Abstract
Precision medicine for cancer is rapidly moving to an approach that integrates multiple dimensions of the biology in order to model mechanisms of cancer progression in each patient. The discovery of multiple drivers per tumor challenges medical decision that faces several treatment options. Drug sensitivity depends on the actionability of the target, its clonal or subclonal origin and coexisting genomic alterations. Sequencing has revealed a large diversity of drivers emerging at treatment failure, which are potential targets for clinical trials or drug repurposing. To effectively prioritize therapies, it is essential to rank genomic alterations based on their proven actionability. Moving beyond primary drivers, the future of precision medicine necessitates acknowledging the intricate spatial and temporal heterogeneity inherent in cancer. The advent of abundant complex biological data will make artificial intelligence algorithms indispensable for thorough analysis. Here, we will discuss the advancements brought by the use of high-throughput genomics, the advantages and limitations of precision medicine studies and future perspectives in this field.
Key words: precision medicine, ESCAT, OncoKB, NGS, high-throughput genomics
Highlights
-
•
Actionable alterations should be prioritized by using validated rankings for treatment tailoring.
-
•
High-throughput genomics could refine drivers and find new targets, mini-drivers and agnostic biomarkers.
-
•
Complex data interpretation should be assisted by machine learning programs and artificial intelligence.
Introduction
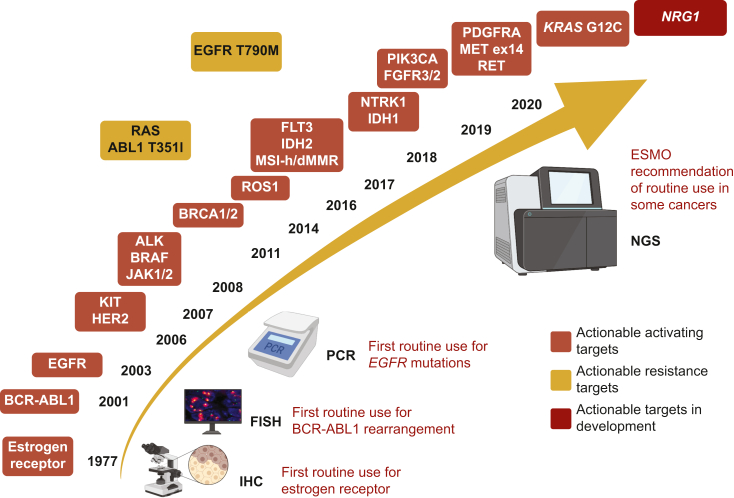
More than a century ago, the physician and scientist Paul Ehrlich described for the first time the concept of finding a ‘magic bullet’ that is able to hit a specific target to kill the disease, while sparing the rest of the body. From the development of endocrine therapy that is the ‘first-generation bullet’ in oncology, we now have reached an era where treatments are designed to selectively target drivers of tumorigenesis, and tailored according to the dynamic tumor evolution. Precision medicine includes the research and validation of predictive, prognostic and resistance biomarkers, but also predictors of toxicity, that help medical decisions and tailor treatment.1 Precision medicine has notably changed the paradigm of treatment in certain cancer types. The discovery of ‘drug–target’ pairs, such as imatinib in chronic myeloid leukemia with BCR-ALB1 fusions,2 gefitinib in epidermal growth factor receptor (EGFR)-mutated non-small-cell lung cancer (NSCLC)3 and vemurafenib in BRAF-mutated melanoma,4 was the first proof that targeted therapy could improve patients’ outcomes and quality of life. Currently, there are >20 biomarkers routinely assessed in clinical practice for the administration of approved targeted therapies (Figure 1).
Figure 1.
History of the implementation of routine evaluation of oncogenic alterations for tailoring treatment with targeted therapy, based on Food and Drug Administration (FDA)/European Medicines Agency (EMA) drug approvals.
ESMO, European Society for Medical Oncology; IHC, immunohistochemistry; NGS, next-generation sequencing.
Over the last decades, there has also been a tremendous progress of molecular techniques, from capillary-based sequencing technologies to next-generation sequencing (NGS), a modern massive parallel sequencing.5,6 High-throughput sequencing enables a rapid assessment of hundreds or thousands of genes from small DNA quantities.7 It may sequence targeted panels of genes (targeted sequencing), the whole exome (whole-exome sequencing, WES) and the whole genome (whole-genome sequencing, WGS) in a few days.6 This has allowed astonishing discoveries in molecular biology, with the first description of an individual genome dating back to 20088 to today’s pan-cancer analyses of whole genomes.9,10 However, the clinical utility of using multigene sequencing is currently modest [European Society for Medical Oncology (ESMO) recommendations] because bioinformatics tools to model cancer biology have not been developed.11
This review will discuss the progress made by high-throughput genomics, the advantages and limitations of precision medicine studies and future perspectives in the field, including data science-based approaches.
Clinical trials of precision medicine investigating the clinical utility of high-throughput genomics to detect genomic drivers
Over the last years, high-throughput sequencing has been studied for its clinical utility in personalized therapy for advanced tumors with actionable molecular alterations. NGS was used to identify genomic alterations, and treatment was customized based on the molecular profile, regardless of tumor histology. Several trials evaluated the outcomes of matched therapy compared to unmatched therapy (Table 1).12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24
Table 1.
Clinical trials of precision medicine where high-throughput genomics was used
| Trial | Technology | Screened | Matched therapy | Outcome |
|||
|---|---|---|---|---|---|---|---|
| N | Type | Matched | Unmatched or prior treatment | P value | |||
| SAFIR 01 (breast)12 | CGH/Sanger seq | 423 | 55 (13%) | OR/SD >4 months | 30% | ||
| SHIVA13 | NGS | 741 | 195 (26%) | HR | 0.88 (95% CI 0.65-1.19) | 0.41 | |
| MDACC14 | NGS (236 genes) | 500 | 122 (24.4%) P1 trials |
Matching score | High | Low | |
| OR/SD ≥6 months | 22% | 9% | |||||
| TTF | HR 0.52 (95% CI 0.36-0.74) | 0.0003 | |||||
| OS | HR 0.65 (95% CI 0.43-1.0) | 0.05 | |||||
| IMPACT/COMPACT15 | Hotspot panel/NGS | 1893 | 84 (5%) | OR | 19% | 9% | <0.026 |
| MOSCATO16 | CGH/NGS/RNAseq | 1035 | 199 (19%) | OR | 11% | ||
| PFS2/PFS1 >1.3 | 33% | ||||||
| OS | 11.9 months | ||||||
| ProfiLER17 | NGS/CGH | 2579 | 163 (6%) | PR | 13% | ||
| IMPACT update18 | NGS (genes ≤182) | 3487 | 711 (20%) | ORR | 16.4% | 5.4% | <0.0001 |
| OR/SD ≥6 months | 35.3% | 20.3% | <0.001 | ||||
| PFS | 4 months | 2.8 months | 0.0001 | ||||
| OS | 9.3 months | 7.3 months | 0.0001 | ||||
| 3-year OS | 15% | 7% | |||||
| 10-year OS | 6% | 1% | |||||
| I-PREDICT19 | NGS (FMI) tissue/ctDNA | 149 | 73 (49%) | Matching score | High | Low | |
| DCR | 50% | 22.4% | 0.028 | ||||
| PFS | 6.5 months | 3.1 months | 0.001 | ||||
| OS | NR | 10.2 months | 0.046 | ||||
| PFS2/PFS1 ≥1.3 | 75% | 36.6% | 0.026 | ||||
| WINTHER20 | DNAseq (236 genes)/RNAseq | 303 | 107 (35%) | OR/SD ≥6 months | 26.2% | ||
| PFS2/PFS1 >1.5 | 22.4% | ||||||
| TARGET21 | NGS on ctDNA (230-641 genes) | 100 | 11 (11%) | PR | 4 pts | 0 | |
| KYT (pancreas)22 | NGS (FMI) | 1856 | 46 (2.5%) | OS | 2.58 years | 1.51 years | 0.0004 |
| HR 0.42 (95% CI 0.26-0.68) | |||||||
| MASTER (young/rare tumors)23 | WGS/WES RNAseq |
1310 | 362 (31.8%) | ORR | 23.9% | 16.3% | |
| DCR | 55.3% | 46.3% | |||||
| PFS2/PFS1 >1.3 | 35.7% | ||||||
| INFORM (pediatric)24 | lcWGS/WES/RNAseq/methylation array | 519 | 20 (3.8%) | PFS | 204 days | 117 days | 0.011 |
| SAFIR02-Breast36 | DNAseq/CGH | 1462 | 157 (10.7%)/238 R∗ | PFS ESCAT I/II Without ESCAT I/II |
9.1 months | 2.8 months | <0.001 0.49 |
| HR 0.41 (90% CI 0.27-0.61) | |||||||
| 2.8 months | 3.1 months | ||||||
| HR 1.15 (95% CI 0.76-1.75) | |||||||
CGH, comparative genomic hybridization; CI, confidence interval; ctDNA, circulating tumor DNA; DCR, disease control rate; ESCAT, ESMO Scale for Clinical Actionability of molecular Targets; FMI, Foundation Medicine; HR, hazard ratio; lcWGS, low-coverage whole-genome sequencing; NGS, next-generation sequencing; NR, not reached; OR, objective response; ORR, objective response rate; OS, overall survival; PFS, progression-free survival; pts, patients; R∗, randomized; RNAseq, RNA sequencing; SD, stable disease; TTF, time to treatment failure; WES, whole-exome sequencing; WGS, whole-genome sequencing.
