Abstract
Exosomal microRNAs (miRNAs/miRs) are potential biomarkers for the diagnosis and treatment of cardiovascular disease, and hyperglycemia serves an important role in the development of atherosclerosis. The present study aimed to investigate the expression profile of serum-derived exosomal miRNAs in coronary heart disease (CHD) with hyperglycemia, and to identify effective biomarkers for predicting coronary artery lesions. Serum samples were collected from eight patients with CHD and hyperglycemia and eight patients with CHD and normoglycemia, exosomes were isolated and differentially expressed miRNAs (DEMIs) were filtered using a human miRNA microarray. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using standard enrichment computational methods for the target genes of DEMIs. Receiver operating characteristic (ROC) curve analysis was applied to evaluate the values of the selected DEMIs in predicting the severity of coronary stenosis. A total of 10 DEMIs, including four upregulated miRNAs (hsa-let-7b-5p, hsa-miR-4313, hsa-miR-4665-3p and hsa-miR-940) and six downregulated miRNAs (hsa-miR-4459, hsa-miR-4687-3p, hsa-miR-6087, hsa-miR-6089, hsa-miR-6740-5p and hsa-miR-6800-5p), were screened in patients with CHD and hyperglycemia. GO analysis showed that the ‘cellular process’, ‘single-organism process’ and ‘biological regulation’ were significantly enriched. KEGG pathway analysis revealed that the ‘mTOR signaling pathway’, ‘FoxO signaling pathway’ and ‘neurotrophin signaling pathway’ were significantly enriched. Among these DEMIs, only hsa-let-7b-5p expression was positively correlated with both hemoglobin A1C levels and Synergy between Percutaneous Coronary Intervention with Taxus and Cardiac Surgery score. ROC curves showed that hsa-let-7b-5p could serve as an effective biomarker for differentiating the severity of coronary stenosis. In conclusion, the present study demonstrated that serum-derived exosomal hsa-let-7b-5p is upregulated in patients with CHD and hyperglycemia, and may serve as a noninvasive biomarker for the severity of coronary stenosis.
Keywords: exosomal miRNAs, hyperglycemia, CHD, hsa-let-7b-5p, coronary stenosis
Introduction
Coronary heart disease (CHD) is a major cardiovascular disease and is the leading cause of mortality worldwide (1). The status of hyperglycemia causes vascular endothelial dysfunction and injury, resulting in the development of atherosclerosis. According to the latest epidemiological data, hyperglycemia is a major risk factor for patients with CHD and affects the prognosis of ~75% of patients (2). Previous studies have suggested that the severity of coronary stenosis in patients with CHD and hyperglycemia is more severe than that in patients with CHD and normoglycemia (3,4). With ongoing progress in the prevention and treatment of CHD, the molecular mechanism underlying the development of CHD and hyperglycemia has become clear. The mechanism is mainly related to increased polyol pathway flux, increased intracellular advanced glycation end product formation, increased hexosamine biosynthesis pathway, protein kinase C activation and mitochondrial production of reactive oxygen species (5); however, there is a lack of clinically effective biomarkers for predicting coronary artery lesions.
MicroRNAs (miRNAs/miRs) are a class of non-coding RNAs 18–25 nucleotides in length, which can accurately regulate gene expression by directly binding to the 3′-untranslated regions of their target mRNAs at transcriptional and post-transcriptional levels. Increasing evidence has suggested that miRNAs are involved in the regulation of multiple physiological and pathological processes, including proliferation, metabolism, inflammation, metastasis and apoptosis (6). Studies have shown that miRNAs from serum or plasma can be used as promising biomarkers for diagnosing and monitoring the progression of cardiovascular diseases, including CHD (7). For example, Bayés-Genis et al (8) demonstrated that circulating miR-1254 and miR-1306-5p are associated with risk of death and hospitalization in patients with heart failure. Based on the results of a meta-analysis, Zhu et al (9) concluded that serum or plasma miR-133a may serve as a diagnostic biomarker for patients with acute myocardial infarction. Wei et al (10) reported that miR-425-5p may serve as novel biomarker of atrial fibrosis that contributes to atrial remodeling in patients with atrial fibrillation. Additionally, Pan et al (11) reported that peripheral blood miR-15a expression is associated with low-density lipoprotein cholesterol (LDL-C) and Gensini score, and may serve as a diagnostic biomarker for patients with coronary artery disease. Recently, Szydelko and Matyjaszek-Matuszek summarized the role of miRNAs as novel biomarkers for the development of diabetic coronary artery disease (12). However, the low content of most miRNAs in serum limits their clinical application.
Exosomes are membrane-bound vesicles 30–150 nm in size, which are present in the majority of bodily fluids and serve key functions in cell-to-cell communication by acting as a delivery cargo shuttle for various molecules (13). Exosomes contain various nucleic acids, including miRNAs, and numerous types of proteins, such as tumor susceptibility gene 101 (TSG101), membrane CD63, membrane-anchored heat-shock 70 (HSP70) and ALG2-interacting protein X. Notably, exosomes have emerged as a promising delivery system for drugs with lower toxicity and high therapeutic efficacy (14). Exosomal miRNAs were first identified in serum, and have also been described in several biological fluids, such as saliva, urine, cerebrospinal fluid and synovial fluid (15). In addition, exosomal miRNAs serve an important role in disease progression by altering cell signal transduction (16). Notably, exosomes can be stably stored under different conditions, indicating that exosomal miRNAs are potential biomarkers for the diagnosis and treatment of diseases, including CHD (17).
Although the expression of miRNAs in patients with CHD has been reported (18), to the best of our knowledge, the relationship between exosomal miRNA expression and the severity of CHD has not been studied. The present study aimed to identify the expression profile of serum-derived exosomal miRNAs in patients with CHD and hyperglycemia, and to identify effective biomarkers for predicting coronary artery lesions. The present study may provide evidence for the use of exosomal miRNAs as novel biomarkers for the severity of coronary stenosis in patients with CHD and hyperglycemia.
Materials and methods
Study population
In the first stage of the present study, eight patients with CHD and hyperglycemia (age range, 39–73 years), and eight age- and sex-matched patients with CHD and normoglycemia (age range, 42–75 years), who had been admitted to The 960th Hospital of the Joint Service Support Force of the People's Liberation Army (Jinan, China) for coronary angiography (CAG) between January 2018 and May 2018 were enrolled to perform a human miRNA microarray analysis. The clinical characteristics of these patients is shown in Table SI. In the second stage, 75 patients with CHD and hyperglycemia (age range, 33–76 years; mean ± SD age, 65.82±8.30 years) and 75 age- and sex-matched patients with CHD and normoglycemia (age range, 30–78 years; mean ± SD age, 63.13±9.85 years) were enrolled from The 960th Hospital of the Joint Service Support Force of the People's Liberation Army between June 2018 and December 2020 for further validation of the miRNA microarray results. The study design is shown in Fig. 1. Hyperglycemia was defined as fasting blood glucose (FBG) ≥6.1 mmol/l. Normoglycemia was defined as FBG <6.0 mmol/l and oral glucose tolerance test (2-h plasma glucose) <7.8 mmol/l. The inclusion criteria of patients with CHD were: At least one major epicardial vessel with >50% stenosis indicated by CAG and presenting with typical angina. The exclusion criteria were as follows: i) Age, <18 years; ii) myocardial infarction within 3 months, cardiac insufficiency, New York Heart Association grade (19) ≥III, cardiomyopathy, congenital heart disease, heart valve disease, autoimmune system disease, hematological system disease, inflammatory disease, lymphatic system disease, active infection, severe liver and kidney insufficiency, and malignant tumor; iii) patients with mental illness that were unable to cooperate with treatments; iv) patients with incomplete medical records. The present study was approved by The 960th Hospital of the Joint Service Support Force of the People's Liberation Army (approval no. 202109) and complied strictly with the 2008 Declaration of Helsinki Principle. Written informed consent was obtained from participants prior to the collection of samples.
