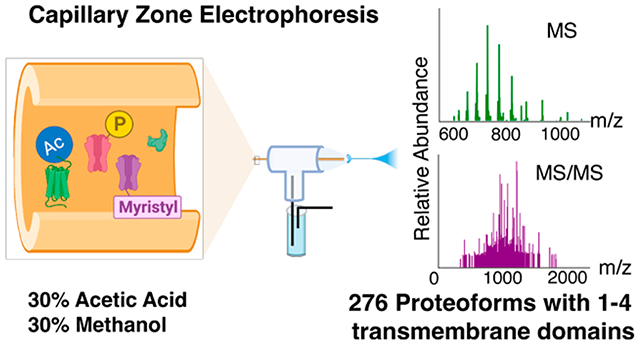
Abstract
Mass spectrometry (MS)-based top-down characterization of integral membrane proteins (IMPs) is crucial for understanding their functions in biological processes. However, it is technically challenging due to their low solubility in typical MS-compatible buffers. In this work, for the first time, we developed an efficient capillary zone electrophoresis (CZE)-tandem MS (MS/MS) method for the top-down proteomics (TDP) of IMPs enriched from mouse brains. Our technique employs a sample buffer containing 30% (v/v) formic acid and 60% (v/v) methanol for solubilizing IMPs and utilizes a separation buffer of 30% (v/v) acetic acid and 30% (v/v) methanol for maintaining the solubility of IMPs during CZE separation. Single-shot CZE-MS/MS identified 51 IMP proteoforms from the mouse brain sample. Coupling size exclusion chromatography (SEC) to CZE-MS/MS enabled the identification of 276 IMP proteoforms from the mouse brain sample containing 1–4 transmembrane domains. This proof-of-concept work demonstrates the high potential of CZE-MS/MS for the large-scale TDP of IMPs.
Graphical Abstract
Integral membrane proteins (IMPs) embedded within the lipid bilayer membranes are fundamental for essential cellular functions as transporters, receptors, channels, and enzymes, and they are also important drug targets.1,2 While membrane protein-coding genes with transmembrane domains (TMDs) were predicted to make up 26%–36% of the human protein-coding genes, IMPs are underrepresented in most proteomic studies due to their relatively low abundance and high hydrophobicity.3–5 Although bottom-up proteomics (BUP) has great success in the identification of IMPs by their peptides, their intact pictures are lost.6–10 Top-down proteomics (TDP) will provide us with a bird’s eye view of intact proteoforms from the same gene. Proteoforms from the same gene due to genetic variations, alternative splicing, and post-translational modifications (PTMs) can have different biological functions.11–15 PTMs of IMPs are related to their stability and functional regulation as well as apoptosis.16,17 Therefore, TDP analysis of IMPs in a proteoform-specific manner will offer a much better understanding of their biological functions.
Top-down MS characterization of IMPs is a hot research topic, and great effort has been made to study well-purified specific IMPs or IMP complexes.18–25 Some studies have been performed on coupling offline or online liquid-phase separation methods (e.g., liquid chromatography (LC)) to MS/MS for proteome-scale studies of IMPs.26–30 The Kelleher group employed a sodium dodecyl sulfate (SDS) buffer for IMP solubilization, followed by gel-eluted liquid fraction entrapment electrophoresis (GELFrEE) fractionation and LC-MS/MS for the identification of IMP proteoforms from human cell lysates.27,28 Whitelegge et al. utilized high-concentration formic acid (FA) to dissolve IMPs, followed by size-exclusion chromatography (SEC) and reversed-phase LC separations before MS and MS/MS.29 The Ge group also used a similar protein solubilization and separation procedure for TDP of IMPs purified from human embryonic kidney cells and cardiac tissue lysates, identifying up to nearly 200 IMPs.30
Capillary zone electrophoresis (CZE)-MS/MS has been proven as a valuable tool for TDP of complex biological samples due to its high separation efficiency and high sensitivity for proteoform separation and detection.31–34 For example, recently our group reported the identification of over 23,000 proteoforms from human cancer cell lysates using CZE-MS/MS, advancing the TDP for global proteoform identifications substantially.33 CZE-MS/MS for TDP analysis of single human cells has also been reported.34 CZE for membrane protein analysis started in the 1990s using background electrolytes containing a high concentration of salts or detergents.35,36 CZE-MS methods have also been developed for BUP of hydrophobic peptides.37,38 However, to the best of our knowledge, there is no report on coupling CZE to MS/MS online for TDP of IMPs.
