Abstract
Neutrophils, until recently, have typically been considered a homogeneous population of terminally differentiated cells with highly conserved functions in homeostasis and disease. In hepatocellular carcinoma (HCC), tumour-associated neutrophils (TANs) are predominantly thought to play a pro-tumour role, promoting all aspects of HCC development and progression. Recent developments in single-cell technologies are now providing a greater insight and appreciation for the level of cellular heterogeneity displayed by TANs in the HCC tumour microenvironment, which we have been able to correlate with other TAN signatures in datasets for gastric cancer, pancreatic ductal adenocarcinoma (PDAC) and non-small cell lung cancer (NSCLC). TANs with classical pro-tumour signatures have been identified as well as neutrophils primed for anti-tumour functions that, if activated and expanded, could become a potential therapeutic approach. In recent years, therapeutic targeting of neutrophils in HCC has been typically focused on impairing the recruitment of pro-tumour neutrophils. This has now been coupled with immune checkpoint blockade with the aim to stimulate lymphocyte-mediated anti-tumour immunity whilst impairing neutrophil-mediated immunosuppression. As a result, neutrophil-directed therapies are now entering clinical trials for HCC. Pharmacological targeting along with ex vivo reprogramming of neutrophils in HCC patients is, however, in its infancy and a greater understanding of neutrophil heterogeneity, with a view to exploit it, may pave the way for improved immunotherapy outcomes. This review will cover the recent developments in our understanding of neutrophil heterogeneity in HCC and how neutrophils can be harnessed to improve HCC immunotherapy.
Keywords: hepatocellular carcinoma, immunotherapy, neutrophils
Introduction
Primary liver cancer is a leading cause of cancer-related death and morbidity, with 905,667 cases and 830,180 related deaths in 2020 [1]. Worryingly, the incidence of primary liver cancer is expected to increase up to 55% by 2040 [2]. Hepatocellular carcinoma (HCC) is the most common form of primary liver cancer making it a significant public health concern. HCC typically develops on the background of chronic liver disease and cirrhosis of various aetiologies. This makes delivering effective therapy challenging due to aetiology-specific immunosuppressive tumour microenvironments and patient frailty due to underlying liver disease and co-morbidities in typically elderly patients. Although there are curative treatments available for early-stage HCC, the majority of HCCs are diagnosed at an advanced stage with limited palliative treatments available and a generally poor prognosis. The IMBrave150 phase III clinical trial resulted in the approval of the immunotherapy combination of Atezolizumab (anti-PD-L1) plus Bevacizumab (anti-VEGF) as a first-line systemic treatment for advanced disease and the first significant change in HCC patient treatment for over a decade [3]. Whilst this immunotherapy combination is highly effective in a subset of patients (one-third responders), the majority of patients fail to clinically respond [4]. Failure to respond to immunotherapy is typically associated with an immunosuppressive HCC tumour microenvironment (TME). Currently approved immunotherapies target immune-inhibitory receptors (e.g. PD-1 and CTLA-4) on T lymphocytes, or their ligands (e.g. PD-L1), aiming to activate T-lymphocyte cytotoxicity and anti-tumour immunity [5]. New therapies targeting immunosuppressive cells in the TME beyond T lymphocytes, alone or in combination with current immunotherapies, might be more effective in a subgroup of HCC patients. Neutrophils are one such target. In health, neutrophils comprise 50–70% of all circulating leukocytes and are the first effector cells to arrive at sites of infection, inflammation and tissue damage [6]. Advancement in single-cell RNA sequencing (scRNAseq) technologies has finally enabled analysis of tissue-derived neutrophils at the single-cell level, allowing us to understand their previously unappreciated plasticity and heterogeneity in chronic disease and cancer. Tumour-associated neutrophils (TANs), through their effector functions; phagocytosis, degranulation, release of neutrophil extracellular traps (NETs) and antigen presentation, have been implicated in a broad range of pro- and anti-tumour activities in HCC, from promoting tumour cell proliferation, immunosuppression, metastasis and angiogenesis to direct tumour killing and activation of anti-tumour immunity, respectively [7–9], and their pro-tumour roles are summarised in Figure 1. Given our improved understanding of the level of cellular heterogeneity and plasticity neutrophils are capable of, future work will need to determine whether the broad range of pro- and anti-tumour roles previously identified for neutrophils are present in either all or discrete populations of TANs, which may have therapeutic implications.
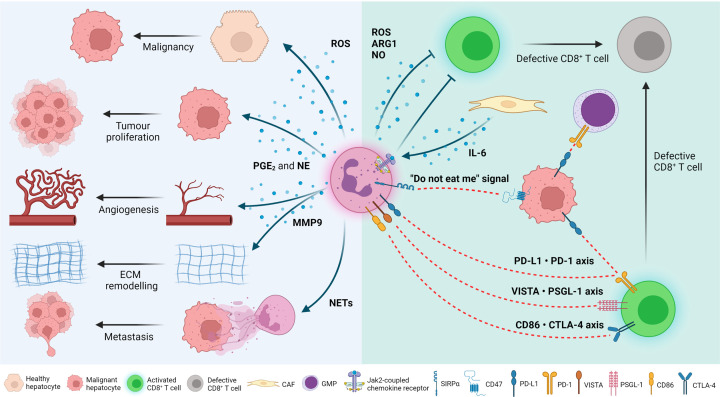
Figure 1. Pro-tumour mechanisms of tumour-associated neutrophils.
Direct effects of neutrophils on cancer cells: Neutrophils release a plethora of bioactive molecules that drive cancer development. Neutrophils secrete reactive oxygen species (ROS) which, through DNA damage, can drive malignant transformation of hepatocytes. They also secrete prostaglandin E2 (PGE2) and neutrophil elastase (NE), which can induce tumour cell proliferation; as well as matrix metalloproteinase 9 (MMP9), which promotes angiogenesis and extracellular matrix (ECM) remodelling in the tumour. Furthermore, release of neutrophil extracellular traps (NETs) promotes metastatic seeding of tumour cells in distal organs. Tumour cells express CD47 which, when bound to its ligand, signal regulatory protein α (SIRPα) found on the surface of neutrophils, impairs their phagocytic capacity; a process commonly known as the ‘do not eat me’ signal. A proportion of tumour cells also express the programmed death-ligand 1 (PD-L1) which, in contact with programmed cell death protein 1 (PD-1) in the surface of CD8+ T cells and granulocyte–monocyte progenitors (GMPs), suppresses their anti-tumour functions. Effects of neutrophils on the immune system: Neutrophils emit ROS, arginase 1 (ARG1) and nitric oxide (NO), which can immunosuppress cytotoxic CD8+ T cells. Moreover, cancer-associated fibroblasts (CAFs) secrete interleukin 6 (IL-6) that is recognised by JAK2-coupled chemokine receptors on the surface of neutrophils, also immunosuppressing T cells. Finally, neutrophils express PD-L1, V-type immunoglobulin domain-containing suppressor of T-cell activation (VISTA) and CD86 which, in contact with their respective receptors/ligands on CD8+ T cells, can impair their cytotoxic ability. Figure created with BioRender.com
This review will cover recent advances in our understanding of neutrophil function and heterogeneity in HCC and discuss how this heterogeneous population of neutrophils may impact the TME and their cellular plasticity can be harnessed to improve immunotherapy in HCC.
Neutrophil heterogeneity in granulopoiesis
Phenotypical and functional heterogeneity in myeloid cells is well established for monocyte and macrophage populations and assures diverse functions that are required to maintain homeostasis, respond to infection and resolve inflammation [10]. On the other hand, neutrophils are traditionally considered as a homogeneous population of terminally differentiated cells with highly conserved function. Recent advances in single-cell technologies are challenging this view and are confirming that human and murine neutrophils are a heterogeneous population of cells that display a range of maturation and polarisation states. This heterogeneity is governed by a multitude of factors including tissue of residence, physiologic state (e.g. homeostasis, inflammation and cancer), circadian rhythms and neutrophil cellular age as well as obesity, sex, tobacco consumption and whole organism aging [11–14].
Initial transcriptomic studies in 2020 [15–17] defined up to eight different neutrophil populations in mice and human, ranging from granulocyte-macrophage progenitor (GMP) precursors to mature circulating neutrophils, with identified clusters unified using the following nomenclature: G0, G1, G2, G3, G4, G5a, G5b and G5c [17]. These clusters have distinct maturation states, a predominant tissue of residence (bone marrow, spleen and peripheral blood), and are present under homeostasis and perturbed during pathogenic infection. G0, G1, G2, G3 and G4 clusters align to GMP, proNeu, preNeu, immNeu and mNeu, respectively, and reside predominantly in the bone marrow. Peripheral blood contained three main neutrophil subsets; G5a, G5b and G5c, which had typical mature neutrophil nuclear morphology. Given the distinct transcriptional signatures of circulating neutrophils, which could not be explained by cellular age, mechanical stress, or insults, it has been proposed that these subsets may be pre-programmed to perform specific homeostatic functions.
