Abstract
Combination therapy with protease (PR) and reverse transcriptase (RT) inhibitors can efficiently suppress human immunodeficiency virus (HIV) replication, but the emergence of drug-resistant variants correlates strongly with therapeutic failure. Here we describe a new method for high-throughput analysis of clinical samples that permits the simultaneous detection of HIV type 1 (HIV-1) phenotypic resistance to both RT and PR inhibitors by means of recombinant virus assay technology. HIV-1 RNA is extracted from plasma samples, and a 2.2-kb fragment containing the entire HIV-1 PR- and RT-coding sequence is amplified by nested reverse transcription-PCR. The pool of PR-RT-coding sequences is then cotransfected into CD4+ T lymphocytes (MT4) with the pGEMT3ΔPRT plasmid from which most of the PR (codons 10 to 99) and RT (codons 1 to 482) sequences are deleted. Homologous recombination leads to the generation of chimeric viruses containing PR- and RT-coding sequences derived from HIV-1 RNA in plasma. The susceptibilities of the chimeric viruses to all currently available RT and/or PR inhibitors is determined by an MT4 cell–3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide-based cell viability assay in an automated system that allows high sample throughput. The profile of resistance to all RT and PR inhibitors is displayed graphically in a single PR-RT-Antivirogram. This assay system facilitates the rapid large-scale phenotypic resistance determinations for all RT and PR inhibitors in one standardized assay.
Within the last decade, many drugs have become available for the treatment of individuals infected with human immunodeficiency virus type 1 (HIV-1). Despite their initial antiretroviral activity, the benefit of treatment with these agents is of limited duration. Complete suppression of HIV-1 replication is rarely achieved with reverse transcriptase (RT) inhibitors either alone or in dual combinations (2). In contrast, treatment with triple drug combinations that include a protease (PR) inhibitor (6, 9, 20) can reduce the virus load in plasma to undetectable levels and provide substantial clinical benefit. Nevertheless, the breakthrough of drug-resistant mutants remains one of the most serious obstacles to sustained suppression of HIV (3, 4, 10, 30, 44). Continuous high-level in vivo replication of HIV-1 and the intrinsic error rate of the RT enzyme are the major driving forces behind the generation of drug-resistant variants (13, 33, 46). When drug pressure is applied to this divergent and rapidly replicating virus population, variants with the appropriate mutation(s) in their genomes will escape the drug inhibition and outgrow the wild-type drug-susceptible viruses.
The inclusion of different RT and PR inhibitors in antiretroviral treatment regimens has resulted in the emergence of many drug-resistant HIV-1 variants (3, 4, 10, 22–24, 30, 34, 36, 41, 43, 44, 47). More than 100 resistance-associated mutations, spanning the HIV-1 RT- and PR-coding regions, have been described (37). In addition, an increasing number of variants carrying multiple or multidrug resistance-associated mutations have been reported (15, 38). Consequently, methods for detecting resistance and cross-resistance are likely to be needed for patient management. Various assays for the genotypic detection of resistance-associated mutations have been developed (11, 18, 42). However, phenotypic assays are needed to determine the effect of complex genotypic mutational patterns on virus drug susceptibility. This is especially the case with viruses having complex combinations of mutations that may result in unpredictable patterns of resistance, cross-resistance, multidrug resistance, or resistance reversal. Phenotypic resistance testing is often performed by peripheral blood mononuclear cell-based methods (16). However, these require freshly isolated donor lymphocytes, isolation of whole virus, and long culture times and are generally considered to be too labor-intensive and expensive for routine use. The prolonged virus culture times have also been shown to select for subpopulations of HIV-1 variants (21) which can influence the drug susceptibility profile. Therefore, the description of the recombinant virus assay by Kellam and Larder (19) generated interest in the development of more rapid and reproducible determinations of the resistance of HIV to RT inhibitors in clinical samples from HIV-1-infected patients (1, 7, 12, 17). With the introduction of combinations of PR and RT inhibitors in antiretroviral treatment regimens, there was clearly a need to extend phenotypic resistance assays. Here we report the development of a phenotypic recombinant virus assay that can determine the susceptibility of HIV-1 to both RT and PR inhibitors.
MATERIALS AND METHODS
Plasma samples.
Plasma samples obtained from HIV-1-infected individuals were shipped with dry ice and stored at −70°C until analysis. Plasma samples used for repeated analyses were thawed no more than two times.
Viral RNA extraction.
Viral RNA was isolated from 200 μl of plasma with the QIAamp Viral RNA Extraction Kit (Qiagen, Hilden, Germany) as instructed by the manufacturer.
Amplification of RT- and PR-coding sequences.
cDNA encoding PR and RT was made with Expand Reverse Transcriptase (Boehringer, Mannheim, Germany). Each reaction mixture (final volume, 20 μl) contained 5 mM MgCl2, 1 mM deoxynucleoside triphosphates (Pharmacia, Uppsala, Sweden), 20 U of RNase inhibitor (Perkin-Elmer, Foster City, Calif.), 2 μl of Expand RT reaction buffer (10×), 4 μl of RNA, 6.5 U of RT enzyme, and 0.75 μM the HIV-1-specific primer OUT3 (see below). The reaction mixture was incubated at 42°C for 30 min to enable cDNA synthesis. The RT enzyme was subsequently inactivated by incubation of the reaction mixture at 99°C for 5 min. All incubations were carried out in a GeneAmp 9600 thermocycler (Perkin-Elmer).
A 2.2-kb PR-RT-coding sequence was amplified from cDNA by nested PCR. The first-round PCR used primers PRTO5 (5′-GCCCCTAGGAAAAAGGGCTGTTGG-3′) and OUT3 (5′-CATTGCTCTCCAATTACTGTGATATTTCTCATG-3′). The reaction mixture contained 2.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, 0.15 μM (each) primer, 5 U of Expand High Fidelity Polymerase mixture (Boehringer), 10 μl of Expand Reaction buffer (10×), and 20 μl of the cDNA mixture. The final volume of the PCR mixture was 100 μl. Ten microliters of the first-round PCR mixture was used for the second-round PCR. All components of the reaction mixture were the same as those used in first-round PCR, but the reaction mixture contained 0.15 μM primers PRTI5 and IN3 (5′-TGAAAGATTGTACTGAGAGACAGG-3′ and 5′-TCTATTCCATCTAAAAATAGTACTTTCCTGATTCC-3′, respectively). All reactions were carried out in a Biometra Uno Thermocycler (Biometra, Göttingen, Germany). PCR conditions for both rounds were 95°C for 3 min and then 30 cycles of 90°C for 1 min, 55°C for 30 s, and 72°C for 2 min, followed by a final 10 min of incubation at 72°C. All amplification products were analyzed by 1% agarose gel electrophoresis. For amplification of sequences encoding only RT, PCR was performed with the primers and under the conditions described previously (19).
Purification of amplified coding sequences.
The PCR product was purified with a QIAquick PCR Purification kit (Qiagen) and was reanalyzed by 1% agarose gel electrophoresis.