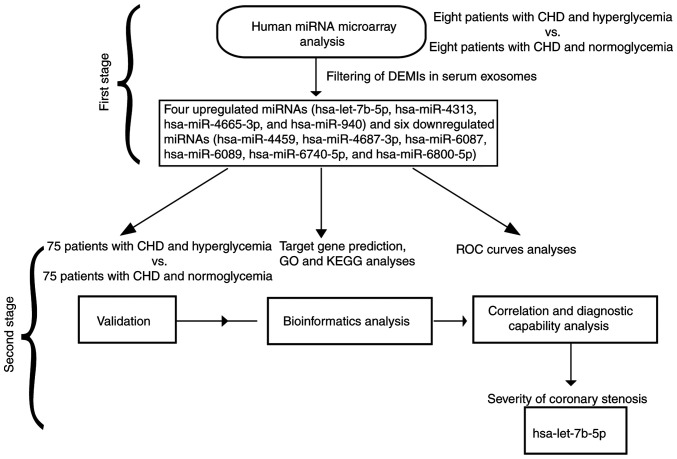
Figure 1.
Flow diagram of the present study. In the first stage, a human miRNA microarray analysis was performed to identify the expression profile of exosomal miRNAs in the serum of patients with CHD and hyperglycemia. In the second stage, validation, bioinformatics analysis, and correlation and diagnostic capability analyses were performed to verify hsa-let-7b-5p as an effective biomarker for differentiating the severity of coronary stenosis. CHD, coronary heart disease; DEMIs, differentially expressed miRNAs; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA/miR, microRNA; ROC, receiver operating characteristic.
Clinical data collection
The clinical baseline data of patients with CHD, including age, sex, body mass index (BMI), systolic blood pressure (SBP), diastolic blood pressure (DBP), smoking, drinking, and history of dyslipidemia and hypertension were collected. The levels of total cholesterol (TC), triglyceride (TG), LDL-C, high-density lipoprotein cholesterol (HDL-C), hemoglobin A1C (Hba1c) and FBG were routinely tested. Based on the Synergy between Percutaneous Coronary Intervention with Taxus and Cardiac Surgery (SYNTAX) score (20), the severity of coronary stenosis in patients with CHD was evaluated, and was divided into the mild lesion group (SYNTAX score <22) and the moderate to severe lesion group (SYNTAX score ≥22).
Isolation of exosomes from serum
Peripheral blood (10 ml) was collected from each participant after fasting for 12 h. The serum was separated by centrifugation at 2,000 × g for 20 min at 37°C, followed by centrifugation at 12,000 × g for 5 min at 4°C. Exosomes were isolated from the serum using an exosome isolation kit (cat. no. EZB-exo1; EZBioscience), according to the manufacturer's instructions. Briefly, 1/3 volume of exosome separation reagent was added to 1 ml serum and was refrigerated at 4°C overnight. The serum and exosome separation reagent were centrifuged together at 15,000 × g at 4°C for 30 min and supernatants were removed by aspiration. The pellet containing the exosomes at the bottom of the tube was resuspended in 200 µl PBS.
Transmission electron microscopy (TEM)
The exosome pellets were fixed with 4% paraformaldehyde at 37°C and then placed in a Formvar/carbon film-coated TEM grid (Thermo Fisher Scientific, Inc.). Subsequently, samples were fixed by incubation with 1% glutaraldehyde for 90 min at 37°C, stained with 1% uranyl acetate for 2 h at 37°C, embedded and polymerized in epoxy resin for 2 h at 37°C, and finally images were captured using a FEI Tecnai T20 transmission electron microscope (Thermo Fisher Scientific, Inc.).
Western blot analysis
Proteins were extracted from exosomes using an exosomal protein extraction kit (EZBioscience), according to the manufacturer's protocol. The concentration of each protein was detected using a BCA Protein Assay Kit (Shanghai Yeasen Biotechnology Co., Ltd.). Proteins (35 µg/lane) were loaded and concentrated on 5% stacking gels, separated by SDS-PAGE on 10% resolving gels, and then transferred to polyvinylidene difluoride membranes (MilliporeSigma). After blocking with 5% bovine serum albumin (Beyotime Institute of Biotechnology) for 2 h at 37°C, the membranes were incubated with primary human anti-HSP70 (cat. no. ab181606; 1:1,000), anti-TSG101 (cat. no. ab133586; 1:750) and anti-CD63 (cat. no. ab134045; 1:2,000) antibodies overnight at 4°C. Subsequently, the membranes were incubated with goat anti-rabbit HRP secondary antibody (cat. no. ab6721; 1:2,500) at 37°C for 60 min. All antibodies were obtained from Abcam. Finally, protein bands were visualized using an ECL kit (Beyotime Institute of Biotechnology), and were further scanned and analyzed using ImageJ version 1.8.0 software (National Institutes of Health).
miRNA microarray analysis
The exosomal miRNAs were extracted using the mirVana™ PARIS™ kit (Ambion; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. For miRNA microarray analysis, extracted miRNAs were labeled with the cyanine 3-cytidine bisphosphate labeling kit (Agilent Technologies, Inc.) according to the manufacturer's protocol, and then hybridized to a human miRNA microarray (cat. no. G4872A; 8×60 K, version 21.0; Agilent Technologies, Inc.) at 55°C with constant rotation at 12 × g for 20 h, according to the manufacturer's instructions. Subsequently, the microarray was rinsed with corresponding wash buffer three times and scanned using an Agilent G2505C microarray scanner (Agilent Technologies, Inc.). The raw data were read and converted to numbers using Feature Extraction Software (version 10.7.1.1; Agilent Technologies, Inc.), and were further analyzed using GeneSpring GX software (version 12.5; Agilent Technologies, Inc.). The signal intensities of the spots on the microarray were normalized to the total signal intensity and shown as percentages. An unpaired Student's t-test was used to determine the statistical significance of differences in microarray signal intensities. The differentially expressed miRNAs (DEMIs) were screened according to the following thresholds: Fold change >2 or <0.5; adjusted P<0.05 (false discovery rate method). Hierarchical clustering analysis of the dataset of the selected DEMIs was performed using ClusterProfiler 3.5 version software packages for Windows (Bioconductor, http://www.bioconductor.org/packages/3.5/bioc/html/clusterProfiler.html) with Ward's method (21).