Our group and others have suggested that neutrophil maturation plays a critical role in determining their phenotype and functionality. Fundamental to this, is the recently proposed ‘neutrotime paradigm’ which aims to provide a framework to understand and describe heterogeneity in neutrophil populations. In this model, neutrophils, rather than existing in discrete maturation states as described previously, follow a developmental continuum, called the ‘neutrotime’ [18]. As neutrophils progress along this continuum and encounter a stimulus they will begin to acquire a phenotype (or polarisation state), that is the product of both time (e.g. maturation stage) and environmental cues (sequence of signals encountered). This mechanism provides an opportunity to generate a highly diverse neutrophil repertoire without a requirement for committed developmental branches or subsets [18]. Neutrophils recruited to sites of inflammation and infection, typically emerge from the mature end of the neutrotime spectrum, containing a full complement of anti-microbial granule proteins allowing them to perform their primary function, clearing pathogens. However, during chronic disease and cancer, where distortions in granulopoiesis are observed, emergence of neutrophils earlier along the neutrotime continuum is common with circulating neutrophils appearing relatively immature. As a result, these cells are phenotypically and functionally distinct from their mature counterparts which have encountered similar environmental cues. Alterations in granulopoiesis in cancer has also led to reports of extended neutrophil lifespan [19], possibly in excess of 5 days for circulating human neutrophils [20], providing further time for phenotype alterations.
Neutrophil heterogeneity in cancer: the age of single-cell technologies
For over a decade, neutrophil heterogeneity in cancer has been dichotomously described with circulating neutrophils labelled based on their density as either high/normal- or low-density neutrophils (HDNs/NDNs and LDNs, respectively) and TANs labelled based on either their identified pro- or anti-tumour functions [21]. Emerging evidence is now confirming that this simple binary characterisation is not sufficient to describe the broad range of neutrophil phenotypes and proposed widespread functionality.
Single-cell transcriptomics has revolutionised our understanding of the tumour microenvironment, providing a wealth of data on nearly all cell types at single-cell resolution. However, until recently, the majority of scRNAseq studies failed to recover a significant number of neutrophils for downstream analysis; this is a feature also prominent in other single-cell analyses performed in solid organs [22–26]. Reasons behind this include the fragile nature of neutrophils during isolation and the relative low abundance of RNA compared with other cell types. Progress, however, has been made, with recent first reports of neutrophil heterogeneity in liver cancer [27] and other solid tumours such as gastric cancer [28], pancreatic ductal adenocarcinoma (PDAC) [29] and non-small cell lung cancer (NSCLC) [30].
Neutrophil heterogeneity in HCC
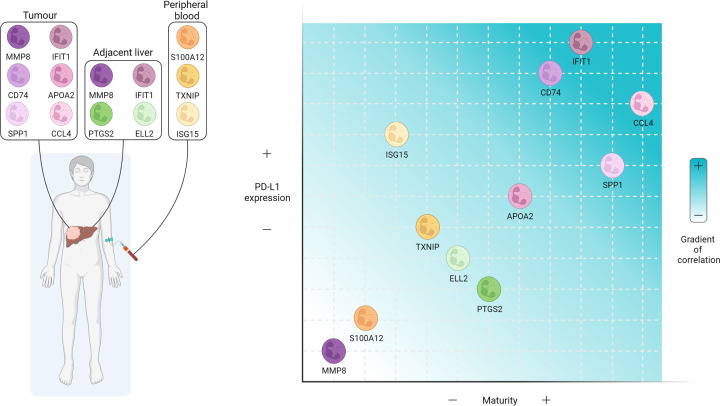
The study of neutrophil subtypes in HCC is still in its infancy. Despite many single-cell transcriptomic analyses of HCC having been performed [31–38] the first description of significant neutrophil heterogeneity in HCC patients and preclinical models was made only in 2022 [27], highlighting the technical difficulty of neutrophil heterogeneity analysis and its emergence as a novel area of research in HCC. Xue et al. analysed 160 samples of 124 treatment-naïve liver cancer patients, of which 100 samples were from 79 HCC patients, comprising a mixture of viral (HBV and HCV) and non-viral HCC. It must be noted that this study was performed on resection samples and as such may not be representative of patients with more advanced HCC, where resections cannot be performed. As such presence of the following neutrophil/TANs will need to be confirmed in biopsy tissue from patients with advanced disease. The authors identified 11 transcriptionally distinct neutrophil clusters in HCC which they termed: Neu_01_MMP8, Neu_02_S100A12, Neu_03_ISG15, Neu_04_TXNIP, Neu_05_ELL2, Neu_06_PTGS2, Neu_07_APOA2, Neu_08_CD74, Neu_09_IFIT1, Neu_10_SPP1 and Neu_11_CCL4. The S100A12+, ISG15+ and TXNIP+ subtypes are found in peripheral blood, the ELL2+ and PTGS2+ mainly in adjacent liver and the MMP8+, APOA2+, CD74+, IFIT1+, SPP1+ and CCL4+ predominantly located in the tumour (Figure 2). These neutrophil clusters were conserved in a genetically engineered mouse model (GEMM), called pTMC (Myc-Δ90Ctnnb1), which is driven by β-catenin and develops HCC tumours. These neutrophil clusters exist along a maturation gradient ranging from the most immature cells present in peripheral blood, the intermediate present in adjacent liver and the most mature cells present in the tumour, with the exception of the MMP8+ cluster, found in the adjacent liver and tumour, having the most immature phenotype (Figure 2). Divergence of neutrophil polarising states arising from both mature and immature neutrophils provides further validity to the ‘neutrotime paradigm’ as an effective model to explain neutrophil heterogeneity in homeostasis and disease.
Figure 2. Neutrophil heterogeneity in liver cancer.
Up to 11 different neutrophil phenotypes have been described in human HCC by scRNAseq [27]. These phenotypes have different predominant tissues of residence, including tumour, adjacent liver and peripheral blood. Moreover, they display a range of maturity stages and PD-L1 expression levels that show a positive correlation. Figure created with BioRender.com
Understanding the biological significance of each of the TAN clusters in HCC development, how they regulate the TME and potential effects on immunotherapy responses are key questions that need to be answered. PD-L1 expression appears to be largely related to maturation in both human and mouse HCC and may provide some insight into the potential pro- or anti-tumour functions of these TANs (Figure 2). In line with this, enrichment of high PD-L1 expressing IFIT1+, SPP1+ and CCL4+ TANs (Figure 2) in HCC tumours correlated with a poorer prognosis indicating a pro-tumour role for these clusters [27]. It is tempting to speculate these HCC tumours would respond to immune checkpoint blockade (anti-PD-L1/anti-PD-1) and those enriched with PD-L1 low TANs will not respond. However, our results suggest that mature TANs express high levels of PD-L1 and correlate with immunotherapy resistance in HCC [39]. Further research needs to be conducted in order to understand this paradox, controlling for the expression of PD-L1 in other cell types including cancer cells, other myeloid cells like macrophages and dendritic cells, and stromal cells, as well as the molecular and immune subtype classification of the tumour, which is known to predict immune checkpoint inhibition response.
The six HCC TAN clusters; MMP8+, APOA2+, CD74+, IFIT1+, SPP1+ and CCL4+ can be characterised by predicted function and role in tumour development. The IFIT1+, SPP1+ and CCL4+ TANs were identified as pro-tumour, were associated with a poorer prognosis and had the highest levels of PD-L1 expression. These TANs were proposed to exert their pro-tumour functions by different mechanisms. IFIT1+ TANs were enriched for genes associated with interferon (IFN) signalling and pathways of type I interferon signalling and response to interferon gamma [40–51]. In addition, IFIT1+ TANs showed the highest expression of PD-L1 and could be the major population of PD-L1+ TANs that inhibits the cytotoxic capacity of CD8+ T cells, as such, these were termed interferon-stimulated immunosuppressive TANs. SPP1+ TANs displayed a gene expression signature similar to pro-angiogenic tumour-associated macrophages (TAMs) [52,53] and were therefore predicted to play a key role in tumour angiogenesis, thus termed angiogenic TANs. The CCL4+ TANs were enriched for chemokine secretion and predicted to promote the recruitment of immunosuppressive myeloid cells [54–56] and thus termed myeloid chemokine-secreting TANs (Figure 3). CD74+ TANs showed high expression of CD74, although protein expression of CD74 in neutrophils has not been reported yet in the bibliography [57], and other MHC-associated genes [58–62] and were termed antigen-presenting TANs. These are predicted to play a role in antigen presentation, which has classically been linked to anti-tumour functions in other cancers like lung cancer [63,64]. However, although they correlate with a better prognosis in patients, their relatively high expression of PD-L1 makes unclear whether these are likely to play a pro- or anti-tumour role (Figure 3). Finally, the MMP8+ and APOA2+ TANs expressed lower levels of PD-L1, correlated with a better prognosis and were therefore predicted to play an anti-tumour role in HCC (Figure 3). MMP8+ TANs were enriched for gene signatures associated with classical neutrophil properties including azurophil and gelatinase granules' secretion, neutrophil activation and phagocytosis, which have all been proposed as potential anti-tumour mechanisms [39,65–68] and were termed immature TANs. The APOA2+ neutrophils were enriched for metabolic pathways such as triglyceride metabolism and regulation of steroid metabolism [69–74] and thus termed hepatic lipid-associated TANs (Figure 3) and hepatic lipid-associated neutrophils or LANs, in the tumour and adjacent liver, respectively. Interestingly, LANs resemble hepatic lipid-associated macrophages (LAMs), which help maintain lipid metabolism in healthy liver and suggestive to hepatoprotective potential, suppressing HCC development [75], implying these neutrophils/TANs may play a similar role in HCC.
