Abstract
The DNA sensor cGAS (cyclic GMP-AMP synthase) and its adaptor protein STING (Stimulator of Interferon Genes) detect the presence of cytosolic DNA as a sign of infection or damage. In cancer cells, this pathway can be activated through persistent DNA damage and chromosomal instability, which results in the formation of micronuclei and the exposure of DNA fragments to the cytosol. DNA damage from radio- or chemotherapy can further activate DNA sensing responses, which may occur in the cancer cells themselves or in stromal and immune cells in the tumour microenvironment (TME). cGAS–STING signalling results in the production of type I interferons, which have been linked to immune cell infiltration in ‘hot’ tumours that are susceptible to immunosurveillance and immunotherapy approaches. However, recent research has highlighted the complex nature of STING signalling, with tumours having developed mechanisms to evade and hijack this signalling pathway for their own benefit. In this mini-review we will explore how cGAS–STING signalling in different cells in the TME can promote both anti-tumour and pro-tumour responses. This includes the role of type I interferons and the second messenger cGAMP in the TME, and the influence of STING signalling on local immune cell populations. We examine how alternative signalling cascades downstream of STING can promote chronic interferon signalling, the activation of the transcription factor nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and the production of inflammatory cytokines, which can have pro-tumour functions. An in-depth understanding of DNA sensing in different cell contexts will be required to harness the anti-tumour functions of STING signalling.
Keywords: cancer, cGAS, DNA sensing, immunology, innate immunity, STING
Introduction: DNA sensing in cancer
Our immune system has the capacity to detect and eliminate cancer cells, despite the fact that they are derived from our own body. This is partly because cancerous and pre-cancerous cells – particularly those with a high mutational burden – express proteins that can be detected as neo-antigens by the cells of our adaptive immune system [1,2]. However, a pre-requisite for effective immune activation is a favourable tumour microenvironment (TME), which relies on the innate immune mechanisms that result in the secretion of interferons, cytokines and chemokines to promote the infiltration and activation of immune cells with anti-cancer properties. One innate immune signalling axis, which can shape the immunological properties of the TME is the signalling pathway involving the adaptor protein STING (Stimulator of Interferon Genes) which is activated by the DNA sensor cGAS (cyclic GMP-AMP synthase) after the detection of cytosolic DNA or DNA damage [3]. Immune cells within the TME, stromal cells and cancer cells themselves can contribute to the DNA sensing response in cancer. Emerging evidence highlights a multi-faceted role of STING signalling in cancer, which can lead to both anti-tumour and pro-tumour signalling outcomes. This review will provide an overview of the recent advances in our understanding of how STING signalling influences anti-tumour immune responses, how this signalling can be evaded by tumours and how it can be subverted in some cancers to aid tumour progression.
Sources of cytosolic DNA in cancer
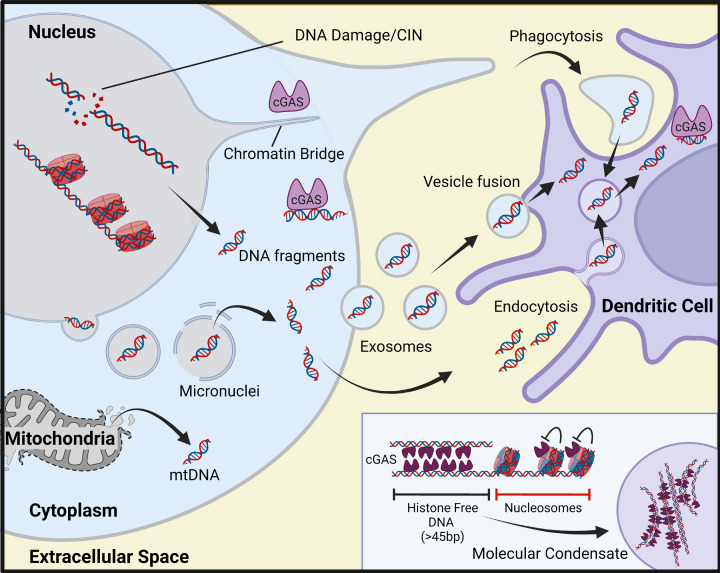
Cytosolic DNA is detected as pathogen-associated molecular pattern (PAMP), during viral infection for instance, but it can also serve as danger- or damage-associated molecular pattern (DAMP) when the cell’s own nuclear or mitochondrial DNA is damaged and gains access to the cytosol. Tumour cells often contain cytosolic DNA, due to persistent DNA damage and chromosomal instability (CIN), which is a hallmark of cancer [4,5], and additional DNA damage is induced by radiotherapy and chemotherapy with DNA damaging agents. The presence of CIN and DNA breaks is associated with the formation of micronuclei – membrane bound structures that form around damaged DNA and lagging/mis-segregated chromosomes during mitotic exit [6]. Upon rupture of their unstable membrane micronuclei expose their DNA content to the cytosol [7–11]. DNA damage and CIN may also lead to the direct leakage of DNA fragments out of the nucleus and the formation of chromatin bridges which can also be detected by cytosolic DNA sensors [12,13] (Figure 1). Furthermore, leakage of DNA from damaged mitochondria can also contribute to the innate immune activation [14].
Figure 1. Sources of immunostimulatory DNA in tumour cells and the tumour microenvironment.
The presence of self-DNA within the cytosol can occur due to chromosomal instability in cancer cells as well as from additional DNA damage caused by cancer therapies. Micronuclei are a source of self-DNA within the cytosol often naturally arising in metastatic and genomically unstable cancer cells as well as from exposure to ionising radiation. Chromatin bridges contain DNA connecting two daughter cells. Mitochondrial (mt) DNA is another potential source of immunostimulatory DNA in the cytosol which arises from mitochondrial damage. cGAS can only bind stretches of unmodified double-stranded DNA greater than 45 bp, its activation is inhibited by nucleosomes. cGAS molecules assemble on histone free DNA forming a ladder-like structure and can form higher order structures which agglomerate into molecular condensates which enhance the production of cGAMP. Nearby immune cells can take up cGAMP or DNA released into the TME by dead or dying tumour cells. Tumour cells can also export DNA into the TME in membrane bound vesicles such as exosomes which are taken up by local immune cells to activate STING signalling.
DNA sensing by cGAS and STING
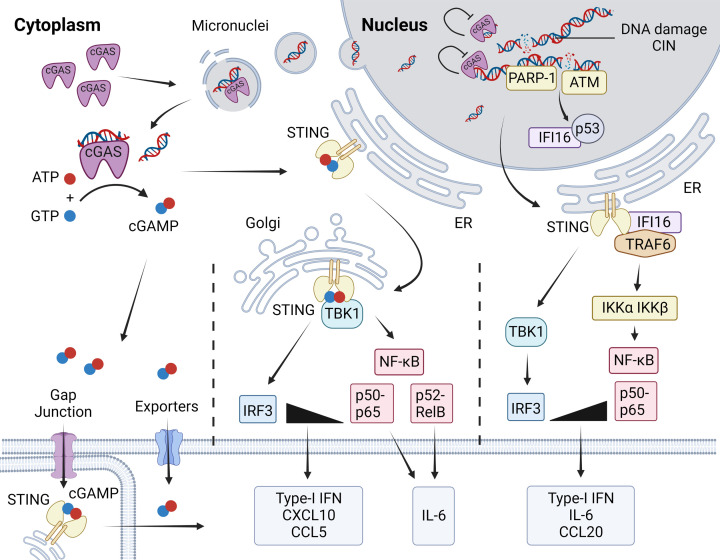
The DNA sensor cGAS and its adaptor protein STING constitute the key signalling axis for sensing dsDNA within the cytosol of many different cell types (Figure 2) [15–19]. cGAS binds double-stranded (ds) DNA in a sequence-independent manner, and cGAS dimers form ladder-like assemblies with dsDNA in higher order signalling domains in the cytosol [20–24]. While cGAS is also present in the nucleus of many cells, its catalytic activity is inhibited by interaction with nucleosomes on chromatin (Figure 1) [25–29]. Independent of its catalytic activity cGAS has also been shown to interact with DNA replication forks, regulate genomic stability as well as inhibit homologous recombination [30–32]. Upon detection of dsDNA in the cytosol, cGAS catalyses the synthesis of the second messenger 2′3′-cyclic GMP-AMP (cGAMP), which then binds STING, a membrane protein residing at the endoplasmic reticulum (ER) [16–18,33–35]. Activated STING dimers translocate through the ER–Golgi intermediate compartments (ERGIC) to the Golgi apparatus [36–39]. STING forms a complex with TANK-binding kinase 1 (TBK1) which phosphorylates STING and the transcription factor interferon regulatory factor (IRF3) as well as activating nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) (Figure 2) [15,39–41]. These transcription factors go on to promote the expression of type I interferons (IFN-I), cytokines and chemokines including CXCL10 and CCL5. STING activation can also induce autophagy, senescence and cell death, depending on signal strength and cell context [42–44]. STING-mediated activation of inflammasome complexes has also been described [45], but other DNA sensors such as AIM2 (Absent in Melanoma 2) may also play an important role in inflammasome activation, depending on cell context [46,47].