The SHIVA study was the first randomized controlled trial that compared matched therapy with chemotherapy in patients failing all standard treatments. Matched therapy consisted of marketed drugs already approved in other indications, which targeted alterations of hormone receptors, phosphatidylinositol 3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) pathways. The study failed to show significant differences in progression-free survival (PFS) and the experimental arm exhibited more grade 3-4 toxicity.13 The MOSCATO 01 study used NGS and comparative genomic hybridization to screen for molecular alterations in fresh tumor biopsies carried out at relapse. It showed positive results with a PFS ratio (ratio between PFS on targeted therapy and PFS on prior therapy) ≥1.3 in 33% of patients receiving matched treatment, but the overall matching rate was low (7% of the total number of patients screened).16 Similarly, a low matching rate (6%) was reported in the ProfiLER study, which enrolled a high number of patients with advanced solid and hematological tumors.17
More recent trials like I-PREDICT and WINTHER, which enrolled fewer patients, reported higher rates of patients receiving matched treatment (49% and 35%, respectively). The I-PREDICT trial targeted tumor heterogeneity and showed improved disease control rates, PFS and overall survival (OS) with higher ‘matching scores’.19 Similarly, the WINTHER trial showed that a higher matching score was associated with a longer PFS and OS. However, the median PFS was only 2 months in all treated patients.20 The NCT/DKTK MASTER trial prospectively investigated the clinical utility of WGS/WES and RNA sequencing (RNAseq) in personalized care in patients with advanced cancers of younger age or with rare tumors. The initial results showed better response rates and PFS with personalized therapy compared to prior therapy, particularly in specific cancer types.23 However, it remains uncertain whether WGS/WES and whole-transcriptome sequencing provide significantly more benefits than large-panel NGS for detecting actionable genomic alterations and guiding available treatments.
The reasons behind failure
Reasons for failure in precision medicine studies can be attributed to several factors (Figure 2). The main cause was the lack of optimal tool to model biology and allocate the right therapy. Treatment choices were often based on experts’ opinions, leading to variability in target and treatment selection criteria across studies. Additionally, inadequate matching algorithms and prioritization of drug availability over target actionability contributed to the limited benefit.
Figure 2.
Lessons learned from precision medicine studies. Heavily pretreated patients with refractory tumors in advanced stages are less likely to significantly benefit from matched treatment. Patient attrition could be diminished by an earlier inclusion of patients in precision medicine trials and shortening the inclusion period. New biopsies should be favored whenever possible over archival specimens. Liquid biopsy could be used as an alternative when tissue is hard to obtain or in case of tissue sampling bias. Molecular alterations should be prioritized for efficient treatment design based on the level of evidence of their actionability and targeted with direct inhibitors. Clinical trials of precision medicine should be carried out in institutions with access to early-phase clinical trials in order to expand the number of patients oriented to a matched treatment.
ctDNA, circulating tumor DNA.
Another reason for failure was the absence of drugs that matched the identified alterations or the use of weak inhibitors. Half of the patients with actionable alterations did not receive a matched treatment in both SHIVA and MOSCATO 01 studies, and >80% in the ProfiLER study. Also, drugs used for matching were not best-in-class, especially in earlier studies. Weak inhibitors were also used, such as afatinib for ERBB2 alterations, multityrosine kinase inhibitors for RET fusions, mammalian target of rapamycin inhibitors for PI3K pathway alterations or MEK inhibitors for KRAS mutations, which are known today to only weakly affect these alterations.13,16 The availability of better drugs today could potentially improve clinical outcomes.
The use of archival samples may result in inadequate quality or quantity of the sample. In the ProfiLER study, 78% of samples were archival and >15% of patients had a premature withdrawal because of inadequate samples.17 In addition, a fresh biopsy should be favored over archival tissue especially in situations when treatment would exert a selective pressure,25,26 in patients who derived initial benefit from targeted therapies, hormone therapies, immunotherapy27,28 or after treatments with mutagenic potential (e.g. temozolomide).29 However, outside these examples, there is limited evolution of the actionable genome of treated metastases over time, which discourages repeated testing in the metastatic setting and supports the use of qualitative archived biopsies for tailoring treatment.28
Molecular analyses that were restricted to DNA analyses might overlook the existence of drivers of tumor progression in the absence of genomic alterations, such as the overexpression of messenger RNA (mRNA) or proteins (e.g. estrogen receptor or androgen receptor). The WINTHER trial included RNAseq in addition to DNA sequencing which resulted in a therapy match for an extra 12% of cases.20
The issue of co-mutations was not taken into account in the majority of trials. For instance, in advanced breast cancer overexpressing ERBB2 and hyperactive PI3K pathway (PIK3CA mutations, PTEN loss or AKT1 mutations), everolimus has been shown to be associated with a decreased hazard of progression when added to trastuzumab.30
These studies have greatly enriched the actual state of knowledge in precision medicine and offered valuable lessons for future studies (Figure 2). They highlighted the importance of choosing the optimal ‘drug–target’ pair,31 with selective, direct inhibitors rather than weak agents used as monotherapy. It also pointed out a need of common guidelines for target prioritization for treatment design, based on clinical evidence.
Ranking actionability of genomic alterations with a validated system
High-throughput sequencing may provide vast molecular information in tumors, revealing an average of 4-5 drivers per tumor.3,9 The challenge for molecular pathologists and physicians lies in assessing the clinical relevance of these data to determine the most suitable treatment.
Targets range from clinically validated to experimental and finally to undruggable alterations, while drugs ranging from direct inhibitors of unique or multiple targets to indirect pathway inhibitors. The missing piece of the puzzle in precision medicine studies was a common algorithm able to prioritize treatment for the alteration with the highest actionability. Several systems have been developed to rank alterations, the most used today being OncoKB (oncokb.org)32 and the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT).33, 34, 35 These systems classify genomic alterations based on the clinical and/or preclinical proof of actionability. Biomarkers with the highest level of evidence are predictive of improved survival endpoints with their matched treatment in prospective clinical trials. In this case, access to the matched treatment should be the standard of care. Investigational biomarkers need additional clinical data as the magnitude of clinical benefit is not clear enough, while hypothetical predictive biomarkers have shown benefit in other tumor types with the same alteration or have preclinical evidence of actionability. The validation of the ESCAT scale was provided by the SAFIR trial in human epidermal growth factor receptor 2 (HER2)-negative metastatic breast cancer. Pooled data from the SAFIR02-BREAST and SAFIR-PI3K studies revealed an improvement in PFS in breast cancer patients who received maintenance therapy matched to targets classified as level I or II according to ESCAT. However, no such benefit was observed in treatments that extended beyond these levels or when treatments were given without ESCAT guidance.36 Also, in a cohort of patients with advanced cholangiocarcinoma, treatment administration according to ESCAT has shown an improved OS for patients with ESCAT I-II alterations who received matched treatment compared to controls who did not.37 Actionable ESCAT tier I-II alterations have been reported in 26% of patients with advanced cholangiocarcinoma,37 39% of patients with urothelial carcinoma38 and 68%-74% of patients with advanced breast cancer.39,40
These ranking systems have high potential, by providing uniformity in the interpretation of molecular reports in the oncological community, and by helping treatment decision for a target with uncertain relevance. It offers a realistic view of the expected benefit and helps to select the most suitable treatment when more genomic alterations are detected.41 As ESCAT evaluates only actionability and not drugs, decision making should also be assisted by tools that evaluate the magnitude of clinical benefit of drugs [e.g. the ESMO-Magnitude of Clinical Benefit Scale (MCBS)].41
The current landscape of high-throughput sequencing in clinical practice
High-throughput sequencing has provided valuable insights in precision medicine, by the detection of targetable drivers, resistance mechanisms or refinement of diagnosis.27,42,43 The ESMO Precision Medicine Working Group recommends routine use of NGS in advanced non-squamous NSCLC, prostate and ovarian cancers due to the identification of specific genomic alterations (e.g. BRCA1/2) that can guide treatment. For other tumor sites, large-scale use of NGS is not yet recommended outside of clinical trials.34 One issue however is the detection of rare tumor-agnostic biomarkers, such as microsatellite instability-high (MSI-H) or NTRK fusion in solid tumors, for which pembrolizumab and TRK inhibitors, respectively, may be administered in the absence of alternative treatments.42 Although immunohistochemistry (IHC) is sufficient for tumors with a high prevalence of these biomarkers,44,45 NGS is more suitable for tumors with lower prevalence and it might also identify other actionable biomarkers, while it may incur additional costs.
