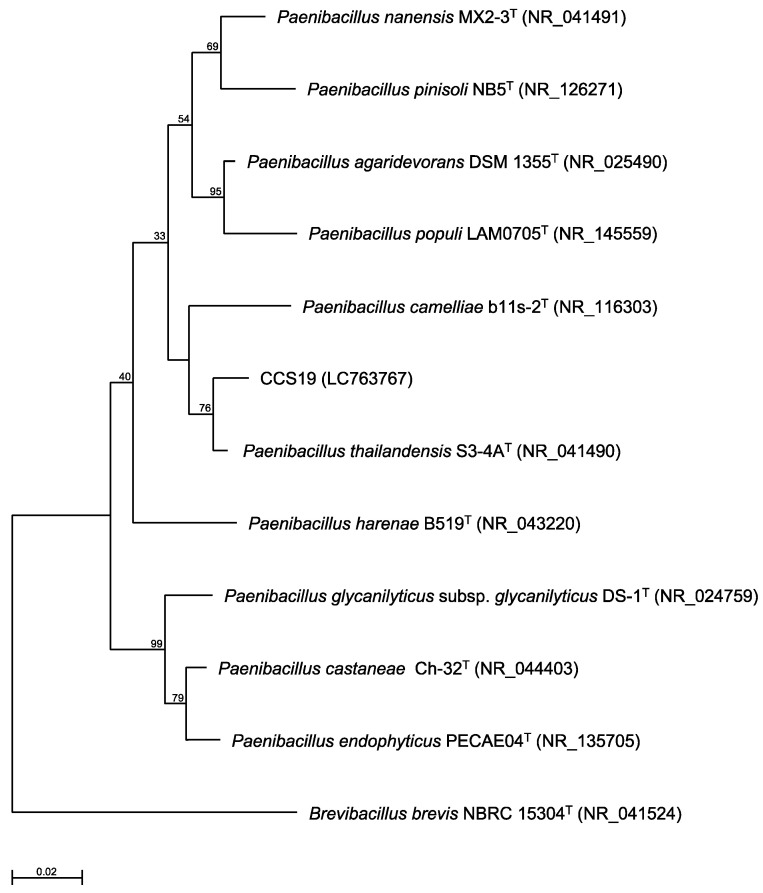
To identify strain CCS19, the 16S rRNA gene sequence was determined and used to construct a phylogenetic tree. Strain CCS19 clustered with members of the genus Paenibacillus (Figure 1). In particular, strain CCS19 exhibited similarities of 98.7%, 98.1%, 97.2%, 96.7%, 96.6%, 96.6%, 96.3%, and 96.3% to its closest relatives, P. thailandensis S3-4AT, P. nanensis MX2-3T, P. agaridevorans DSM1355T, P. castaneae Ch-32T, P. harenae B519T, P. populi LAM0705T, P. camelliae b11s-2T, and P. pinisoli NB5T, respectively. Based on these results, we identified strain CCS19 as Paenibacillus sp. CCS19.
Figure 1.
Phylogenetic tree constructed from analysis of 16S rRNA gene sequences and showing the relationships between strain CCS19 and related Paenibacillus type strains. The bar indicates a 0.02% nucleotide substitution rate. The tree was rooted using Brevibacillus brevis NBRC 15304T as the outgroup.
Most type strains in the genus Paenibacillus are characterized as gram-positive or -negative, aerobic or facultative anaerobic, rod-shaped, motile bacteria. Currently, more than 300 species and 6 subspecies of Paenibacillus have been identified based on 16S rRNA gene sequence homology as well as physiological and chemotaxonomic characteristics (https://www.bacterio.net/genus/paenibacillus). Moreover, Paenibacillus were found to produce novel enzymes exhibiting substrate specificity different from that of known enzymes such as alcohol oxidase 14 and (2R,3R)-2,3-butanediol dehydrogenase 15. To enable the use of enzymes produced by Paenibacillus sp. CCS19, the draft genome sequence was determined. The raw data after genome sequencing using a MiSeq sequencer yielded 64,617 reads with 105-fold coverage. The assembled genome sequence of Paenibacillus sp. CCS19 contained 107 contigs, which consisted of 6,816,589 bp and GC content of 51.5%. Moreover, within the draft genome sequence of Paenibacillus sp. CCS19, a total of 5,935 predicted coding sequences were identified.