Herein, we demonstrate the first example of CZE-MS/MS for the TDP of IMP proteoforms purified from mouse brains. Briefly, the IMPs were extracted from the mouse brain and purified by an alkaline and urea wash following procedures in the literature.39,40 After chloroform–methanol precipitation, 120 μg of IMPs was dissolved in a buffer containing 30% (v/v) FA and 60% (v/v) methanol, followed by SEC fractionation using a mobile phase (5% FA, 60% methanol, v/v). Each SEC fraction was analyzed by CZE-MS/MS with a BGE containing 30% (v/v) acetic acid (AA) and 30% (v/v) methanol, Figure 1A. The proteoform identification was performed via database search against a UniProt mouse database (UP000000589, 55315 entries) using TopPIC software (TOP-down mass spectrometry-based proteoform identification and characterization, versions 1.6.0 and 1.6.2).41 The proteoform identification was filtered by a proteoform-level false discovery rate (FDR) of less than 5%. The number of transmembrane domains (TMDs) of identified proteoforms was predicted by Deep TMHMM, which is a deep learning model for transmembrane topology prediction.28 TMHMM has been widely used to predict the transmembrane helices with a high accuracy since the first version in 2001.42 The detailed experimental procedures are described in Supporting Information I.
Figure 1.
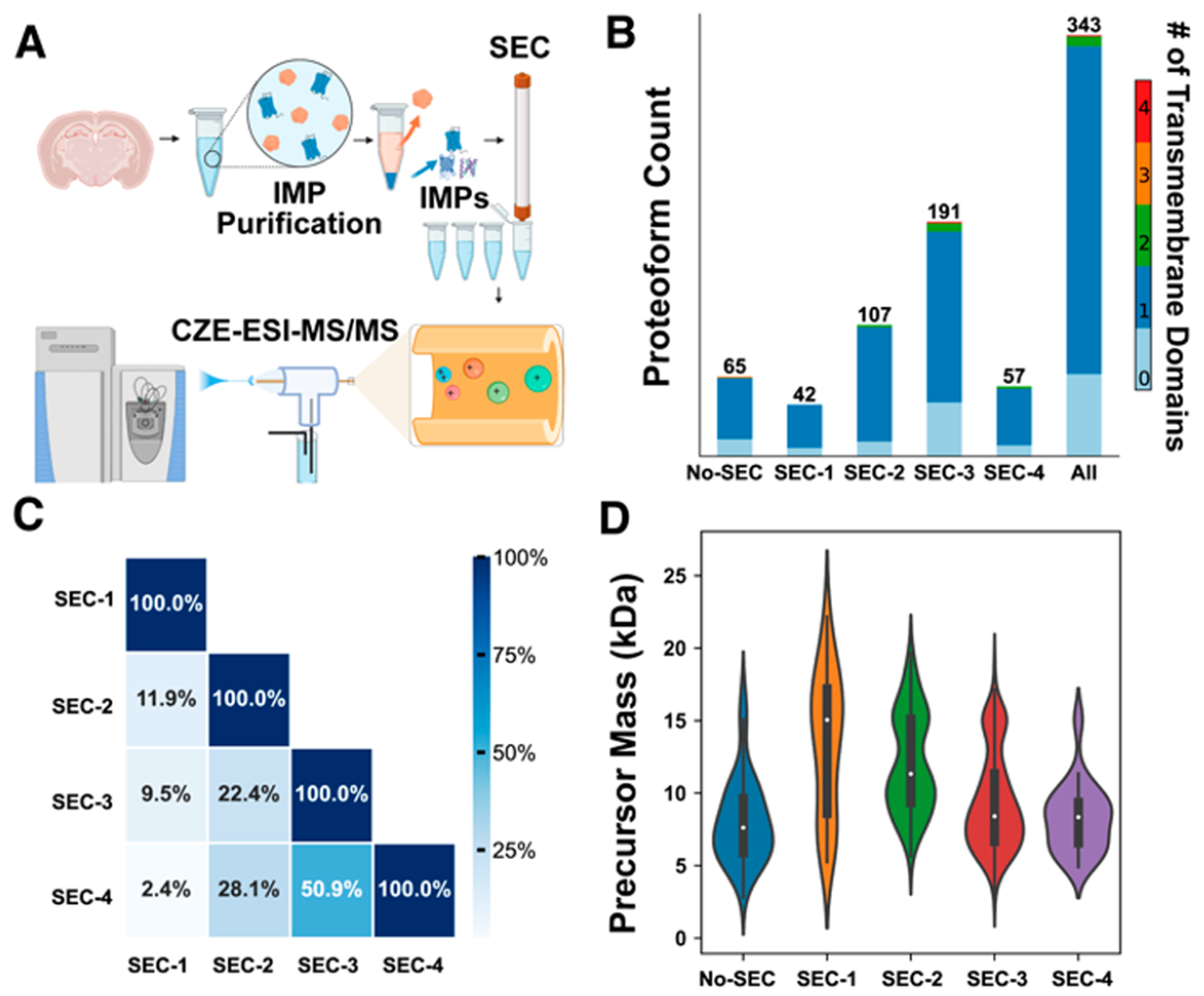