Construction of the proviral clone pGEMT3ΔPRT.
The previously described (19) proviral molecular clone pHIVΔRTBstEII from which the sequence for RT was deleted was used as starting material for the construction of a proviral clone from which the sequences for PR and RT were deleted. The construct was obtained from the Medical Research Council AIDS Directed Programme Reagent Project (repository reference ADP206) and contains a 12.5-kb XbaI insert of HXB2 (HIV-IIIB) and flanking cellular sequences. The nucleotide and amino acid positions used in this paper are those of the GenBank HIVHXB2CG sequence (sequence identification no. 327742; accession no. KØ3455). To generate the proviral clone from which the sequences for PR and RT were deleted, pHIVΔRTBstEII was digested with XbaI, and the resulting full-length HIV-1 proviral RT-deleted fragment was subcloned into the XbaI site of pGEM9zf(−) (Promega, Madison, Wis.). After BstEII linearization of this intermediate construct (pGEM9HIVΔRT), the reaction mixture was treated with DNA polymerase I (Klenow fragment) and was further digested with ApaI to remove a 750-bp ApaI-BstEII fragment. In parallel, the original molecular clone pHIVΔRTBstEII was linearized with AhdI, incubated with DNA polymerase I, and subsequently treated with ApaI. The resulting 270-bp ApaI-AhdI fragment was then subcloned into the recipient vector, pGEM7zf(−). A 270-bp ApaI-XmaI fragment was recovered from this clone (pGEM7Apa/Sma) by consecutive XmaI digestion, treatment with DNA polymerase I, and ApaI digestion. Finally, the 750-bp ApaI-BstEII fragment, removed from the vector pGEM9HIVΔRT, was replaced by the 270-bp ApaI-XmaI fragment recovered from the construct pGEM7Apa/Sma. Restriction analysis and PCR with the appropriate primers (see above) were used to confirm the identity and length of the new construct, pGEMT3ΔPRT. PR- and RT-coding sequences were thus deleted from the HIV-1 proviral genome starting from the AhdI cleavage site (nucleotide position 2280 of the PR sequence; amino acid 9 of the PR sequence) to nucleotide position 4115 (amino acid 483) of the RT sequence. The resulting construct was transformed into Escherichia coli JM109. For use in recombination experiments, large-scale plasmid DNA preparations (Qiagen) were linearized by BstEII digestion, purified with phenol-chloroform, precipitated with ethanol, and resuspended in water. The pHIVΔRTBstEII construct was also used for all comparative validation experiments.
Cotransfection of PR-RT-coding sequences with pGEMT3ΔPRT.
MT4 cells were subcultured at a density of 250,000 cells/ml on the day before transfection. Cells were pelleted and resuspended in phosphate-buffered saline at a concentration of 3.1 × 106 cells/ml. A 0.8-ml portion (2.5 × 106 cells/ml) was used for each transfection. Transfections were performed with the Bio-Rad Gene pulser (Bio-Rad, Hercules, Calif.) with 0.4-cm electrode cuvettes (Bio-Rad). Cells were electroporated with 10 μg of BstEII-linearized pGEMT3ΔPRT and approximately 5 μg of purified PR-RT-PCR product at 250 μF and 300 V, followed by a 30-min incubation at room temperature. Ten milliliters of fresh culture medium was then added to the suspension of transfected cells, and incubation was performed at 37°C in a humidified atmosphere with 5% CO2. Cell cultures were monitored for the appearance of viral cytopathic effect (CPE). Culture supernatants were typically harvested by centrifugation at 8 to 10 days after transfection and were stored at −70°C for subsequent infectivity and drug susceptibility determinations. Infectivity was determined by the viral CPE assay described below by using a 50% endpoint method (50% cell culture infectious dose).
Drug susceptibility assays.
HIV-1 drug susceptibility was determined by an MT4 cell–3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (MT4-MTT)-based CPE protection assay (32). MT4 cells were infected with 200 50% cell culture infective doses of recombinant viruses in the presence of fivefold dilutions of different antiretroviral drugs. In general, the susceptibilities of the viruses to zidovudine (AZT; 3′-azido-3′-deoxythymidine), lamivudine (3TC; β-l-2′,3′-dideoxy-3′-thiacytidine), didanosine (ddI; 2′,3′-dideoxyinosine), zalcitabine (ddC; 2′,3′-dideoxycytidine), stavudine (d4T; 2′,3′-didehydro-3′-deoxythymidine), loviride (R89439), nevirapine, tivirapine (8CI-TIBO, R91767), saquinavir (Ro-31-8959), indinavir (MK-639), and ritonavir (ABT-538) were determined in one assay. Four replicate determinations were performed in duplicate plates for each concentration of antiretroviral drug. Four wild-type recombinant viruses derived from HIV-1 IIIB/LAI RNA were generated and tested in parallel with clinical samples for each assay. Fold resistance values were calculated by dividing the mean 50% inhibitory concentration (IC50) for a recombinant virus from a patient by the mean IC50 for recombinant wild-type viruses.
Dideoxynucleotide-based sequence analysis (genotyping).
A 785-bp fragment containing the first 260 codons of the RT sequence was amplified by PCR with primers A(35) (5′-TTGGTTGCACTTTAAATTTTCCCATTAGTCCTATT-3′) and NE-1(35) (5′-CCTACTAACTTCTGTATGTCATTGACAGTCCAGCT-3′) (25). The PCR product was treated with shrimp alkaline phosphatase and exonuclease I (reagent pack for PCR product pretreatment; Amersham) and analyzed by cycle sequencing (Thermo Sequenase fluorescent labelled primer cycle sequencing kit with 7-deaza-dGTP; Amersham) with the following fluorescein isothiocyanate-labelled primers: RTGSS01 (5′-TTAGCCCTATTGAGACTGTACC-3′) and RTGSS02 (5′-TACTGGATGTGGGTGATGCATA-3′) in the sense direction and RTGAS03 (5′-TCCCTGTGGAAGCACATTG-3′) and RTGAS04 (5′-GTTCATAACCCATCCAAAG-3′) in the antisense direction. The reaction products were analyzed with an ALF automated sequencer (Pharmacia). DNA sequences were aligned to the HXB2 reference sequence with GeneWorks software, version 2.5 (Oxford Molecular). For PR, a 400-bp fragment containing all 99 codons of protease plus gag- and RT-flanking sequences was amplified with primers DP10 (5′-CAACTCCCTCTCAGAAGCAGGAGCCG-3′) and DP11 (5′-CCATTCCTGGCTTTAATTTTACTGGTA-3′) (26). The primers used for sequence analysis were FH1P1802 (5′-CAAATCACTCTTTGGCAACGACC-3′) and FH1P2055A (5′-AATCTGAGTCAACAGATTTCTTCC-3′).
Viral load measurements.
HIV-1 RNA levels in plasma were quantified with the Amplicor HIV-1 Monitor Kit (Roche), as instructed by the manufacturer.
RESULTS
Construction of proviral clone pGEMT3ΔPRT from which sequences for PR and RT were deleted.