Figure 2. Innate immune signalling in response to cytosolic DNA and DNA damage.
On detecting cytosolic DNA, cGAS synthesises the second messenger cGAMP which can be exported to adjacent cells or the extracellular environment using gap junctions and exporters. Within the cell, cGAMP can go on to bind STING anchored to the endoplasmic reticulum (ER). STING is subject to numerous posttranslational modifications, then transported from the ER to the Golgi where it forms a complex with TBK1 and is phosphorylated by TBK1 at Serine 366. This serves to recruit the transcription factor IRF3 to the complex, which is also phosphorylated by TBK1. IRF3 and NF-κB p65 then promote the production of IFN-Is and chemokines including CXCL10 and CCL5. An alternative non-canonical STING signalling pathway has been identified in human epithelial cells. This pathway does not require the activity of cGAS, but the DNA damage sensing proteins PARP-1 and ATM, as well as the DNA binding protein IFI16 which shuttles between the nucleus and cytosol. cGAS-independent DNA damage sensing favours the activation of NF-κB over IRF3, generating a more pro-inflammatory cytokine profile that is distinct from canonical STING signalling. In cancer cells, canonical STING activation can be re-wired to induce the activation of NF-κB p52, which also induces a pro-inflammatory cytokine profile that may aid tumour progression.
cGAS-STING signalling is tightly regulated by post-translational modifications, including ubiquitylation with K48-, K63-, K27- and K11-linked ubiquitin chains, modification with ubiquitin-like proteins as well modification with metabolites and lipids [45,47]. Additional DNA binding proteins have also been shown to synergise with cGAS in the activation of STING. This includes Interferon-γ-Inducible Protein 16 (IFI16), the helicase DDX41, the DNA damage response kinase DNA-PK and the Z DNA-binding protein ZBP1 [51–57]. It remains to be elucidated whether additional DNA binding proteins contribute specificity to the sensing of DNA ligands, or whether they represent additional fail-safe mechanisms for the full activation of DNA sensing responses only when appropriate. Antagonistic and synergistic cross-talk between DNA sensing pathways such as cGAS-STING signalling and inflammasome activation has been reported [58–60], which further influences downstream signalling outcomes.
STING can also be activated independently of cGAS, such as following membrane fusion or after uptake of cGAMP from neighbouring cells [61,58]. Bacterial cyclic di-nucleotides (CDNs) can also activate STING directly in response to infection [59]. While cGAMP sensing by STING bypasses the DNA sensing role of cGAS in bystander cells, a non-canonical involvement of cGAS in the detection of extracellular cGAMP has also been described [62]. cGAS-independent STING activation following the detection of nuclear DNA damage has also been described in human epithelial cells, which may be of particular relevance to cancer. This alternative STING activation pathway involves the DNA binding protein IFI16, as well as the DNA damage sensing factors ATM, PARP1 and p53 [60]. The cGAS-independent activation of STING links the nuclear DNA damage response to innate immune activation prior to the formation of micronuclei and results in the expression of a different – more pro-inflammatory – set of cytokines and chemokines [60].
Anti-tumour functions of STING signalling
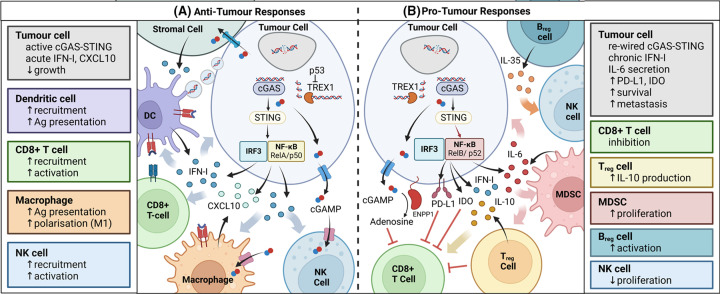
cGAS–STING signalling has been shown to have potent anti-cancer activities in a variety of mouse tumour models (Figure 3A). The production of STING-induced IFN-Is and chemokines by immune cells in the TME and by the cancer cells themselves facilitates further immune cell infiltration and the promotion of adaptive anti-cancer immune responses [9,11,63–66]. Furthermore the induction of cGAS–STING signalling in cancer treatments such as radiotherapy, is also thought to enable the generation of spontaneous adaptive anti-tumour immunity [66–70]. This has led to a significant interest in harnessing cGAS–STING signalling in cancer therapy and the development of STING agonists [63,65,66,71–76].
Figure 3. Pro-tumour and anti-tumour responses of STING signalling in cancer.
(A) Anti-tumour responses. The cGAS-STING signalling axis with IFN-I and CXCL10 secretion as signalling output co-ordinates multiple anti-tumour functions. The tumour suppressor p53 negatively regulates TREX1, making cytosolic DNA available for detection by cGAS. Tumour DNA released into the TME can be taken up by DCs promoting their activation. cGAMP can also be transported via gap junctions directly to adjacent cells or into the TME where it is taken up by local immune cells including NK cells and macrophages. The uptake of cGAMP by the importers of these cells can trigger their own STING signalling cascades and the production of IFN-Is. IFN-I responses enhance the activity of cytotoxic T-cells, DCs and NK cells as well as promote M2 to M1 macrophage polarisation. The activation of DCs is particularly important for generating adaptive CD8+ T-cell responses. (B) Pro-tumour responses. Chronic IFN-β signalling can promote tumour growth and immunosuppression. This signalling can trigger the expression of a subset of ISGs known as the IFN-related DNA damage resistance signature (IRDS) with potentially pro-tumour functions. IFN-I and IFN-II signalling has been linked to the up-regulation of PD-L1/2 and IDO which have immunosuppressive properties. STING signalling can also be subverted in some cancers to promote the activation of the non-canonical family of NF-κB transcription factors (p52/RelB) which result in the expression of IL-6. IL-6 is important for cancer cell survival and metastasis, and a pro-inflammatory TME can suppress anti-tumour immune responses through the recruitment of MDSC and regulatory T cells. cGAMP can be exported into the TME where it can be broken down and converted to immunosuppressive adenosine by ENPP1. cGAMP can trigger apoptosis and impair proliferation in T-cells which express high levels of STING. STING signalling in regulatory B cells can also impair NK cell function via IL-35 secretion.
STING signalling in cancer cells
The first potential point of activation of cGAS–STING pathway is within the cancer cells themselves following detection of cytosolic DNA from micronuclei for instance [7,8]. This may be a result of inherent CIN or further DNA damage from cancer therapy. Cancer cells can contribute to the anti-tumour responses generated against them through the secretion of IFN-Is and chemokines into the TME [9,11,63,77–79]. In that way, the STING signalling capacity of tumour cells themselves may shape their TME, with IFN-I signalling being associated with ‘hot’ tumours containing infiltrating DCs and T cells. Moreover, following cell-intrinsic DNA sensing, cancer cells can also export the second messenger cGAMP directly into neighbouring immune and stromal cells in the TME. Gap junctions between cells have been shown to facilitate the transfer of cGAMP between tumour and stromal cells to promote IFN-I responses in neighbouring cells [58,80,81]. cGAMP can also be secreted into the extracellular space. Several transporters, channels and pumps which shuttle cGAMP between the extracellular environment and the cytosol have recently been identified and may contribute to the propagation of STING signalling in the TME [62,82–87].
