Abstract
Erythromycin resistance among streptococci is commonly due to target site modification by an rRNA-methylating enzyme, which results in coresistance to macrolide, lincosamide, and streptogramin B antibiotics (MLSB resistance). Genes belonging to the ermAM (ermB) gene class are the only erythromycin resistance methylase (erm) genes in Streptococcus pyogenes with MLSB resistance that have been sequenced so far. We identified a novel erm gene, designated ermTR, from an erythromycin-resistant clinical strain of S. pyogenes (strain A200) with an inducible type of MLSB resistance. The nucleotide sequence of ermTR is 82.5% identical to ermA, previously found, for example, in Staphylococcus aureus and coagulase-negative staphylococci. Our finding provides the first sequence of an erm gene other than ermAM that mediates MLSB resistance in S. pyogenes.
Three principal mechanisms have so far been found to be responsible for the acquired erythromycin resistance in bacteria: target site modification, enzymatic inactivation of erythromycin, and active efflux of erythromycin (20, 21). In streptococci as well as in many other gram-positive bacteria, target site modification is a common resistance mechanism (45). It is due to the presence of an rRNA methylase that mono- or dimethylates the N6 amino group of an adenine residue in 23S rRNA. Methylation probably results in a conformational change in the ribosome, leading to reduced binding of macrolide, lincosamide, and streptogramin B (MLSB) antibiotics to their target site in the 50S ribosomal subunit. MLSB resistance can be divided into constitutive resistance, when the methylating enzyme is produced continuously, and inducible resistance, when the presence of an inducing antibiotic is required for production of the enzyme. Different types of erythromycin resistance methylases, encoded by erm genes, are produced by different bacteria. The erm genes have been divided into at least 12 different classes on the basis of hybridization studies and sequence comparisons (45).
In addition to MLSB resistance, active efflux has recently been shown to be a common mechanism of erythromycin resistance, at least in Streptococcus pyogenes and Streptococcus pneumoniae (41). In some of these bacteria, resistance to 14- and 15-membered macrolides is due to the mefA gene, which encodes a membrane-associated protein (9).
In streptococci MLSB resistance has commonly been due to genes belonging to the ermAM (ermB) gene class. The ermAM gene was first sequenced from plasmid pAM77 of Streptococcus sanguis (13). Thereafter, genes of the same class have been sequenced, for example, from S. pneumoniae (43), S. pyogenes (7, 8), and Streptococcus agalactiae (5). In S. pyogenes, genes belonging to the ermAM class are actually the only erm genes that have been sequenced. In this study, we have characterized a novel erm gene, designated ermTR, from an erythromycin-resistant clinical strain of S. pyogenes isolated in Finland. Our data provide the first sequence of an erm gene other than ermAM that mediates MLSB resistance in S. pyogenes.
MATERIALS AND METHODS
Bacterial strains and determination of antimicrobial resistance.
S. pyogenes A200 of serotype T11 is an erythromycin-resistant clinical skin isolate. Its erythromycin resistance phenotype was determined by the double-disk test and MIC determinations (36). The MICs of different antimicrobial agents were determined by the plate dilution method (30).
S. agalactiae 90-30-591 was used as a positive control for the ermAM gene in PCR. S. pyogenes 13 234 containing the 17.5-MDa MLSB resistance plasmid pERL1 (25) was used as a control strain for plasmid isolation.
DNA extraction and dot blot hybridization.
Streptococcal DNA was extracted by the cell lysis method described by Anderson and McKay (1), with the following modifications. The cells were grown in 3 ml of Todd-Hewitt broth (BBL, Cockeysville, Md.) with 1% yeast extract (Oxoid, Basingstoke, United Kingdom). Instead of lysozyme alone, a combination of lysozyme (10 mg/ml) and mutanolysin (300 U/ml) (Sigma, St. Louis, Mo.) was used to enhance cell lysis during an incubation for 2 h at 37°C. After cell lysis DNA was extracted once with 1 volume of phenol and once with 1 volume of chloroform, and after the addition of 1/10 volume of 2 M sodium acetate (pH 6.2), the DNA was precipitated with 2 volumes of ethanol. The DNA was dissolved in 20 μl of water, and 1 μl of the solution was used as template in PCRs.