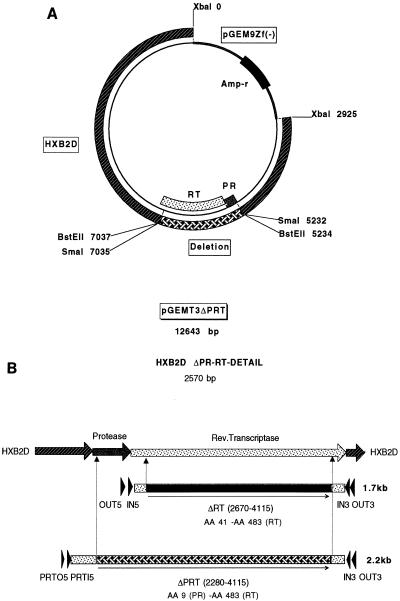
Proviral clone pHIVΔRTBstEII from which the sequence for RT was deleted (19) was used to construct proviral clone pGEMT3ΔPRT from which the sequences for PR and RT were deleted (Fig. 1A). PR- and RT-coding sequences located upstream from the RT deletion in pHIVΔRTBstEII were deleted starting with the AhdI cleavage site (nucleotide position 2280; amino acid 9 in the PR gene) (Fig. 1B). Cotransfection of MT4 cells with the BstEII-linearized proviral construct pGEMT3ΔPRT from which the sequences for PR and RT were deleted and the appropriate PR-RT-coding sequences, amplified from viral RNA in plasma, resulted in the homologous recombination and the generation of infectious virus. An extensive HIV-induced CPE became apparent within 8 to 10 days of transfection. After harvesting and titration of recombinant viruses, drug susceptibility assays were performed to determine the resistance profile of the virus in any given patient plasma sample (see below). Population-based sequencing confirmed there were no major differences in the RT mutational patterns between HIV RNA in the original sample and the RNA of the corresponding recombinant viruses (Table 1).
FIG. 1.
(A) Proviral clone pGEMT3ΔPRT from which the sequences encoding PR and RT were deleted was constructed by removing a 750-bp ApaI-BstEII fragment upstream of the RT deletion of the proviral clone pHIVΔRTBstEII and replacing it with a 270-bp ApaI-XmaI fragment recovered from the intermediate construct pGEM7Apa/Sma (see Materials and Methods). This resulted in a clone from which coding sequences starting from amino acid position 10 in the HIV-1 PR gene to amino acid position 484 in the HIV-1 RT gene have been removed. (B) Nested PCR is used to amplify either RT-only coding sequences (1.7 kb) (primer pairs OUT3-OUT5 and IN3-IN5) or combined PR-RT-coding sequences (2.2 kb) (primer pairs OUT3-PRTO5 and IN3-PRTI5). RT-only coding sequences encompass all resistance-associated amino acid changes between residues 41 and 483 of the HIV-1 RT gene. Combined PR- and RT-coding sequences encode for all resistance-associated mutations between amino acid residue 9 of the PR gene and residue 483 of the RT gene.
TABLE 1.
Comparison of amino acid substitutions in RT between HIV-1 from plasma and the corresponding recombinant virusa
| Sample no. | Virus source | Amino acid at the following position:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M41 | D67 | K70 | A98 | K101 | K103 | E138 | Y181 | M184 | Y188 | G190 | H208 | T215 | K219 | K238 | ||
| 100 | Plasma | L | N | R | N | V | Y | F | T | T | ||||||
| Recombinant | L | N | R | N | V | Y | F | Q | T | |||||||
| 200 | Plasma | L | Y | |||||||||||||
| Recombinant | L | Y | N/K | |||||||||||||
| 300 | Plasma | E | N/K | A | A/G | ND | ND | |||||||||
| Recombinant | E/K | N/K | A | |||||||||||||
| 400 | Plasma | N | V | |||||||||||||
| Recombinant | N | V | ||||||||||||||
| 500 | Plasma | Q | ND | |||||||||||||
| Recombinant | Q | |||||||||||||||
| r1 BEL4 16/0 | Plasma | N | R | G | C | F | Q | |||||||||
| Recombinant | N | R | G | C | F | Q | ||||||||||
| r9 BEL5 15/4 | Plasma | R | H | Y | ||||||||||||
| Recombinant | R | L | Y | |||||||||||||
Viral RNA was extracted from either 200 μl of plasma or 200 μl of culture supernatant containing the corresponding chimeric virus. The RT-coding sequences were amplified, and a population-based sequence analysis was performed with both isolates (see Materials and Methods). ND, not determined.
PCR amplification of full-length RT- and combined PR- and RT-coding sequences.
Following cDNA synthesis, nested PCR amplification of cDNA was performed to generate either a 1.7-kb DNA amplicon encompassing the full-length RT-coding sequence or the combined 2.2-kb PR- and RT-coding sequence (Fig. 1B). To determine the sensitivity of the reverse transcription-PCR procedure, two approaches were followed. First, the amplification efficiency was determined with a serially diluted stock of a laboratory HIV strain (IIIB/LAI) containing 108 to 10 RNA copies/ml (data not shown). Second, 10- and 100-fold serial dilutions of RNA isolated from patient plasma samples with known copy numbers per milliliter (Roche Amplicor) were tested. Reverse transcription-PCR was performed with both undiluted and diluted RNAs. These experiments showed that the RT-only sequences (1.7 kb) could be amplified from as few as 50 copies of viral RNA (250 copies/ml). Amplification of the combined PR- and RT-coding sequences (2.2 kb) required at least 200 copies of viral RNA (1,000 copies/ml) (Table 2). The difference in sensitivity between the two different reverse transcription-PCRs is caused by the differences in the lengths of the amplicons that need to be generated. Similar results were obtained with serially diluted IIIB/LAI culture supernatants (data not shown).
TABLE 2.
Sensitivity of reverse transcription-PCR amplification of RT-coding sequence (1.7 kb) versus that of amplification of PR-RT-coding sequence (2.2 kb)a
| Viral load (no. of RNA copies/ml) | % Samples positive by PCR (no. positive/total no. tested)
|
|
|---|---|---|
| RT-coding sequence (1.7 kb) | PR-RT-coding sequence (2.2 kb) | |
| <1,000 | 67 (10/15) | 13 (2/15) |
| 1–5,000 | 76 (16/21) | 71 (15/21) |
| 5–10,000 | 95 (20/21) | 86 (18/21) |
| 10–50,000 | 93 (28/30) | 90 (27/30) |
| 50–100,000 | 100 (21/21) | 100 (21/21) |
| 100–500,000 | 100 (23/23) | 100 (23/23) |
| >500,000 | 100 (22/22) | 100 (22/22) |
PCR amplification of RT-only or combined PR- and RT-coding sequences was performed with plasma samples (n = 160) with viral loads ranging from 100 to >1,000,000 copies of viral RNA/ml. Results are presented as the percentage or number of samples included in each viral load category giving a positive PCR result for either RT-only (1.7 kb) or combined PR- and RT (2.2 kb)-coding sequences.