Prediction of miRNA target genes and enrichment analysis
The target genes of DEMIs were predicted using TargetScan8.0 (https://www.targetscan.org/vert_80/), miRDB (https://mirdb.org/miRDB/), and Tarbase7.0 (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index) databases. For Gene Ontology (GO; http://geneontology.org/) enrichment analysis, the target genes of the selected DEMIs were mapped to GO terms (biological process, cellular component and molecular function) in the dataset, and the gene numbers for every term were calculated. Subsequently, hypergeometric test was performed to identify significantly enriched GO terms in the input gene list, and P<0.05 was set as the cut-off criterion. For Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/) pathway enrichment analysis, the significantly enriched metabolic or signal transduction pathways with P<0.05 from target genes of the selected DEMIs were identified when compared with the whole genome background.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from exosomes using an exosome RNA purification kit (EZBioscience), according to the manufacturer's protocol. RNA was reverse transcribed into cDNA using a miRNA First Strand cDNA Synthesis (Stem-loop Method) kit (Sangon Biotech Co., Ltd.), according to the manufacturer's instructions. For the quantification of miRNA expression levels, qPCR was performed according to the instructions of the SYBR Green One™ miRNAs qPCR Detection Kit (BioTeke Corporation), with U6 small nuclear RNA used as the endogenous reference. The total reaction system of qPCR was 25 µl, including master mix, primers, cDNA and RNase-free deionized water, and the thermocycling conditions were as follows: One cycle at 95°C for 15 min, followed by 40 reaction cycles at 94°C for 20 sec and 60°C for 34 sec. The primer sequences are shown in Table I. The relative expression levels of miRNAs were normalized against the expression of U6 using the 2−∆∆Cq method (22).
Table I.
Primers used for reverse transcription-quantitative PCR.
| miRNA | Forward primer, 5′-3′ | Reverse primer, 5′-3′ |
|---|---|---|
| hsa-let-7b-5p | TGAGGTAGTAGGTTGTGTGGTT | CGCAGGGTCCGAGGTATTC |
| hsa-miR-4313 | AGCCCCCTGGCCCCAAACCC | CGCAGGGTCCGAGGTATTC |
| hsa-miR-4665-3p | CTCGGCCGCGGCGCGTAGCCCCCGCC | CGCAGGGTCCGAGGTATTC |
| hsa-miR-940 | AAGGCAGGGCCCCCGCTCCCC | CGCAGGGTCCGAGGTATTC |
| hsa-miR-4459 | CCAGGAGGCGGAGGAGGTGGAG | CGCAGGGTCCGAGGTATTC |
| hsa-miR-4687-3p | TGGCTGTTGGAGGGGGCAGGC | CGCAGGGTCCGAGGTATTC |
| hsa-miR-6087 | TGAGGCGGGGGGGCGAGC | CGCAGGGTCCGAGGTATTC |
| hsa-miR-6089 | GGAGGCCGGGGTGGGGCGGGGCGG | CGCAGGGTCCGAGGTATTC |
| hsa-miR-6740-5p | AGTTTGGGATGGAGAGAGGAGA | CGCAGGGTCCGAGGTATTC |
| hsa-miR-6800-5p | GTAGGTGACAGTCAGGGGCGG | CGCAGGGTCCGAGGTATTC |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
miRNA/miR, microRNA.
Statistical analysis
Statistical analysis was performed using SPSS software (version 27.0; IBM Corp.). The Kolmogorov-Smirnov test was used to test the normal distribution of all variables. Data conforming to normal distribution were expressed as the mean ± standard deviation, and unpaired Student's t-test was used to assess the significant difference. Data conforming to non-normal distribution were expressed as the median (interquartile range), and Mann-Whitney U test was used to assess the significant difference. Categorical variables were expressed as frequency and percentage, and were analyzed by χ2 test or Fisher's exact test. Correlation coefficients between the expression of selected DEMIs and the levels of biochemical parameters in patients with CHD and hyperglycemia were determined by Spearman correlation analysis. The receiver operating characteristic (ROC) curve and area under the curve (AUC) were analyzed to assess the possibility of using exosomal miRNAs as diagnostic biomarkers for the severity of coronary stenosis. P<0.05 was considered to indicate a statistically significant difference.
Results
Characterization of serum exosomes
Firstly, the isolated exosomes were assessed by TEM and western blot analysis. TEM showed that isolated exosomes exhibited a round-shaped appearance within 30–150 nm, corresponding to the expected size range of exosomes (Fig. S1A and B). Western blot analysis showed that the marker proteins, CD63, TSG101 and HSP70, were enriched in the exosomes, further confirming the successful isolation of exosomes from serum (Fig. S1C).
Profile of exosomal miRNA expression in the serum of patients with CHD and hyperglycemia
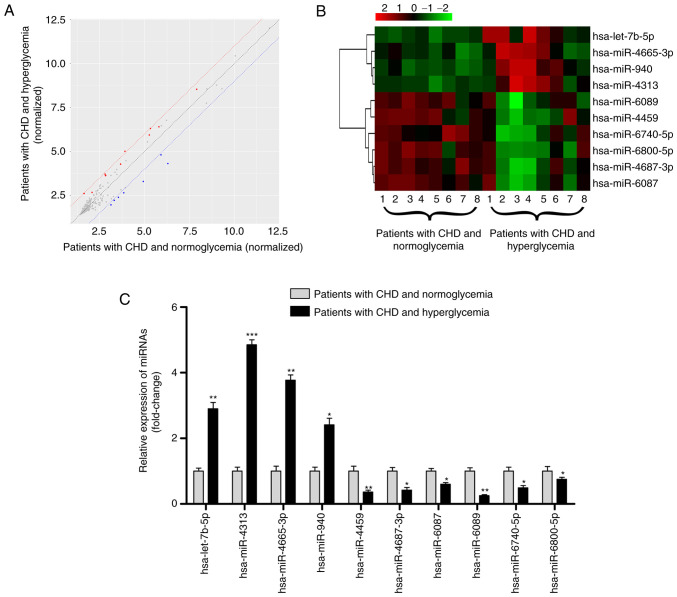
The miRNA transcript profile was characterized using a microarray analysis in 16 human serum exosome samples, eight from patients with CHD and hyperglycemia compared with eight from patients with CHD and normoglycemia. A total of 10 DEMIs were identified according to the criteria (Fig. 2A). The 10 DEMIs are shown in a heatmap (Fig. 2B); among them, hsa-let-7b-5p, hsa-miR-4313, hsa-miR-4665-3p and hsa-miR-940 were upregulated, and hsa-miR-4459, hsa-miR-4687-3p, hsa-miR-6087, hsa-miR-6089, hsa-miR-6740-5p and hsa-miR-6800-5p were downregulated (Table II; all P<0.05).
Figure 2.