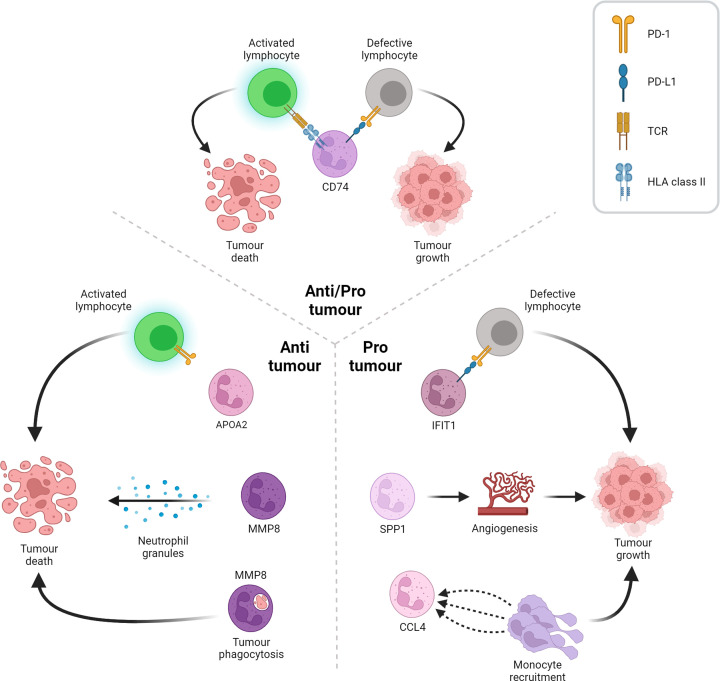
Figure 3. Role of tumour-associated neutrophils in the HCC microenvironment.
The six TANs found in the scRNAseq dataset are predicted to play diverse roles in the HCC TME. On the left side, the APOA2 subtype expresses low levels of PD-L1, which avoids the suppression of T lymphocytes that act against the tumour. Moreover, MMP8 shows an elevated secretory profile of intracellular enzyme-loaded granules that results in cell toxicity for the tumour. On the right side, the IFIT1 subtype that, although characterised for an intense interferon signalling, possesses the highest PD-L1 expression, which turns the T lymphocytes defective upon binding to their PD-1 receptor. Furthermore, the SPP1 subtype, which expresses pro-angiogenic factors, enhances angiogenic programs in the TME, resulting in tumour fuelling. The third pro-tumour TAN, the CCL4 subtype, secretes myeloid chemoattractant peptides that potentiates the infiltration of immunosuppressive monocytes to the TME. Finally, on the upper side, the CD74 subtype, which is able to activate T lymphocyte-dependant tumour death programs through its antigen-presenting activity, at the same time that possesses the second most elevated PD-L1 expression, which suppresses T lymphocytes and reinvigorates tumour growth. Figure created with BioRender.com
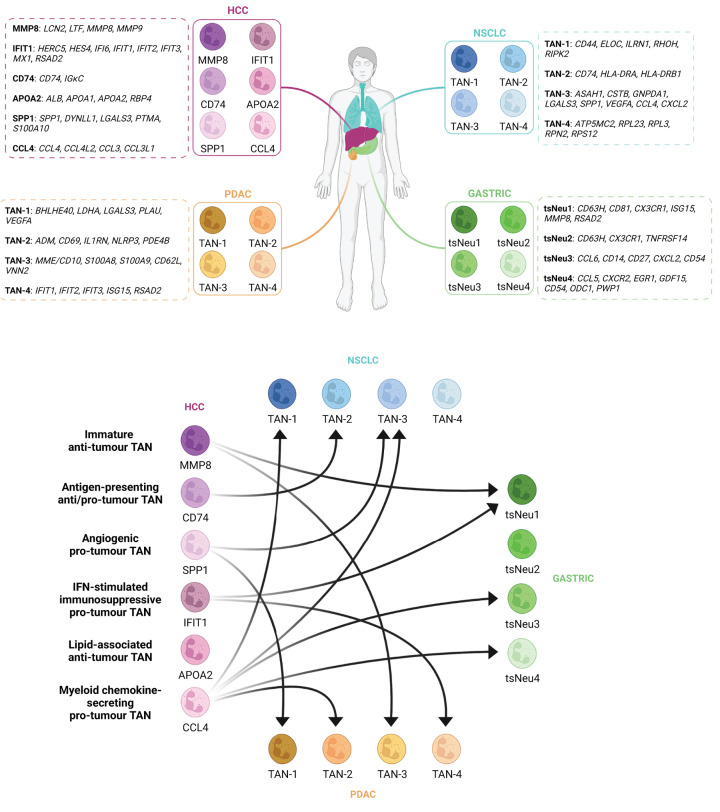
Interestingly, the HCC TAN clusters [27] can be identified in scRNAseq analysis of other cancers including gastric cancer [28], pancreatic ductal adenocarcinoma (PDAC) [29] and non-small cell lung cancer (NSCLC) [30] by comparison of most differentially expressed marker genes, with the exception of APOA2+ lipid-associated TANs which so far appear to be specific to the liver. Several TAN subtypes were found to be present in at least three of the four scRNAseq neutrophil analyses. Firstly, pro-tumour TAN subtypes were identified in all four datasets. Angiogenic TANs were identified in PDAC (TAN-1), NSCLC (TAN-3) and HCC (SPP1+). IFN-stimulated immunosuppressive TANs were identified in PDAC (TAN-4), gastric cancer (tsNeu1) and HCC (IFIT1+). Interestingly, variations of myeloid chemokine-secreting TANs were identified in all four cancers including two types identified in gastric cancer (tsNeu3 and tsNeu4), NSCLC (TAN-3) and HCC (CCL4+), all of which have elevated levels of myeloid chemokines. Similar TANs identified in NSCLC (TAN-1) and PDAC (TAN-2) are predicted to be pro-inflammatory and are therefore likely to contribute to myeloid cell recruitment. Antigen-presenting TANs were identified in NSCLC (TAN-2) and HCC (CD74+). Finally, immature TANs that are likely to play anti-tumour roles were identified in PDAC (TAN-3), gastric cancer (tsNeu1) and HCC (MMP8+). Interestingly, tsNeu1 TANs identified in gastric cancer expressed genes associated with immature and IFN-stimulated immunosuppressive TANs, as such, it is difficult to hypothesise a role for these TANs in tumour progression. Given our current understanding of neutrophil heterogeneity across multiple cancers, therapies that expand intratumoural MMP8+ and APOA2+ TANs whilst supressing IFIT1+, SPP1+ and CCL4+ TANs may be effective in improving immunotherapy responses in HCC (Figure 4).
Figure 4. Functional correlation of neutrophil subtypes between multiple cancers.
The upper part of the figure shows the different neutrophil subtypes found in HCC [27], gastric cancer [28], PDAC [29] and NSCLC [30] by scRNAseq along with the list of the most differentially expressed genes for each one. The bottom part of the figure represents the functional correlation between the diverse neutrophil subtypes in the four cancers studied. Hence, the MMP8 subtype in HCC, which possess an immature anti-tumour profile, correlates with tsNeu1 gastric cancer and TAN-3 PDAC. Secondly, the CD74 subtype in HCC, with an antigen-presenting anti/pro-tumour potential, correlates with TAN-2 NSCLC. Thirdly, the SPP1 subtype in HCC, characterised for secreting pro-angiogenic factors, correlates with TAN-3 NSCLC and TAN-1 PDAC. Fourthly, the IFIT1 subtype in HCC, with an interferon-stimulated immunosuppressive pro-tumour profile, correlates with tsNeu1 gastric cancer and TAN-4 PDAC. Fifthly, the APOA2 subtype in HCC, with a lipid-associated anti-tumour role remains specific for HCC, as TAN-4 NSCLC and tsNeu2 gastric cancer do. Finally, the CCL4 subtype in HCC, with a myeloid chemokine-secreting pro-tumour profile, correlates with TAN-1 and TAN-3 NSCLC, tsNeu3 and tsNeu4 gastric cancer and TAN-2 PDAC. Figure created with BioRender.com
Harnessing neutrophils for immunotherapy in HCC
TANs are predominantly thought to be immunosuppressive in HCC favouring tumour progression [7], with preclinical pan-neutrophil depletion experiments shown to be effective in reducing tumour burden in rodents [27,76]. Therapies leading to neutropenia such as traditional chemotherapy agents, are typically poorly tolerated in HCC patients, who often have cirrhosis and portal hypertension. This coupled with the high risk of severe bacterial infection makes complete clearance of neutrophils in the absence of direct tumour cytotoxic effects not an appealing therapeutic strategy. Given the recent advances in our understanding of neutrophil heterogeneity in the HCC tumour microenvironment it may be possible to exploit their cellular plasticity allowing for the specific targeting or reprogramming of TANs with pro-tumour functions, without affecting the circulating pool of functional anti-microbial neutrophils.