Reproducibility of phenotypic drug susceptibility testing.
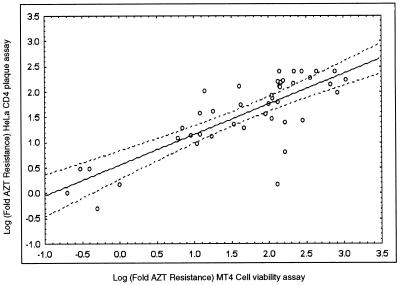
The RT susceptibility profiles obtained with plasma-derived recombinants from patients treated with AZT and 3TC were generated by the HeLa CD4 plaque reduction assay (19) and were compared with the susceptibility profiles obtained by the MT4 cell viability-based assay (32). The fold resistance values for AZT (fold increase in IC50 relative to the IC50 for the wild type) obtained by the two methods are presented in Fig. 2 and were strongly correlated (n = 43; r = 0.8; P < 0.00001). However, the MT4-MTT method was more amenable to automation and thus was the method of choice for high sample throughput. To evaluate the reproducibility of the susceptibility assay, the entire procedure including RNA extraction, reverse transcription-PCR amplification, cotransfection, and susceptibility testing was repeated four times with plasma samples with known HIV-1 drug resistance profiles. All independently generated recombinant viruses (carrying plasma-derived sequences encoding RT only or PR and RT) were harvested and tested for drug susceptibility by the MT4-MTT method. The results (IC50s and the corresponding fold resistance values) of four independent susceptibility determinations with a sample with documented AZT and 3TC and nonnucleoside RT inhibitor (NNRTI) resistance and a sample with documented PR inhibitor resistance are presented in Table 3 and demonstrate that the respective phenotypic resistance profiles can be reproducibly detected in these plasma samples.
FIG. 2.
Phenotypic AZT resistance was determined either by the HeLa CD4 plaque reduction assay (y axis) or the MT4-MTT cell viability assay (x axis). The correlation between AZT resistance data obtained by both methods and presented as log fold AZT resistance values (fold increase in the mean IC50 relative to the mean wild-type IC50 for the wild type; the mean IC50 for both patient and wild-type viruses is derived from two to four separate susceptibility determinations) was statistically determined (n = 43; r = 0.8; P = < 0.00001).
TABLE 3.
Reproducibility of phenotypic resistance testing in the MT4-MTT cell viability assaya
| Sample no. and drug | IC50 (μM)
|
Fold resistanceb
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Determin. 1 | Determin. 2 | Determin. 3 | Determin. 4 | Mean | SEM (n = 4)c | CVd | Determin. 1 | Determin. 2 | Determin. 3 | Determin. 4 | Mean | SEM (n = 4) | CV | |
| 10 | ||||||||||||||
| AZT | 13.6 | 3.70 | 6.50 | 12.5 | 9.1 | 2.38 | 0.26 | 211 | 79 | 137 | 192 | 155 | 30 | 0.19 |
| Tivirapine | 0.47 | 0.32 | 0.44 | 0.51 | 0.4 | 0.04 | 0.09 | 30 | 23 | 31 | 37 | 30 | 3 | 0.09 |
| Loviride | 0.31 | 0.28 | 0.36 | 0.41 | 0.3 | 0.03 | 0.08 | 8 | 9 | 11 | 8 | 9 | 1 | 0.08 |
| ddI | 8.10 | 8.20 | 8.70 | 12.8 | 9.5 | 1.12 | 0.12 | 1 | 2 | 2 | 2 | 2 | 0.3 | 0.14 |
| ddC | 7.50 | 8.30 | 8.90 | 9.20 | 8.5 | 0.37 | 0.04 | 2 | 2 | 3 | 3 | 3 | 0.3 | 0.12 |
| d4T | 10.0 | 3.20 | 5.40 | 9.20 | 7.0 | 1.60 | 0.23 | 2 | 2 | 3 | 3 | 3 | 0.3 | 0.12 |
| 3TC | >100 | >100 | >100 | >100 | >100 | NDe | ND | >17 | >10 | >10 | >13 | >12 | ND | |
| 20 | ||||||||||||||
| Indinavir | 0.15 | 0.63 | 0.42 | 0.26 | 0.4 | 0.10 | 0.29 | 18 | 8 | 12 | 6 | 11 | 3 | 0.24 |
| Ritonavir | 0.56 | 1.71 | 1.72 | 0.38 | 1.1 | 0.36 | 0.33 | 22 | 20 | 21 | 10 | 18 | 3 | 0.15 |
| Saquinavir | 0.23 | 0.37 | 0.40 | 0.15 | 0.3 | 0.06 | 0.20 | 82 | 37 | 71 | 21 | 53 | 14 | 0.27 |
The PR and RT inhibitor resistance profiles were determined for four different recombinant viruses separately generated from the same plasma sample (sample 10 or 20). The profiles of the susceptibilities of the respective viruses to seven RT inhibitors (sample 10) and three PR inhibitors (sample 20) are expressed as IC50s and as the means of these determinations (Determin.). The same results are also expressed as fold resistance values.
Fold increase in the mean IC50 relative to the mean IC50 for the wild type. The mean IC50 for both viruses from patients and wild-type viruses is derived from two to four separate susceptibility determinations by the MT4-MTT assay.
SEM, standard error of the mean.
CV, coefficient of variation.
ND, not determined.
Comparison of susceptibility data obtained with recombinant viruses carrying plasma-derived sequences encoding RT only with data obtained with recombinant viruses carrying plasma-derived sequences encoding PR and RT.
To evaluate whether similar drug susceptibility data are obtained with recombinant viruses carrying plasma-derived sequences encoding PR and RT and recombinant viruses carrying plasma-derived sequences encoding only RT, HIV-1 RNA was extracted from 35 plasma samples and both recombinant viruses (those carrying plasma-derived sequences encoding RT only and PR and RT) were produced, and virus susceptibilities to five HIV-1 RT inhibitors were determined in one MT4-MTT-based assay. There were no significant differences in susceptibility results obtained with recombinant viruses carrying plasma-derived sequences encoding RT only and recombinant viruses carrying plasma-derived sequences encoding PR and RT. Representative results (IC50s and the corresponding fold resistance values) are given in Table 4.
TABLE 4.