Profile of exosomal miRNAs in the serum of patients with CHD and hyperglycemia. (A) Scatter plot showing the expression profile of exosomal miRNAs between eight patients with hyperglycemia and eight patients with normoglycemia; miRNAs above and below the border lines (red and blue) exhibited a >2 fold-change difference in expression. (B) Clustered heatmap for the 10 DEMIs, including hsa-let-7b-5p, hsa-miR-4313, hsa-miR-4665-3p, hsa-miR-940, hsa-miR-4459, hsa-miR-4687-3p, hsa-miR-6087, hsa-miR-6089, hsa-miR-6740-5p and hsa-miR-6800-5p. (C) Reverse transcription-quantitative PCR validation of the 10 DEMIs in 75 patients with CHD and hyperglycemia and 75 patients with CHD and normoglycemia. Data are expressed as the mean ± standard deviation. *P<0.05, **P<0.01, ***P<0.001 vs. patients with CHD and normoglycemia. CHD, coronary heart disease; DEMIs, differentially expressed miRNAs; miRNA/miR, microRNA.
Table II.
List of the 10 differentially expressed miRNAs in the serum of patients with coronary heart disease and hyperglycemia.
| miRNA | Regulation | Fold change | P-value |
|---|---|---|---|
| hsa-let-7b-5p | Up | 2.47 | 0.007a |
| hsa-miR-4313 | Up | 2.60 | 0.015b |
| hsa-miR-4665-3p | Up | 2.38 | 0.027b |
| hsa-miR-940 | Up | 3.60 | 0.012b |
| hsa-miR-4459 | Down | 0.48 | 0.039b |
| hsa-miR-4687-3p | Down | 0.52 | 0.006a |
| hsa-miR-6087 | Down | 0.50 | 0.013b |
| hsa-miR-6089 | Down | 0.46 | 0.020b |
| hsa-miR-6740-5p | Down | 0.50 | 0.013b |
| hsa-miR-6800-5p | Down | 0.47 | 0.007a |
miRNA/miR, microRNA.
P<0.01,
P<0.05.
Validation of exosomal miRNA profile identified by microarray
The exosomal miRNA profile was further confirmed by RT-qPCR using samples from 75 patients with CHD and hyperglycemia and 75 patients with CHD and normoglycemia. The baseline characteristics of these patients are shown in Table III. The levels of Hba1c and FBG, the number of stenotic vessels, severity of stenosis and SYNTAX score were significantly higher in patients with CHD and hyperglycemia than with those in patients with CHD and normoglycemia (all P<0.05). There was no significant difference in other baseline data (all P>0.05), including age, sex, BMI, SBP, DBP, smoking, drinking, history of dyslipidemia and hypertension, TC, TG, LDL-C and HDL-C. As shown in Fig. 2C, RT-qPCR results were consistent with the miRNA microarray analysis results, as increased expression levels of hsa-let-7b-5p, hsa-miR-4313, hsa-miR-4665-3p and hsa-miR-940, and decreased expression levels of hsa-miR-4459, hsa-miR-4687-3p, hsa-miR-6087, hsa-miR-6089, hsa-miR-6740-5p and hsa-miR-6800-5p were observed in patients with CHD and hyperglycemia (all P<0.05), thus demonstrating the reliability of this miRNA profile.
Table III.
Clinical baseline data of patients with CHD and hyperglycemia or normoglycemia.
| Characteristic | Patients with CHD and hyperglycemia (n=75) | Patients with CHD and normoglycemia (n=75) | P-value |
|---|---|---|---|
| Age, years | 65.82±8.30 | 63.13±9.85 | 0.725 |
| Sex, female/male | 28/47 | 29/46 | 0.866 |
| BMI, kg/m2 | 27.16±4.68 | 26.32±4.16 | 0.402 |
| SBP, mmHg | 147.91±14.45 | 148.04±15.41 | 0.331 |
| DBP, mmHg | 83.64±7.76 | 81.57±13.14 | 0.681 |
| Smoking, n (%) | 21 (28.0) | 24 (32.0) | 0.593 |
| Drinking, n (%) | 13 (17.33) | 17 (22.67) | 0.414 |
| Dyslipidemia, n (%) | 20 (26.67) | 27 (36.0) | 0.218 |
| Hypertension, n (%) | 52 (69.33) | 45 (60.0) | 0.232 |
| TC, mmol/l | 4.32±1.09 | 4.16±1.27 | 0.609 |
| TG, mmol/l | 1.47±0.86 | 1.87±1.35 | 0.055 |
| LDL-C, mmol/l | 2.51±0.85 | 2.40±0.93 | 0.252 |
| HDL-C, mmol/l | 1.02±0.21 | 1.14±0.20 | 0.711 |
| Hba1c, % | 8.41±1.06 | 4.19±1.57 | <0.001a |
| FBG, mmol/l | 8.78±2.10 | 4.49±1.26 | <0.001a |
| Number of stenotic vessels | 2.90±0.30 | 2.30±0.80 | 0.046b |
| Severity of stenosis, % | 78.02±14.15 | 56.42±23.17 | 0.013b |
| SYNTAX score | 29.50 (24.0, 31.50) | 24.0 (21.50, 27.0) | <0.001a |
P<0.001,
P<0.05. SYNTAX score is expressed as the median (interquartile range) and other data are presented as the mean ± standard deviation. BMI, body mass index; DBP, diastolic blood pressure; FBG, fasting blood glucose; Hba1c, hemoglobin A1C; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; SBP, systolic blood pressure; SYNTAX, Synergy between Percutaneous Coronary Intervention with Taxus and Cardiac Surgery; TC, total cholesterol; TG, triglyceride.
Functional enrichment and pathway analysis of target genes of the 10 DEMIs
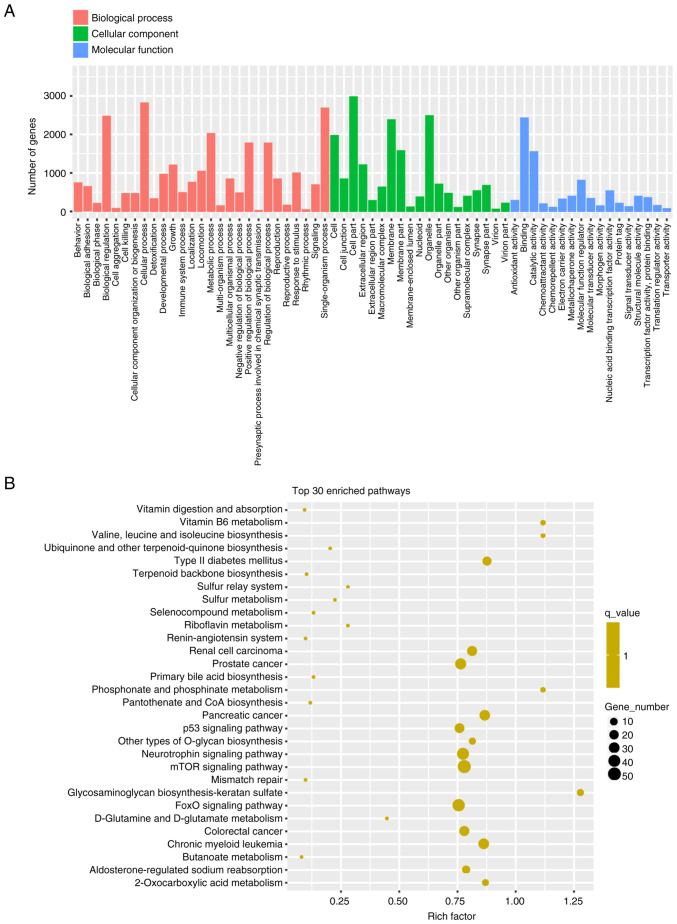
For the GO enrichment analysis, ‘cellular process’, ‘single-organism process’ and ‘biological regulation’ were the top three enriched terms in biological process; ‘cell part’, organelle’ and ‘membrane’ were the top three enriched terms in cellular component; and ‘binding’, ‘catalytic activity’ and ‘molecular function regulator’ were the top three enriched terms in molecular function (Fig. 3A). The results of KEGG pathway analysis are shown in Fig. 3B. Several enriched pathways were identified, including ‘mTOR signaling pathway’, ‘FoxO signaling pathway’, ‘neurotrophin signaling pathway’, ‘p53 signaling pathway’ and ‘type 2 diabetes mellitus’, which were related to the 10 DEMIs.