Profiling relapsed disease might tailor drugs for resistance alterations.27 For instance, in EGFR-mutated NSCLC failing osimertinib, on-target alterations are found in 6%-10% of cases, with EGFR C797S being the most frequent mutation,46 for which fourth-generation EGFR inhibitors are undergoing evaluation in phase I/II studies47 (NCT04862780). The MET amplification, which occurs in 7%-15% of cases, could be targeted with a MET tyrosine kinase inhibitor (TKI), while continuing the EGFR inhibition with osimertinib.46,48,49 Gene fusions, such as ALK, RET, NTRK and BRAF fusions, occur in 1%-8% of cases. The dual inhibition of EGFR and of the acquired alteration showed tumor responses in several case reports, but the magnitude of benefit is currently unknown for most targets.50, 51, 52, 53, 54 The development of clinical trials in this setting is challenged by the diversity and rarity of certain acquired alterations. Moreover, more than one acquired resistance alteration frequently co-exists at TKI failure27,55 and polyclonal resistance with different aberrations within the same tumor or in different metastatic sites increases the complexity of strategies to overcome resistance. For these situations and beyond, investigational genotype-directed therapies for the treatment of resistance are now challenged by molecular-agnostic approaches. For instance, patritumab-deruxtecan, an antibody-drug conjugate (ADC) targeting HER3, which is overexpressed in most EGFR-mutated NSCLC, has recently shown promising results in patients failing EGFR inhibitors in the phase I HERTHENA trial (objective response rate 39%, median PFS 8.2 months), irrespective of the post-TKI mechanism of resistance.56 It is clear that further refinement is needed for the optimal decision making in the treatment of resistance, possibly by recommending personalized post-progression therapies to those patients with a single, targetable mechanism of resistance and agents with less specificity in patients with complex resistance.
As for immunotherapy, to date, there are no good genomic predictors of immunotherapy response or resistance,57 while more extensive prediction systems that integrate genomics, tumor microenvironment and patients’ characteristics are now investigated.27
Beyond genomic alterations, homologous recombination deficiency (HRD) is found in nearly half of high-grade serous ovarian tumors,58 including tumors without BRCA alterations. In the phase III PAOLA-1 trial, tumor HRD has been shown to be a predictive biomarker of the efficacy of olaparib when used as a maintenance in combination with bevacizumab in patients with newly diagnosed advanced ovarian cancer.59 The Myriad MyChoice HRD score is based on whole-genome tumor loss of heterozygosity, telomeric allelic imbalance and large-scale state transition scores.60 It is a Food and Drug Administration (FDA)- and European Medicines Agency (EMA)-recognized biomarker, whose assessment is necessary for the prescription of olaparib–bevacizumab as a maintenance treatment in the first-line treatment of advanced ovarian cancer without BRCA mutations.
The promise of high-throughput sequencing beyond target detection
Prediction of prognosis and duration of response from baseline clonal heterogeneity analyses
Intratumoral clonal heterogeneity has been linked to poor clinical outcome across multiple cancer types.61, 62, 63, 64, 65, 66 This has been shown to be a key player in tumor evolution and treatment resistance, being found even in treatment-naïve tumors. In the TRACERx cohort of 100 operated NSCLC patients, subclonal events were detected by multiregion WES, with a median of 30% and 48% of the identified somatic mutations and copy number alterations, respectively. However, there was an increased variability in intratumoral heterogeneity among tumors [e.g. copy number alterations (CNAs) ranging from 0.06% to 81%]. Of note, patients with ≥48% subclonal CNAs had lower relapse-free survival (RFS) than those with lower proportions (hazard ratio 3.7 in a multivariate analysis), while for patients with high proportions of subclonal somatic mutations there were no significant associations with RFS.65 Similarly, in the renal cohort of TRACERx, the most chromosomally rearranged renal tumors were associated with the highest propensity to metastasize. The authors identified three types of cancer evolution, including tumors with low chromosomal complexity and low diversity between tumor regions (the lowest metastatic potential), high chromosomal complexity and high diversity (intermediate metastatic potential, subclonal somatic CNAs, high driver heterogeneity) and high chromosomal complexity with low diversity (high metastatic potential, clonal CNAs).66
The presence of de novo subclonal resistance mutations may impact the duration of response to standard treatment. For instance, in a study on 2774 lung cancer patients with available routine molecular testing, 2% (n = 20) of EGFR-mutant NSCLC tumors had de novo EGFR T790M mutations that predict resistance to first- and second-generation EGFR inhibitors. In 13 of these patients treated with erlotinib, the response rate was only 8%, the median PFS was 2 months and the median OS was 16 months.67 As these subclones may have a frequency below current standard detection thresholds,61 the use of molecular techniques with increased sensitivity such as single-cell analyses68 should better identify such de novo subclonal alterations, but it will be important to assess if this strategy has a clinical utility for the prediction of tumor relapse or progression that justifies its costs.
Resistance by clonal evolution over time
Over the past decade, the development of sequencing technologies accompanied by advances in bioinformatics resulted in a rise of clonal evolution studies. These studies aimed at rebuilding the global architecture of multiple cancer cell populations within a single patient; they allowed a better understanding of clonal heterogeneity, clonal evolution and cancer drivers with the definition of truncal or branchial genetic alterations.69 The trajectory of cancer characterized by WGS analyses in 2658 cancers by the Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium revealed a mutational spectrum that undergoes substantial changes during the course of disease progression. While early oncogenesis features are limited to a subset of driver genes and certain copy number gains, there is a nearly fourfold increase in driver alterations in later stages, with increased genomic instability.70 In 5% of cases, there were no identified drivers, suggesting that further research in this field is still needed.9 While data on driver discovery are mainly available on coding genome regions, the non-coding genome will need to also be explored for novel driver candidates to be identified.10
Classically, driver alterations have been considered to strongly impact tumor progression, while passenger alterations are inconsequential. However, there is an additional class of mutations with a weak tumor-promoting effect, which have been described as ‘mini-drivers’ or ‘putative passenger alterations with medium impact’.71,72 Thus, putative passenger alterations range from low-impact passengers to medium-impact passengers, which could act as weak drivers or deleterious alterations. Their molecular impact correlates with temporal evolution (early versus late events) and different genomic signatures. Across multiple cancer cohorts, signatures 1, 6, 7, 19, 28 and 35 had a higher contribution toward high-impact passengers. Similarly, tumors featuring MSI-H harbored more impactful passengers as compared to microsatellite stable tumors. Moreover, impact could change over time, from neutral function to impactful role after treatment or cell seeding in another organ.72 However, up to now, these data remained mainly descriptive and difficult to integrate in daily clinical practice for cancer treatment choices.
Persister cells
Studies have highlighted the fact that some rare cancer cells, called ‘persister cells’, are ‘waiting’ in a drug-tolerant state that allows them to survive treatment until they acquire the additional genetic alteration that will drive the full, classically described, resistance to therapy causing eventually patient relapse.73, 74, 75 Epigenetic adaptation is likely responsible for this dormant, quiescent, epithelial–mesenchymal transition-like or stemness cancer cell phenotype and DNA repair defects are expected to be responsible for the acquisition of resistant mutations and chromosomal rearrangements.76, 77, 78 Being able to eradicate those cancer cells early in cancer treatment could block or drastically postpone the development of resistance and maybe turn those refractory cancers into a curable disease (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2023.101642). Identifying and characterizing these cells require the holistic integration of whole-genome, -transcriptome and -epigenome profiling at the single-cell level on repeated sequential cancer samples. The recent advances in high-throughput NGS technologies such as RNAseq, WGS, ATACseq or MedIPseq at the single cell now allow this level of characterization. The scientists and clinicians are however dealing with huge amount of data that cannot be integrated by a single person and bioinformatics is also reaching here its limitation in terms of clinical decision-making support.
Developing new technologies to detect drivers
Beyond high-throughput genomics: protein-based assays and epigenetics
Genomic analyses offer only a glimpse into the complexity of cancer. Connecting genotype to phenotype is an essential challenge of precision medicine, with the deployment of proteomic, metabolomic, epigenetic and microRNA-based profiling in clinical practice.79,80 This would answer several challenges: (i) improvement of driver detection (e.g. estrogen, androgen, c-KIT receptors are often overexpressed in the absence of genomic alterations, or conversely, not all genomic alterations are stably expressed as proteins); (ii) intercepting resistance from early adaptive changes that occur via feedback loops or epigenetic modifications; (iii) better characterize acquired resistance in the absence of genomic determinants of progression (e.g. cellular plasticity, epigenetic changes); (iv) identify post-translational changes, especially phosphorylation, with functional consequences. This might improve the prioritization of target inhibition when more candidates are concurrently present.