STING signalling in endothelial cells
cGAMP production and export by tumour cells can activated STING signalling in neighbouring endothelial cells, to induce further IFN-β and CXCL10 production in the TME. Activation of STING within the endothelium was also shown to be accompanied by increased expression of factors associated with T-cell adhesion to the endothelium and the infiltration of lymphocytes into the TME. The activation of STING in endothelial cells in the tumour vasculature is also thought to protect against angiogenesis and promote vascular normalisation in tumours [63–65,88].
STING signalling in immune cells
Immune cells in the TME can respond to extracellular tumour-derived DNA and activate the DNA sensing pathway. The uptake of tumour DNA has been shown to be particularly important for activating DCs in the TME [68,69,89]. How precisely the uptake of DNA from the TME in these instances occurs is uncertain. Extracellular tumour DNA may be available following cancer cell death, and uptake may be facilitated by association with chromatin modifying proteins such as HMGB1, or through fusion of DNA-containing extracellular vesicles [68,69,89–95]. Following the uptake of extracellular DNA in the cytosol of myeloid cells, cGAS is activated leading to IFN-I production. In mice CD11c+ DCs were found to be one of the main producers of STING-dependent IFN-Is in the TME [68–70]. STING-induced type I interferons (IFN-Is) can further stimulate tumour infiltrating dendritic cells (DCs) and promote the presentation of tumour antigens to CD8+ T cells [96,97]. Anti-tumour adaptive immune responses are lost in mouse hosts lacking the IFN-I receptor or STING in DCs, rendering them unable to reject implanted tumours [70,97,97]. Conversely the intra-tumoral injection of STING agonists leads to immune-mediated clearance of tumours in mice, showing that activation of STING signalling in both tumour cells and immune cells may contribute to tumour clearance [81,98,99]. STING signalling has also shown to play an important role in regulating the polarisation of macrophages to an M1 anti-tumour phenotype in some tumours [99,100]. The human importer SLC46A2 facilitates the uptake of cGAMP by human macrophages and monocytes where it activates STING signalling [84]. Similarly, STING-dependent IFN-β production has been shown to be important for promoting NK activity in mouse models of glioblastoma, lymphoma and melanoma tumour [101,102]. STING signalling is also thought to reduce numbers of myeloid-derived suppressor cells in the TME [103,104], although this may not be the case in all tumours [105,106]. The STING-induced production of chemokines such as CXCL9, CXCL10 and CCL5 promote further recruitment of immune cells into the TME, and high levels of STING expression in tumours has been linked to overall higher immune cell infiltration in cancers [107].
STING agonists as anti-tumour agents
Activation of STING signalling has been shown to be a feature of ‘hot’ tumours, which show infiltration of DCs and cytotoxic T cells – a pre-requisite for detection of tumour antigens by adaptive immune responses and for effective immunotherapy [108]. Thus, there has been considerable interest in utilising STING agonists as potential anti-cancer therapies. Numerous studies have investigated using STING agonists such as cGAMP, other cyclic di-nucleotides or small molecule activators of STING in mouse tumour models [71–76,99,109]. There is a large body of evidence that STING agonists have dramatic anti-tumour effects in these systems, enabling tumour clearance and often protecting against subsequent challenge with tumour cells. However, so far none of these successes have translated into an effective therapy in clinical trials [110]. While the lack of success with an early STING agonist, DMXAA, was later shown to be due to its specificity for murine STING [111,112], it is unclear what limits the efficacy of other STING agonists in patients. It remains to be investigated whether this is due to limitations in agonist activity and bio-availability (which could be overcome through advances in drug design and delivery) or due to more fundamental mechanistic limitations which would be more difficult to address. Underlying reasons may be mouse–human species differences in STING function or the dynamic re-wiring of STING signalling in human tumours.
Evasion of STING signalling during tumour progression
Due to of the anti-tumour effects of STING signalling, it was originally proposed that the pathway must be dysfunctional or epigenetically suppressed in cancer cells [113]. Although this has been demonstrated in several cancer cell lines, the genes for either cGAS or STING are rarely mutated in cancers [4,113–115]. Similarly, while STING and cGAS expression have been found to be epigenetically silenced in some tumours, many cancers show normal or elevated cGAS and STING expression [114–118]. The sensing of cytosolic DNA can also be attenuated through the over-expression of the cytosolic DNase TREX1 or extracellular DNases, and the inhibition of DNA uptake into DCs [79,94,118–120]. Analogously, cGAMP in the extracellular environment can be broken down by ectonucleotide pyrophosphate phosphodiesterase 1 (ENPP1) found on cell surfaces [83,121]. It has been shown that ENPP1 is increasingly up-regulated as cancer cells become metastatic, and this enhances cancer cell migration in a cGAS-dependent manner [122]. In addition to preventing immune cell activation by removal of extracellular cGAMP, the breakdown of cGAMP increases levels of adenosine which further inhibits anti-tumour immune responses [122,123]. This has generated interest in ENPP1 as therapeutic target in its own right [124].
Pro-tumour functions of STING signalling
In addition to its potent anti-tumour activities, STING signalling has also been shown to contribute to cancer cell survival and tumour progression in some contexts. The balance of downstream STING signalling outputs may be differentially regulated in tumour cells, so that anti-tumour IFN-I and cell death responses are limited, while pro-inflammatory and pro-survival programs are promoted.
While short-lived interferon responses due to acute STING stimulation have clear anti-tumour effects mediated by IFN-Is, it has been shown that low levels of chronic IFN-β signalling can promote cancer cell survival (Figure 3B) [125–128]. Mechanistically, prolonged low levels of IFN-β signalling cause the activation of an unphosphorylated ISGF3 (U-ISGF3) complex, which promotes the expression of a subset of interferon-stimulated genes with pro-tumour functions [127–129]. Chronic IFN-I signalling and low-grade inflammation can have additional effects on the immune composition of the TME, with the recruitment of myeloid-derived suppressor cells contributing to radioresistance and tumour progression [105]. In addition to this, both type-I and -II IFNs up-regulate the expression of programmed death ligand (PD-L1/2) in cancer cells [78,130]. PD-L1 can interfere with STING-mediated IFN production and IFN-mediated cytotoxicity in cancer cells alongside its ability to repress T cell responses [128,131]. The expression of the enzyme indoleamine-2,3-dioxygenase (IDO), which promotes an immune-suppressive TME has also been linked to STING signalling [106,132,133].
STING-dependent NF-κB activation in tumour cells
The STING-dependent expression of pro-inflammatory cytokines, and particularly IL-6, can also promote inflammation-driven tumour progression. STING-deficient mice have been found to be resistant to the development of skin tumours induced by the mutagen DMBA, which drives tumorigenesis through IL-6-mediated inflammation [134]. Analysis of transcriptomics data from the Cancer Cell Line Encyclopedia (CCLE) and The Cancer Genome Atlas (TCGA) of primary tumour samples (including pancreatic adenocarcinoma, cutaneous melanoma, prostate adenocarcinoma and breast adenocarcinoma) has also shown that higher levels of STING expression is associated with higher levels of pro-inflammatory gene expression [10]. IL-6 in particular is a key enhancer of survival and metastasis in cancer [135–138]. In triple negative breast cancer cells with CIN, the depletion of cGAS–STING resulted in a reduction in IL-6 production and impaired cancer cell survival [139].
The expression of pro-inflammatory cytokines such as IL-6 is driven by the NF-kB family of transcription factors. In most cells, canonical cGAS–STING signalling induces only modest levels of NF-κB p65 activation, through signalling by TBK1 which favours the activation of the transcription factor IRF3 [41]. However, an alternative cGAS-independent mode of STING activation by nuclear DNA damage results in preferential NF-κB p65 activation and a more pro-inflammatory cytokine profile than conventional DNA sensing [60]. Thus, it is possible that this pathway contributes the production of pro-inflammatory cytokines during chronic DNA damage or CIN in cancer.