Dot blot hybridization was performed by standard techniques. The probes used are shown in Table 1. The probes were labelled with two 32P-labelled nucleotide triphosphates and two unlabelled nucleotides by nick translation as described previously (40). Filters underwent three 30-min washes at 52°C in 0.1% sodium dodecyl sulfate, 0.015 M NaCl, and 0.0015 M sodium citrate followed by three 15-min washes at 52°C in 0.015 M NaCl and 0.0015 M sodium citrate. Under these high-stringency conditions, none of the probes cross-hybridized. The plasmids that are the original sources of the genes were used as controls (Table 1).
TABLE 1.
Probes and controls used in dot blot hybridization to detect known erm genes in S. pyogenes A200
PCR and sequencing experiments.
The oligonucleotide primers used in PCR and sequencing are shown in Table 2. Primers III1 and III2 were designed from the sequence of ermAM found in S. pyogenes (8), S. sanguis (13), and S. pneumoniae (43). Primers III9 and III13 were directly from the sequence of ermA of S. aureus (28). The rest of the primers were from newly sequenced regions of strain A200. Primers III14 and III15 were used in the ligation PCR after self-ligation of A200 DNA digested with TaqI restriction endonuclease. TaqI was chosen because there were no TaqI recognition sites in the newly sequenced DNA. The oligonucleotides were synthesized by the Applied Biosystems 391 DNA synthesizer (PCR-Mate, Foster City, Calif.). DNA amplification was performed by using a DNA thermocycler (HB-TR1; Hybaid Ltd., Middlesex, United Kingdom). The PCR mixture of 100 μl contained 50 mM KCl, 10 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2, 0.1% (vol/vol) Triton X-100, 200 μM (each) deoxyribonucleotides, 20 pmol of oligonucleotides, 1 U of DynaZyme DNA polymerase (Finnzymes Oy, Espoo, Finland), and approximately 50 ng of template DNA extracted from A200. A total of 35 cycles, with denaturation at 93°C for 30 s, annealing at temperatures adjusted for each primer pair for 1 min, and extension at 72°C for 90 s, were carried out in the thermal reactor. A 20-μl volume of the reaction mixture was run in a 1.0% agarose gel (FMC BioProducts, Rockland, Maine). A 100-bp ladder (Gibco-BRL, Gaithersburg, Md.) was run in the gel as a size marker. After staining with ethidium bromide, the PCR products were visualized under UV light and were photographed with type 667 Polaroid film.
TABLE 2.
Oligonucleotide primers used in PCR and sequencing
| Primer | Sequence (5′ to 3′)a | Gene used for primer design | Position in ermTR | Primer used with primer |
|---|---|---|---|---|
| III1 | GAA ATT GG(A/C) ACA GGT AAA GGG CA | ermAM | 316–338 | III2b |
| III2b | AAA (C/T)TG ATT TTT AGT AAA | ermAM | 828–846 | III1 |
| III6 | GAA GTT TAG CTT TCC TAA | ermTR | 468–485 | III2b |
| III7 | TGC TGT TAA TGG TGG AAA | ermTR | 641–658 | III2b |
| III8b | GCA TGA CAT AAA CCT TCA | ermTR | 393–410 | III9 |
| III9 | ACA TAA GGA GGT TTC AAT | ermA | 1–18 | III8b |
| III10 | AGG TTA TAA TGA AAC AGA | ermTR | 204–221 | III8b |
| III13b | TTA GTG AAA CAA TTT GTA | ermA | 925–942 | III7 |
| III14 | TCT CCT TGC CGG TTA TAA | ermTR | 186–203 | III15b |
| III15 | ATC AAT TAA GAC AGG TGC TGA AGC | ermTR | 839–862 | III14b |
Nucleotides in parentheses indicate that during synthesis either of the nucleotides was incorporated into the oligonucleotide.
The primer was biotinylated at the 5′ end for sequencing purposes.
Sequencing was done directly from the PCR products. Purification of the PCR products and sequencing according to Sanger’s dideoxynucleotide chain termination method were performed as described previously (39). The sequences of the regions corresponding to the positions (Table 2) of the primers derived from the ermAM and ermA genes (primers III1, III2, III9, and III13) were determined from PCR products that were produced with other primers.
Comparison of sequences.
The nucleotide and amino acid sequence similarities of different erm genes and their predicted products were determined by the programs GAP and PILEUP of the Genetics Computer Group Program Package (11). The output of PILEUP includes a dendrogram, constructed by the neighbor joining method, and an ordered gapped listing of sequences.
Plasmid isolation experiments.