The gap between genomics data and the phenotype of cancer cells can be primarily explained by the multilevel regulatory processes of gene expression. In cancer cells, for example, studies integrating genomics, transcriptomics and proteomics data (proteogenomics) have revealed the uncoupling between mRNA and protein expression levels in cancer cells.81, 82, 83 Alterations of cancer cells post-transcriptional machinery, especially protein synthesis and degradation, may account for this discrepancy.81,82 Therefore, proteomic data integration can contribute to identify actionable cancer-associated events not apparent within transcriptomic data and help interpret ambiguous genomic alterations. For example, proteogenomic approaches can point to a gene within an amplified region that is relevant for cancer cell phenotype and hence nominate precisely a therapeutic target, bridging genomic alteration to phenotype.
Protein activity is not always mirrored by its abundance. The post-translational modifications (PTMs) act as a switch that can regulate protein activity. Amongst the many PTMs, phosphorylation can be pharmacologically controlled by a large number of small molecule inhibitors and is accessible to omic-type analysis by mass spectrometry (MS)-based approaches. Phosphoproteomics opens a window on cellular protein activity and, associated to bioinformatics analysis tools, enables kinase activity scores to be calculated and dominant signaling processes to be identified.80,84, 85, 86, 87
The mammalian genome contains not only coding regions but also instructions for gene expression through epigenetic regulations. Epigenetic control switches chromatin states between inaccessible heterochromatin and accessible euchromatin, influencing the cell’s phenotypic space. Alterations in epigenetic regulators play a significant role in tumorigenesis, tumor progression and resistance.88,89 The non-coding genome, rarely studied in cancer, may hold important cis-acting alterations associated with oncogenesis, necessitating epigenetic scrutiny to understand their effects on cancer cells and expand the catalog of driver alterations. Laboratory initiatives have enriched genomic data with tumor samples’ epigenetic profiles using approaches like ATACseq.90 Integrating epigenetic data with multi-omics information uncovers new cis-regulatory elements and cancer subtype-specific enhancers, revealing potential driver transcriptional factors and genetic risk loci for cancer predisposition. This integration enhances our understanding of non-coding genomic alterations’ role in cancer predisposition, refines molecular subtyping and identifies driver transcriptional regulations.
Application of protein analyses for potential ADC tailoring
Along with increased knowledge of cancer biology and clonal evolution, and the need for more selective and potent anticancer therapies, the field of ADCs has rapidly expanded over the past decade.91, 92, 93, 94, 95, 96 The mechanism of action of ADCs is complex and not yet well elucidated, requiring drug internalization, intracellular trafficking and payload release.97,98 Currently, it remains unclear whether biomarker expression would improve drug efficacy, which raises several points. Firstly, the main modality used to determine target expression was IHC and several cut-offs have been used without a clear rationale. More sensitive technologies seeking to detect target expression below the standard threshold such as RNAseq could help to address this question. Innovative technologies such as NanoString, which looks at the spatial distribution of gene expression, or Imaging Mass Cytometry used to examine protein expression at the single-cell level and their spatial localization with precision could aid to better understand the mechanism of action and toxicity of ADCs.97,98 Some of these approaches are ongoing in the translational axis from the DAISY trial, which assesses trastuzumab deruxtecan (T-DXd) in patients with metastatic breast cancer, with results expected by the end of 2023 (NCT04132960).99 Secondly, it has been demonstrated in vivo that the efficacy of certain ADCs depends on the ‘bystander effect’, the capacity to kill neighboring cells irrespective of target antigen expression.100 Finally, target-independent ADC uptake such as pinocytosis or via binding to Fc receptors was also described.101
Moreover, biomarker characterization in patients with tumors harboring oncogenic driver mutations needs further exploration. In lung cancer cell lines and patient-derived xenograft models, it was demonstrated that clinical activity of anti-HER2 ADCs such as T-DXd in cells with ERBB2-activating mutation was favored by target ubiquitination and internalization facilitating endocytosis, regardless of HER2 expression.102 These data underline that ADCs’ efficacy is independent of their capability to inhibit oncogenic driver signaling. On the contrary, EGFR-mutated NSCLC expresses high levels of HER3 without detection of ERBB3 genomic alterations presenting a meaningful benefit from HER3-DXd.56
On the basis of this evidence, biomarkers of response and resistance to ADCs detected through tumor or plasma are an unmet medical need and essential for the suitable use of these drugs.
The use of organoids for cancer modeling
Cellular approaches, particularly organoids grown from stem/progenitor cells, offer promising therapeutic strategies in cancer modeling and precision medicine. Grown from stem and/or progenitor cells, organoids are multicellular structures that self-organize ex vivo in simple tridimensional matrix-based cultures. Despite them lacking most of their natural microenvironment, they recapitulate the main structural and functional properties of the organ they are originating from. This new generation of experimental models bridges a gap between cell lines and animal models, providing versatile but relevant tools to study most organs, from the brain to the gut.103,104
Over the past 10 years, organoids have been used in stem cell biology and for the comprehension of organogenesis. More recently, they have been engineered to modulate gene expression and model the appearance and progression of monogenetic diseases such as cystic fibrosis (CF) or cancer.104,105 As an example, the APC/KRAS/TP53 sequence identified by Fearon and Vogelstein in colorectal cancer in the 1990s has been introduced in normal gut organoids to demonstrate the causative role of these driver mutations in tumorigenesis.106, 107, 108 The culture of pathological organoids can also be initiated from the adult stem cells contained in patient biopsies or surgical specimens (Supplementary Figure S2, available at https://doi.org/10.1016/j.esmoop.2023.101642). Virtually immortal and genetically stable over time, patient-derived organoids (PDOs) provide unique tools to decrypt tumor phylogeny and intratumoral heterogeneity while large PDO collections (or living biobanks) recapitulate intertumoral diversity.
Now referred to as ‘avatars’, organoids display a remarkable clinical applicability. PDO collections are great preclinical tools for drug discovery while their normal counterparts can be used to assess compound toxicities. Organoids derived from rectal biopsies are already exploited by clinicians in precision medicine strategies to identify the best therapeutic option for CF patients with uncommon mutations in cystic fibrosis transmembrane conductance regulator (CFTR).109 Several publications have made clear that PDOs established from digestive, ovarian and bladder cancers corroborate patient responses to treatment with impressive positive and negative predictive values.110, 111, 112, 113, 114, 115 Clinical trials should now assess the power of organoids in the management of cancer patients. PDO-based functional assays could identify chemotherapies for tumors with no/low identified actionable oncogenic alterations. Furthermore, bridging functional and molecular precision medicine for each patient could reduce uncertainty in the decision-making process by validating the efficacy of targeted therapies or anticipating resistance. Engineering more complex organoid co-cultures with stromal components, including endothelial and immune cells, should also open the possibility to assess individual response to anti-angiogenics or immune checkpoint inhibitors.116, 117, 118 Thus, modeling patients’ tumors ex vivo using organoids and cell-based assays could be exploited to tailor individual treatment, prevent toxicities and anticipate future lines of therapies.
Modeling immune reaction
Cancer heterogeneity, with genetic and epigenetic modifications over time and space,119, 120, 121, 122 presents challenges in understanding its impact on the cancer immunopeptidome. Neoantigens, arising from point mutations, have been confirmed to influence antitumor responses.123, 124, 125 A significant proportion of neoantigens can also originate from non-mutated and non-exonic sequences, such as introns, intergenic regions or alternative reading frames.126, 127, 128, 129, 130, 131, 132, 133 The therapeutic potential of an antigen depends on its expression level, proportion of tumor cells expressing it and immunogenicity. Predictive algorithms aid in identifying immunogenic epitopes, but limitations exist, necessitating further refinement.
Given the heterogeneity of cancers, and the source of neoantigens (exon, intron, intergenic, 5′ and 3′ untranslated region sequences), it is still very difficult to model the cancer immune response accurately. A recent study showed that there is heterogeneity in the exonic immunopeptidome between four organoid clones derived from the same colorectal cancer patient.134 The use of NGS technologies, RNAseq, proteogenomics, bioinformatics and immunopeptide tools based on MS analysis has already allowed the identification of neoantigens from a biopsy of patient’s tumor tissue within ∼3weeks (Figure 3). Nevertheless, in the near future, in order to model the anticancer immune response, it will be important to accelerate the identification of large-scale cancer immunopeptidomes. To achieve this, considerable improvements are needed in (i) the process of immunopurification of tumor neoantigens from biopsies or organoids, (ii) the sensitivity of MS analyses, (iii) better predictive algorithms for major histocompatibility complex class I/peptide binding and also in terms of neoantigen immunogenicity and (iv) the development of ‘in situ’ platforms. Such a platform using biopsies or organoids derived from these biopsies could make possible the analysis of the immune reactivity of cancer infiltrates in primary tumors to the different neoantigens previously identified by the analysis of cancerous immunopeptidomes.
Figure 3.