Comparison of RT susceptibility data obtained with recombinant viruses with sequences encoding RT only or with recombinant viruses with sequences encoding PR and RTa
| Patient no. | Construct | IC50 (μM) (fold resistanceb)
|
||||
|---|---|---|---|---|---|---|
| AZT | 3TC | ddI | Loviride | Nevirapine | ||
| 3 | ΔRT | 1.91 (44) | >100 (>67) | 9.15 (2) | 2.41 (50) | 16.4 (214) |
| ΔPR/RT | 4.54 (49) | >100 (>13) | 12.3 (2) | 3.49 (46) | 94.7 (622) | |
| 11 | ΔRT | 0.16 (13) | >100 (>13) | 10.1 (2) | 0.09 (2) | NDc |
| ΔPR/RT | 1.49 (16) | >100 (>13) | 13.1 (2) | 0.10 (1) | 0.53 (3) | |
| 12 | ΔRT | 0.10 (2) | 9.25 (1) | 7.02 (3) | 0.08 (1) | ND |
| ΔPR/RT | 0.09 (1) | 9.76 (1) | 9.52 (1) | 0.14 (2) | 0.31 (2) | |
| 13 | ΔRT | 1.51 (19) | >100 (>48) | 56.4 (4) | 0.08 (1) | 0.28 (2) |
| ΔPR/RT | 1.87 (17) | >100 (>42) | 53.7 (5) | 0.08 (1) | 0.14 (1) | |
| 14 | ΔRT | 11.6 (142) | >100 (>48) | 47.4 (4) | 0.18 (3) | 1.08 (8) |
| ΔPR/RT | 6.68 (60) | >100 (>42) | 36.1 (4) | 0.21 (3) | ND | |
RT susceptibility data for chimeric viruses with RT only (ΔRT) were compared with similar data obtained with recombinant viruses with PR and RT (ΔPR/RT). Plasma samples from 35 patients (70 recombinant viruses) were investigated. Data for five samples with profiles indicating resistance to NRTI and NNRTI are presented. Resistance data are given as IC50s and as fold resistance.
Fold increase in the mean IC50 relative to the mean IC50 for the wild type. The mean IC50s for both patient and wild-type viruses were derived from two to four separate susceptibility determinations by the MT4-MTT assay.
ND, not determined.
Simultaneous measurement of HIV-1 susceptibility to PR and RT inhibitors with recombinant viruses carrying the plasma-derived sequences encoding PR and RT.
To investigate combined phenotypic PR and RT resistance testing with recombinant viruses carrying plasma-derived PR- and RT-coding sequences, a selection of samples from patients with documented PR and RT inhibitor therapy was analyzed as outlined above. Representative results are given in Table 5 and are discussed below.
TABLE 5.
Simultaneous determination of resistance to HIV-1 PR and RT inhibitors by recombinant viruses with plasma-derived sequences encoding PR and RTa
| Drug | IC50 (μM) (fold resistanceb)
|
||||
|---|---|---|---|---|---|
| Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | |
| AZT | 0.32 (5) | 66.6 (998) | 5.60 (57) | 73.6 (460) | 31.9 (426) |
| 3TC | 7.77 (1) | 35.7 (5) | >100 (>13) | >100 (>11) | 18.42 (4) |
| ddI | 2.34 (1) | 10.1 (1) | 12.4 (2) | 1.98 (1) | 7.62 (1) |
| ddC | 1.94 (1) | 3.52 (1) | 9.75 (3) | 9.21 (3) | 2.06 (1) |
| d4T | 1.95 (1) | 7.68 (1) | 7.21 (1) | 7.73 (2) | 7.15 (4) |
| Loviride | 0.047 (1) | 0.09 (1) | 3.49 (46) | 0.02 (1) | >100 (>1700) |
| Nevirapine | ND | 0.32 (2) | 94.7 (622) | ND | >100 (>758) |
| Indinavir | 0.023 (3) | 0.11 (7) | 0.39 (21) | 0.63 (8) | 1.95 (104) |
| Saquinavir | 0.046 (15) | 0.003 (1) | 0.05 (8) | 0.36 (37) | 3.50 (579) |
| Ritonavir | 0.092 (4) | 1.15 (23) | 7.71 (120) | 1.71 (20) | 11.3 (175) |
| Viral loadc | 26,000 | 191,000 | 269,000 | 400,000 | NA |
| CD4 count | 224 | 75 | 54 | 314 | NA |
The combined PR-RT resistance pattern (10 drugs) was determined for different samples. The results for five samples with different patterns of resistance to all inhibitors are presented. Resistance to nevirapine was not determined (ND) for samples 1 and 4. Results are expressed as IC50s and fold resistance. Viral load data and CD4 count (numbers of cells per milliliter) were not available (NA) for sample 5.
Fold-increase in the mean IC50 relative to the mean IC50 for the wild type. The mean IC50s for both patient and wild-type viruses were derived from two to four separate susceptibility determinations by the MT4-MTT assay.
Number of HIV-1 RNA copies per milliliter of plasma as determined by the Roche Amplicor HIV-1 Monitor assay.
(i) Sample 1.
The patient from whom sample 1 was obtained had been treated with AZT, ddC, and saquinavir for 1 year and harbored a virus with decreased susceptibility to AZT and saquinavir. This susceptibility profile is in agreement with the therapeutic history and other clinical manifestations. Viral load was 26,000 RNA copies/ml, while the CD4 count had dropped from 301 to 224 cells/ml over a period of 1 year.
(ii) Sample 2.
Sample 2 was from a patient with documented nucleoside RT inhibitor (NRTI) treatment that was extended with 6 months of ritonavir therapy and harbored a virus with resistance to AZT and 3TC in combination with resistance to ritonavir and cross-resistance to indinavir. After an initial response to ritonavir (the viral load fell to 3,000 RNA copies/ml), the patient started failing therapy (viral load, 191,000 RNA copies/ml), most probably because PR-RT-resistant virus had emerged.
(iii) Sample 3.
Sample 3 was from a patient with documented NRTI and NNRTI treatment that was extended with 1 year of ritonavir therapy and harbored a virus with a susceptibility profile that is in agreement with therapy and viral load status (269,000 RNA copies/ml). AZT resistance, high-level NNRTI resistance or cross-resistance, and ritonavir resistance accompanied by indinavir cross-resistance were observed.
(iv) Sample 4.
The patient from whom sample 4 was obtained was treated with all available NRTIs as well as with the PR inhibitors ritonavir, indinavir, and saquinavir and had a viral load of 400,000 RNA copies/ml. The combined PR-RT susceptibility determination demonstrated the presence of dual resistance to AZT and 3TC in combination with resistance to all three PR inhibitors tested (ritonavir, indinavir, and saquinavir).
(v) Sample 5.
The patient from whom sample 5 was obtained was treated with all available NRTIs in combination with PR inhibitors. Recent combinations administered consisted of saquinavir-ddC, saquinavir-ddI, ritonavir-3TC, ritonavir-saquinavir-ddC, and indinavir-saquinavir-AZT-3TC. Resistance to AZT, NNRTIs, and all PR inhibitors tested was detected.
Comparative analysis of PR resistance phenotype data with PR genotype and treatment history.
PR-coding regions were sequenced to assess the relation between the protease resistance phenotype of recombinant viruses with plasma-derived sequences encoding PR and RT and their genotypes. Table 6 summarizes a subset of the results from this analysis. No single pattern of amino acid substitutions in PR was required for resistance to PR inhibitors. Although amino acid substitutions at position 48 (G to V) and position 90 (L to M) were associated with saquinavir resistance and substitutions at position 82 were indicative of indinavir and ritonavir resistance, the level of resistance varied and appeared to be the result of the combined effects of multiple combinations of amino acid changes. The highest levels of phenotypic resistance to PR inhibitors were found in samples containing the most amino acid changes in the sequence encoding PR.