To facilitate the screening and isolation of novel enzymes, in the present study, we isolated and determined the draft genome sequence of Paenibacillus sp. CCS19. Analysis of the draft genome sequence of Paenibacillus sp. CCS19 could reveal novel enzymes suitable for industrial use.
Nucleotide Sequence Accession Number
The draft genome sequence of Paenibacillus sp. CCS19 was deposited in the DDBJ/EMBL/GenBank databases under accession numbers BTCK01000001 to BTCK01000158. The raw sequence reads were deposited in DDBJ under BioProject number PRJDB15611 and BioSample number SAMD00590491.
Acknowledgments
We are grateful to all members of the Department of Basic Science at our institute (Nihon University) for their technical assistance and valuable discussions. This research was supported by JSPS KAKENHI Fund for the Promotion of Joint International Research (Fostering Joint International Research (A)) Grant Number 20KK0343.
References
- 1.Akita H, Fujino Y, Doi K, Ohshima T. Highly stable meso-diaminopimelate dehydrogenase from an Ureibacillus thermosphaericus strain A1 isolated from a Japanese compost: purification, characterization and sequencing. AMB Express. 2011;1:43. doi: 10.1186/2191-0855-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akita H, Hayashi J, Sakuraba H, Ohshima T. Artificial thermostable D-amino acid dehydrogenase: creation and application. Front Microbiol. 2018;9:1760. doi: 10.3389/fmicb.2018.01760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akita H, Itoiri Y, Ihara S, Takeda N, Matsushika A, Kimura ZI. Deinococcus kurensis sp. nov, isolated from pond water collected in Japan. Arch Microbiol. 2020;202:1757–1762. doi: 10.1007/s00203-020-01845-8. [DOI] [PubMed] [Google Scholar]
- 4.Akita H, Matsushika A, Kimura ZI. Enterobacter oligotrophica sp. nov, a novel oligotroph isolated from leaf soil. Microbiologyopen. 2019;8:e00843. doi: 10.1002/mbo3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akita H, Kimura ZI, Hoshino T. Pseudomonas humi sp. nov, isolated from leaf soil. Arch Microbiol. 2019;201:245–251. doi: 10.1007/s00203-018-1588-x. [DOI] [PubMed] [Google Scholar]
- 6.Akita H, Itoiri Y, Takeda N, Matsushika A, Kimura ZI. Paenibacillus glycanilyticus subsp. hiroshimensis subsp. nov, isolated from leaf soil collected in Japan. Arch Microbiol. 2021;203:1787–1793. doi: 10.1007/s00203-020-02166-6. [DOI] [PubMed] [Google Scholar]
- 7.Lane DJ, 16S/23S rRNA sequencing. Nucleic acid techniques in bacterial systematics. Stackebrandt, E, & Goodfellow, M. eds, New York. 1991. pp. 115–175.
- 8.Turner S, Pryer KM, Miao VP, Palmer JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. 1999;46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 11.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seemann T. Shovill: faster SPAdes assembly of Illumina reads. https://github.com/tseemann/shovill.
- 13.Tanizawa Y, Fujisawa T, Nakamura Y. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018;34:1037–1039. doi: 10.1093/bioinformatics/btx713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isobe K, Kato A, Sasaki Y, Suzuki S, Kataoka M, Ogawa J, Iwasaki A, Hasegawa J, Shimizu S. Purification and characterization of a novel alcohol oxidase from Paenibacillus sp. AIU 311. J Biosci Bioeng. 2007;104:124–128. doi: 10.1263/jbb.104.124. [DOI] [PubMed] [Google Scholar]
- 15.Yu B, Sun J, Bommareddy RR, Song L, Zeng AP. Novel (2R,3R)-2,3-butanediol dehydrogenase from potential industrial strain Paenibacillus polymyxa ATCC 12321. Appl Environ Microbiol. 2011;77:4230–4233. doi: 10.1128/AEM.02998-10. [DOI] [PMC free article] [PubMed] [Google Scholar]