Targeting neutrophils: combination therapies and the ‘recruitment paradox’
To date, most neutrophil-based therapies have aimed to either suppress recruitment of TANs or modulate their immunosuppressive nature to elicit an anti-tumour response. Recruitment and modulation of neutrophils in HCC and their associated targeted therapies have been reviewed elsewhere in detail [7]. In this section, we will discuss recent advances in our understanding of combination immunotherapies in HCC, how this impacts neutrophil recruitment and tumour burden, and how therapies that elicit neutrophil-based anti-tumour responses can improve anti-PD-1 immunotherapy.
The effectiveness of targeting neutrophil chemokine receptors in combination with immune checkpoint blockade, chemotherapy and hormone therapy has been reported for nearly a decade, in various cancers, including pancreas, lung, prostate, breast, bladder, nasopharyngeal carcinoma, ovary [77–85] and, in 2022, the liver [39]. These studies aimed to limit the recruitment of pro-tumour immunosuppressive neutrophils, largely through targeting the neutrophil specific chemokine receptor CXCR2, which would allow a robust anti-tumour response to be generated. In GEMMs of intrahepatic cholangiocarcinoma (ICC) [86], Haining Liu and colleagues demonstrated that co-inhibition of both CXCR2 and METTL1, a protein directly involved in the expression of the soluble chemo attractants CXCL5/CXCL8, in combination with anti-PD-1, achieved a complete response, with a survival rate of 100% during 60 days of tracking [87]. This therapeutic response was associated with a significant reduction in infiltrating tumour neutrophils (referred to as polymorphonuclear myeloid-derived suppressor cells). Interestingly, however, we have demonstrated that the pharmacological inhibition of both CXCR2 and PD-1 sensitises immunotherapy-resistant non-alcoholic steatohepatitis-associated HCC (NASH-HCC) mouse models to immune checkpoint blockade, resulting in an influx of intratumoural neutrophils that promote CD8+ T cell and CD103+XCR1+ cDC1 dendritic cell activation, leading to a decrease in tumour burden (Figure 5) [39]. These recruited neutrophils were of a lactoferrin-positive immature anti-tumour phenotype and were organised in immune hubs, which we propose act to establish and maintain anti-tumour responses in the NASH-HCC immunotherapy-resistant niche [39]. This ‘recruitment paradox’ of combination immunotherapies in NASH-HCC, we suggest, could be the result of several mechanisms. By their very nature, mature neutrophils express the highest levels of surface CXCR2 and would therefore be preferentially repressed by a CXCR2 antagonist, leading to the potential accumulation of immature neutrophils. Furthermore, synergy of CXCR2 and anti-PD-1 inhibition led to the expansion of an intratumoural immature neutrophil population originating from either a neutrophil progenitor (NeP) or tumour-seeded hematopoietic stem cell. This therapeutic synergy is also responsible for the acquisition of an anti-tumour phenotype characterised by elevated neutrophil activation and phagocytosis as well as antigen presentation capability. Indeed, a remarkably similar population of tumour-associated immature neutrophils (MMP8+ subtype) was identified by scRNAseq analysis and expressed genes associated with the early neutrotime signature [27]. Interestingly, this immature neutrophil population had the lowest expression of PD-L1 (CD274) among all neutrophil subsets identified and could, if expanded, represent an interesting mechanism to stimulate anti-tumour immunity. Therefore, it is clear that the therapeutic combination of CXCR2 inhibition and immune checkpoint immunotherapy overcomes a significant clinical problem faced with anti-PD-1 resistance in NASH-HCC [88] and a phase I/II clinical trial for advanced HCC [89] is currently underway targeting the CXCR2 and PD-L1/PD-1 axes.
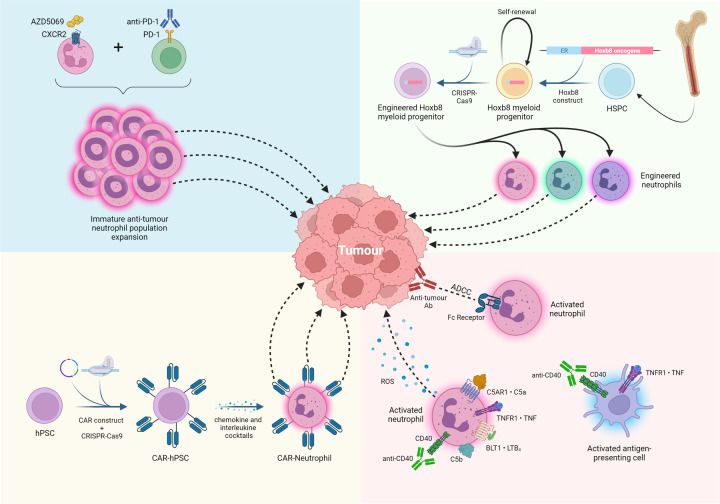
Figure 5. Current clinical and preclinical neutrophil therapies in cancer.
Top left corner: the combination of a CXCR2 small molecule inhibitor (AZD5069) plus anti-PD-1 expanded an immature anti-tumour neutrophil population, resulting in a reduction in tumour burden and an increase in survival [39]; clinical trial ongoing targeting these axes in advanced HCC [89]. Top right corner: Hoxb8 myeloid progenitors provide a continuous pool of neutrophils for genetic modification, enabling the identification of novel neutrophil-specific therapeutic targets to improve immunotherapy. Bottom left corner: the recent development of human CAR-neutrophils from human pluripotent stem cells (hPSCs) showed high efficacy in a model of glioblastoma [96] and may be suitable for use in HCC. Bottom right corner: Further immunotherapy approaches, combining TNF and monoclonal antibodies anti-CD40 and anti-tumour-associated antigen, result in a high infiltration of activated anti-tumour neutrophils and antigen-presenting cells’ activation [91]. Figure created with BioRender.com
Given that neutrophils display a range of anti-tumour functions, we propose that due to their cellular plasticity it may be possible to reprogram TANs to promote anti-tumour immunity and that this may not necessarily be specific to targeting a chemokine receptor as we have demonstrated. Esteban-Fabró and colleagues showed that combining anti-PD-1 therapy with Cabozantinib, a small molecule tyrosine kinase inhibitor largely selective for c-Met and VEGFR2, increased anti-tumour efficacy compared with monotherapies [90]. This therapeutic effect was associated with a neutrophil-based immune response, which, when used to stratify human HCC patients, represented those with favourable molecular and clinical features [90]. In melanoma models, immunotherapies combining tumour necrosis factor (TNF), anti-CD40 and anti-tumour-associated antigen antibody have also been shown to polarise neutrophils to an anti-tumour phenotype promoting tumour clearance via ROS secretion and improved antibody-dependant cellular cytotoxicity (ADCC). This provides further evidence that neutrophils have a substantial cellular plasticity which could be therapeutically exploited in HCC (Figure 5) [91].
Due to our recent improvements in the understanding of intratumoural neutrophil populations it is now clearer than ever the importance of understanding patient-specific neutrophil heterogeneity. As more neutrophil specific therapies are developed and tested in combination with immune checkpoint immunotherapies, it will be critical to understand how these drugs impact each neutrophil population and how the abundance of each one in the patients' tumours impacts therapy outcome.