TABLE 6.
Comparative analysis of PR phenotypes and genotypes of recombinant viruses containing plasma-derived sequences for PR and RTa
| Sample no. | Fold resistanceb
|
PR mutational patternc | PR inhibitor treatment history | ||
|---|---|---|---|---|---|
| RTV | IDV | SQV | |||
| 1 | 4 | 3 | 15 | 10I, 19Q, 35D, 36I, 37N, 48V, 63P, 69Y/H, 71T, 90M, 93L | SQV, 1 yr |
| 2 | 23 | 7 | 1 | 14R, 15V, 32I, 36I, 37N, 46I, 82A | RTV, 6 mo |
| 3 | 120 | 21 | 8 | 10I, 20R, 36I, 37D, 54V, 57K, 60E, 61R, 63P, 71V, 72V, 82A, 90M, 93L | RTV, 1 yr |
| 4 | 20 | 8 | 37 | 36I, 37N, 48V, 54V, 60E, 62V, 63P, 82A | RTV-IDV-SQVd |
| 5 | 175 | 104 | 579 | 10I, 13V, 36I, 37D, 48V, 54V, 60E, 61E, 62V, 64V, 71V, 82A, 90M, 93L | RTV-IDV-SQVd |
| 6 | 8 | 18 | 1 | 32I, 37N, 46I, 63P, 82A, 93L | IDV, 6 mo |
| 7 | 38 | 15 | 1 | 34K, 37N, 41K, 43R, 54V, 62V, 63I, 71V, 74S/T, 82A, 90M | RTV, 18 mo |
| 8 | 3 | 4 | 21 | 10I, 35D, 36I, 37N, 48V, 60E, 63P, 69Y | RTV, 1 mo; SAQ, 7 mo |
| 9 | 23 | 45 | 121 | 13V/I, 14R/V, 20M, 35D/N, 36I/M, 37G/D, 45R/K, 63P, 69K/Q/N/H, 71V/A, 84V/I, 89P/M/T/L | IDV, 8 mo |
The phenotypic resistance profiles of viruses isolated from the plasma of patients (samples 1 to 9) not responding to PR inhibitor therapy were determined and were compared with the genotypic mutation patterns of the corresponding viruses. Phenotypic resistance data are given as fold resistance. The sequences were aligned with the corresponding sequence of HXB2D to identify mutations and polymorphisms present in the viruses from patients. RTV, ritonavir; IDV, indinavir; SQV, saquinavir.
Fold increase in the mean IC50 relative to the mean IC50 for the wild type. The mean IC50s for both patient and wild-type viruses were derived from two to four separate susceptibility determinations by the MT4-MTT assay.
Boldface indicates primary resistance-associated mutations.
The three PR inhibitors consecutively included in treatment regimen.
DISCUSSION
Serial measurement of HIV RNA levels in plasma can be used to monitor the antiviral activities of drugs and drug combinations (8, 29, 31, 35, 45). Shortly after the initiation of antiretroviral treatment, the levels of HIV RNA in a patient’s plasma usually decrease. The order of magnitude of this reduction greatly depends on the type and combination of antiretroviral drugs used. After an initial decline, HIV RNA levels may increase, indicating drug failure. Drug therapy may fail for several reasons including drug potency, noncompliance, pharmacological factors, and the emergence of drug-resistant virus strains (3, 4, 10, 22–24, 30, 34, 36, 39, 41, 43, 44, 47). Therefore, drug susceptibility testing is becoming increasingly important. Current methods for the detection of drug resistance include phenotypic and genotypic assays. Genotypic assays (11, 18, 41) are relatively rapid and particularly useful when a strong correlation between a specific single mutation and drug resistance exists. Interpretation of genotypic information is much more difficult when complex mutational patterns that can interact to cause resistance, cross-resistance, or resistance reversal are identified.
The development of the recombinant virus assay (19) and modifications thereof (1, 7, 12, 17) opened the way for rapid, reproducible, and large-scale phenotypic analysis of drug resistance. Phenotypic assays directly measure the ability of HIV to grow in the presence of each drug. As a result, they can provide information on cross-resistance, multidrug resistance, or resistance reversal. Phenotypic resistance testing was initially described for the analysis of resistance to HIV-1 RT inhibitors. However, inclusion of PR inhibitors into treatment regimens also requires testing for susceptibility to these drugs. Here we report the first recombinant virus assay that allows the simultaneous testing of HIV-1 susceptibility to PR and RT inhibitors in one standardized assay system (PR-RT Antivirogram). Amplification of the combined PR-RT (2.2-kb)-coding sequences was possible from as few as 1,000 viral RNA copies/ml of plasma from a patient. Higher plasma HIV RNA levels, associated with the emergence of drug-resistant virus strains, will result in the most efficient amplification. The assay has been optimized for the detection of HIV-1 subtype B virus strains, and although amplification of other subtypes is possible, the adaptation of the assay for efficient PCR amplification (2.2 kb) of other subtypes and samples with a viral load near the detection limit is under investigation.
After transfection of MT4 cells, homologous recombination of patient-derived coding sequences with proviral constructs from which the sequences encoding PR and RT were deleted results in recombinant viruses which are isogenic except for their PR and/or RT genes. Except for the first 9 amino acid residues of the sequence for PR, the inserted PR-RT amplicon contains the coding information for all currently described resistance-conferring amino acid substitutions (37). As a consequence, changes in the susceptibilities of these viruses compared to the susceptibilities of wild-type recombinant strains should result from amino acid substitutions within the inserted PR or RT genes. Most resistance profiles (>90%) in the patient population phenotypically tested for PR inhibitor resistance so far (n = 500) could be explained by retrospective analysis of treatment schedules. The potential influence of compensatory amino acid changes around the p7/p1 and p1/p6 PR cleavage sites on susceptibility to PR inhibitors and viral fitness is being investigated with new genetic constructs and amplicons that will include these sites.
When the genotypes of HIV RT genes in viruses from plasma were compared with those of the genes present in recombinant viruses, both viruses had nearly identical mutational patterns (Table 1). Hence, the in vitro phenotypic resistance pattern should be a good reflection of the resistance pattern of circulating virus in vivo. Moreover, the reduction of virus culture time for recombinant virus to 8 to 10 days posttransfection, compared with 3 to 4 weeks for standard virus isolation in peripheral blood mononuclear cells, lowers the potential for the selection of minority viral species (21). Nevertheless, a detailed analysis of virus mixtures is needed to define the threshold proportion that minority species must exceed before they can be detected either phenotypically or genotypically. Detection of minor populations of resistant viruses in previously treated individuals may be important. A clonal analysis (27, 40), making use of ultrasensitive PCR and phenotypic resistance testing, is most suitable for this purpose. Unfortunately, clonal analysis is not yet amenable to high-throughput analysis of patient samples.