Two methods were used to detect plasmids in strain A200. The procedure of Anderson and McKay (1) was used, but with the modifications in the cell lysis procedures described earlier in this paper. In addition, a 125-ml overnight culture (Todd-Hewitt broth with 1% yeast extract) was analyzed with the Wizard Plus Maxipreps DNA Purification System (Promega, Madison, Wis.) according to the instructions of the manufacturer, except that half of the volumes were used and mutanolysin (300 U/ml) and pronase (500 μg/ml) were added in the cell resuspension solution and the mixture was incubated for 1 h at 37°C.
Nucleotide sequence accession number.
The nucleotide sequence of ermTR and its leader sequence has been assigned GenBank accession number AF002716.
RESULTS AND DISCUSSION
Resistance phenotype and antimicrobial susceptibilities of S. pyogenes A200.
Strain A200 expressed an inducible type of MLSB resistance. This was indicated by resistance to erythromycin and susceptibility to clindamycin in MIC determinations (the MICs were 8 and 0.25 μg/ml, respectively) and by a reduced clindamycin inhibition zone proximal to the erythromycin disk in the double disk test. A200 was susceptible to all other antimicrobial agents tested; the MICs were as follows: penicillin, 0.016 μg/ml; cephalothin, 0.125 μg/ml; tetracycline, 0.5 μg/ml; chloramphenicol, 4 μg/ml; ciprofloxacin, 0.5 μg/ml; and vancomycin, 0.5 μg/ml.
Dot blot hybridization.
Hybridization of strain A200 gave negative results with the probes listed in Table 1, indicating that the ermA, ermBP (which belongs to the ermAM gene class), ermC, ermF, or ermQ gene was not present in A200.
Sequence analysis of ermTR and upstream sequences and comparison to other erm genes.
To identify the resistance gene in strain A200 a PCR-based approach was used. Primers III1 and III2 successfully amplified an approximately 530-bp region of the methylase gene from the genome of S. agalactiae 90-30-591 used as a positive control. A PCR product of the same size was also amplified from A200. The nucleotide sequence of the 530-bp PCR product of A200 was determined. Unexpectedly, the sequence that was obtained shared 82% homology with the ermA gene of S. aureus. The ermA gene has not previously been found in S. pyogenes. Therefore, in sequencing of the A200 gene, the sequence of ermA was used to help the primer design, when applicable (Table 2).
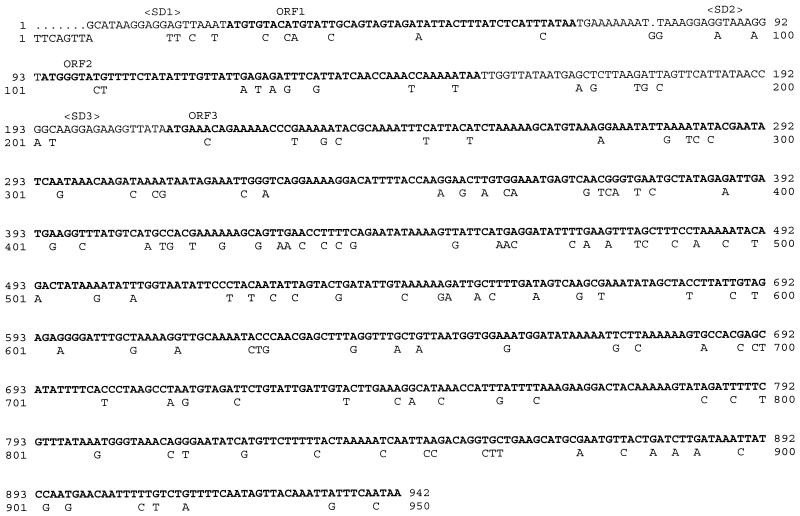
The completely sequenced 942-bp region of strain A200 aligned with the S. aureus ermA gene region is presented in Fig. 1. Three potential open reading frames (ORFs) were detected. The longest ORF consists of the same number of nucleotides as is present in the ermA gene, that is, 732 nucleotides. The sequence of this ORF is 82.5% identical to the ermA sequence, and it is predicted that it encodes a polypeptide of 243 amino acids (Fig. 2). This strongly suggests that the polypeptide is also a methylase conferring resistance to erythromycin by methylating an adenine residue in the 23S rRNA. The structural gene for the methylase was designated ermTR.
FIG. 1.