Intact integral membrane protein (IMP) analysis by CZE-MS/MS. (A) Flowchart of SEC-CZE-ESI-MS/MS for profiling IMPs of mouse brains, created with Biorender. (B) The identification number of proteoforms from each SEC fraction and the combined result from the four fractions. Different colors represent the number of predicted transmembrane domains (TMDs) based on the proteoform sequence by DeepTMHMM. (C) Heatmap of proteoform overlaps between any two SEC fractions. (D) Violin plots of mass distributions of identified proteoforms from the four SEC fractions and CZE-MS/MS alone (No-SEC).
Single-shot CZE-MS/MS analysis identified 21 proteins and 65 proteoforms from the mouse brain. Of the proteoforms identified, 51 (79%) have at least one TMD predicted by Deep TMHMM. The data demonstrate that our CZE-MS/MS method is efficient for IMP proteoform identification. We also checked the mass shifts of identified proteoforms for potential formylation modifications [+28 Da] due to the high concentration of FA that was used for the solubilization of IMPs before CZE-MS/MS. We did not see any 28-Da mass shifts in the identified proteoforms. We suspect the lack of artifactual formylation is due both to our storage of the FA solution at −20 °C before use and to the rapid dilution after solubilization.43 We noted that all of the identified IMP proteoforms were smaller than 18 kDa. As shown in Figure S1, the mouse brain IMP proteoforms migrated through the CZE capillary within the time window of 40–90 min. The earlier peaks during 40–50 min correspond to proteoforms smaller than 18 kDa. The later peaks during 50–90 min are most likely relatively large proteoforms, and there are no clear charge state distributions in those mass spectra under our instrument conditions.