All susceptibility measurements described here were performed by a standardized MT4-MTT-based cell viability assay (32). This assay has the advantage over other assays in that it does not require expensive reagents and can be automated. This method, in combination with the proviral construct from which the sequence encoding PR and RT is deleted and the recombinant viruses that were produced also provides the option for in vitro analysis of the development of resistance to new antiretroviral drugs at early stages. Furthermore, no changes in the technology will be required when new drugs such as other PR or RT inhibitors (e.g., 1592U89, DMP-266, and VX-478) become approved for use in antiretroviral therapy. At this time, the combined PR-RT assay (Antivirogram) is being used for the simultaneous determination of resistance to 12 currently available antiretroviral drugs. Resistance to single or multiple RT and/or PR inhibitors can be reproducibly detected with the assay system described here. A high degree of automation at all steps involved in the entire procedure makes the assay amenable to high sample throughput, with a current capacity of more then 300 complete (12 drugs) phenotyping assays per week.
It has been demonstrated that any ongoing HIV replication, even below the detection limits of the current assays, provides resistant virus opportunities to emerge (14). Nevertheless, circumstantial evidence suggests that pharmacokinetic factors might also be responsible for the clinical failure of patients on combination therapy including PR inhibitors. These factors include poor drug absorption, increased drug metabolism via P-450 enzyme induction, and noncompliance with the drug regimen (28). Testing of the resistance of viral isolates from individual patients can also be considered a tool for the detection of transmission of resistant virus populations (5) and, simultaneously, as the most valuable technology for defining from the start of antiretroviral therapy the most potent drug combination.
Ideally, optimal antiretroviral therapy should provide durable suppression of HIV replication. Potent combination therapy may temporarily delay resistance, but drug-resistant virus will probably be selected by any regimen that does not completely suppress virus replication. Therefore, combined high-throughput genotypic and phenotypic monitoring of resistance needs to be included in clinical trials to assess the usefulness of resistance testing as a prognostic marker of treatment outcome or failure. Moreover, the information generated by the recombinant virus assay described here and graphically presented in the Antivirogram assay should not only help to detect the resistance present in clinical samples but should also identify the most effective therapeutic strategies active against susceptible and/or resistant viral populations.
ACKNOWLEDGMENTS
This work was supported in part by European Community grant ERB3514 PL 950995 and grant FWO G.0134.97 of the Fonds voor Wetenschappelijk Onderzoek (FWO).
We thank Koen Andries and Marc Moeremans for fruitful discussions and John Mellors for helpful comments.
REFERENCES
- 1.Boucher C, Keulen W, Van Bommel T, Nijhuis M, De Jong D, De Jong M, Schipper P, Back N. Human immunodeficiency virus type 1 drug susceptibility determination by using recombinant viruses generated from patient sera tested in a cell killing assay. Antimicrob Agents Chemother. 1996;40:2404–2409. doi: 10.1128/aac.40.10.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter C, Fischi M, Hammer S, Hirsch M, Jacobsen D, Katzenstein D, Montaner J, Richman D, Saag M, Schooley R, Thompson M, Vella S, Yeni P, Volberding P. Antiretroviral therapy for HIV infection. JAMA. 1996;276:146–154. [PubMed] [Google Scholar]
- 3.Condra J, Holder D, Schleif W, Blahy O, Danovich R, Gabryelski L, Graham D, Laird D, Quintero J, Rhodes A, Robbins H, Roth E, Shivaprakash M, Yang T, Chodakewitz J, Deutsch P, Leavitt R, Massari F, Mellors J, Squires K, Steigbigel R, Teppler H, Emini E. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condra J, Schlief W, Blahy O, Gabryelski L, Graham D, Quintero J, Rhodes A, Robbins H, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K, Deutsch P, Emini E. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;375:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 5.Conway B, Montessori V, Rouleau D, Montaner J, Fransen S, Shillington A, Mayers D. Program and abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication. 1997. Primary lamivudine resistance in acute HIV infection, abstr. 103; p. 68. [Google Scholar]
- 6.Craig J, Duncan I, Hockley D, Grief C, Roberts N, Mills J. Antiviral properties of Ro 31-8959, an inhibitor of human immunodeficiency virus (HIV) proteinase. Antivir Res. 1991;16:295–305. doi: 10.1016/0166-3542(91)90045-s. [DOI] [PubMed] [Google Scholar]
- 7.de Béthune, M. P., K. Hertogs, K. Andries, P. Stoffels, J. Van Roey, P. Schel, A. Van Cauwenberge, C. Van den Eynde, M. De Brabander, J. De Cree, and R. Pauwels. 1996. Monitoring of anti HIV-1 therapy by drug resistance phenotyping (RT-Antivirogram™) and plasma viral load determination: a case study. Antivir. Ther. 1(Suppl. 1):42.
- 8.Dewar R, Highbarger H, Sarmiento M, Todd J, Vasudevachari M, Davey R, Kovacs J, Salzman N, Lane H, Urdea M. Application of branched DNA signal amplification to monitor human immunodeficiency virus type 1 burden in human plasma. J Infect Dis. 1994;170:1172–1179. doi: 10.1093/infdis/170.5.1172. [DOI] [PubMed] [Google Scholar]
- 9.Dorsey B, Levin R, McDaniel S, Vacca J, Guare J, Darke P, Zugay J, Emini E, Schleif W, Quintero J, Lin J, Chen I, Holloway M, Fitzgerald P, Axel M, Ostovic D, Anderson P, Huff J. L-735,524—the design of a potent and orally bioavailable HIV protease inhibitor. J Med Chem. 1994;37:3443–3451. doi: 10.1021/jm00047a001. [DOI] [PubMed] [Google Scholar]
- 10.Eberle J, Bechowsky B, Rose D, Hauser U, Vonderhelm K, Gurtler L, Nitschko H. Resistance of HIV type-1 to proteinase inhibitor Ro 31-8959. AIDS Res Hum Retroviruses. 1995;11:671–676. doi: 10.1089/aid.1995.11.671. [DOI] [PubMed] [Google Scholar]
- 11.Gingeras, T., G. Mamtora, N. Shen, J. Drenkow, M. Winters, and T. Merigan. 1996. Genetic analysis of HIV-1 plasma using high density oligonucleotide arrays and dideoxynucleotide sequencing. Antivir. Ther. 1(Suppl. 1):42.
- 12.Hertogs, K., M. Conant, P. Schel, A. Van Cauwenberge, M. P. de Béthune, and R. Pauwels. 1996. The RT-Antivirogram™: a rapid and accurate method to determine phenotypic (multi-) drug resistance in plasma of patients treated with various HIV-1 RT inhibitors. Antivir. Ther. 1(Suppl. 1):40.