Neutrophil engineering: ex vivo reprogramming
Transfusion of exogenously-reprogrammed immune cells, such as CAR-T and CAR-NK therapies, are beginning to revolutionise how haematological cancers, such as lymphoma or adult and childhood leukaemias are treated. In these cancers, T and NK cells are manipulated to express recombinant chimeric antigen receptors (CARs), which significantly enhances their anti-tumour potential. However, to date, efficacy in solid tumours has been limited [92], although big efforts are made to improve this [93]. Moreover, clinical trials, based on preclinical studies in NOD scid γ (NSG) mice [94], are ongoing for advanced HCC with CAR-T cells targeting glypican-3, a cancer neoantigen widely expressed on the surface of the malignant hepatocytes [95]. Similarly, CAR-neutrophils are in the early stages of development and have shown promising preclinical results in glioblastoma [96]. This study genetically engineered human pluripotent stem cells (hPSCs) in order to create anti-glioblastoma CAR-neutrophils, which had enhanced anti-tumour activity both in vitro and in vivo. Mechanistically, the CAR-neutrophils were shown to be phenotypically similar to hPSC derived neutrophils but had greater anti-tumour functionality as assessed by neutrophil-tumour immune synapse formation, phagocytosis and ROS-mediated tumour killing (Figure 5) [96]. In line with this, we propose that CAR-neutrophils may be effective in HCC. Compared to CAR-T cells, neutrophils express matrix metalloproteinases which give them a superior ability to infiltrate liver tumours with a dense stroma. Furthermore, the short lifespan of CAR-neutrophils may reduce toxicities, a recurrent clinical issue experienced by patients receiving CAR-T therapy. In addition, neutrophils could also be genetically modified in order modulate specific pro-tumour and anti-tumour functions. To do so there is a unique pre-clinical model of immortalised myeloid progenitors the can be maintained, modified and expanded ex vivo, called Hoxb8-conditional myeloid progenitor cells. Neutrophils derived from this system are phenotypically similar to mouse primary neutrophils [97,98] and display neutrophil functions when administered in vivo (Figure 5) [99]. Furthermore, given the success of combination therapies in preclinical models in reprogramming neutrophils in vivo to drive anti-tumour immunity, transfusion of exogenously-stimulated neutrophils may be sufficient to drive an anti-cancer response. As a proof of concept, preclinical work has shown that transfusion of immature neutrophils isolated from LPS-treated mice is sufficient to reactivate anti-tumour immunity in immunotherapy-resistant mouse models of NASH-HCC [39]. Currently, granulocyte infusion for the treatment of refractory neutropenic sepsis is the only approved neutrophil cellular therapy [100]. In theory, granulocytes used for this purpose could be polarised to an anti-tumour phenotype ex vivo prior to infusion in order to stimulate anti-tumour immune responses. A fast-acting stimulus which robustly polarises neutrophils to an anti-tumour phenotype whilst not over activating them would need to be identified. Moreover, granulocyte transfusions carry significant risk which would need to be carefully considered, especially when genetically modifying or stimulating them. The main immediate risks being febrile reactions and pulmonary toxicity [100].
Conclusion and outlook
Interrogation of TAN biology has finally entered the era of scRNAseq allowing for the first time the analysis of neutrophil heterogeneity and the identification of TAN subtypes, many of which appear to be conserved across cancers. However, this is just the start, and this cutting-edge technology is not without its limitations. Firstly, scRNAseq read depth is much lower than that of conventional bulk RNAseq and, as such, only a superficial picture of the neutrophil transcriptome is provided [101]. When this is coupled with the low abundance of RNA and high abundance of RNAses present in neutrophils, caution must be taken when analysing and interpreting scRNAseq from TANs. Secondly, bioinformatical clustering of scRNAseq data can lead to over clustering, and the fallacy of affirming the existence of more neutrophil subtypes than may exist. Ultimately, future work needs to focus on identifying surface protein markers that will allow for functional analysis and validation of proposed TAN subtypes and enable the identification of a suitable homogeneous and universal nomenclature going forward.
From a therapeutic perspective, neutrophils have conventionally been regarded as promoting tumorigenesis. As a result, immunotherapeutic strategies targeting neutrophils in cancer have largely focused on inhibiting the recruitment of neutrophils to the tumour. Recent data, however, suggest that neutrophils in the HCC tumour microenvironment can be programmed with anti-tumour functions if provided with the right exogenous stimulus and could even overcome immunotherapy resistance in NASH-HCC patients. Finally, neutrophil-based cell therapies may provide an exciting new therapeutic avenue and an area of research for improving our understanding of neutrophil-mediated anti-tumour immunity and improving HCC immunotherapy outcomes. Altogether, recent developments in our understanding of neutrophil biology in cancer has shed light on the cellular heterogeneity and plasticity of a once considered ‘inert’ cell. Future work should aim to exploit these recent findings to boost the efficacy of existent treatments and lead to the development of new immunotherapies for patients with cancer.
Summary
Neutrophils are a heterogeneous population of cells that can display a range of phenotypes in homeostasis, disease and cancer.
Tumour-associated neutrophil (TAN) subtypes functionally correlate between cancers.
Neutrophils exhibit cellular plasticity and can undergo reprogramming in the HCC tumour microenvironment.
Reprogrammed neutrophils, in situ or ex vivo, represent a viable anti-cancer target.
Abbreviations
- Ab
Antibody
- ADCC
Antibody-dependant cellular cytotoxicity
- APOA2
Apolipoprotein A-II
- ARG1
Arginase 1
- BLT1
Leukotriene B4 receptor 1
- CAF
Cancer-associated fibroblast
- CAR
Chimeric antigen receptor
- Cas9
CRISPR-associated protein 9
- CCL4
C-C motif chemokine ligand 4
- CD
Cluster of differentiation
- cDC1
Conventional dendritic cell 1
- c-MET
Tyrosine kinase MET
- CRISPR
Clustered regularly interspaced short palindromic repeats
- CTLA-4
Cytotoxic T-lymphocyte-associated protein 4
- CXCL5
CXC motif chemokine ligand 5
- CXCL8
CXC motif chemokine ligand 8
- CXCR2
CXC motif chemokine receptor 2
- C5a
Complement component C5a
- C5AR1
Complement component C5a receptor 1
- C5b
Complement component C5b
- ECM
Extracellular matrix
- ELL2
Elongation factor for RNA polymerase II 2
- GEMM
Genetically engineered mouse model
- GMP
Granulocyte-monocyte progenitor
- HBV
Hepatitis B virus
- HCC
Hepatocellular carcinoma
- HCV
Hepatitis C virus
- HDN
High-density neutrophil
- HLA class II
Human leukocyte antigen class II
- Hoxb8
Homeobox B8
- hPSC
Human pluripotent stem cell
- HSPC
Hematopoietic stem and progenitor cell
- ICC
Intrahepatic cholangiocarcinoma
- IFIT1
Interferon induced protein with tetratricopeptide repeats 1
- IFN
Interferon
- IL-6
Interleukin 6
- ISG15
Interferon-stimulated gene 15
- JAK2
Janus kinase 2
- LAM
Lipid-associated macrophage
- LAN
Lipid-associated neutrophil
- LDN
Low-density neutrophil
- LPS
Lipopolysaccharide
- LTB4
Leukotriene B4
- METTL1
tRNA (guanine-N(7)-)-methyltransferase
- MHC
Major histocompatibility complex
- MMP8
Matrix metalloproteinase 8
- MMP9
Matrix metalloproteinase 9
- NASH
Non-alcoholic steatohepatitis
- NDN
Normal-density neutrophil
- NE
Neutrophil elastase
- NeP
Neutrophil progenitor
- NET
Neutrophil extracellular trap
- NK
Natural killer
- NO
Nitric oxide
- NSCLC
Non-small cell lung cancer
- NSG
NOD scid γ
- PDAC
Pancreatic ductal adenocarcinoma
- PD-1
Programmed cell death protein 1
- PD-L1
Programmed death-ligand 1
- PGE2
Prostaglandin E2
- PSGL-1
P-selectin glycoprotein ligand-1
- PTGS2
Prostaglandin-endoperoxide synthase 2
- ROS
Reactive oxigen species
- scRNAseq
Single-cell RNA sequencing
- SIRPα
Signal regulatory protein α
- SPP1
Osteopontin 1
- S100A12
S100 calcium-binding protein A12
- TAM
Tumour-associated macrophage
- TAN
tumour-associated neutrophil
- TCR
T cell receptor
- TME
Tumour microenvironment
- TNF
Tumour necrosis factor
- TNFR1
Tumour necrosis factor receptor 1
- TXNIP
Thioredoxin interacting protein
- VEGF
Vascular endothelial growth factor
- VEGFR2
Vascular endothelial growth factor receptor 2
- VISTA
V-type immunoglobulin domain-containing suppressor of T-cell activation
- XCR1
X-C motif chemokine receptor 1
Competing Interests
J.L. is a shareholder in FibroFind Limited.
Funding
J.L. is supported by a JGW Patterson Foundation grant and CRUK program grants [grant numbers C18342/A23390 and DRCRPG-Nov22/100007] and an MRC program grant [grant number MR/R023026/1]. E.R.G. is funded by the W.E. Harker Foundation. D.G. is supported by the Newcastle CRUK Clinical Academic Training Programme.
Open Access
Open access for this article was enabled by the participation of Newcastle University in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with JISC.