Nucleotide sequence of the ermTR gene of S. pyogenes A200 and its leader sequence. The ribosome-binding sites (Shine-Dalgarno sequence) and the ORFs (boldface) are indicated. The ermTR gene has been aligned with the ermA gene, and only those nucleotides of ermA that differ from the ermTR sequence are shown.
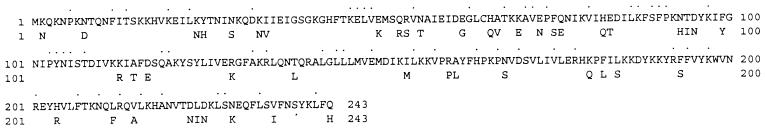
FIG. 2.
Predicted amino acid sequence of the polypeptide encoded by the ermTR gene of S. pyogenes A200. The sequence has been aligned with the predicted amino acid sequence of the polypeptide encoded by the ermA gene, and only those amino acids that differ are shown below the sequence. The 56 identical amino acids that are encoded by different codons are indicated by black spots above the sequence.
The G+C content of the coding sequence of ermTR is 30% and that of ermA is 32.5%; these are equally close to the G+C contents of 32 to 36% and 34.5 to 38.5% of the chromosomes of S. aureus and S. pyogenes, respectively (38). It is possible that these genes share a common origin and have relatively recently diverged from each other. The native gene may have been a staphylococcal or a streptococcal gene, or it may have been transferred to these species from another gram-positive organism. In addition to S. aureus, the ermA gene has been found in coagulase-negative staphylococci and in the gram-negative organisms Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae (32, 42, 44).
Upstream of ermA, there is a leader sequence with two ORFs (28). One is predicted to encode a 15-amino-acid peptide and the other is predicted to encode a 19-amino-acid peptide. This situation resembles that of the ermTR gene, in which a leader sequence potentially encodes two small peptides of the same size (Fig. 1), with the identity to the ermA ORFs being 87.5 and 85.0%, respectively. The regulation of the expression of ermA in S. aureus has been shown to depend on the leader sequence upstream of ermA (28). It has been suggested that in the presence of inducing concentrations of erythromycin, translation of peptide 1 by a ribosome that has bound to erythromycin would result in a ribosome stall, allowing translation of peptide 2. In turn, stalling of ribosomes translating peptide 2 would lead to the translation of ermA. On the basis of the similarities also found between the leader sequences of ermA and ermTR (Fig. 1), regulation of the expression of ermTR probably occurs via a similar mechanism.
Comparison of the ermTR gene to other erm gene classes indicated that the sequence of ermTR is about 61 to 64% identical to the sequences of ermC, ermG, and ermGT, 58% identical to the sequence of ermAM (ermB), which is the only erm gene in S. pyogenes previously sequenced, 56% identical to the sequence of ermQ, and 48 to 49% identical to the sequences of ermF and ermD (which is similar to those of ermJ and ermK). The conserved regions encoding conserved amino acid motifs of the rRNA-methylating enzymes that are present in the different erm genes were also found within the ermTR gene sequence (data not shown).
Comparison of amino acid sequences.
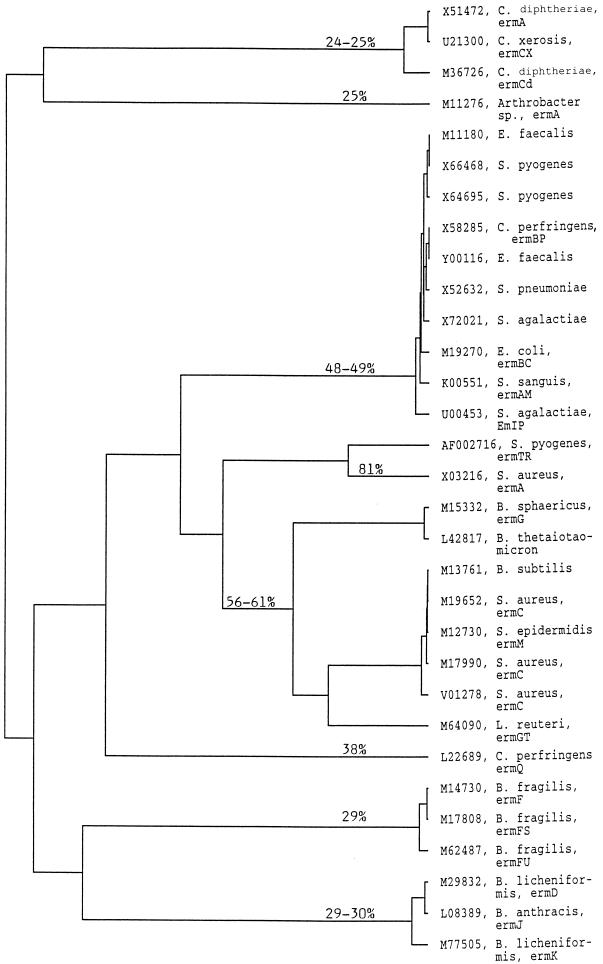
The alignment showing the similarities between the predicted amino acid sequences of the ermTR and ermA gene products is shown in Fig. 2. Altogether, 197 (81.1%) of the encoded 243 amino acids are identical, but the similarity between the two gene products reaches 90.1%. A dendrogram showing the potential evolutionary relationships between the methylase encoded by ermTR and other rRNA-methylating enzymes is shown in Fig. 3.