To identify the immunopeptidome of each cancer, tumor biopsies are collected from the patient for whole-exome sequencing (WES) and RNA sequencing (RNAseq) in order to evaluate the repertoire of canonical and non-canonical antigens which are susceptibly produced in order to build a personalized peptide sequence database for each patient. In parallel, for the same tumor biopsies, the major histocompatibility complex class I (MHC-I) immunopeptidome is isolated by immunoprecipitation with antibodies that recognize the different human leukocyte antigen (HLA) molecules. The isolated MHC-I immunopeptidome is then analyzed by liquid chromatography-mass spectrometry (LC-MS)/MS. The MS/MS spectra of each canonical and non-canonical polypeptide found at the tumor cell surface are then searched against the established personalized peptide sequence database. The identified candidate epitopes are then selected using different algorithms to predict their affinity for MHC-I complexes. Then, the immunogenicity of the candidate neoantigens is finally obtained by in vitro immunological tests using autologous T cells or peripheral blood mononuclear cells (PBMCs).
MIP, MHC I-associated peptides; NGS, next-generation sequencing; UTR, untranslated region.
Big data management and sharing
Cohort-based discovery of drivers
Cancers accumulate multiple somatic mutations in their genomes, but only a small fraction of these mutations drive tumorigenesis. The discovery of driver mutations in cancer-related genes became an endeavor of cancer genomics in recent years, resulting in the development of numerous sophisticated bioinformatic algorithms. The majority of current methods model the gene-specific background mutation rate based on selectively neutral synonymous mutations and local epigenomic characteristics.3,135,136 Furthermore, background mutational models consider the nucleotide contexts of the mutations per tumor type and the direction of gene transcription.3,137
Positive selection of mutations in a driver gene may cause anomalies in its mutational properties and underlying deviation from the background mutational profile. Various mutation features of a gene are employed to predict its driver role in tumorigenesis. Most commonly used is an excess of functional mutations, causing amino acid changes (non-synonymous mutations), premature truncation of the proteins (stop mutations, frameshift indels) or anomalies of splicing (splice-site mutations).3,137 Moreover, scoring systems assessing the deleteriousness of functional mutations which integrate multiple annotations including conservation and functional impact are utilized to detect driver genes and prioritize driver mutations.9,138 Such mutations may also be localized at functionally relevant regions of genes, for example, rendering the genes constitutively active139 or impeding protein degradation.140 The resulting non-random distribution of mutations across the gene causes mutational recurrence and clusterization and it is utilized as a signature of positive selection by some algorithms.9,141,142 Furthermore, it was noted and successfully implemented in the driver-detection pipeline that nucleotide contexts of driver mutations may be different from the characteristic nucleotide contexts of passenger mutations in a given tumor type.143 At the same time the most comprehensive catalog of driver genes can be obtained through a combination of multiple prediction tools.144
As current algorithms depend on the cohort-based estimates of background mutational rate and profile, precision and sensitivity of these estimates depend on the cohort size and variation of mutational landscapes within the cohort. For example, the mutational landscape of a subset of tumors in a cohort may be predominantly driven by a specific mutational process, such as APOBEC-related, mismatch repair deficiency or mutant polymerase ε.145, 146, 147 This introduces heterogeneity in the cohort in terms of mutation rates, spectrums and distribution across the genome hampering the accuracy of estimation of the background mutagenesis and affecting the detection of driver genes.148 This aspect is particularly relevant for the detection of drivers in tumor relapses after a mutagenic treatment, which is frequently associated with the mutator phenotype and specific mutational profile,149,150 and urges the need of coordinated multicentric sequencing effort for rare and pretreated aggressive tumors.
Merge information in a single device to model cancer biology
Current strategies to stratify patients or to design new therapeutic approaches are based on the segmented analysis of tumor molecular traits and rarely consider tumor biology as a whole. The reason lies in the complexity of such endeavor. The resistance observed in the clinic arises from virtually unique combinations of genomic alterations that result in epigenetic, transcriptional, translation as well as post-translational perturbations. To be efficient, the design of therapeutic approaches needs to encompass this multilayered reality.
There are two main paths to model this level of complexity. The first strategy relies on ex vivo avatar of individual patient tumors. A second approach coalesces comprehensive molecular information into a single device (Figure 4). Data science can now feed in to generate in silico modeling of tumor phenotypic and evolutionary space and vulnerabilities on a per patient mode. Tumor models need to integrate oncogenic drivers in their full diversity. Considering the variability of individual cancer evolution and the various consequences of treatment selection pressure, there is a compelling need to model biology on an individual and temporal scale. Oncogenic drivers represent the source of therapeutic targets but only at a given time and possibly for only a subset of tumor cells. Accurate models need to take into account the ability of tumor cells to evolve individually within a tumor, giving rise to clonal heterogeneity. The determination of the clonal makeup of a tumor can rely on genetic subclones tracking through longitudinally collected biopsies or alternatively involve circulating tumor DNA serial sampling. Beside the well-documented clonal, genetic-based tumor heterogeneity, tumor cells of the same genetic makeup can display phenotypic variability, the so-called phenotypic heterogeneity which stems from combinations of environmental inputs and stochastic events that eventually impact gene expression and transcriptional programs controlled by epigenetic regulations. Of note, epigenetic processes have been shown to enable tumor cells to acquire a persistent cell phenotype, from which full therapy resistance can arise.151 The phenotypic plasticity of tumor cells also needs to be delineated in order to feed integrative tumor models able to predict tumor evolution upon exposure to therapy. Finally, the tumor microenvironment is also key to understand treatment outcome. In particular, response to immunotherapy must be faithfully modeled in the current anticancer therapeutic landscape. Cancer models should therefore integrate the determinants of tumor immune status and immunotherapy response discussed in the preceding text.
Figure 4.
From the understanding of cancer progression to the identification of targets for treatment personalization.
ICB, immune checkpoint blockade.
It seems likely that only artificial intelligence approaches would be powerful enough to integrate massive data with spatial and temporal timepoints and holistic analysis aiming at providing clinical support on a daily basis. How artificial intelligence models would be able to define signature biomarkers early in cancer treatment to predict resistance and anticipate treatment tailoring is now the challenge for the coming years.
Conclusion
The examples of success in precision medicine are based on the discovery of truncal drivers and the use of an optimally matched drug. Resistance alterations and mini-drivers have also been gaining field over the last years, becoming promising drug targets. The rank of genomic alterations based on their level of evidence of actionability is essential for drug-matched prioritization. Beyond drivers, further advancements in the field of precision medicine should consider the multilayered reality of the spatial and temporal cancer heterogeneity. With the increasing availability of complex biological data, artificial intelligence algorithms have already become an indispensable tool for analysis. On a clinical perspective, integrative analysis of multiple omics data gives a focus to functional targets but also identifies master regulatory processes, both being targetable and hence highly valuable for clinical translation. On a more basic point of view, integrative data analysis enables the oncogenic processes to be comprehensively interrogated, leading to a broader understanding of tumor biology and opening the way to integrative tumor model development.
Acknowledgements
Figures were created by Mihaela Aldea with BioRender.com.
Funding
None declared.
Disclosure
MA: expenses: Sandoz; advisory board: Viatris; funding for academic research: Sandoz, Amgen, AstraZeneca. JCS has been a full-time employee of AstraZeneca between September 2017 and January 2020. He is a shareholder of AstraZeneca and Gritstone and currently Amgen’s senior vice president of Oncology. FA: travel/accommodation/expenses from AstraZeneca, GlaxoSmithKline, Novartis and Roche, and his institution has received research funding from AstraZeneca, Lilly, Novartis, Pfizer and Roche. Consultant/advisory fees from Amgen, Astellas, Astra Zeneca, Bayer, BeiGene, BMS, Celgene, Debiopharm, Genentech, Ipsen, Janssen, Lilly, MedImmune, MSD, Novartis, Pfizer, Roche, Sanofi, Orion. Principal/sub-investigator of clinical trials for AbbVie, Aduro, Agios, Amgen, Argen-x, Astex, AstraZeneca, Aveo pharmaceuticals, Bayer, Beigene, Blueprint, BMS, Boehringer Ingelheim, Celgene, Chugai, Clovis, Daiichi Sankyo, Debiopharm, Eisai, Eos, Exelixis, Forma, Gamamabs, Genentech, Gortec, GSK, H3 biomedecine, Incyte, InnatePharma, Janssen, Kura Oncology, Kyowa, Lilly, Loxo, Lysarc, Lytix Biopharma, Medimmune, Menarini, Merus, MSD, Nanobiotix, Nektar Therapeutics, Novartis, Octimet, Oncoethix, Oncopeptides AB, Orion, Pfizer, Pharmamar, Pierre Fabre, Roche, Sanofi, Servier, Sierra Oncology, Taiho, Takeda, Tesaro, Xencor. All other authors have declared no conflicts of interest.