Figure 3.
Functional enrichment and pathway analyses of the target genes of the 10 DEMIs. (A) Enrichment scores in GO enrichment analysis on target genes of the 10 DEMIs. (B) Top 30 enrichment scores in the KEGG pathway analysis of the target genes. DEMIs, differentially expressed miRNAs; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA/miR, microRNA.
Correlation between expression of the 10 DEMIs and biochemical parameters in patients with CHD and hyperglycemia
The correlation between expression of the 10 DEMIs and biochemical parameters is shown in Table SII. Results indicated that hsa-let-7b-5p expression showed a strong positive correlation with HbA1c levels and SYNTAX score in patients with CHD and hyperglycemia (all P<0.001). In addition, hsa-miR-940 expression was positively related to HbA1c level, and hsa-miR-6087 and hsa-miR-6800-5p expression was negatively related to HbA1c level (all P<0.01). Furthermore, hsa-miR-4459 expression was positively related to SYNTAX score (P<0.01). Notably, no correlation was observed between the expression of the other DEMIs and HbA1c level or SYNTAX score (all P>0.05). These data suggested that exosomal hsa-let-7b-5p may be associated with coronary stenosis in patients with CHD and hyperglycemia.
Diagnostic potential of the 10 DEMIs for the severity of coronary stenosis
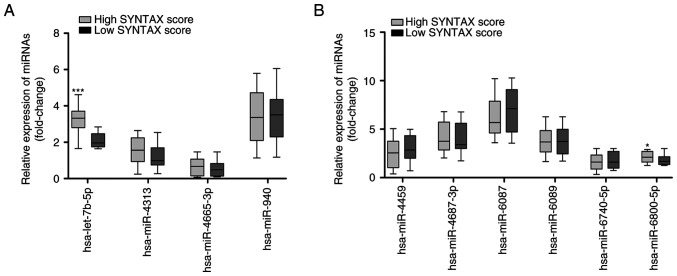
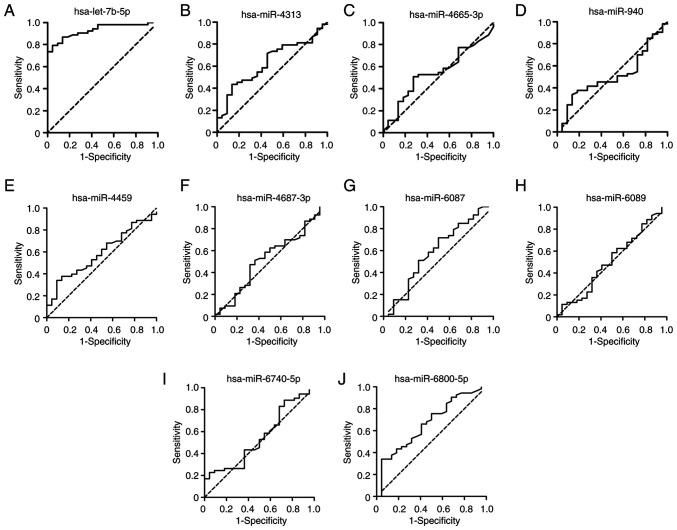
Based on the SYNTAX score, the 75 patients with CHD and hyperglycemia were divided into two groups, including 22 patients with low SYNTAX score and 53 patients with high SYNTAX score. RT-qPCR results showed that the expression levels of hsa-let-7b-5p were significantly increased in the patients with high SYNTAX score compared with those in the patients with low SYNTAX score (Fig. 4A; P<0.001). In addition, the expression levels of hsa-miR-6800-5p were significantly increased in patients with high SYNTAX score compared with those in the patients with low SYNTAX score (Fig. 4B; P<0.05). ROC curves of the 10 DEMIs in the diagnosis of the severity of coronary stenosis are shown in Fig. 5. The results showed that hsa-let-7b-5p yielded the highest diagnostic accuracy among the 10 DEMIs in discriminating high SYNTAX score from low SYNTAX score in patients with CHD and hyperglycemia (Table SIII; P<0.001). These data demonstrated that exosomal hsa-let-7b-5p has a promising potential as a biomarker for the diagnosis of the severity of coronary stenosis.
Figure 4.
Expression of the 10 differentially expressed miRNAs in patients with CHD and hyperglycemia split into two groups according to SYNTAX score. (A) RT-qPCR analysis of the expression levels of hsa-let-7b-5p, hsa-miR-4313, hsa-miR-4665-3p and hsa-miR-940 in 22 patients with low SYNTAX score and 53 patients with high SYNTAX score. (B) RT-qPCR analysis of the expression levels of hsa-miR-4459, hsa-miR-4687-3p, hsa-miR-6087, hsa-miR-6089, hsa-miR-6740-5p and hsa-miR-6800-5p in 22 patients with low SYNTAX score and 53 patients with high SYNTAX score. Data are presented as box and whisker plots with the minimum, median and maximum values. *P<0.05, ***P<0.001 vs. low SYNTAX score. CHD, coronary heart disease; miRNA/miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; SYNTAX, Synergy between Percutaneous Coronary Intervention with Taxus and Cardiac Surgery.
Figure 5.
Diagnostic value of the 10 differentially expressed miRNAs for the severity of coronary stenosis. Receiver operating characteristic curves were plotted using the expression levels of (A) hsa-let-7b-5p, (B) hsa-miR-4313, (C) hsa-miR-4665-3p, (D) hsa-miR-940, (E) hsa-miR-4459, (F) hsa-miR-4687-3p, (G) hsa-miR-6087, (H) hsa-miR-6089, (I) hsa-miR-6740-5p and (J) hsa-miR-6800-5p for distinguishing high SYNTAX score from low SYNTAX score in patients with coronary heart disease and hyperglycemia. miR/miRNA, microRNA; SYNTAX, Synergy between Percutaneous Coronary Intervention with Taxus and Cardiac Surgery.