As an additional immune evasion strategy, tumour cells can also re-wire cGAS-mediated STING signalling towards increased non-canonical NF-κB activation in cancer cells through the NF-κB subunits p52 and RelB [118,140]. This drives the production of pro-inflammatory cytokines, rather than IFN-Is following DNA sensing, and thus could be a reason why cancers retain cGAS–STING signalling and subvert it to a different outcome. Non-canonical NF-κB signalling through p52 can in turn interfere with conventional STING–TBK1–IFN signalling, and this has been shown to further impair the anti-tumour effects of radiotherapy [140]. Similar inhibitory effects on STING signalling have also been attributed to IL-6 production in prostate cancer [141]. Thus, the collective evidence points towards a model where STING signalling is not restricted to interferon responses with anti-tumour functions, but rather can be adapted dynamically to produce both anti- and pro-tumour signalling outcomes during tumour progression.
Pro-tumour functions of STING signalling in immune cells
It is becoming increasingly clear that the ultimate outcome of STING signalling is also highly sensitive to cell context and differs between different cell types. For instance, while cGAMP transport to DCs can stimulate potent anti-tumour responses, its transport into astrocytes can promote brain metastasis through the production of cytokines that are beneficial for cancer cell growth and survival [80]. Activation of STING signalling in adaptive immune cells themselves can also negatively impact anti-cancer immune responses. For instance, regulatory B cells can make use of STING–IRF3 signalling to induce the production of IL-35, which impairs NK cell-mediated anti-tumour activity [142]. Activation of STING signalling can also induce apoptosis in T cells, which may negatively impact anti-tumour responses [44,133,143–151]. Adding to the complexity, the outcomes of STING signalling in T cells are also influenced by the engagement of the T-cell receptor [133,147,151]. It has been proposed that these negative impacts of STING signalling may be overcome by the use of lower doses of STING agonists or the specific targeting to tumour cells [149,151]. However, given that activation thresholds and cell type specificities may also differ between mouse models and individual cancer patients, there is an urgent need to understand how downstream STING signalling effects are regulated in different human cell contexts.
Conclusions and outlook
The intense study of DNA sensing pathways in the last decade or so has revealed a crucial role of the cGAS–STING–interferon signalling axis in shaping immune responses during infection, autoinflammation and cancer. However, despite the fact that the anti-tumour role of STING signalling is well documented in mouse tumour models and multiple STING agonists have been tested in clinical trials, these approaches have not yet shown efficacy as anti-tumour agents in patients. Indeed, STING signalling has been shown to have both pro- and anti-tumour functions in cancer cells, stromal cells and the immune cells in the TME. While the activation of canonical, acute STING signalling that results in IFN-I induction has been studied in great molecular detail and clearly possess anti-tumour functions, we know much less about how low-grade, chronic STING signalling is induced in response to DNA damage. We will also need to understand how different kinds of DNA damage induced by radio- and chemotherapy regimens affect DNA sensing pathways, so that we can avoid potential pro-tumour effects. There are many open questions about how STING activation drives different cellular outcomes, from the predominant activation of IRF3 to canonical or non-canonical NF-κB activation, as well as inflammasome activation and cell death or senescence pathways. In the future, it may be possible to therapeutically influence the balance of STING signalling outputs, rather than STING activation per se, to promote the anti-tumour functions of DNA sensing in cancer, while limiting adverse effects on tumour progression.
Summary
The detection of cytosolic DNA and DNA damage results in the activation of the innate immune adaptor STING in cancer cells, immune cells or endothelial cells within the tumour microenvironment. All these cell types can contribute to the production of type I interferons which have anti-tumour functions.
If the STING signalling response is evaded or diverted towards a more pro-inflammatory cytokine profile, this can instead promote the establishment of an immune-suppressive microenvironment and can aid tumour cell survival and metastasis.
A deeper understanding of how different cell contexts and regulatory mechanisms influence signalling outputs downstream of STING will be important for the development of STING-targeting immunotherapies.
Abbreviations
- CCLE
Cancer Cell Line Encyclopedia
- CIN
chromosomal instability
- cGAMP
cyclic GMP-AMP
- cGAS
cyclic GMP-AMP synthase
- DAMP
damage-associated molecular pattern
- DC
dendritic cell
- ENPP1
ectonucleotide pyrophosphate phosphodiesterase 1
- ER
endoplasmic reticulum
- ERGIC
ER–Golgi intermediate compartment
- IFI16
interferon gamma inducible protein 16
- IFN-I
type I interferon
- IL
interleukin
- IRF3
interferon regulatory factor
- ISG
interferon-stimulated gene
- NF-κB
nuclear factor κ-light-chain-enhancer of activated B cell
- NK
natural killer
- PAMP
pathogen-associated molecular pattern
- PARP
poly(ADP-ribose) polymerase
- PD-L1/2
programmed death ligand
- STING
STimulator of Interferon Genes
- TBK1
TANK-binding kinase 1
- TCGA
The Cancer Genome Atlas
- TME
tumour microenvironment
- TREX1
three prime repair exonuclease
- ZBP-1
Z DNA binding protein 1
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Open Access
Open access for this article was enabled by the participation of Lancaster University in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with JISC.
References
- 1.Schumacher T.N., Scheper W. and Kvistborg P. (2019) Cancer neoantigens. Ann. Rev. Immunol. 37, 173–200 10.1146/annurev-immunol-042617-053402 [DOI] [PubMed] [Google Scholar]
- 2.Jardim D.L., Goodman A., de Melo Gagliato D. and Kurzrock R. (2021) The challenges of tumour mutational burden as an immunotherapy biomarker. Cancer Cell 39, 154–173 10.1016/j.ccell.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hopfner K.P. and Hornung V. (2020) Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 21, 501–521 10.1038/s41580-020-0244-x [DOI] [PubMed] [Google Scholar]
- 4.Bakhoum S.F. and Cantley L.C. (2018) The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174, 1347–1360 10.1016/j.cell.2018.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drews R.M., Hernando B., Tarabichi M., Haase K., Lesluyes T., Smith P.S.et al. (2022) A pan-cancer compendium of chromosomal instability. Nature 606, 976–983 10.1038/s41586-022-04789-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krupina K., Goginashvili A. and Cleveland D.W. (2021) Causes and consequences of micronuclei. Curr. Opin. Cell Biol. 70, 91–99 10.1016/j.ceb.2021.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harding S.M., Benci J.L., Irianto J., Discher D.E., Minn A.J. and Greenberg R.A. (2017) Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 10.1038/nature23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacKenzie K.J., Carroll P., Martin C.A., Murina O., Fluteau A., Simpson D.J.et al. (2017) CGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 10.1038/nature23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Y.J., LeBert N., Chitre A.A., Koo C.X.E., Nga X.H., Ho S.S.W.et al. (2015) Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells. Cell Rep. 11, 460–473 10.1016/j.celrep.2015.03.041 [DOI] [PubMed] [Google Scholar]
- 10.Dou Z., Ghosh K., Vizioli M.G., Zhu J., Sen P., Wangensteen K.J.et al. (2017) Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 10.1038/nature24050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho S.S.W., Zhang W.Y.L., Tan N.Y.J., Khatoo M., Suter M.A., Tripathi S.et al. (2016) The DNA structure-specific endonuclease MUS81 mediates DNA sensor STING-dependent host rejection of prostate cancer cells. Immunity 44, 1177–1189 10.1016/j.immuni.2016.04.010 [DOI] [PubMed] [Google Scholar]