FIG. 3.
Dendrogram of methylases encoded by different erm genes. The source of the amino acid sequence is indicated by the GenBank accession number, the bacterial species, and the name of the gene, if given. The percent identity of the predicted amino acid sequences of the polypeptides encoded by different erm genes to the predicted amino acid sequence of the polypeptide encoded by the ermTR gene of S. pyogenes A200 is marked on the dendrogram. Note that the most remote methylases associated with resistance to MLSB antibiotics were not included in this analysis.
Plasmid isolation experiments.
Attempts to detect extrachromosomal DNA in strain A200 were unsuccessful, although the methods did detect the control plasmid in S. pyogenes 13 234. In general, antibiotic resistance genes in streptococci are carried by the chromosome, and they are often associated with conjugative transposons (12, 14, 35, 44). However, plasmids carrying determinants for MLSB resistance have previously been isolated from S. pyogenes (2, 7, 8, 10, 22, 24–27, 34). Most streptococcal plasmids carrying antibiotic resistance genes are conjugative and have been shown to transfer by conjugation between streptococcal species (4, 6, 15, 17) and especially among S. pyogenes strains by transduction (16, 23).
Epidemiology of the ermTR gene.
We have carried out epidemiological investigations to study the distribution of different erm genes, including ermTR, in clinical isolates of streptococci collected from different parts of Finland by use of a methodology consisting of PCR and digestion of the PCR products (18, 19). Twenty-four S. pyogenes isolates representing five different serotypes and 29 group G streptococcal isolates, all expressing the inducible type of MLSB resistance, were studied, and all contained the ermTR gene (18, 19). Although the drug efflux gene mefA is at present a predominant erythromycin resistance determinant in S. pyogenes and probably also in other streptococci (41), these studies suggest that ermTR may be widely distributed among streptococci with MLSB resistance.
We conclude that ermTR, characterized in this study, is the first sequenced erm gene that mediates MLSB resistance in S. pyogenes but that does not belong to the ermAM gene class.
ACKNOWLEDGMENTS
This work was supported in part by the Sigrid Juselius Foundation (funds to H. Seppälä and P. Huovinen), the Finnish Academy (funds to H. Seppälä and M. Skurnik), the Maud Kuistila Foundation (funds to H. Seppälä), and the National Institutes of Health (grant DE 10913; funds to M. C. Roberts).
We are indebted to Tuula Randell for expert technical assistance. We are grateful to Claude Mabilat for S. agalactiae 90-30-591 and Horst Malke for S. pyogenes 13 234.