Supplementary data
References
- 1.Yates L.R., Seoane J., Le Tourneau C., et al. The European Society for Medical Oncology (ESMO) precision medicine glossary. Ann Oncol. 2018;29:30–35. doi: 10.1093/annonc/mdx707. [DOI] [PubMed] [Google Scholar]
- 2.Druker B.J., Talpaz M., Resta D.J., et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 3.Martincorena I., Raine K.M., Gerstung M., et al. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171:1029–1041.e1021. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman P.B., Hauschild A., Robert C., et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caspar S.M., Dubacher N., Kopps A.M., et al. Clinical sequencing: from raw data to diagnosis with lifetime value. Clin Genet. 2018;93:508–519. doi: 10.1111/cge.13190. [DOI] [PubMed] [Google Scholar]
- 7.Zehir A., Benayed R., Shah R.H., et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler D.A., Srinivasan M., Egholm M., et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 9.Consortium ITP-CAoWG Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rheinbay E., Nielsen M.M., Abascal F., et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature. 2020;578:102–111. doi: 10.1038/s41586-020-1965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swanton C., Soria J.C., Bardelli A., et al. Consensus on precision medicine for metastatic cancers: a report from the MAP conference. Ann Oncol. 2016;27:1443–1448. doi: 10.1093/annonc/mdw192. [DOI] [PubMed] [Google Scholar]
- 12.Andre F., Bachelot T., Commo F., et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15:267–274. doi: 10.1016/S1470-2045(13)70611-9. [DOI] [PubMed] [Google Scholar]
- 13.Le Tourneau C., Delord J.P., Goncalves A., et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16:1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 14.Wheler J.J., Janku F., Naing A., et al. Cancer therapy directed by comprehensive genomic profiling: a single center study. Cancer Res. 2016;76:3690–3701. doi: 10.1158/0008-5472.CAN-15-3043. [DOI] [PubMed] [Google Scholar]
- 15.Stockley T.L., Oza A.M., Berman H.K., et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med. 2016;8:109. doi: 10.1186/s13073-016-0364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massard C., Michiels S., Ferte C., et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7:586–595. doi: 10.1158/2159-8290.CD-16-1396. [DOI] [PubMed] [Google Scholar]
- 17.Tredan O., Wang Q., Pissaloux D., et al. Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: analysis from the ProfiLER trial. Ann Oncol. 2019;30:757–765. doi: 10.1093/annonc/mdz080. [DOI] [PubMed] [Google Scholar]
- 18.Tsimberidou A.M., Hong D.S., Wheler J.J., et al. Long-term overall survival and prognostic score predicting survival: the IMPACT study in precision medicine. J Hematol Oncol. 2019;12:145. doi: 10.1186/s13045-019-0835-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sicklick J.K., Kato S., Okamura R., et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med. 2019;25:744–750. doi: 10.1038/s41591-019-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodon J., Soria J.C., Berger R., et al. Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat Med. 2019;25:751–758. doi: 10.1038/s41591-019-0424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothwell D.G., Ayub M., Cook N., et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25:738–743. doi: 10.1038/s41591-019-0380-z. [DOI] [PubMed] [Google Scholar]
- 22.Pishvaian M.J., Blais E.M., Brody J.R., et al. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: a retrospective analysis of the Know Your Tumor registry trial. Lancet Oncol. 2020;21:508–518. doi: 10.1016/S1470-2045(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horak P., Heining C., Kreutzfeldt S., et al. Comprehensive genomic and transcriptomic analysis for guiding therapeutic decisions in patients with rare cancers. Cancer Discov. 2021;11:2780–2795. doi: 10.1158/2159-8290.CD-21-0126. [DOI] [PubMed] [Google Scholar]
- 24.van Tilburg C.M., Pfaff E., Pajtler K.W., et al. The pediatric precision oncology INFORM registry: clinical outcome and benefit for patients with very high-evidence targets. Cancer Discov. 2021;11:2764–2779. doi: 10.1158/2159-8290.CD-21-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aftimos P., Oliveira M., Irrthum A., et al. Genomic and transcriptomic analyses of breast cancer primaries and matched metastases in AURORA, the Breast International Group (BIG) molecular screening initiative. Cancer Discov. 2021;11:2796–2811. doi: 10.1158/2159-8290.CD-20-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertucci F., Ng C.K.Y., Patsouris A., et al. Author Correction: Genomic characterization of metastatic breast cancers. Nature. 2019;572:E7. doi: 10.1038/s41586-019-1380-3. [DOI] [PubMed] [Google Scholar]
- 27.Aldea M., Andre F., Marabelle A., et al. Overcoming resistance to tumor-targeted and immune-targeted therapies. Cancer Discov. 2021;11:874–899. doi: 10.1158/2159-8290.CD-20-1638. [DOI] [PubMed] [Google Scholar]
- 28.van de Haar J., Hoes L.R., Roepman P., et al. Limited evolution of the actionable metastatic cancer genome under therapeutic pressure. Nat Med. 2021;27:1553–1563. doi: 10.1038/s41591-021-01448-w. [DOI] [PubMed] [Google Scholar]
- 29.Pietrantonio F., Morano F., Lonardi S., et al. 383O MAYA trial: temozolomide (TMZ) priming followed by combination with low-dose ipilimumab and nivolumab in patients with microsatellite stable (MSS), MGMT silenced metastatic colorectal cancer (mCRC) Ann Oncol. 2021;32:S530–S531. doi: 10.1200/JCO.21.02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andre F., Hurvitz S., Fasolo A., et al. Molecular alterations and everolimus efficacy in human epidermal growth factor receptor 2-overexpressing metastatic breast cancers: combined exploratory biomarker analysis from BOLERO-1 and BOLERO-3. J Clin Oncol. 2016;34:2115–2124. doi: 10.1200/JCO.2015.63.9161. [DOI] [PubMed] [Google Scholar]
- 31.Tsimberidou A.M., Kurzrock R. Precision medicine: lessons learned from the SHIVA trial. Lancet Oncol. 2015;16:e579–e580. doi: 10.1016/S1470-2045(15)00397-6. [DOI] [PubMed] [Google Scholar]
- 32.Chakravarty D., Gao J., Phillips S.M., et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mateo J., Chakravarty D., Dienstmann R., et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) Ann Oncol. 2018;29:1895–1902. doi: 10.1093/annonc/mdy263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosele F., Remon J., Mateo J., et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31:1491–1505. doi: 10.1016/j.annonc.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Condorelli R., Mosele F., Verret B., et al. Genomic alterations in breast cancer: level of evidence for actionability according to ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) Ann Oncol. 2019;30:365–373. doi: 10.1093/annonc/mdz036. [DOI] [PubMed] [Google Scholar]
- 36.Andre F., Filleron T., Kamal M., et al. Genomics to select treatment for patients with metastatic breast cancer. Nature. 2022;610:343–348. doi: 10.1038/s41586-022-05068-3. [DOI] [PubMed] [Google Scholar]
- 37.Verdaguer H., Sauri T., Acosta D.A., et al. ESMO Scale for Clinical Actionability of Molecular Targets driving targeted treatment in patients with cholangiocarcinoma. Clin Cancer Res. 2022;28:1662–1671. doi: 10.1158/1078-0432.CCR-21-2384. [DOI] [PubMed] [Google Scholar]
- 38.Necchi A., Madison R., Pal S.K., et al. Comprehensive genomic profiling of upper-tract and bladder urothelial carcinoma. Eur Urol Focus. 2021;7:1339–1346. doi: 10.1016/j.euf.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 39.van Geelen C.T., Savas P., Teo Z.L., et al. Clinical implications of prospective genomic profiling of metastatic breast cancer patients. Breast Cancer Res. 2020;22:91. doi: 10.1186/s13058-020-01328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hempel D., Ebner F., Garg A., et al. Real world data analysis of next generation sequencing and protein expression in metastatic breast cancer patients. Sci Rep. 2020;10 doi: 10.1038/s41598-020-67393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gyawali B., Kesselheim A.S. The promise of ESCAT: a new system for evaluating cancer drug-target pairs. Nat Rev Clin Oncol. 2019;16:147–148. doi: 10.1038/s41571-018-0110-3. [DOI] [PubMed] [Google Scholar]
- 42.Le D.T., Durham J.N., Smith K.N., et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aldea M., Cerbone L., Bayle A., et al. Detection of additional occult malignancy through profiling of ctDNA in late-stage cancer patients. Ann Oncol. 2021;32:1642–1645. doi: 10.1016/j.annonc.2021.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Hause R.J., Pritchard C.C., Shendure J., Salipante S.J. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016;22:1342–1350. doi: 10.1038/nm.4191. [DOI] [PubMed] [Google Scholar]
- 45.Cocco E., Scaltriti M., Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15:731–747. doi: 10.1038/s41571-018-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leonetti A., Sharma S., Minari R., et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer. 2019;121:725–737. doi: 10.1038/s41416-019-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eno M.S., Brubaker J.D., Campbell J.E., et al. Discovery of BLU-945, a reversible, potent, and wild-type-sparing next-generation EGFR mutant inhibitor for treatment-resistant non-small-cell lung cancer. J Med Chem. 2022;65:9662–9677. doi: 10.1021/acs.jmedchem.2c00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sequist L.V., Han J.Y., Ahn M.J., et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol. 2020;21:373–386. doi: 10.1016/S1470-2045(19)30785-5. [DOI] [PubMed] [Google Scholar]