- 12.Glück S., Guey B., Gulen M.F., Wolter K., Kang T.W., Schmacke N.A.et al. (2017) Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19, 1061–1070 10.1038/ncb3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flynn P.J., Koch P.D. and Mitchison T.J. (2021) Chromatin bridges, not micronuclei, activate cGAS after drug-induced mitotic errors in human cells. Proc. Natl. Acad. Sci. U.S.A. 118, e2103585118 10.1073/pnas.2103585118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamazaki T., Kirchmair A., Sato A., Buqué A., Rybstein M., Petroni G.et al. (2020) Mitochondrial DNA drives abscopal responses to radiation that are inhibited by autophagy. Nat. Immunol. 21, 1160–1171 10.1038/s41590-020-0751-0 [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa H. and Barber G.N. (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Civril F., Deimling T., de Oliveira Mann C.C., Ablasser A., Moldt M., Wittie G.et al. (2013) Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337 10.1038/nature12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X., Shu C., Yi G., Chaton C.T., Shelton C.L., Diao J.et al. (2013) Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39, 1019–1031 10.1016/j.immuni.2013.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun L., Wu J., Du F., Chen X. and Chen Z.J. (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type i interferon pathway. Science 339, 786–791 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun W., Li Y., Chen L., Chen H., You F., Zhou X.et al. (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U.S.A. 106, 8653–8658 10.1073/pnas.0900850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou W., Whiteley A.T., de Oliveira Mann C.C., Morehouse B.R., Nowak R.P., Fischer E.C.et al. (2018) Structure of the human cGAS-DNA complex reveals enhanced control of immune surveillance. Cell 174, 300–311 10.1016/j.cell.2018.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreeva L., Hiller B., Kostrewa D., Lässig C., De Oliveira Mann C.C., Jan Drexler D.et al. (2017) CGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 549, 394–398 10.1038/nature23890 [DOI] [PubMed] [Google Scholar]
- 22.Xie W., Lama L., Adura C., Tomita D., Glickman J.F., Tuschl T.et al. (2019) Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation. Proc. Natl. Acad. Sci. U.S.A. 116, 11946–11955 10.1073/pnas.1905013116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Wu J., Du F., Xu H., Sun L., Chen Z.et al. (2014) The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 6, 421–430 10.1016/j.celrep.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du M. and Chen Z.J. (2018) DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 10.1126/science.aat1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyer J.A., Spangler C.J., Strauss J.D., Cesmat A.P., Liu P., Mcginty R.K.et al. (2020) Structural basis of nucleosome-dependent cGAS inhibition. Science 370, 450–454 10.1126/science.abd0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kujirai T., Zierhut C., Takizawa Y., Kim R., Negishi L., Uruma N.et al. (2020) Structural basis for the inhibition of cGAS by nucleosomes. Science 370, 455–458 10.1126/science.abd0237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pathare G.R., Decout A., Glück S., Cavadini S., Makasheva K., Hovius R.et al. (2020) Structural mechanism of cGAS inhibition by the nucleosome. Nature 587, 668–672 10.1038/s41586-020-2750-6 [DOI] [PubMed] [Google Scholar]
- 28.Zhao B., Xu P., Rowlett C.M., Jing T., Shinde O., Lei Y.et al. (2020) The molecular basis of tight nuclear tethering and inactivation of cGAS. Nature 587, 673–677 10.1038/s41586-020-2749-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michalski S., de Oliveira Mann C.C., Stafford C.A., Witte G., Bartho J., Lammens K.et al. (2020) Structural basis for sequestration and autoinhibition of cGAS by chromatin. Nature 587, 678–682 10.1038/s41586-020-2748-0 [DOI] [PubMed] [Google Scholar]
- 30.Chen H., Zhang J., Wang Y., Simoneau A., Yang H., Levine A.S.et al. (2020) cGAS suppresses genomic instability as a decelerator of replication forks. Sci. Adv. 6, eabb8941 10.1126/sciadv.abb8941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang H., Xue X., Panda S., Kawale A., Hooy R.M., Liang F.et al. (2019) Chromatin‐bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO 38, e102718 10.15252/embj.2019102718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H., Zhang H., Wu X., Ma D., Wu J., Wang L.et al. (2018) Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563, 131–136 10.1038/s41586-018-0629-6 [DOI] [PubMed] [Google Scholar]
- 33.Wu J., Sun L., Chen X., Du F., Shi H., Chen C.et al. (2013) Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Röhl I.et al. (2013) CGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 10.1038/nature12306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X., Shi H., Wu J., Zhang X., Sun L., Chen C.et al. (2013) Cyclic GMP-AMP containing mixed Phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235 10.1016/j.molcel.2013.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa H., Ma Z. and Barber G.N. (2009) STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 10.1038/nature08476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobbs N., Burnaevskiy N., Chen D., Gonugunta V.K., Alto N.M. and Yan N. (2015) STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 18, 157–168 10.1016/j.chom.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukai K., Konno H., Akiba T., Uemura T., Waguri S., Kobayashi T.et al. (2016) Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 7, 1–10 10.1038/ncomms11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S., Cai X., Wu J., Cong Q., Chen X., Li T.et al. (2015) Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, 1217–1231 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- 40.Ma X., Helgason E., Phung Q.T., Quan C.L., Iyer R.S., Lee M.W.et al. (2012) Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc. Natl. Acad. Sci. U.S.A. 109, 9378–9383 10.1073/pnas.1121552109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abe T. and Barber G.N. (2014) Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol. 88, 5328–5341 10.1128/JVI.00037-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gui X., Yang H., Li T., Tan X., Shi P., Li M.et al. (2019) Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266 10.1038/s41586-019-1006-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C. and Xu P. (2023) Cellular functions of cGAS-STING signaling. Trends Cell Biol. 33, 630–648 10.1016/j.tcb.2022.11.001 [DOI] [PubMed] [Google Scholar]
- 44.Gulen M.F., Koch U., Haag S.M., Schuler F., Apetoh L., Villunger A.et al. (2017) Signalling strength determines proapoptotic functions of STING. Nat. Commun. 8, 1–10 10.1038/s41467-017-00573-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaidt M.M., Ebert T.S., Chauhan D., Ramshorn K., Pinci F., Zuber S.et al. (2017) The DNA inflammasome in human myeloid cells is initiated by a STING-Cell death program upstream of NLRP3. Cell 171, 1110–1124 10.1016/j.cell.2017.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han C., Godfrey V., Liu Z., Han Y., Liu L., Peng H.et al. (2021) The AIM2 and NLRP3 inflammasomes trigger IL-1-mediated antitumor effects during radiation. Sci. Immunol. 6, eabc6998 10.1126/sciimmunol.abc6998 [DOI] [PubMed] [Google Scholar]
- 47.Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R.et al. (2009) AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 10.1038/nature07725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang J., Wu J., Liu Q., Wu X., Zhao Y. and Ren J. (2022) Post-translational modifications of STING: a potential therapeutic target. Front Immunol. 13, 1–19 10.3389/fimmu.2022.888147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ablasser A. and Hur S. (2020) Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat. Immunol. 21, 17–29 10.1038/s41590-019-0556-1 [DOI] [PubMed] [Google Scholar]
- 50.Li W., Li Y., Kang J., Jiang H., Gong W., Chen L.et al. (2023) 4-octyl itaconate as a metabolite derivative inhibits inflammation via alkylation of STING. Cell Rep. 42, 1–18 10.1016/j.celrep.2023.112145 [DOI] [PubMed] [Google Scholar]