REFERENCES
- 1.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behnke D, Golubkov V I, Malke H, Boitsov A S, Totolian A A. Restriction endonuclease analysis of group A streptococcal plasmids determining resistance to macrolides, lincosamides and streptogramin-B antibiotics. FEMS Microbiol Lett. 1979;6:5–9. [Google Scholar]
- 3.Berryman D I, Rood J I. Cloning and hybridization analysis of ermP, a macrolide-lincosamide-streptogramin B resistance determinant from Clostridium perfringens. Antimicrob Agents Chemother. 1989;33:1346–1353. doi: 10.1128/aac.33.8.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bougueleret L, Bieth G, Horodniceanu T. Conjugative R plasmids in group C and G streptococci. J Bacteriol. 1981;145:1102–1105. doi: 10.1128/jb.145.2.1102-1105.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brantl S, Kummer C, Behnke D. Complete nucleotide sequence of plasmid pGB3631, a derivative of the Streptococcus agalactiae plasmid pIP501. Gene. 1994;142:155–156. doi: 10.1016/0378-1119(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 6.Buu-Hoi A, Bieth G, Horaud T. Broad host range of streptococcal macrolide resistance plasmids. Antimicrob Agents Chemother. 1984;25:289–291. doi: 10.1128/aac.25.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceglowski P, Alonzo J C. Gene organization of the Streptococcus pyogenes plasmid pDB101: sequence analysis of the orfη-orfS region. Gene. 1994;145:33–39. doi: 10.1016/0378-1119(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 8.Ceglowski P, Boitsov A, Chai S, Alonso J C. Analysis of the stabilization system of pSM19035-derived plasmid pBT233 in Bacillus subtilis. Gene. 1993;136:1–12. doi: 10.1016/0378-1119(93)90441-5. [DOI] [PubMed] [Google Scholar]
- 9.Clancy J, Petitpas J, Dib-Hajj F, Yuan W, Cronan M, Kamath A V, Bergeron J, Retsema J A. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 10.Clewell D B, Franke A E. Characterization of a plasmid determining resistance to erythromycin, lincomycin, and vernamycin B in a strain of Streptococcus pyogenes. Antimicrob Agents Chemother. 1974;5:534–537. doi: 10.1128/aac.5.5.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genetics Computer Group. Program manual for the Wisconsin package, version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 12.Horaud T, De Cespedes G, Clermont D, David F, Delbos F. Variability of chromosomal genetic elements in streptococci. In: Dunny G M, Cleary P P, McKay L L, editors. Genetics and molecular biology of streptococci, lactococci, and enterococci. Washington, D.C: American Society for Microbiology; 1991. pp. 16–20. [Google Scholar]
- 13.Horinouchi S, Byeon W-H, Weisblum B. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J Bacteriol. 1983;154:1252–1262. doi: 10.1128/jb.154.3.1252-1262.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horodniceanu T, Bougueleret L, Bieth G. Conjugative transfer of multiple-antibiotic resistance markers in beta-hemolytic group A, B, F, and G streptococci in the absence of extrachromosomal deoxyribonucleic acid. Plasmid. 1981;5:127–137. doi: 10.1016/0147-619x(81)90014-7. [DOI] [PubMed] [Google Scholar]
- 15.Horodniceanu T, Buu-Hoi A, Le Bouguenec C, Bieth G. Narrow host range of some streptococcal R plasmids. Plasmid. 1982;8:199–206. doi: 10.1016/0147-619x(82)90057-9. [DOI] [PubMed] [Google Scholar]
- 16.Hyder S L, Streitfeld M M. Inducible and constitutive resistance to macrolide antibiotics and lincomycin in clinically isolated strains of Streptococcus pyogenes. Antimicrob Agents Chemother. 1973;4:327–331. doi: 10.1128/aac.4.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kataja, J., P. Huovinen, M. Skurnik, and H. Seppälä. Unpublished data.
- 19.Kataja, J., H. Seppälä, M. Skurnik, H. Sarkkinen, and P. Huovinen. Unpublished data.