Author Contribution
E.R.G and J.L. contributed to all aspects of the article. D.G. contributed significantly to the writing and review or editing of the manuscript before submission.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A.et al. (2021) Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71, 209–249 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Rumgay H., Arnold M., Ferlay J., Lesi O., Cabasag C.J., Vignat J.et al. (2022) Global burden of primary liver cancer in 2020 and predictions to 2040. J. Hepatol. 77, 1598–1606 10.1016/j.jhep.2022.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finn R.S., Qin S., Ikeda M., Galle P.R., Ducreux M., Kim T.Y.et al. (2020) Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 382, 1894–1905 10.1056/NEJMoa1915745 [DOI] [PubMed] [Google Scholar]
- 4.Cheng A.L., Qin S., Ikeda M., Galle P.R., Ducreux M., Kim T.Y.et al. (2022) Updated efficacy and safety data from IMbrave150: Atezolizumab plus bevacizumab vs. sorafenib for unresectable hepatocellular carcinoma. J. Hepatol. 76, 862–873 10.1016/j.jhep.2021.11.030 [DOI] [PubMed] [Google Scholar]
- 5.Llovet J.M., Castet F., Heikenwalder M., Maini M.K., Mazzaferro V., Pinato D.J.et al. (2022) Immunotherapies for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 19, 151–172 10.1038/s41571-021-00573-2 [DOI] [PubMed] [Google Scholar]
- 6.Borregaard N. (2010) Neutrophils, from marrow to microbes. Immunity 33, 657–670 10.1016/j.immuni.2010.11.011 [DOI] [PubMed] [Google Scholar]
- 7.Geh D., Leslie J., Rumney R., Reeves H.L., Bird T.G. and Mann D.A. (2022) Neutrophils as potential therapeutic targets in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 19, 257–273 10.1038/s41575-021-00568-5 [DOI] [PubMed] [Google Scholar]
- 8.Jaillon S., Ponzetta A., Di Mitri D., Santoni A., Bonecchi R. and Mantovani A. (2020) Neutrophil diversity and plasticity in tumour progression and therapy. Nat. Rev. Cancer 20, 485–503 10.1038/s41568-020-0281-y [DOI] [PubMed] [Google Scholar]
- 9.Hedrick C.C. and Malanchi I. (2022) Neutrophils in cancer: heterogeneous and multifaceted. Nat. Rev. Immunol. 22, 173–187 10.1038/s41577-021-00571-6 [DOI] [PubMed] [Google Scholar]
- 10.Gordon S. and Taylor P.R. (2005) Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5, 953–964 10.1038/nri1733 [DOI] [PubMed] [Google Scholar]
- 11.Ballesteros I., Rubio-Ponce A., Genua M., Lusito E., Kwok I., Fernández-Calvo G.et al. (2020) Co-option of neutrophil fates by tissue environments. Cell 183, 1282.e1218–1297.e1218 10.1016/j.cell.2020.10.003 [DOI] [PubMed] [Google Scholar]
- 12.Casanova-Acebes M., Pitaval C., Weiss L.A., Nombela-Arrieta C., Chèvre R., N A.G.et al. (2013) Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell 153, 1025–1035 10.1016/j.cell.2013.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adrover J.M., Del Fresno C., Crainiciuc G., Cuartero M.I., Casanova-Acebes M., Weiss L.A.et al. (2019) A neutrophil timer coordinates immune defense and vascular protection. Immunity 50, 390.e310–402.e310 10.1016/j.immuni.2019.01.002 [DOI] [PubMed] [Google Scholar]
- 14.Quail D.F., Amulic B., Aziz M., Barnes B.J., Eruslanov E., Fridlender Z.G.et al. (2022) Neutrophil phenotypes and functions in cancer: A consensus statement. J. Exp. Med. 219, e20220011 10.1084/jem.20220011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muench D.E., Olsson A., Ferchen K., Pham G., Serafin R.A., Chutipongtanate S.et al. (2020) Mouse models of neutropenia reveal progenitor-stage-specific defects. Nature 582, 109–114 10.1038/s41586-020-2227-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwok I., Becht E., Xia Y., Ng M., Teh Y.C., Tan L.et al. (2020) Combinatorial single-cell analyses of granulocyte-monocyte progenitor heterogeneity reveals an early uni-potent neutrophil progenitor. Immunity 53, 303.e305–318.e305 10.1016/j.immuni.2020.06.005 [DOI] [PubMed] [Google Scholar]
- 17.Xie X., Shi Q., Wu P., Zhang X., Kambara H., Su J.et al. (2020) Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat. Immunol. 21, 1119–1133 10.1038/s41590-020-0736-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grieshaber-Bouyer R., Radtke F.A., Cunin P., Stifano G., Levescot A., Vijaykumar B.et al. (2021) The neutrotime transcriptional signature defines a single continuum of neutrophils across biological compartments. Nat. Commun. 12, 2856 10.1038/s41467-021-22973-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ocana A., Nieto-Jiménez C., Pandiella A. and Templeton A.J. (2017) Neutrophils in cancer: prognostic role and therapeutic strategies. Mol. Cancer 16, 137 10.1186/s12943-017-0707-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillay J., den Braber I., Vrisekoop N., Kwast L.M., de Boer R.J., Borghans J.A.et al. (2010) In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood 116, 625–627 10.1182/blood-2010-01-259028 [DOI] [PubMed] [Google Scholar]
- 21.Sagiv J.Y., Michaeli J., Assi S., Mishalian I., Kisos H., Levy L.et al. (2015) Phenotypic diversity and plasticity in circulating neutrophil subpopulations in cancer. Cell Reports 10, 562–573 10.1016/j.celrep.2014.12.039 [DOI] [PubMed] [Google Scholar]
- 22.Ramachandran P., Dobie R., Wilson-Kanamori J.R., Dora E.F., Henderson B.E.P., Luu N.T.et al. (2019) Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 10.1038/s41586-019-1631-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bischoff P., Trinks A., Obermayer B., Pett J.P., Wiederspahn J., Uhlitz F.et al. (2021) Single-cell RNA sequencing reveals distinct tumor microenvironmental patterns in lung adenocarcinoma. Oncogene 40, 6748–6758 10.1038/s41388-021-02054-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Narayanan S.P., Mannan R., Raskind G., Wang X., Vats P.et al. (2021) Single-cell analyses of renal cell cancers reveal insights into tumor microenvironment, cell of origin, and therapy response. Proc. Natl. Acad. Sci. U.S.A. 118, e2103240118 10.1073/pnas.2103240118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen K., Wang Q., Li M., Guo H., Liu W., Wang F.et al. (2021) Single-cell RNA-seq reveals dynamic change in tumor microenvironment during pancreatic ductal adenocarcinoma malignant progression. EBioMedicine 66, 103315 10.1016/j.ebiom.2021.103315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G., Qiu M., Xing X., Zhou J., Yao H., Li M.et al. (2022) Lung cancer scRNA-seq and lipidomics reveal aberrant lipid metabolism for early-stage diagnosis. Sci. Transl. Med. 14, eabk2756 10.1126/scitranslmed.abk2756 [DOI] [PubMed] [Google Scholar]
- 27.Xue R., Zhang Q., Cao Q., Kong R., Xiang X., Liu H.et al. (2022) Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature 612, 141–147 10.1038/s41586-022-05400-x [DOI] [PubMed] [Google Scholar]
- 28.Nie P., Zhang W., Meng Y., Lin M., Guo F., Zhang H.et al. (2022) A YAP/TAZ-CD54 axis is required for CXCR2-CD44- tumor-specific neutrophils to suppress gastric cancer. Protein Cell 14, 513–531 10.1093/procel/pwac045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L., Liu Y., Dai Y., Tang X., Yin T., Wang C.et al. (2022) Single-cell RNA-seq analysis reveals BHLHE40-driven pro-tumour neutrophils with hyperactivated glycolysis in pancreatic tumour microenvironment. Gut 72, 958–971 10.1136/gutjnl-2021-326070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salcher S., Sturm G., Horvath L., Untergasser G., Kuempers C., Fotakis G.et al. (2022) High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell. 40, 1503.e1508–1520.e1508 10.1016/j.ccell.2022.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R.et al. (2019) Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 179, 829.e820–845.e820 10.1016/j.cell.2019.10.003 [DOI] [PubMed] [Google Scholar]
- 32.Sharma A., Seow J.J.W., Dutertre C.A., Pai R., Blériot C., Mishra A.et al. (2020) Onco-fetal Reprogramming of Endothelial Cells Drives Immunosuppressive Macrophages in Hepatocellular Carcinoma. Cell 183, 377.e321–394.e321 10.1016/j.cell.2020.08.040 [DOI] [PubMed] [Google Scholar]
- 33.Ma L., Wang L., Khatib S.A., Chang C.W., Heinrich S., Dominguez D.A.et al. (2021) Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J. Hepatol. 75, 1397–1408 10.1016/j.jhep.2021.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma L., Hernandez M.O., Zhao Y., Mehta M., Tran B., Kelly M.et al. (2019) Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell. 36, 418.e416–430.e416 10.1016/j.ccell.2019.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y., Wu L., Zhong Y., Zhou K., Hou Y., Wang Z.et al. (2021) Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell 184, 404.e416–421.e416 10.1016/j.cell.2020.11.041 [DOI] [PubMed] [Google Scholar]
- 36.Zheng C., Zheng L., Yoo J.K., Guo H., Zhang Y., Guo X.et al. (2017) Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169, 1342.e1316–1356.e1316 10.1016/j.cell.2017.05.035 [DOI] [PubMed] [Google Scholar]