- 51.Almine J.F., O'Hare C.A.J., Dunphy G., Haga I.R., Naik R.J., Atrih A.et al. (2017) IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat. Commun. 8, 1–15 10.1038/ncomms14392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jønsson K.L., Laustsen A., Krapp C., Skipper K.A., Thavachelvam K., Hotter D.et al. (2017) IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Commun. 8, 1–17 10.1038/ncomms14391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z., Yuan B., Bao M., Lu N., Kim T. and Liu Y.J. (2011) The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12, 959–965 10.1038/ni.2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh R.S., Vidhyasagar V., Yang S., Arna A.B., Yadav M., Aggarwal A.et al. (2022) DDX41 is required for cGAS-STING activation against DNA virus infection. Cell Rep. 39, 1–17 10.1016/j.celrep.2022.110856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferguson B.J., Mansur D.S., Peters N.E., Ren H. and Smith G.L. (2012) DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife 1, e00047 10.7554/eLife.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taffoni C., Marines J., Chamma H., Guha S., Saccas M., Bouzid A.et al. (2023) DNA damage repair kinase DNA-PK and cGAS sunergize to induce cancer-related inflammation in glioblastoma. EMBO J. 42, e111961 10.15252/embj.2022111961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y., Wu M., Cao D., Yang C., Jin J., Wu L.et al. (2021) ZBP1-MLKL necroptotic signaling potentiates radiation-induced antitumor immunity via intratumoral STING pathway activation. Sci Adv. 7, 1–16 10.1126/sciadv.abf6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ablasser A., Schmid-Burgk J.L., Hemmerling I., Horvath G.L., Schmidt T., Latz E.et al. (2013) Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503, 530–534 10.1038/nature12640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burdette D.L., Monroe K.M., Sotelo-Troha K., Iwig J.S., Eckert B., Hyodo M.et al. (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 10.1038/nature10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunphy G., Flannery S.M., Almine J.F., Connolly D.J., Paulus C., Jønsson K.L.et al. (2018) Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol. Cell 71, 745–760 10.1016/j.molcel.2018.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holm C.K., Jensen S.B., Jakobsen M.R., Cheshenko N., Horan K.A., Moeller H.B.et al. (2012) Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat. Immunol. 13, 737–743 10.1038/ni.2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maltbaek J.H., Cambier S., Snyder J.M. and Stetson D.B. (2022) ABCC1 transporter exports the immunostimulatory cyclic dinucleotide cGAMP. Immunity 55, 1799–1812 10.1016/j.immuni.2022.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Campisi M., Sundararaman S.K., Shelton S.E., Knelson E.H., Mahadevan N.R., Yoshida R.et al. (2020) Tumor-derived cGAMP regulates activation of the vasculature. Front Immunol. 11, 1–16 10.3389/fimmu.2020.02090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang H., Lee W.S., Kong S.J., Kim C.G., Kim J.H., Chang S.K.et al. (2019) STING activation reprograms tumor vasculatures and synergizes with VEGFR2 blockade. J. Clin. Invest. 129, 4350–4364 10.1172/JCI125413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Demaria O., De Gassart A., Coso S., Gestermann N., Di Domizio J., Flatz L.et al. (2015) STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc. Natl. Acad. Sci. U.S.A. 112, 15408–15413 10.1073/pnas.1512832112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sen T., Rodriguez B.L., Chen L., Della Corte C.M., Morikawa N., Fujimoto J.et al. (2019) Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov. 9, 646–661 10.1158/2159-8290.CD-18-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tian J., Zhang D., Kurbatov V., Wang Q., Wang Y., Fang D.et al. (2021) 5‐Fluorouracil efficacy requires anti‐tumor immunity triggered by cancer‐cell‐intrinsic STING. EMBO 40, e106065 10.15252/embj.2020106065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deng L., Liang H., Xu M., Yang X., Burnette B., Arina A.et al. (2014) STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41, 843–852 10.1016/j.immuni.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woo S.R., Fuertes M.B., Corrales L., Spranger S., Furdyna M.J., Leung M.Y.K.et al. (2014) STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41, 830–842 10.1016/j.immuni.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burnette B.C., Liang H., Lee Y., Chlewicki L., Khodarev N.N., Weichselbaum R.R.et al. (2011) The efficacy of radiotherapy relies upon induction of type I interferon-dependent innate and adaptive immunity. Cancer Res. 71, 2488–2496 10.1158/0008-5472.CAN-10-2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corrales L., Glickman L.H., McWhirter S.M., Kanne D.B., Sivick K.E., Katibah G.E.et al. (2015) Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030 10.1016/j.celrep.2015.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baird J.R., Friedman D., Cottam B., Dubensky T.W., Kanne D.B., Bambina S.et al. (2016) Radiotherapy combined with novel STING-targeting oligonucleotides results in regression of established tumors. Cancer Res. 76, 50–61 10.1158/0008-5472.CAN-14-3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H., Hu S., Chen X., Shi H., Chen C., Sun L.et al. (2017) cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc. Natl. Acad. Sci. U.S.A. 114, 1637–1642 10.1073/pnas.1621363114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen J., Zhao W., Ju Z., Wang L., Peng Y., Labrie M.et al. (2019) PARPI triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCANEss. Cancer Res. 79, 311–319 10.1158/0008-5472.CAN-18-1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Conde E., Vercher E., Soria-Castellano M., Suarez-Olmos J., Mancheno U., Elizalde E.et al. (2021) Epitope spreading driven by the joint action of CART cells and pharmacological STING stimulation counteracts tumor escape via antigen-loss variants. J. Immunother. Cancer 9, e00335 10.1136/jitc-2021-003351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ding L., Wang Q., Martincuks A., Kearns M.J., Jiang T., Lin Z.et al. (2023) STING agonism overcomes STAT3-mediated immunosuppression and adaptive resistance to PARP inhibition in ovarian cancer. J. Immunother. Cancer 11, 1–15 10.1136/jitc-2022-005627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Song J.X., Villagomes D., Zhao H. and Zhu M. (2022) cGAS in nucleus: The link between immune response and DNA damage repair. Front. Immunol. 13, 1–16 10.3389/fimmu.2022.1076784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu Y., Manasrah B.K., McGregor S.M., Lera R.F., Norman R.X., Tucker J.B.et al. (2021) Paclitaxel induces micronucleation and activates pro-inflammatory cGAS-STING signaling in triple-negative breast cancer. Mol. Cancer Ther. 20, 2553–2567 10.1158/1535-7163.MCT-21-0195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vanpouille-Box C., Alard A., Aryankalayil M.J., Sarfraz Y., Diamond J.M., Schneider R.J.et al. (2017) DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat. Commun. 8, 1–15 10.1038/ncomms15618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Q., Boire A., Jin X., Valiente M., Er E.E., Lopez-Soto A.et al. (2016) Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 533, 493–498 10.1038/nature18268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schadt L., Sparano C., Schweiger N.A., Silina K., Cecconi V., Lucchiari G.et al. (2019) Cancer-cell-intrinsic cGAS expression mediates tumor immunogenicity. Cell Rep. 29, 1236–1248 10.1016/j.celrep.2019.09.065 [DOI] [PubMed] [Google Scholar]
- 82.Lahey L.J., Mardjuki R.E., Wen X., Hess G.T., Ritchie C., Carozza J.A.et al. (2020) LRRC8A:C/E heteromeric channels are ubiquitous transporters of cGAMP. Mol. Cell 80, 578–591 10.1016/j.molcel.2020.10.021 [DOI] [PubMed] [Google Scholar]
- 83.Carozza J.A., Böhnert V., Nguyen K.C., Skariah G., Shaw K.E., Brown J.A.et al. (2020) Extracellular cGAMP is a cancer-cell-produced immunotransmitter involved in radiation-induced anticancer immunity. Nat. Cancer 1, 184–196 10.1038/s43018-020-0028-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cordova A.F., Ritchie C., Böhnert V. and Li L. (2021) Human SLC46A2 is the dominant cGAMP importer in extracellular cGAMP-sensing macrophages and monocytes. ACS Cent Sci. 7, 1073–1088 10.1021/acscentsci.1c00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ritchie C., Cordova A.F., Hess G.T., Bassik M.C. and Li L. (2019) SLC19A1 is an importer of the immunotransmitter cGAMP. Mol. Cell 75, 372–381 10.1016/j.molcel.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luteijn R.D., Zaver S.A., Gowen B.G., Wyman S.K., Garelis N.E., Onia L.et al. (2019) SLC19A1 transports immunoreactive cyclic dinucleotides. Nature 573, 434–438 10.1038/s41586-019-1553-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou Y., Fei M., Zhang G., Liang W.C., Lin W.Y., Wu Y.et al. (2020) Blockade of the phagocytic receptor MerTK on tumor-associated macrophages enhances P2X7R-dependent STING activation by tumor-derived cGAMP. Immunity 52, 357–373 10.1016/j.immuni.2020.01.014 [DOI] [PubMed] [Google Scholar]