- 20.Leclercq R, Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob Agents Chemother. 1991;35:1267–1272. doi: 10.1128/aac.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leclercq R, Courvalin P. Intrinsic and unusual resistance to macrolide, lincosamide, and streptogramin antibiotics in bacteria. Antimicrob Agents Chemother. 1991;35:1273–1276. doi: 10.1128/aac.35.7.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malke H. Genetics of resistance to macrolide antibiotics and lincomycin in natural isolates of Streptococcus pyogenes. Mol Gen Genet. 1974;135:349–367. doi: 10.1007/BF00271149. [DOI] [PubMed] [Google Scholar]
- 23.Malke H. Transfer of a plasmid mediating antibiotic resistance between strains of Streptococcus pyogenes in mixed cultures. Z Allg Mikrobiol. 1975;15:645–649. doi: 10.1002/jobm.3630150810. [DOI] [PubMed] [Google Scholar]
- 24.Malke H. Conjugal transfer of plasmid determining resistance to macrolides, lincosamides and streptogramin-B type antibiotics among group A, B, D and H streptococci. FEMS Microbiol Lett. 1979;5:335–338. [Google Scholar]
- 25.Malke H, Jacob H E, Störl K. Characterization of the antibiotic resistance plasmid ERL1 from Streptococcus pyogenes. Mol Gen Genet. 1976;144:333–338. doi: 10.1007/BF00341732. [DOI] [PubMed] [Google Scholar]
- 26.Malke H, Reichard W, Hartmann M, Walter F. Genetic study of plasmid-associated zonal resistance to lincomycin in Streptococcus pyogenes. Antimicrob Agents Chemother. 1981;19:91–100. doi: 10.1128/aac.19.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsuhashi S, Inoue M, Saito K, Nakae M. Drug resistance in Streptococcus pyogenes strains isolated in Japan. In: Schlessinger D, editor. Microbiology—1982. Washington, D.C: American Society for Microbiology; 1982. pp. 151–154. [Google Scholar]
- 28.Murphy E. Nucleotide sequence of ermA, a macrolide-lincosamide-streptogramin B determinant in Staphylococcus aureus. J Bacteriol. 1985;162:633–640. doi: 10.1128/jb.162.2.633-640.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy E, Löfdahl S. Transposition of Tn554 does not generate a target duplication. Nature. 1984;307:292–294. doi: 10.1038/307292a0. [DOI] [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, vol. 10, no. 8. M7-A2. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1990. [Google Scholar]
- 31.Rinckel L A, Savage D C. Characterization of plasmids and plasmid-borne macrolide resistance from Lactobacillus sp. strain 100-33. Plasmid. 1990;23:119–125. doi: 10.1016/0147-619x(90)90030-g. [DOI] [PubMed] [Google Scholar]
- 32.Roe D E, Weinberg A, Roberts M C. Mobile rRNA methylase genes in Actinobacillus actinomycetemcomitans. J Antimicrob Chemother. 1996;37:457–464. doi: 10.1093/jac/37.3.457. [DOI] [PubMed] [Google Scholar]
- 33.Rood J I, Cole S T. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol Rev. 1991;55:621–648. doi: 10.1128/mr.55.4.621-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schalen C, Gebreselassie D, Stahl S. Characterization of an erythromycin resistance (erm) plasmid in Streptococcus pyogenes. APMIS. 1995;103:59–68. doi: 10.1111/j.1699-0463.1995.tb01080.x. [DOI] [PubMed] [Google Scholar]
- 35.Scott R J D, Naidoo J, Lightfoot N F, George R C. A community outbreak of group A beta-haemolytic streptococci with transferable resistance to erythromycin. Epidemiol Infect. 1989;102:85–91. doi: 10.1017/s095026880002971x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seppälä H, Nissinen A, Yu Q, Huovinen P. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J Antimicrob Chemother. 1993;32:885–891. doi: 10.1093/jac/32.6.885. [DOI] [PubMed] [Google Scholar]
- 37.Smith C J. Nucleotide sequence analysis of Tn4551: use of ermFS operon fusion to detect promoter activity in Bacteroides fragilis. J Bacteriol. 1987;169:4589–4596. doi: 10.1128/jb.169.10.4589-4596.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. [Google Scholar]
- 39.Soini H, Böttger E C, Viljanen M K. Identification of mycobacteria by PCR-based sequence determination of the 32-kilodalton protein gene. J Clin Microbiol. 1994;32:2944–2947. doi: 10.1128/jcm.32.12.2944-2947.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiegel C A, Roberts M. Mobiluncus gen. nov., Mobiluncus curtisii subspecies curtisii sp. nov., Mobiluncus curtisii subspecies holmesii subsp. nov., and Mobiluncus mulieris sp. nov., curved rods from the human vagina. Int J Syst Bacteriol. 1984;34:177–184. [Google Scholar]
- 41.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thakker-Varia S, Jenssen W D, Moon-McDermott L, Weinstein M P, Dubin D T. Molecular epidemiology of macrolides-lincosamides-streptogramin B resistance in Staphylococcus aureus and coagulase-negative staphylococci. Antimicrob Agents Chemother. 1987;31:735–743. doi: 10.1128/aac.31.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trieu-Cuot P, Poyart-Salmeron C, Carlier C, Courvalin P. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 1990;18:3660. doi: 10.1093/nar/18.12.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wasteson Y D, Roe E, Falk K, Roberts M C. Characterization of antibiotic resistance in Actinobacillus pleuropneumoniae. Vet Microbiol. 1996;48:41–50. doi: 10.1016/0378-1135(95)00130-1. [DOI] [PubMed] [Google Scholar]
- 45.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]