Discussion
The incidence of CHD has exhibited an increase in recent decades, which may be due to the prevalence of obesity and lifestyle changes (23). CHD is considered to be caused by atherosclerosis-induced arterial stenosis and hyperglycemia can exacerbate the progression of arterial stenosis. An increasing number of studies have demonstrated that accurate diagnosis of coronary artery lesions is key to the treatment of CHD (24,25). CAG is considered the gold standard for detecting coronary stenosis in routine clinical practice; however, it is expensive and some patients cannot be tested due to contraindications, such as hemophilia, severe heart failure, and allergy to iodine and contrast agents. Therefore, identification of noninvasive biomarkers for evaluating the severity of coronary stenosis in CHD is required (26). It has been suggested that exosomal miRNAs participate in a wide range of biological processes and their dysregulated levels are associated with complex phenotypes of human disease (27). In the present study, serum samples were collected from patients with CHD and hyperglycemia, exosomes were isolated and DEMIs were detected using human miRNA microarray analysis. Subsequently, the target mRNAs of the selected DEMIs were analyzed by bioinformatics analysis. Furthermore, the correlations and diagnostic values of the selected DEMIs regarding the severity of coronary stenosis in patients with CHD and hyperglycemia were analyzed. To the best of our knowledge, the present study was the first to reveal that upregulated exosomal hsa-let-7b-5p expression may serve as a noninvasive biomarker the severity of coronary stenosis, which could provide a novel insight into the diagnostic capacity of exosomal miRNAs in coronary artery lesions.
miRNAs are transported mainly through exosomes in the circulatory system. The transfer of miRNAs in these exosomes prevents them from being degraded, causing miRNAs to circulate in vivo and act on target cells. Numerous studies have determined the expression profiles of exosomal miRNAs in human disease (28). For example, Liu et al (29) reported 66 DEMIs from exosomes in the serum of patients with esophageal squamous cell carcinoma and Su et al (30) identified 13 DEMIs from exosomes in the serum of patients with acute myocardial infarction. In serum exosomes from patients with endometriosis, 24 DEMIs were identified by a miRNA microarray (31). By contrast, investigations into exosomal miRNAs in patients with CHD and hyperglycemia are rare and limited (32,33).
In the present study, using a human miRNA microarray analysis, 10 DEMIs, including four upregulated miRNAs (hsa-let-7b-5p, hsa-miR-4313, hsa-miR-4665-3p and hsa-miR-940) and six downregulated miRNAs (hsa-miR-4459, hsa-miR-4687-3p, hsa-miR-6087, hsa-miR-6089, hsa-miR-6740-5p and hsa-miR-6800-5p), were identified in the serum exosomes of patients with CHD and hyperglycemia. These results were consistent with those of previous reports regarding serum exosomes in esophageal squamous cell carcinoma, acute myocardial infarction and endometriosis (29–31), suggesting the credibility of the present data. The expression levels of 10 DEMIs were further validated in 75 patients with CHD and hyperglycemia. Consistent with a previous study (34), the results revealed that the SYNTAX score was significantly higher in patients with CHD and hyperglycemia than in patients with CHD and normoglycemia, thus confirming the promotion of coronary stenosis by hyperglycemia. A KEGG pathway enrichment analysis was subsequently conducted, and the 10 DEMI target genes were significantly enriched in the ‘mTOR signaling pathway’, ‘FoxO signaling pathway’ and ‘neurotrophin signaling pathway’. The 10 DEMIs may participate in several biological processes, such as ‘cellular process’, ‘single-organism process’ and ‘biological regulation’. These data suggested that the 10 DEMIs may be involved in the pathogenesis of patients with CHD and hyperglycemia.
As a member of the hsa-let-7 family of miRNAs, hsa-let-7b-5p acts as a crucial regulator of developmental processes in human diseases, such as Parkinson's disease, multiple sclerosis, asthenozoospermia and Crohn disease (35–38). In the current study, serum-derived exosomal hsa-let-7b-5p was shown to be upregulated in patients with CHD and hyperglycemia, which is consistent with a previous study by Zhang et al (39). Subsequently, the correlations between expression levels of the 10 DEMIs and biochemical parameters were analyzed. Only hsa-let-7b-5p expression was found to have a strong positive correlation with HbA1c levels and SYNTAX score in patients with CHD and hyperglycemia. Furthermore, increased hsa-let-7b-5p expression was detected in patients with high SYNTAX score. Based on these findings, it was hypothesized that exosomal hsa-let-7b-5p may be associated with coronary stenosis in patients with CHD and hyperglycemia.
Exosome-derived miRNAs can easily be isolated from various bodily fluids, indicating potential opportunities for clinical translation. Exosomal miRNAs have emerged as promising biomarkers for diagnosis, risk stratification and prognosis prediction (27). Current research on the relationships of coronary stenosis and miRNAs is increasing (40). Ling et al (41) suggested that upregulated miR-122-5p has the ability to predict the severity of coronary lesions. Li et al (42) demonstrated that miR-34a may be a novel biomarker in assistance of the diagnosis of coronary stenosis. Similarly, miR-221/222 have been reported to serve as promising biomarkers for the diagnosis of ≥50% coronary stenosis and the occurrence of acute coronary syndrome (43). Notably, the present study found that hsa-let-7b-5p yielded the highest diagnostic accuracy among the 10 DEMIs in discriminating high SYNTAX score from low SYNTAX score in patients with CHD and hyperglycemia. Similarly, previous studies have reported that plasma hsa-let-7b-5p serves as a biomarker for the diagnosis of heroin use disorders, non-small cell lung and multiple sclerosis (36,44,45). These data demonstrated that exosomal hsa-let-7b-5p could be used as a promising biomarker for differentiating the severity of coronary stenosis. However, the methods used for exosome isolation critically impact subsequent analyses due to lack of internationally standardized methodologies. It is thus strongly recommended to standardize the isolation procedure before integrating studies across different laboratories. Similar to all other biomarkers in CHD, before exosomal hsa-let-7b-5p can be translated to the clinic, it must be validated in large cohort studies and accredited by the International Organization for Standardization.
The present study has some limitations. Firstly, a relatively small sample size of patients with CHD and hyperglycemia was assessed and selection bias may exist, thus more investigations with large clinical samples are required for further validation of the diagnostic power of exosomal hsa-let-7b-5p for differentiating the severity of coronary stenosis. Secondly, there was a lack of continuously monitoring disease progression and follow-up results, and the relationship between exosomal hsa-let-7b-5p and prognosis needs further study using regression analysis. Thirdly, the association of exosomal hsa-let-7b-5p and hyperglycemia requires further confirmation. Finally, the effects and related signaling pathways of exosomal hsa-let-7b-5p in coronary stenosis were not investigated, which could be explored in patients with CHD in further studies.