- 88.Chelvanambi M., Fecek R.J., Taylor J.L. and Storkus W.J. (2021) STING agonist-based treatment promotes vascular normalization and tertiary lymphoid structure formation in the therapeutic melanoma microenvironment. J. Immunother. Cancer 9, e001906 10.1136/jitc-2020-001906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu M.M., Pu Y., Han D., Shi Y., Cao X., Liang H.et al. (2017) Dendritic cells but not macrophages sense tumor mitochondrial DNA for cross-priming through signal regulatory protein α signaling. Immunity 47, 363–373 10.1016/j.immuni.2017.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mender I., Zhang A., Ren Z., Han C., Deng Y., Siteni S.et al. (2020) Telomere stress potentiates STING-dependent anti-tumor immunity. Cancer Cell 38, 400–411 10.1016/j.ccell.2020.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fang C., Mo F., Liu L., Du J., Luo M., Men K.et al. (2021) Oxidized mitochondrial DNA sensing by STING signaling promotes the antitumor effect of an irradiated immunogenic cancer cell vaccine. Cell Mol. Immunol. 18, 2211–2223 10.1038/s41423-020-0456-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Klarquist J., Hennies C.M., Lehn M.A., Reboulet R.A., Feau S. and Janssen E.M. (2014) STING-mediated DNA Sensing promotes antitumor and autoimmune responses to dying cells. J. Immunol. 193, 6124–6134 10.4049/jimmunol.1401869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kitai Y., Kawasaki T., Sueyoshi T., Kobiyama K., Ishii K.J., Zou J.et al. (2017) DNA-containing exosomes derived from cancer cells treated with topotecan activate a STING-dependent pathway and reinforce antitumor immunity. J. Immunol. 198, 1649–1659 10.4049/jimmunol.1601694 [DOI] [PubMed] [Google Scholar]
- 94.Diamond J.M., Vanpouille-Box C., Spada S., Rudqvist N.P., Chapman J.R., Ueberheide B.M.et al. (2018) Exosomes shuttle TREX1-sensitive IFN-stimulatory dsDNA from irradiated cancer cells to DCs. Cancer Immunol. Res. 6, 910–920 10.1158/2326-6066.CIR-17-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Mingo Pulido Á., Hänggi K., Celias D.P., Gardner A., Li J., Batista-Bittencourt B.et al. (2021) The inhibitory receptor TIM-3 limits activation of the cGAS-STING pathway in intra-tumoral dendritic cells by suppressing extracellular DNA uptake. Immunity 54, 1154–1167 10.1016/j.immuni.2021.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Diamond M.S., Kinder M., Matsushita H., Mashayekhi M., Dunn G.P., Archambault J.M.et al. (2011) Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med. 208, 1989–2003 10.1084/jem.20101158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fuertes M.B., Kacha A.K., Kline J., Woo S.R., Kranz D.M., Murphy K.M.et al. (2011) Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8α+ dendritic cells. J. Exp. Med. 208, 2005–2016 10.1084/jem.20101159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baird J.R., Friedman D., Cottam B., Dubensky T.W., Kanne D.B., Bambina S.et al. (2016) Radiotherapy combined with novel STING-targeting oligonucleotides results in regression of established tumors. Cancer Res. 76, 50–61 10.1158/0008-5472.CAN-14-3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Downey C.M., Aghaei M., Schwendener R.A. and Jirik F.R. (2014) DMXAA causes tumor site-specific vascular disruption in murine non-small cell lung cancer, and like the endogenous non-canonical cyclic dinucleotide STING agonist, 2939-cGAMP, induces M2 macrophage repolarization. PloS ONE 9, e99988 10.1371/journal.pone.0099988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Q., Bergholz J.S., Ding L., Lin Z., Kabraji S.K., Hughes M.E.et al. (2022) STING agonism reprograms tumor-associated macrophages and overcomes resistance to PARP inhibition in BRCA1-deficient models of breast cancer. Nat. Commun. 13, 1–17 10.1038/s41467-022-30568-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marcus A., Mao A.J., Lensink-Vasan M., Wang L.A., Vance R.E. and Raulet D.H. (2018) Tumor-derived cGAMP triggers a STING-mediated interferon response in non-tumor cells to activate the NK cell response. Immunity 49, 754–763 10.1016/j.immuni.2018.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Berger G., Knelson E.H., Jimenez-Macias J.L., Nowicki M.O., Han S., Panagioti E.et al. (2022) STING activation promotes robust immune response and NK cell-mediated tumor regression in glioblastoma models. Proc. Natl. Acad. Sci. U.S.A. 119, e2111003119 10.1073/pnas.2111003119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang C.-X., Ye S.-B., Ni J.-J., Cai T.-T., Liu Y.-N., Huang D.-J.et al. (2019) STING signaling remodels the tumor microenvironment by antagonizing myeloid-derived suppressor cell expansion. Cell Death Differ. 26, 2314–2328 10.1038/s41418-019-0302-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheng H., Xu Q., Lu X., Yuan H., Li T., Zhang Y.et al. (2020) Activation of STING by cGAMP Regulates MDSCs to Suppress Tumor Metastasis via Reversing Epithelial-Mesenchymal Transition. Front. Oncol. 10, 1–14 10.3389/fonc.2020.00896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liang H., Deng L., Hou Y., Meng X., Huang X., Rao E.et al. (2017) Host STING-dependent MDSC mobilization drives extrinsic radiation resistance. Nat. Commun. 8, 1–10 10.1038/s41467-017-01566-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lemos H., Mohamed E., Huang L., Ou R., Pacholczyk G., Arbab A.S.et al. (2016) STING promotes the growth of tumors characterized by low antigenicity via IDO activation. Cancer Res. 76, 2076–2081 10.1158/0008-5472.CAN-15-1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.An X., Zhu Y., Zheng T., Wang G., Zhang M., Li J.et al. (2019) An analysis of the expression and association with immune cell infiltration of the cGAS/STING pathway in Pan-Cancer. Mol. Ther. Nucleic Acids 14, 80–89 10.1016/j.omtn.2018.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fridman W.H., Zitvogel L., Sautès-Fridman C. and Kroemer G. (2017) The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 14, 717–734 10.1038/nrclinonc.2017.101 [DOI] [PubMed] [Google Scholar]
- 109.Corrales L., Glickman L.H., McWhirter S.M., Kanne D.B., Sivick K.E., Katibah G.E.et al. (2015) Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor Regression and Immunity. Cell Rep. 11, 1018–1030 10.1016/j.celrep.2015.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Le Naour J., Zitvogel L., Galluzzi L., Vacchelli E. and Kroemer G. (2020) Trial watch: STING agonists in cancer therapy. Oncoimmunology 9, 1–12 10.1080/2162402X.2020.1777624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Conlon J., Burdette D.L., Sharma S., Bhat N., Thompson M., Jiang Z.et al. (2013) Mouse, but not Human STING, Binds and Signals in Response to the Vascular Disrupting Agent 5,6-Dimethylxanthenone-4-Acetic Acid. J. Immunol. 190, 5216–5225 10.4049/jimmunol.1300097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gao P., Ascano M., Zillinger T., Wang W., Dai P., Serganov A.A.et al. (2013) Structure-function analysis of STING activation by c[G(2′,5′) pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 154, 748–762 10.1016/j.cell.2013.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xia T., Konno H., Ahn J. and Barber G.N. (2016) Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 14, 282–297 10.1016/j.celrep.2015.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Konno H., Yamauchi S., Berglund A., Putney R.M., Mulé J.J. and Barber G.N. (2018) Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene 37, 2037–2051 10.1038/s41388-017-0120-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xia T., Konno H. and Barber G.N. (2016) Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis. Cancer Res. 76, 6747–6759 10.1158/0008-5472.CAN-16-1404 [DOI] [PubMed] [Google Scholar]
- 116.Lee K.M., Lin C.C., Servetto A., Bae J., Kandagatla V., Ye D.et al. (2022) Epigenetic repression of STING by MYC promotes immune evasion and resistance to immune checkpoint inhibitors in triple-negative breast cancer. Cancer Immunol. 10, 829–843 10.1158/2326-6066.CIR-21-0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kitajima S., Tani T., Springer B.F., Campisi M., Osaki T., Haratani K.et al. (2022) MPS1 inhibition primes immunogenicity of KRAS-LKB1 mutant lung cancer. Cancer Cell 40, 1128–1144 10.1016/j.ccell.2022.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bakhoum S.F., Ngo B., Laughney A.M., Cavallo J.A., Murphy C.J., Ly P.et al. (2018) Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 10.1038/nature25432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tusseau M., Lovšin E., Samaille C., Pescarmona R., Mathieu A.L., Maggio M.C.et al. (2022) DNASE1L3 deficiency, new phenotypes, and evidence for a transient type I IFN signaling. J. Clin. Immunol. 42, 1310–1320 10.1007/s10875-022-01287-5 [DOI] [PubMed] [Google Scholar]