- 37.Zhang M., Yang H., Wan L., Wang Z., Wang H., Ge C.et al. (2020) Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J. Hepatol. 73, 1118–1130 10.1016/j.jhep.2020.05.039 [DOI] [PubMed] [Google Scholar]
- 38.Aizarani N., Saviano A., Sagar X.X.X., Mailly L., Durand S., Herman J.S.et al. (2019) A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204 10.1038/s41586-019-1373-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leslie J., Mackey J.B.G., Jamieson T., Ramon-Gil E., Drake T.M., Fercoq F.et al. (2022) CXCR2 inhibition enables NASH-HCC immunotherapy. Gut 71, 2093–2106 10.1136/gutjnl-2021-326259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wrage M., Hagmann W., Kemming D., Uzunoglu F.G., Riethdorf S., Effenberger K.et al. (2015) Identification of HERC5 and its potential role in NSCLC progression. Int. J. Cancer 136, 2264–2272 10.1002/ijc.29298 [DOI] [PubMed] [Google Scholar]
- 41.Lewis M.W., Wisniewska K., King C.M., Li S., Coffey A., Kelly M.R.et al. (2022) Enhancer RNA transcription is essential for a novel CSF1 enhancer in triple-negative breast cancer. Cancers 14, 1852 10.3390/cancers14071852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheriyath V., Kaur J., Davenport A., Khalel A., Chowdhury N. and Gaddipati L. (2018) G1P3 (IFI6), a mitochondrial localised antiapoptotic protein, promotes metastatic potential of breast cancer cells through mtROS. Br. J. Cancer 119, 52–64 10.1038/s41416-018-0137-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z., Gu S., Lu T., Wu K., Li L., Dong C.et al. (2020) IFI6 depletion inhibits esophageal squamous cell carcinoma progression through reactive oxygen species accumulation via mitochondrial dysfunction and endoplasmic reticulum stress. J. Exp. Clin. Cancer Res. 39, 144 10.1186/s13046-020-01646-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McManus M., Kleinerman E., Yang Y., Livingston J.A., Mortus J., Rivera R.et al. (2017) Hes4: A potential prognostic biomarker for newly diagnosed patients with high-grade osteosarcoma. Pediatric Blood Cancer 64, 10.1002/pbc.26318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aljohani A.I., Joseph C., Kurozumi S., Mohammed O.J., Miligy I.M., Green A.R.et al. (2020) Myxovirus resistance 1 (MX1) is an independent predictor of poor outcome in invasive breast cancer. Breast Cancer Res. Treat. 181, 541–551 10.1007/s10549-020-05646-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gong W., Donnelly C.R., Heath B.R., Bellile E., Donnelly L.A., Taner H.F.et al. (2021) Cancer-specific type-I interferon receptor signaling promotes cancer stemness and effector CD8+ T-cell exhaustion. Oncoimmunology 10, 1997385 10.1080/2162402X.2021.1997385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li T.H., Zhao B.B., Qin C., Wang Y.Y., Li Z.R., Cao H.T.et al. (2021) IFIT1 modulates the proliferation, migration and invasion of pancreatic cancer cells via Wnt/β-catenin signaling. Cell. Oncol. (Dordrecht) 44, 1425–1437 10.1007/s13402-021-00651-8 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Pidugu V.K., Pidugu H.B., Wu M.M., Liu C.J. and Lee T.C. (2019) Emerging functions of human IFIT proteins in cancer. Front. Mol. Biosci. 6, 148 10.3389/fmolb.2019.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakamoto S., Inoue H., Kohda Y., Ohba S.I., Mizutani T. and Kawada M. (2020) Interferon-induced transmembrane protein 1 (IFITM1) promotes distant metastasis of small cell lung cancer. Int. J. Mol. Sci. 21, 4934 10.3390/ijms21144934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sari I.N., Yang Y.G., Phi L.T., Kim H., Baek M.J., Jeong D.et al. (2016) Interferon-induced transmembrane protein 1 (IFITM1) is required for the progression of colorectal cancer. Oncotarget 7, 86039–86050 10.18632/oncotarget.13325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su W., Xiao W., Chen L., Zhou Q., Zheng X., Ju J.et al. (2019) Decreased IFIT2 expression in human non-small-cell lung cancer tissues is associated with cancer progression and poor survival of the patients. OncoTargets Ther. 12, 8139–8149 10.2147/OTT.S220698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moorman H.R., Poschel D., Klement J.D., Lu C., Redd P.S. and Liu K. (2020) Osteopontin: a key regulator of tumor progression and immunomodulation. Cancers 12, 3379 10.3390/cancers12113379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y., Liu H., Zhao Y., Yue D., Chen C., Li C.et al. (2021) Tumor-associated macrophages (TAMs)-derived osteopontin (OPN) upregulates PD-L1 expression and predicts poor prognosis in non-small cell lung cancer (NSCLC). Thoracic Cancer 12, 2698–2709 10.1111/1759-7714.14108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kodama T., Koma Y.-i., Arai N., Kido A., Urakawa N., Nishio M.et al. (2020) CCL3–CCR5 axis contributes to progression of esophageal squamous cell carcinoma by promoting cell migration and invasion via Akt and ERK pathways. Lab. Invest. 100, 1140–1157 10.1038/s41374-020-0441-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L., Liu Y.D., Zhan Y.T., Zhu Y.H., Li Y., Xie D.et al. (2018) High levels of CCL2 or CCL4 in the tumor microenvironment predict unfavorable survival in lung adenocarcinoma. Thoracic Cancer 9, 775–784 10.1111/1759-7714.12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geng Y., Feng J., Huang H., Wang Y., Yi X., Wei S.et al. (2022) Single-cell transcriptome analysis of tumor immune microenvironment characteristics in colorectal cancer liver metastasis. Ann. Transl. Med. 10, 1170 10.21037/atm-22-5270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pantouris G., Syed M.A., Fan C., Rajasekaran D., Cho T.Y., Rosenberg E.M. Jret al. (2015) An analysis of MIF structural features that control functional activation of CD74. Chem. Biol. 22, 1197–1205 10.1016/j.chembiol.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foers A.D., Dagley L.F., Chatfield S., Webb A.I., Cheng L., Hill A.F.et al. (2020) Proteomic analysis of extracellular vesicles reveals an immunogenic cargo in rheumatoid arthritis synovial fluid. Clin. Transl. Immunol. 9, e1185 10.1002/cti2.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt M., Edlund K., Hengstler J.G., Heimes A.-S., Almstedt K., Lebrecht A.et al. (2021) Prognostic impact of immunoglobulin Kappa C (IGKC) in early breast cancer. Cancers 13, 3626 10.3390/cancers13143626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt M., Micke P., Gehrmann M. and Hengstler J.G. (2012) Immunoglobulin kappa chain as an immunologic biomarker of prognosis and chemotherapy response in solid tumors. Oncoimmunology 1, 1156–1158 10.4161/onci.21653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schindler L., Zwissler L., Krammer C., Hendgen-Cotta U., Rassaf T., Hampton M.B.et al. (2021) Macrophage migration inhibitory factor inhibits neutrophil apoptosis by inducing cytokine release from mononuclear cells. J. Leukoc. Biol. 110, 893–905 10.1002/JLB.3A0420-242RRR [DOI] [PubMed] [Google Scholar]
- 62.Fukuda Y., Bustos M.A., Cho S.-N., Roszik J., Ryu S., Lopez V.M.et al. (2022) Interplay between soluble CD74 and macrophage-migration inhibitory factor drives tumor growth and influences patient survival in melanoma. Cell Death Dis. 13, 117 10.1038/s41419-022-04552-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singhal S., Bhojnagarwala P.S., O'Brien S., Moon E.K., Garfall A.L., Rao A.S.et al. (2016) Origin and Role of a Subset of Tumor-Associated Neutrophils with Antigen-Presenting Cell Features in Early-Stage Human Lung Cancer. Cancer Cell. 30, 120–135 10.1016/j.ccell.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eruslanov E.B., Bhojnagarwala P.S., Quatromoni J.G., Stephen T.L., Ranganathan A., Deshpande C.et al. (2014) Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J. Clin. Invest. 124, 5466–5480 10.1172/JCI77053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Németh T., Sperandio M. and Mócsai A. (2020) Neutrophils as emerging therapeutic targets. Nat. Rev. Drug Discovery 19, 253–275 10.1038/s41573-019-0054-z [DOI] [PubMed] [Google Scholar]
- 66.Rees D.J.v., Bouti P., Klein B., Verkuijlen P.J.H., Houdt M.v., Schornagel K.et al. (2022) Cancer cells resist antibody-mediated destruction by neutrophils through activation of the exocyst complex. J. Immunotherapy Cancer 10, e004820 10.1136/jitc-2022-004820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Behrens L.M., van Egmond M. and van den Berg T.K. (2023) Neutrophils as immune effector cells in antibody therapy in cancer. Immunol. Rev. 314, 280–301 10.1111/imr.13159 [DOI] [PubMed] [Google Scholar]