- 13.Ho D, Neumann A, Perelson A, Chen W, Leonard J, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 14.Imamichi H, Zhang Y-M, Lane H C, Fallon J, Salzman N P. Program and abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication. 1997. Continued evolution of HIV-1 during combination therapy despite levels of HIV-1 RNA <500 copies/ml, abstr. 63; p. 41. [Google Scholar]
- 15.Iversen A, Shafer R, Wehrly K, Winters M, Mullins J, Chesebro B, Merigan T. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Beckett L A, Arduino J-M, Lane J, Black R J, Reichelderfer P S, D’Aquila R T, Crumpacker C S the RV-43 Study Group; the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jellinger R, Shafer R, Merigan T. A novel approach to assessing the drug susceptibility and replication of human immunodeficiency virus type 1 isolates. J Infect Dis. 1997;175:561–566. doi: 10.1093/infdis/175.3.561. [DOI] [PubMed] [Google Scholar]
- 18.Kaye S, Loveday C, Tedder R. A microtiter format point mutation assay: application to the detection of drug resistance in human immunodeficiency virus type-1 infected patients treated with zidovudine. J Med Virol. 1992;37:241–246. doi: 10.1002/jmv.1890370402. [DOI] [PubMed] [Google Scholar]
- 19.Kellam P, Larder B. Recombinant virus assay: a rapid, phenotypic assay for assessment of drug susceptibility of human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1994;38:23–30. doi: 10.1128/aac.38.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kempf D, Marsh K, Denissen J, McDonald E, Vasavanonda S, Flentge C, Green B, Fino L, Park C, Kong X, Wideburg N, Saldivar A, Ruiz L, Kati W, Sham H, Robins T, Stewart K, Hsu A, Plattner J, Leonard J, Norbeck D. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc Natl Acad Sci USA. 1995;92:2484–2488. doi: 10.1073/pnas.92.7.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusumi K, Conway B, Cunningham S, Berson A, Evans C, Iversen A, Colvin D, Gallo M, Coutre S, Shpaer E, Faulkner D, De Ronde A, Volkman S, Williams C, Hirsch M, Mullins J. Human immunodeficiency virus type 1 envelope gene structure and diversity in vivo and after cocultivation in vitro. J Virol. 1992;66:875–885. doi: 10.1128/jvi.66.2.875-885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larder B, Darby G, Richman D. HIV with reduced sensitivity to zidovudine isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 23.Larder B, Kemp S. Multiple mutations in HIV-1 reverse transcriptase confer high level resistance to zidovudine. Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 24.Larder B, Kemp S, Harrigan R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1991;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 25.Larder B, Kohli A, Kellam P, Kemp S, Kronick M, Henfrey R. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature. 1993;365:671–673. doi: 10.1038/365671a0. [DOI] [PubMed] [Google Scholar]
- 26.Luiz M, Janini L, Pieniazek D, Peralta J, Schechter M, Tanuri A, Vicente A, Della Torre N, Pieniazek N, Luo C, Kalish M, Schochetman G, Rayfield M. Identification of single and dual infections with distinct subtypes of human immunodeficiency virus type 1 by using restriction fragment length polymorphism analysis. Virus Genes. 1996;13:69–81. doi: 10.1007/BF00576981. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Picado J, Sutton L, Hoes Helfant A, Savara A, Kaplan J, D’Aquila R. Program and abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication. 1997. Novel clonal analyses of resistance to HIV-1 RT and protease inhibitors, abstr. 45; p. 30. [Google Scholar]
- 28.Mayers D, Gallahan D, Martin G, Emmons W, Chung R, Spooner K, Newton J, Aronson N, Weislow O. Program and abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication. 1997. Drug resistance genotypes from plasma virus of HIV-1 infected patients failing combination drug therapy, abstr. 80; p. 52. [Google Scholar]
- 29.Mellors J, Rinaldo C, Gupta P, White R, Todd J, Kingsley L. Prognosis in HIV-1 infection predicted by the quantity of virus in the plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 30.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P, Mo H, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G, Ho D, Boucher C, Leonard J, Norbeck D, Kempf D. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 31.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantification of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauwels R, Balzarini J, Baba M, Snoeck R, Schols D, Herdewijn P, Desmyter J, De Clercq E. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods. 1988;20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 33.Perelson A, Neumann A, Markowitz M, Leonard J, Ho D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 34.Richman D, Havlir D, Corbeil J, Looney D, Ignacio C, Spector S, Sullivan J, Cheeseman S, Barringer K, Pauletti D. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1994;68:1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saag M, Shaw G, Richman D, Volberding P. HIV viral load markers in clinical practice. Nat Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 36.Sardana V, Emini E, Gotlib L, Graham D, Lineberger D, Long W, Schlabach A, Wolfgang J, Condra J. Functional analysis of HIV-1 reverse transcriptase amino acids involved in resistance to multiple nonnucleoside inhibitors. J Biol Chem. 1992;267:17526–17530. [PubMed] [Google Scholar]
- 37.Schinazi R, Larder B, Mellors J. Mutations in retroviral genes associated with drug resistance. Int Antivir News. 1996;4:95–107. [Google Scholar]
- 38.Schmit J C, Cogniaux J, Hermans P, Van Vaeck C, Sprecher S, Van Remoortel B, Witvrouw M, Balzarini J, Desmyter J, De Clercq E, Vandamme A. Multiple drug resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors in an efficiently replicating human immunodeficiency virus type 1 patient strain. J Infect Dis. 1996;174:962–968. doi: 10.1093/infdis/174.5.962. [DOI] [PubMed] [Google Scholar]
- 39.Shafer R, Iversen A, Winters M, Aguiniga E, Katzenstein D, Merigan T. Drug resistance and heterogeneous long-term virologic responses of human immunodeficiency virus type 1-infected subjects to zidovudine and didanosine combination therapy. J Infect Dis. 1995;172:70–78. doi: 10.1093/infdis/172.1.70. [DOI] [PubMed] [Google Scholar]
- 40.Shi C, Mellors J. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob Agents Chemother. 1997;41:2781–2785. doi: 10.1128/aac.41.12.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.St. Clair M, Martin J, Tudor-Williams G, Bach M, Vavro L, King D, Kellam P, Kemp S, Larder B. Resistance to didanosine and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 42.Stuyver L, Wyseur A, Rombout A, Louwagie J, Scarcez T, Verhofstede C, Rimland D, Schinazi R, Rossau R. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tisdale M, Kemp S, Perry N, Larder B. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tisdale M, Meyers R, Maschera B, Parry N, Oliver N, Blair E. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob Agents Chemother. 1995;39:1704–1710. doi: 10.1128/aac.39.8.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Gemen B, van Beuningen R, Nabbe A, van Strijp D, Jurriaans S, Lens P, Kievits T. A one-tube quantitative RNA NASBA nucleic acid amplification assay using electrochemiluminescent (ECL) labelled probes. J Virol Methods. 1994;49:157–167. doi: 10.1016/0166-0934(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 46.Wei X, Ghosh S, Taylor M, Johnson V, Emini E, Deutsch P, Lifsin J, Bonhoeffer S, Nowak M, Hahn B, Saag M, Shaw G. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;375:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 47.Zhang D, Caliendo A, Eron J, DeVore K, Kaplan J, Hirsch M, D’Aquila R. Resistance to 2′,3′-dideoxycytidine conferred by a mutation in codon 65 of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1994;38:1428–1432. doi: 10.1128/aac.38.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]