- 49.Oxnard G.R., Yang J.C., Yu H., et al. TATTON: a multi-arm, phase Ib trial of osimertinib combined with selumetinib, savolitinib, or durvalumab in EGFR-mutant lung cancer. Ann Oncol. 2020;31:507–516. doi: 10.1016/j.annonc.2020.01.013. [DOI] [PubMed] [Google Scholar]
- 50.Piotrowska Z., Isozaki H., Lennerz J.K., et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov. 2018;8:1529–1539. doi: 10.1158/2159-8290.CD-18-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang W., He Q., Chen Y., et al. Metastatic EML4-ALK fusion detected by circulating DNA genotyping in an EGFR-mutated NSCLC patient and successful management by adding ALK inhibitors: a case report. BMC Cancer. 2016;16:62. doi: 10.1186/s12885-016-2088-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schrock A.B., Zhu V.W., Hsieh W.S., et al. Receptor tyrosine kinase fusions and BRAF kinase fusions are rare but actionable resistance mechanisms to EGFR tyrosine kinase inhibitors. J Thorac Oncol. 2018;13:1312–1323. doi: 10.1016/j.jtho.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 53.Sun M., Wang X., Xu Y., et al. Combined targeting of EGFR and BRAF triggers regression of osimertinib resistance by using osimertinib and vemurafenib concurrently in a patient with heterogeneity between different lesions. Thorac Cancer. 2021;13:514–516. doi: 10.1111/1759-7714.14295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rotow J., Patel J., Hanley M., et al. FP14.07 Combination osimertinib plus selpercatinib for EGFR-mutant non-small cell lung cancer (NSCLC) with acquired RET fusions. J Thorac Oncol. 2021;16:S230. [Google Scholar]
- 55.Roper N., Brown A.-L., Wei J.S., et al. Clonal evolution and heterogeneity of osimertinib acquired resistance mechanisms in EGFR mutant lung cancer. Cell Rep Med. 2020;1 doi: 10.1016/j.xcrm.2020.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Janne P.A., Baik C., Su W.C., et al. Efficacy and safety of patritumab deruxtecan (HER3-DXd) in EGFR inhibitor-resistant, EGFR-mutated non-small cell lung cancer. Cancer Discov. 2022;12:74–89. doi: 10.1158/2159-8290.CD-21-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Domingues I., Cedres S., Callejo A., et al. Long duration of immunotherapy in a STK11 mutated/KRAS wild-type non-small cell lung cancer patient. Pulmonology. 2020;26:49–50. doi: 10.1016/j.pulmoe.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 58.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ray-Coquard I., Pautier P., Pignata S., et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. 2019;381:2416–2428. doi: 10.1056/NEJMoa1911361. [DOI] [PubMed] [Google Scholar]
- 60.Hodgson D.R., Dougherty B.A., Lai Z., et al. Candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. Br J Cancer. 2018;119:1401–1409. doi: 10.1038/s41416-018-0274-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chowell D., Napier J., Gupta R., et al. Modeling the subclonal evolution of cancer cell populations. Cancer Res. 2018;78:830–839. doi: 10.1158/0008-5472.CAN-17-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J., Fujimoto J., Zhang J., et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andor N., Graham T.A., Jansen M., et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pereira B., Chin S.F., Rueda O.M., et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7 doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jamal-Hanjani M., Wilson G.A., McGranahan N., et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 66.Turajlic S., Xu H., Litchfield K., et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell. 2018;173:595–610.e511. doi: 10.1016/j.cell.2018.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu H.A., Arcila M.E., Hellmann M.D., et al. Poor response to erlotinib in patients with tumors containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol. 2014;25:423–428. doi: 10.1093/annonc/mdt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Velazquez-Villarreal E.I., Maheshwari S., Sorenson J., et al. Single-cell sequencing of genomic DNA resolves sub-clonal heterogeneity in a melanoma cell line. Commun Biol. 2020;3:318. doi: 10.1038/s42003-020-1044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGranahan N., Favero F., de Bruin E.C., et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerstung M., Jolly C., Leshchiner I., et al. The evolutionary history of 2,658 cancers. Nature. 2020;578:122–128. doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castro-Giner F., Ratcliffe P., Tomlinson I. The mini-driver model of polygenic cancer evolution. Nat Rev Cancer. 2015;15:680–685. doi: 10.1038/nrc3999. [DOI] [PubMed] [Google Scholar]
- 72.Kumar S., Warrell J., Li S., et al. Passenger mutations in more than 2,500 cancer genomes: overall molecular functional impact and consequences. Cell. 2020;180:915–927.e916. doi: 10.1016/j.cell.2020.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hata A.N., Niederst M.J., Archibald H.L., et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22:262–269. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hangauer M.J., Viswanathan V.S., Ryan M.J., et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247–250. doi: 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramirez M., Rajaram S., Steininger R.J., et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun. 2016;7 doi: 10.1038/ncomms10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niveditha D., Sharma H., Sahu A., et al. Drug tolerant cells: an emerging target with unique transcriptomic features. Cancer Inform. 2019;18 doi: 10.1177/1176935119881633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vallette F.M., Olivier C., Lezot F., et al. Dormant, quiescent, tolerant and persister cells: four synonyms for the same target in cancer. Biochem Pharmacol. 2019;162:169–176. doi: 10.1016/j.bcp.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 78.Raoof S., Mulford I.J., Frisco-Cabanos H., et al. Targeting FGFR overcomes EMT-mediated resistance in EGFR mutant non-small cell lung cancer. Oncogene. 2019;38:6399–6413. doi: 10.1038/s41388-019-0887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang B., Whiteaker J.R., Hoofnagle A.N., et al. Clinical potential of mass spectrometry-based proteogenomics. Nat Rev Clin Oncol. 2019;16:256–268. doi: 10.1038/s41571-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang B., Wang J., Wang X., et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clark D.J., Dhanasekaran S.M., Petralia F., et al. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell. 2019;179:964–983.e931. doi: 10.1016/j.cell.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dou Y., Kawaler E.A., Cui Zhou D., et al. Proteogenomic characterization of endometrial carcinoma. Cell. 2020;180:729–748.e726. doi: 10.1016/j.cell.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gillette M.A., Satpathy S., Cao S., et al. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell. 2020;182:200–225.e235. doi: 10.1016/j.cell.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Petralia F., Tignor N., Reva B., et al. Integrated proteogenomic characterization across major histological types of pediatric brain cancer. Cell. 2020;183:1962–1985.e1931. doi: 10.1016/j.cell.2020.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mertins P., Mani D.R., Ruggles K.V., et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rivero-Hinojosa S., Lau L.S., Stampar M., et al. Proteomic analysis of medulloblastoma reveals functional biology with translational potential. Acta Neuropathol Commun. 2018;6:48. doi: 10.1186/s40478-018-0548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang H., Liu T., Zhang Z., et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell. 2016;166:755–765. doi: 10.1016/j.cell.2016.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cancer Genome Atlas Research Network. Weinstein J.N., Collisson E.A., et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bertucci F., Ng C.K.Y., Patsouris A., et al. Genomic characterization of metastatic breast cancers. Nature. 2019;569:560–564. doi: 10.1038/s41586-019-1056-z. [DOI] [PubMed] [Google Scholar]
- 90.Corces M.R., Granja J.M., Shams S., et al. The chromatin accessibility landscape of primary human cancers. Science. 2018;362 doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tsuchikama K., An Z. Antibody-drug conjugates: recent advances in conjugation and linker chemistries. Protein Cell. 2018;9:33–46. doi: 10.1007/s13238-016-0323-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bardia A., Tolaney S.M., Punie K., et al. Biomarker analyses in the phase III ASCENT study of sacituzumab govitecan versus chemotherapy in patients with metastatic triple-negative breast cancer. Ann Oncol. 2021;32:1148–1156. doi: 10.1016/j.annonc.2021.06.002. [DOI] [PubMed] [Google Scholar]
- 93.Cortes J., Kim S.B., Chung W.-P., et al. LBA1 Trastuzumab deruxtecan (T-DXd) vs trastuzumab emtansine (T-DM1) in patients (Pts) with HER2+ metastatic breast cancer (mBC): results of the randomized phase III DESTINY-Breast03 study. Ann Oncol. 2021;32:S1287–S1288. [Google Scholar]
- 94.Modi S., Park H., Murthy R.K., et al. Antitumor activity and safety of trastuzumab deruxtecan in patients with HER2-low-expressing advanced breast cancer: results from a phase Ib study. J Clin Oncol. 2020;38:1887–1896. doi: 10.1200/JCO.19.02318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smit E., Nakagawa K., Nagasaka M., et al. Trastuzumab deruxtecan (T-DXd; DS-8201) in patients with HER2-mutated metastatic non-small cell lung cancer (NSCLC): interim results of DESTINY-Lung01. J Clin Oncol. 2020;38:9504. 9504. [Google Scholar]
- 96.Nakagawa K., Nagasaka M., Felip E., et al. OA04.05 Trastuzumab deruxtecan in HER2-overexpressing metastatic non-small cell lung cancer: interim results of DESTINY-Lung01. J Thorac Oncol. 2021;16:S109–S110. [Google Scholar]