In conclusion, the present study screened 10 DEMIs in the serum of patients with CHD and hyperglycemia using a human miRNA microarray analysis. Among the DEMIs, exosomal hsa-let-7b-5p was upregulated in patients with CHD and hyperglycemia, and was positively correlated with both HbA1c levels and SYNTAX score. Further analysis showed that exosomal hsa-let-7b-5p could potentially serve as a noninvasive biomarker for the severity of coronary stenosis. The present study provides a novel insight into the diagnostic capacity of exosomal miRNAs in coronary artery lesions.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- AUC
area under the curve
- CHD
coronary heart disease
- DEMIs
differentially expressed miRNAs
- GO
Gene Ontology
- HSP70
heat-shock 70
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LDL-C
low-density lipoprotein cholesterol
- miRNAs
microRNAs
- ROC
receiver operating characteristic
- RT-qPCR
reverse transcription-quantitative PCR
- SYNTAX
Synergy between Percutaneous Coronary Intervention with Taxus and Cardiac Surgery
- TEM
transmission electron microscopy
- TSG101
tumor susceptibility gene 101
Funding Statement
This work was supported by the Medical and Health Science and Technology Development Planning Project of Shandong Province (grant no. 202103011061).
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due to the institutional policy that prohibits data sharing, but are available from the corresponding author on reasonable request.
Authors' contributions
QJ participated in the study conception and design. SFH participated in the sample collection, experiments, data collection, statistical analysis and funding support. JF, LLY, BL, YHH, RMC, CYL, CXZ, JYL, YNW, YQG and HT assisted in the sample collection, experiments, data collection and statistical analysis. QJ and SFH prepared the draft of the manuscript. QJ and SFH confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by The 960th Hospital of the Joint Service Support Force of the People's Liberation Army (Jinan, China; approval no. 202109) and complied strictly with the 2008 Declaration of Helsinki Principle. Written informed consent was obtained from the participants prior to the collection of samples.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Katta N, Loethen T, Lavie CJ, Alpert MA. Obesity and coronary heart disease: Epidemiology, pathology, and coronary artery imaging. Curr Probl Cardiol. 2021;46:100655. doi: 10.1016/j.cpcardiol.2020.100655. [DOI] [PubMed] [Google Scholar]
- 2.Carneiro AV. Coronary heart disease in diabetes mellitus: Risk factors and epidemiology. Rev Port Cardiol. 2004;23:1359–1366. (In English, Portuguese) [PubMed] [Google Scholar]
- 3.Goodarzi MO, Rotter JI. Genetics Insights in the relationship between type 2 diabetes and coronary heart disease. Circ Res. 2020;126:1526–1548. doi: 10.1161/CIRCRESAHA.119.316065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schutt K, Muller-Wieland D, Marx N. Diabetes mellitus and the heart. Exp Clin Endocrinol Diabetes. 2019;127((S 01)):S102–S104. doi: 10.1055/a-1018-9065. [DOI] [PubMed] [Google Scholar]
- 5.Battault S, Renguet E, Van Steenbergen A, Horman S, Beauloye C, Bertrand L. Myocardial glucotoxicity: Mechanisms and potential therapeutic targets. Arch Cardiovasc Dis. 2020;113:736–748. doi: 10.1016/j.acvd.2020.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234:5451–5465. doi: 10.1002/jcp.27486. [DOI] [PubMed] [Google Scholar]
- 7.Kalayinia S, Arjmand F, Maleki M, Malakootian M, Singh CP. MicroRNAs: Roles in cardiovascular development and disease. Cardiovasc Pathol. 2021;50:107296. doi: 10.1016/j.carpath.2020.107296. [DOI] [PubMed] [Google Scholar]
- 8.Bayés-Genis A, Lanfear DE, de Ronde MWJ, Lupon J, Leenders JJ, Liu Z, Zuithoff NPA, Eijkemans MJC, Zamora E, De Antonio M, et al. Prognostic value of circulating microRNAs on heart failure-related morbidity and mortality in two large diverse cohorts of general heart failure patients. Eur J Heart Fail. 2018;20:67–75. doi: 10.1002/ejhf.984. [DOI] [PubMed] [Google Scholar]
- 9.Zhu L, Liu F, Xie H, Feng J. Diagnostic performance of microRNA-133a in acute myocardial infarction: A meta-analysis. Cardiol J. 2018;25:260–267. doi: 10.5603/CJ.a2017.0126. [DOI] [PubMed] [Google Scholar]
- 10.Wei F, Ren W, Zhang X, Wu P, Fan J. miR-425-5p is negatively associated with atrial fibrosis and promotes atrial remodeling by targeting CREB1 in atrial fibrillation. J Cardiol. 2022;79:202–210. doi: 10.1016/j.jjcc.2021.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Pan X, He Y, Ling S, Chen Z, Yan G. MiR-15a functions as a diagnostic biomarker for coronary artery disease. Clin Lab. 2020;66 doi: 10.7754/Clin.Lab.2020.191138. [DOI] [PubMed] [Google Scholar]
- 12.Szydelko J, Matyjaszek-Matuszek B. MicroRNAs as biomarkers for coronary artery disease related to type 2 diabetes mellitus-from pathogenesis to potential clinical application. Int J Mol Sci. 2022;24:616. doi: 10.3390/ijms24010616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li SP, Lin ZX, Jiang XY, Yu XY. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol Sin. 2018;39:542–551. doi: 10.1038/aps.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Zhou T, Chen J, Li R, Chen H, Luo S, Chen D, Cai C, Li W. The role of Exosomal miRNAs in cancer. J Transl Med. 2022;20:6. doi: 10.1186/s12967-021-03215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaac R, Reis FCG, Ying W, Olefsky JM. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021;33:1744–1762. doi: 10.1016/j.cmet.2021.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu M, Yuan S, Li S, Li L, Liu M, Wan S. The exosome-derived biomarker in atherosclerosis and its clinical application. J Cardiovasc Transl Res. 2019;12:68–74. doi: 10.1007/s12265-018-9796-y. [DOI] [PubMed] [Google Scholar]
- 18.Wang SS, Wu LJ, Li JJ, Xiao HB, He Y, Yan YX. A meta-analysis of dysregulated miRNAs in coronary heart disease. Life Sci. 2018;215:170–181. doi: 10.1016/j.lfs.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Caraballo C, Desai NR, Mulder H, Alhanti B, Wilson FP, Fiuzat M, Felker GM, Piña IL, O'Connor CM, Lindenfeld J, et al. Clinical Implications of the New York Heart Association Classification. J Am Heart Assoc. 2019;8:e014240. doi: 10.1161/JAHA.119.014240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi KH, Lee JM, Koo BK, Nam CW, Shin ES, Doh JH, Rhee TM, Hwang D, Park J, Zhang J, et al. Prognostic implication of functional incomplete revascularization and residual functional SYNTAX score in patients with coronary artery disease. JACC Cardiovasc Interv. 2018;11:237–245. doi: 10.1016/j.jcin.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Ward JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–244. doi: 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Dalen JE, Alpert JS, Goldberg RJ, Weinstein RS. The epidemic of the 20(th) century: Coronary heart disease. Am J Med. 2014;127:807–812. doi: 10.1016/j.amjmed.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 24.Yang F, Dong J, Wang W, Wang X, Fu X, Kumar NC, Zhang T. Evaluation of stenosis severity of coronary calcified lesions using transluminal attenuation gradient: Clinical application of 320-row volume CT. Minerva Med. 2017;108:305–316. doi: 10.23736/S0026-4806.17.04862-5. [DOI] [PubMed] [Google Scholar]