- 120.Ghosh M., Saha S., Li J., Montrose D.C. and Martinez L.A. (2023) p53 engages the cGAS/STING cytosolic DNA sensing pathway for tumor suppression. Mol. Cell 83, 266–280 10.1016/j.molcel.2022.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li L., Yin Q., Kuss P., Maliga Z., Millán J.L., Wu H.et al. (2014) Hydrolysis of 2’3’-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol. 10, 1043–1048 10.1038/nchembio.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li J., Duran M.A., Dhanota N., Chatila W.K., Bettigole S.E., Kwon J.et al. (2021) Metastasis and immune evasion from extracellular cGAMP hydrolysis. Cancer Discov. 11, 1212–1227 10.1158/2159-8290.CD-20-0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vijayan D., Young A., Teng M.W.L. and Smyth M.J. (2017) Targeting immunosuppressive adenosine in cancer. Nat. Rev. Cancer 17, 709–724 10.1038/nrc.2017.86 [DOI] [PubMed] [Google Scholar]
- 124.Goswami A., Deb B., Goyal S., Gosavi A., Mali M., Martis A.M.et al. (2022) AVA-NP-695 selectively inhibits ENPP1 to activate STING pathway and abrogate tumor metastasis in 4T1 breast cancer syngeneic mouse model. Molecules 27, 1–19 10.3390/molecules27196721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cheon H., Holvey-Bates E.G., Schoggins J.W., Forster S., Hertzog P., Imanaka N.et al. (2013) IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO 32, 2751–2763 10.1038/emboj.2013.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Khodarev N.N., Beckett M., Labay E., Darga T., Roizman B. and Weichselbaum R.R. (2004) STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc. Natl. Acad. Sci. U.S.A. 101, 1714–1719 10.1073/pnas.0308102100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Weichselbaum R.R., Ishwaran H., Yoon T., Nuyten D.S.A., Baker S.W., Khodarev N.et al. (2008) An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl. Acad. Sci. U.S.A. 105, 18490–18495 10.1073/pnas.0809242105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cheon H., Holvey-Bates E.G., Mcgrail D.J., Stark G.R., Diaz L.A. and Hertzog P.J. (2021) PD-L1 sustains chronic, cancer cell-intrinsic responses to type I interferon, enhancing resistance to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 118, 1–10 10.1073/pnas.2112258118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Erdal E., Haider S., Rehwinkel J., Harris A.L. and McHugh P.J. (2017) A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 31, 353–369 10.1101/gad.289769.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Garcia-Diaz A., Shin D.S., Moreno B.H., Saco J., Escuin-Ordinas H., Rodriguez G.A.et al. (2017) Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 19, 1189–1201 10.1016/j.celrep.2017.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gato-Cañas M., Zuazo M., Arasanz H., Ibañez-Vea M., Lorenzo L., Fernandez-Hinojal G.et al. (2017) PDL1 signals through conserved sequence motifs to overcome interferon-mediated cytotoxicity. Cell Rep. 20, 1818–1829 10.1016/j.celrep.2017.07.075 [DOI] [PubMed] [Google Scholar]
- 132.Lemos H., Ou R., McCardle C., Lin Y., Calver J., Minett J.et al. (2020) Overcoming resistance to STING agonist therapy to incite durable protective antitumor immunity. J. Immunother Cancer 8, e001182 10.1136/jitc-2020-001182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Quaney M.J., Pritzl C.J., Luera D., Newth R.J., Knudson K.M., Saxena V.et al. (2023) STING controls T cell memory fitness during infection through T cell-intrinsic and IDO-dependent mechanisms. Proc. Natl. Acad. Sci. U. S. A. 120, 1–10 10.1073/pnas.2205049120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ahn J., Xia T., Konno H., Konno K., Ruiz P. and Barber G.N. (2014) Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 5, 1–9 10.1038/ncomms6166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li S., Wang N. and Brodt P. (2012) Metastatic cells can escape the proapoptotic effects of TNF-α through increased autocrine IL-6/STAT3 signaling. Cancer Res. 72, 865–875 10.1158/0008-5472.CAN-11-1357 [DOI] [PubMed] [Google Scholar]
- 136.Yun U.J., Park S.E., Jo Y.S., Kim J. and Shin D.Y. (2012) DNA damage induces the IL-6/STAT3 signaling pathway, which has anti-senescence and growth-promoting functions in human tumors. Cancer Lett. 323, 155–160 10.1016/j.canlet.2012.04.003 [DOI] [PubMed] [Google Scholar]
- 137.Jones S.A. and Jenkins B.J. (2018) Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat. Rev. Immunol. 18, 773–789 10.1038/s41577-018-0066-7 [DOI] [PubMed] [Google Scholar]
- 138.Taniguchi K. and Karin M. (2014) IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin. Immunol. 26, 54–74 10.1016/j.smim.2014.01.001 [DOI] [PubMed] [Google Scholar]
- 139.Hong C., Schubert M., Tijhuis A.E., Requesens M., Roorda M., van den Brink A.et al. (2022) cGAS-STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature 607, 366–373 10.1038/s41586-022-04847-2 [DOI] [PubMed] [Google Scholar]
- 140.Hou Y., Liang H., Rao E., Zheng W., Huang X., Deng L.et al. (2018) Non-canonical NF-κB antagonizes STING sensor-mediated DNA sensing in radiotherapy. Immunity 49, 490–503 10.1016/j.immuni.2018.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Suter M.A., Tan N.Y., Thiam C.H., Khatoo M., MacAry P.A., Angeli V.et al. (2021) cGAS-STING cytosolic DNA sensing pathway is suppressed by JAK2-STAT3 in tumor cells. Sci. Rep. 11, 1–13 10.1038/s41598-021-86644-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li S., Mirlekar B., Johnson B.M., Brickey W.J., Wrobel J.A., Yang N.et al. (2022) STING-induced regulatory B cells compromise NK function in cancer immunity. Nature 610, 373–380 10.1038/s41586-022-05254-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tang C.H.A., Zundell J.A., Ranatunga S., Lin C., Nefedova Y., Del Valle J.R.et al. (2016) Agonist-mediated activation of STING induces apoptosis in malignant B cells. Cancer Res. 76, 2137–2152 10.1158/0008-5472.CAN-15-1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cerboni S., Jeremiah N., Gentili M., Gehrmann U., Conrad C., Stolzenberg M.C.et al. (2017) Intrinsic antiproliferative activity of the innate sensor STI NG in T lymphocytes. J. Exp. Med. 214, 1769–1785 10.1084/jem.20161674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Larkin B., Ilyukha V., Sorokin M., Buzdin A., Vannier E. and Poltorak A. (2017) Cutting Edge: activation of STING in T cells induces type I IFN responses and cell death. J. Immunol. 199, 397–402 10.4049/jimmunol.1601999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sivick K.E., Desbien A.L., Glickman L.H., Reiner G.L., Corrales L., Surh N.H.et al. (2018) Magnitude of therapeutic STING activation determines CD8+ T cell-mediated anti-tumor immunity. Cell Rep. 25, 3074–3085 10.1016/j.celrep.2018.11.047 [DOI] [PubMed] [Google Scholar]
- 147.Imanishi T., Unno M., Kobayashi W., Yoneda N., Matsuda S., Ikeda K.et al. (2019) Reciprocal regulation of STING and TCR signaling by mTORC1 for T-cell activation and function. Life Sci Alliance 2, e201800282 10.26508/lsa.201800282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yum S., Li M., Fang Y. and Chen Z.J. (2021) TBK1 recruitment to STING activates both IRF3 and NF-κB that mediate immune defense against tumors and viral infections. Proc. Natl. Acad. Sci. U.S.A. 118, 1–9 10.1073/pnas.2100225118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wu J., Chen Y.J., Dobbs N., Sakai T., Liou J., Miner J.J.et al. (2019) STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J. Exp. Med. 216, 867–883 10.1084/jem.20182192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wu J., Dobbs N., Yang K. and Yan N. (2020) Interferon-independent activities of mammalian STING mediate antiviral response and tumor immune evasion. Immunity 53, 115–126 10.1016/j.immuni.2020.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kuhl N., Linder A., Philipp N., Nixdorf D., Fischer H., Veth S.et al. (2023) STING agonism turns human T cells into interferon‐producing cells but impedes their functionality. EMBO 24, e55536 10.15252/embr.202255536 [DOI] [PMC free article] [PubMed] [Google Scholar]