- 97.Schlam I., Church S.E., Hether T.D., et al. The tumor immune microenvironment of primary and metastatic HER2- positive breast cancers utilizing gene expression and spatial proteomic profiling. J Transl Med. 2021;19:480. doi: 10.1186/s12967-021-03113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bianchini G., Wang X.Q., Danenberg E., et al. Abstract GS1-00: Single-cell spatial analysis by imaging mass cytometry and immunotherapy response in triple-negative breast cancer (TNBC) in the NeoTRIPaPDL1 trial. Cancer Res. 2022;82 [Google Scholar]
- 99.Mosele M.F., Lusque A., Dieras V., et al. LBA1 Unraveling the mechanism of action and resistance to trastuzumab deruxtecan (T-DXd): biomarker analyses from patients from DAISY trial. Ann Oncol. 2022;33:S123. [Google Scholar]
- 100.Ogitani Y., Hagihara K., Oitate M., et al. Bystander killing effect of DS-8201a, a novel anti-human epidermal growth factor receptor 2 antibody-drug conjugate, in tumors with human epidermal growth factor receptor 2 heterogeneity. Cancer Sci. 2016;107:1039–1046. doi: 10.1111/cas.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mahalingaiah P.K., Ciurlionis R., Durbin K.R., et al. Potential mechanisms of target-independent uptake and toxicity of antibody-drug conjugates. Pharmacol Ther. 2019;200:110–125. doi: 10.1016/j.pharmthera.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 102.Li B.T., Michelini F., Misale S., et al. HER2-mediated internalization of cytotoxic agents in ERBB2 amplified or mutant lung cancers. Cancer Discov. 2020;10:674–687. doi: 10.1158/2159-8290.CD-20-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim J., Koo B.K., Knoblich J.A. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 2020;21:571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tuveson D., Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 105.Dekkers J.F., Wiegerinck C.L., de Jonge H.R., et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 106.Fearon E.R., Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 107.Matano M., Date S., Shimokawa M., et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 108.Drost J., van Jaarsveld R.H., Ponsioen B., et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 109.Berkers G., van Mourik P., Vonk A.M., et al. Rectal organoids enable personalized treatment of cystic fibrosis. Cell Rep. 2019;26:1701–1708.e1703. doi: 10.1016/j.celrep.2019.01.068. [DOI] [PubMed] [Google Scholar]
- 110.Pauli C., Hopkins B.D., Prandi D., et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vlachogiannis G., Hedayat S., Vatsiou A., et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee S.H., Hu W., Matulay J.T., et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 2018;173:515–528.e517. doi: 10.1016/j.cell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hill S.J., Decker B., Roberts E.A., et al. Prediction of DNA repair inhibitor response in short-term patient-derived ovarian cancer organoids. Cancer Discov. 2018;8:1404–1421. doi: 10.1158/2159-8290.CD-18-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tiriac H., Belleau P., Engle D.D., et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8:1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ooft S.N., Weeber F., Dijkstra K.K., et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aay2574. [DOI] [PubMed] [Google Scholar]
- 116.Neal J.T., Li X., Zhu J., et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988.e1916. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dijkstra K.K., Cattaneo C.M., Weeber F., et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174:1586–1598.e1512. doi: 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Takebe T., Sekine K., Enomura M., et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 119.Khurana E., Fu Y., Chakravarty D., et al. Role of non-coding sequence variants in cancer. Nat Rev Genet. 2016;17:93–108. doi: 10.1038/nrg.2015.17. [DOI] [PubMed] [Google Scholar]
- 120.Ji Z., Song R., Regev A., Struhl K. Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015;4 doi: 10.7554/eLife.08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kassiotis G., Stoye J.P. Immune responses to endogenous retroelements: taking the bad with the good. Nat Rev Immunol. 2016;16:207–219. doi: 10.1038/nri.2016.27. [DOI] [PubMed] [Google Scholar]
- 122.Verdegaal E.M., de Miranda N.F., Visser M., et al. Neoantigen landscape dynamics during human melanoma-T cell interactions. Nature. 2016;536:91–95. doi: 10.1038/nature18945. [DOI] [PubMed] [Google Scholar]
- 123.van Rooij N., van Buuren M.M., Philips D., et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Parkhurst M., Gros A., Pasetto A., et al. Isolation of T-cell receptors specifically reactive with mutated tumor-associated antigens from tumor-infiltrating lymphocytes based on CD137 expression. Clin Cancer Res. 2017;23:2491–2505. doi: 10.1158/1078-0432.CCR-16-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Robbins P.F., Lu Y.C., El-Gamil M., et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Apcher S., Millot G., Daskalogianni C., et al. Translation of pre-spliced RNAs in the nuclear compartment generates peptides for the MHC class I pathway. Proc Natl Acad Sci U S A. 2013;110:17951–17956. doi: 10.1073/pnas.1309956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Laumont C.M., Daouda T., Laverdure J.P., et al. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat Commun. 2016;7 doi: 10.1038/ncomms10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Laumont C.M., Perreault C. Exploiting non-canonical translation to identify new targets for T cell-based cancer immunotherapy. Cell Mol Life Sci. 2018;75:607–621. doi: 10.1007/s00018-017-2628-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pearson H., Daouda T., Granados D.P., et al. MHC class I-associated peptides derive from selective regions of the human genome. J Clin Invest. 2016;126:4690–4701. doi: 10.1172/JCI88590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shastri N., Cardinaud S., Schwab S.R., et al. All the peptides that fit: the beginning, the middle, and the end of the MHC class I antigen-processing pathway. Immunol Rev. 2005;207:31–41. doi: 10.1111/j.0105-2896.2005.00321.x. [DOI] [PubMed] [Google Scholar]
- 131.Granados D.P., Sriranganadane D., Daouda T., et al. Impact of genomic polymorphisms on the repertoire of human MHC class I-associated peptides. Nat Commun. 2014;5:3600. doi: 10.1038/ncomms4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bassani-Sternberg M., Braunlein E., Klar R., et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun. 2016;7 doi: 10.1038/ncomms13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schuster H., Peper J.K., Bosmuller H.C., et al. The immunopeptidomic landscape of ovarian carcinomas. Proc Natl Acad Sci U S A. 2017;114:E9942–E9951. doi: 10.1073/pnas.1707658114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Demmers L.C., Kretzschmar K., Van Hoeck A., et al. Single-cell derived tumor organoids display diversity in HLA class I peptide presentation. Nat Commun. 2020;11:5338. doi: 10.1038/s41467-020-19142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Supek F., Minana B., Valcarcel J., et al. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156:1324–1335. doi: 10.1016/j.cell.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 136.Lawrence M.S., Stojanov P., Polak P., et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Alexandrov L.B., Nik-Zainal S., Wedge D.C., et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mularoni L., Sabarinathan R., Deu-Pons J., et al. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 2016;17:128. doi: 10.1186/s13059-016-0994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Davies H., Bignell G.R., Cox C., et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 140.Martínez-Jiménez F., Muiños F., López-Arribillaga E., et al. Systematic analysis of alterations in the ubiquitin proteolysis system reveals its contribution to driver mutations in cancer. Nat Cancer. 2020;1:122–135. doi: 10.1038/s43018-019-0001-2. [DOI] [PubMed] [Google Scholar]
- 141.Tamborero D., Gonzalez-Perez A., Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics. 2013;29:2238–2244. doi: 10.1093/bioinformatics/btt395. [DOI] [PubMed] [Google Scholar]
- 142.Bonilla X., Parmentier L., King B., et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016;48:398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 143.Dietlein F., Weghorn D., Taylor-Weiner A., et al. Identification of cancer driver genes based on nucleotide context. Nat Genet. 2020;52:208–218. doi: 10.1038/s41588-019-0572-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bailey M.H., Tokheim C., Porta-Pardo E., et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385.e318. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Roberts S.A., Lawrence M.S., Klimczak L.J., et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Supek F., Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–84. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Church D.N., Briggs S.E., Palles C., et al. DNA polymerase epsilon and delta exonuclease domain mutations in endometrial cancer. Hum Mol Genet. 2013;22:2820–2828. doi: 10.1093/hmg/ddt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nik-Zainal S., Davies H., Staaf J., et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Boot A., Huang M.N., Ng A.W.T., et al. In-depth characterization of the cisplatin mutational signature in human cell lines and in esophageal and liver tumors. Genome Res. 2018;28:654–665. doi: 10.1101/gr.230219.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Angus L., Smid M., Wilting S.M., et al. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat Genet. 2019;51:1450–1458. doi: 10.1038/s41588-019-0507-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sharma S.V., Lee D.Y., Li B., et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.