- 25.Sirtori CR, Labombarda F, Castelnuovo S, Perry R. The use of echocardiography for the non-invasive evaluation of coronary artery disease. Ann Med. 2017;49:134–141. doi: 10.1080/07853890.2016.1243801. [DOI] [PubMed] [Google Scholar]
- 26.Cao RY, Yang J, Zheng Y, Li H, Zhao Q, Ding Y, Li Q, Liu S, Wang L, Zheng H. The potential value of Copeptin and Pentraxin3 for evaluating the severity of coronary stenosis in patients with coronary artery disease. Clin Biochem. 2021;87:32–38. doi: 10.1016/j.clinbiochem.2020.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Mori MA, Ludwig RG, Garcia-Martin R, Brandao BB, Kahn CR. Extracellular miRNAs: From biomarkers to mediators of physiology and disease. Cell Metab. 2019;30:656–673. doi: 10.1016/j.cmet.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He X, Kuang G, Wu Y, Ou C. Emerging roles of exosomal miRNAs in diabetes mellitus. Clin Transl Med. 2021;11:e468. doi: 10.1002/ctm2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S, Lin Z, Zheng Z, Rao W, Lin Y, Chen H, Xie Q, Chen Y, Hu Z. Serum exosomal microRNA-766-3p expression is associated with poor prognosis of esophageal squamous cell carcinoma. Cancer Sci. 2020;111:3881–3892. doi: 10.1111/cas.14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su J, Li J, Yu Q, Wang J, Li X, Yang J, Xu J, Liu Y, Xu Z, Ji L, et al. Exosomal miRNAs as potential biomarkers for acute myocardial infarction. IUBMB Life. 2020;72:384–400. doi: 10.1002/iub.2189. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Li H, Yuan M, Li D, Sun C, Wang G. Serum exosomal MicroRNAs as potential circulating biomarkers for endometriosis. Dis Markers. 2020;2020:2456340. doi: 10.1155/2020/2456340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng D, Huo M, Li B, Wang W, Piao H, Wang Y, Zhu Z, Li D, Wang T, Liu K. The role of exosomes and exosomal MicroRNA in cardiovascular disease. Front Cell Dev Biol. 2021;8:616161. doi: 10.3389/fcell.2020.616161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aghabozorgi AS, Ahangari N, Eftekhaari TE, Torbati PN, Bahiraee A, Ebrahimi R, Pasdar A. Circulating exosomal miRNAs in cardiovascular disease pathogenesis: New emerging hopes. J Cell Physiol. 2019;234:21796–21809. doi: 10.1002/jcp.28942. [DOI] [PubMed] [Google Scholar]
- 34.Zhang YX, Zeng RR, Yang Y, Shen Y. Application of SYNTAX and its derivative scores in the selection of revascularization strategies for complex coronary heart disease. Chin Med Sci J. 2022;37:340–348. doi: 10.24920/004085. [DOI] [PubMed] [Google Scholar]
- 35.Huang Y, Liu Y, Huang J, Gao L, Wu Z, Wang L, Fan L. Let-7b-5p promotes cell apoptosis in Parkinson's disease by targeting HMGA2. Mol Med Rep. 2021;24:820. doi: 10.3892/mmr.2021.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandolesi G, Rizzo FR, Balletta S, Stampanoni Bassi M, Gilio L, Guadalupi L, Nencini M, Moscatelli A, Ryan CP, Licursi V, et al. The microRNA let-7b-5p Is negatively associated with inflammation and disease severity in multiple sclerosis. Cells. 2021;10:330. doi: 10.3390/cells10020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou R, Zhang Y, Du G, Han L, Zheng S, Liang J, Huang X, Qin Y, Wu W, Chen M, et al. Down-regulated let-7b-5p represses glycolysis metabolism by targeting AURKB in asthenozoospermia. Gene. 2018;663:83–87. doi: 10.1016/j.gene.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 38.Gong L, Xiao J, Yi J, Lu F, Liu X. Immunomodulatory effect of serum exosomes from crohn disease on macrophages via Let-7b-5p/TLR4 Signaling. Inflamm Bowel Dis. 2022;28:96–108. doi: 10.1093/ibd/izab132. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Yin B, Shu B, Liu Z, Ding H, Jia C. Differential expression of microRNA let-7b-5p regulates burn-induced hyperglycemia. Oncotarget. 2017;8:72886–72892. doi: 10.18632/oncotarget.20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berkan O, Arslan S, Lalem T, Zhang L, Sahin NO, Aydemir EI, Korkmaz O, Egilmez HR, Cekin N, Devaux Y. Regulation of microRNAs in coronary atherosclerotic plaque. Epigenomics. 2019;11:1387–1397. doi: 10.2217/epi-2019-0036. [DOI] [PubMed] [Google Scholar]
- 41.Ling H, Guo Z, Du S, Liao Y, Li Y, Ding C, Song C. Serum exosomal miR-122-5p is a new biomarker for both acute coronary syndrome and underlying coronary artery stenosis. Biomarkers. 2020;25:539–547. doi: 10.1080/1354750X.2020.1803963. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Chen M, Feng Q, Zhu L, Bai Z, Wang B, Guo Z, Hou A. MicroRNA-34a in coronary heart disease: Correlation with disease risk, blood lipid, stenosis degree, inflammatory cytokines, and cell adhesion molecules. J Clin Lab Anal. 2022;36:e24138. doi: 10.1002/jcla.24138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu X, Xu JF, Song M, Zhang L, Li YH, Han L, Tang MX, Zhang W, Zhong M, Wang ZH. Associations of circulating microRNA-221 and 222 with the severity of coronary artery lesions in acute coronary Syndrome patients. Angiology. 2022;73:579–587. doi: 10.1177/00033197211034286. [DOI] [PubMed] [Google Scholar]
- 44.Liu H, Xu W, Feng J, Ma H, Zhang J, Xie X, Zhuang D, Shen W, Zhou W. Increased expression of plasma miRNA-320a and let-7b-5p in heroin-dependent patients and its clinical significance. Front Psychiatry. 2021;12:679206. doi: 10.3389/fpsyt.2021.679206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vadla GP, Daghat B, Patterson N, Ahmad V, Perez G, Garcia A, Manjunath Y, Kaifi JT, Li G, Chabu CY. Combining plasma extracellular vesicle Let-7b-5p, miR-184 and circulating miR-22-3p levels for NSCLC diagnosis and drug resistance prediction. Sci Rep. 2022;12:6693. doi: 10.1038/s41598-022-10598-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due to the institutional policy that prohibits data sharing, but are available from the corresponding author on reasonable request.