- 68.Gruijs M., Sewnath C.A.N. and van Egmond M. (2021) Therapeutic exploitation of neutrophils to fight cancer. Semin. Immunol. 57, 101581 10.1016/j.smim.2021.101581 [DOI] [PubMed] [Google Scholar]
- 69.Maras J.S., Das S., Bhat A., Kumar Vyas A., Yadav G., Chaudhary S.et al. (2019) Dysregulated Lipid Transport Proteins Correlate With Pathogenesis and Outcome in Severe Alcoholic Hepatitis. Hepatol. Commun. 3, 1598–1625 10.1002/hep4.1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie H., Wei L., Liu M., Liang Y., Yuan G., Gao S.et al. (2022) Neutrophil-albumin ratio as a biomarker for postoperative complications and long-term prognosis in patients with colorectal cancer undergoing surgical treatment. Front. Nutrition 9, 976216 10.3389/fnut.2022.976216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mahfouz M.H., Assiri A.M. and Mukhtar M.H. (2016) Assessment of Neutrophil Gelatinase-Associated Lipocalin (NGAL) and Retinol-Binding Protein 4 (RBP4) in Type 2 Diabetic Patients with Nephropathy. Biomarker Insights 11, 31–40 10.4137/BMI.S33191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li M., Wang Z., Zhu L., Shui Y., Zhang S. and Guo W. (2021) Down-regulation of RBP4 indicates a poor prognosis and correlates with immune cell infiltration in hepatocellular carcinoma. Biosci. Rep. 41, BSR20210328 10.1042/BSR20210328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Georgila K., Vyrla D. and Drakos E. (2019) Apolipoprotein A-I (ApoA-I), immunity, inflammation and cancer. Cancers 11, 1097 10.3390/cancers11081097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ai J., Tan Y., Ying W., Hong Y., Liu S., Wu M.et al. (2006) Proteome analysis of hepatocellular carcinoma by laser capture microdissection. Proteomics 6, 538–546 10.1002/pmic.200500257 [DOI] [PubMed] [Google Scholar]
- 75.Hu B., Lin J.Z., Yang X.B. and Sang X.T. (2020) Aberrant lipid metabolism in hepatocellular carcinoma cells as well as immune microenvironment: A review. Cell Prolif. 53, e12772 10.1111/cpr.12772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson C.L., Jurk D., Fullard N., Banks P., Page A., Luli S.et al. (2015) NFκB1 is a suppressor of neutrophil-driven hepatocellular carcinoma. Nat. Commun. 6, 6818 10.1038/ncomms7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steele C.W., Karim S.A., Leach J.D.G., Bailey P., Upstill-Goddard R., Rishi L.et al. (2016) CXCR2 inhibition profoundly suppresses metastases and augments immunotherapy in pancreatic ductal adenocarcinoma. Cancer Cell. 29, 832–845 10.1016/j.ccell.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng Y., Mo F., Li Q., Han X., Shi H., Chen S.et al. (2021) Targeting CXCR2 inhibits the progression of lung cancer and promotes therapeutic effect of cisplatin. Mol. Cancer 20, 62 10.1186/s12943-021-01355-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y., He Y., Butler W., Xu L., Chang Y., Lei K.et al. (2019) Targeting cellular heterogeneity with CXCR2 blockade for the treatment of therapy-resistant prostate cancer. Sci. Transl. Med. 11, eaax0428 10.1126/scitranslmed.aax0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang G., Lu X., Dey P., Deng P., Wu C.C., Jiang S.et al. (2016) Targeting YAP-dependent MDSC infiltration impairs tumor progression. Cancer Discovery 6, 80–95 10.1158/2159-8290.CD-15-0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ghallab A.M., Eissa R.A. and El Tayebi H.M. (2022) CXCR2 small-molecule antagonist combats chemoresistance and enhances immunotherapy in triple-negative breast cancer. Front. Pharmacol. 13, 862125 10.3389/fphar.2022.862125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang H., Ye Y.L., Li M.X., Ye S.B., Huang W.R., Cai T.T.et al. (2017) CXCL2/MIF-CXCR2 signaling promotes the recruitment of myeloid-derived suppressor cells and is correlated with prognosis in bladder cancer. Oncogene 36, 2095–2104 10.1038/onc.2016.367 [DOI] [PubMed] [Google Scholar]
- 83.Liu X., Lan T., Mo F., Yang J., Wei Y. and Wei X. (2021) Antitumor and radiosensitization effects of a CXCR2 inhibitor in nasopharyngeal carcinoma. Front. Cell Developmental Biol. 9, 689613 10.3389/fcell.2021.689613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Henriques T.B., Dos Santos D.Z., Dos Santos Guimarães I., Tessarollo N.G., Lyra P.C.M. Jr, Mesquita P.et al. (2021) Inhibition of CXCR2 plays a pivotal role in re-sensitizing ovarian cancer to cisplatin treatment. Aging 13, 13405–13420 10.18632/aging.203074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Devapatla B., Sharma A. and Woo S. (2015) CXCR2 inhibition combined with sorafenib improved antitumor and antiangiogenic response in preclinical models of ovarian cancer. PLoS ONE 10, e0139237 10.1371/journal.pone.0139237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dai Z., Liu H., Liao J., Huang C., Ren X., Zhu W.et al. (2021) N7-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol. Cell. 81, 3339.e3338–3355.e3338 10.1016/j.molcel.2021.07.003 [DOI] [PubMed] [Google Scholar]
- 87.Liu H., Zeng X., Ren X., Zhang Y., Huang M., Tan L.et al. (2022) Targeting tumour-intrinsic N(7)-methylguanosine tRNA modification inhibits MDSC recruitment and improves anti-PD-1 efficacy. Gut 72, 1555–1567 10.1136/gutjnl-2022-327230 [DOI] [PubMed] [Google Scholar]
- 88.Pfister D., Núñez N.G., Pinyol R., Govaere O., Pinter M., Szydlowska M.et al. (2021) NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature 592, 450–456 10.1038/s41586-021-03362-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Evans T.R.J., Basu B., Hubner R., Ma Y.T., Meyer T., Palmer D.H.et al. (2023) A phase I/II study of the CXCR2 inhibitor, AZD5069, in combination with durvalumab, in patients (pts) with advanced hepatocellular carcinoma (HCC). J. Clin. Oncol. 41, TPS631–TPS631 10.1200/JCO.2023.41.4_suppl.TPS631 [DOI] [Google Scholar]
- 90.Esteban-Fabró R., Willoughby C.E., Piqué-Gili M., Montironi C., Abril-Fornaguera J., Peix J.et al. (2022) Cabozantinib enhances anti-PD1 activity and elicits a neutrophil-based immune response in hepatocellular carcinoma. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 28, 2449–2460 10.1158/1078-0432.CCR-21-2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Linde I.L., Prestwood T.R., Qiu J., Pilarowski G., Linde M.H., Zhang X.et al. (2023) Neutrophil-activating therapy for the treatment of cancer. Cancer Cell. 41, 356–372.e10 10.1016/j.ccell.2023.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sterner R.C. and Sterner R.M. (2021) CAR-T cell therapy: current limitations and potential strategies. Blood Cancer J. 11, 69 10.1038/s41408-021-00459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zannikou M., Duffy J.T., Levine R.N., Seblani M., Liu Q., Presser A.et al. (2023) IL15 modification enables CAR T cells to act as a dual targeting agent against tumor cells and myeloid-derived suppressor cells in GBM. J. Immunotherapy Cancer 11, e006239 10.1136/jitc-2022-006239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li D., Li N., Zhang Y.F., Fu H., Feng M., Schneider D.et al. (2020) Persistent polyfunctional chimeric antigen receptor T cells that target glypican 3 eliminate orthotopic hepatocellular carcinomas in mice. Gastroenterology 158, 2250.e2220–2265.e2220 10.1053/j.gastro.2020.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. NCT05003895 . (2021) ClinicalTrials.gov [Google Scholar]
- 96.Chang Y., Syahirah R., Wang X., Jin G., Torregrosa-Allen S., Elzey B.D.et al. (2022) Engineering chimeric antigen receptor neutrophils from human pluripotent stem cells for targeted cancer immunotherapy. Cell Rep. 40, 111128 10.1016/j.celrep.2022.111128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khoyratty T.E., Ai Z., Ballesteros I., Eames H.L., Mathie S., Martín-Salamanca S.et al. (2021) Distinct transcription factor networks control neutrophil-driven inflammation. Nat. Immunol. 22, 1093–1106 10.1038/s41590-021-00968-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang G.G., Calvo K.R., Pasillas M.P., Sykes D.B., Häcker H. and Kamps M.P. (2006) Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat. Methods 3, 287–293 10.1038/nmeth865 [DOI] [PubMed] [Google Scholar]
- 99.Cohen J.T., Danise M., Hinman K.D., Neumann B.M., Johnson R., Wilson Z.S.et al. (2022) Engraftment, fate, and function of HoxB8-conditional neutrophil progenitors in the unconditioned murine host. Front. Cell Developmental Biol. 10, 840894 10.3389/fcell.2022.840894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gea-Banacloche J. (2017) Granulocyte transfusions: A concise review for practitioners. Cytotherapy 19, 1256–1269 10.1016/j.jcyt.2017.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li X. and Wang C.Y. (2021) From bulk, single-cell to spatial RNA sequencing. Int. J. Oral Sci. 13, 36 10.1038/s41368-021-00146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]