To improve the identification of large proteoforms, we further fractionated the mouse brain IMP fraction dissolved in 30% (v/v) FA and 60% (v/v) methanol by SEC into four fractions to reduce the sample complexity. The mobile phase of SEC was kept at a high concentration of FA (5%) and methanol (60%) to maintain the solubility of IMPs. Each SEC fraction was analyzed by CZE-MS/MS in a technical duplicate. We identified 42, 107, 191, and 57 proteoforms from SEC fractions 1, 2, 3, and 4, respectively, Figure 1B. In total, 65 proteins and 343 proteoforms were identified from the mouse brain sample using SEC-CZE-MS/MS. Out of the identified proteoforms, 276 proteoforms are IMP proteoforms and had 1–4 TMDs, including proteoforms from enzymes (e.g., ATP synthase, Cytochrome c oxidase, and NADH dehydrogenase), channels (i.e., voltage-dependent anion-selective channel protein 1), and receptors (i.e., mitochondrial import receptor). The number of IMP proteoforms from SEC-CZE-MS/MS is improved by more than 5-fold compared to that from single-shot CZE-MS/MS (276 vs 51). We need to highlight that SEC-CZE-MS/MS identified 13 proteoforms higher than 18 kDa and that single-shot CZE-MS/MS did not identify any proteoforms larger than 18 kDa. The data clearly indicate that our SEC-CZE-MS/MS technique could be a useful tool for the proteome-scale characterization of IMP proteoforms. The proteoforms identified by SEC-CZE-MS/MS are listed in Supporting Information II. The MS raw files have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD042298.44 We noted that most of the identified IMP proteoforms had one TMD, which is consistent across all of the SEC fractions, Figure 1B. The phenomenon is most likely due to the relatively small proteoforms identified in this study. As shown in Figure S2, CZE-MS/MS alone identified proteoforms in the mass range of 3–17 kDa, with the majority in the range of 5–10 kDa. SEC-CZE-MS/MS identified proteoforms in a mass range of 3–22 kDa, producing much more proteoforms larger than 10 kDa compared to CZE-MS/MS alone. Identification of large proteoforms (i.e., >30 kDa) from complex proteomes by TDP is challenging due to their low signal-to-noise ratios caused by wide charge state distributions and due to the limited mass resolution of commonly used mass spectrometers in TDP studies.30,32,33 Figure S3 shows a Venn diagram about the proteoforms identified by CZE-MS/MS and SEC-CZE-MS/MS. Interestingly, although SEC-CZE-MS/MS identified many more proteoforms than CZE-MS/MS alone (343 vs 65), only less than 50% of proteoforms from CZE-MS/MS alone are covered by SEC-CZE-MS/MS.
We further studied the proteoform overlaps between any two SEC fractions, as shown in Figure 1C. The proteoform overlap ranges from 2% to 51% and becomes smaller when the two fractions have a bigger elution time difference. This suggests that SEC can fractionate the mouse brain IMP proteoforms efficiently. Figure 1D shows the violin plots of mass distributions of identified proteoforms from CZE-MS/MS alone (no-SEC) and the four SEC fractions. It is clear that the median proteoform mass gradually decreases from fraction 1 (SEC-1, ~15 kDa) to fraction 3 (SEC-3, ~8 kDa), indicating a reasonably good separation of SEC for the proteoforms by their size. Fractions 3 and 4 (SEC-3 and SEC-4) have comparable mass distributions, although substantially more proteoforms were identified in F3 compared to F4 (191 vs 57). SEC-CZE-MS/MS improved the identification of relatively large proteoforms compared to that of CZE-MS/MS alone (no-SEC).
Figure 2 shows the representative electropherogram of CZE-MS/MS analysis of SEC fraction 2 including three examples of IMP proteoforms identified in the CZE-MS/MS run at different migration times. These three proteoforms were identified with low E-values ranging from 8.3 × 10−24 to 7.3 × 10−38, indicating high-confidence identifications. These three proteoforms all contain one TMD, and good backbone cleavage coverages were achieved for the TMDs using the higher energy collision dissociation (HCD) method. Electropherograms of SEC fractions 1, 3, and 4 as well as some example proteoforms identified in those fractions are shown in Figures S4–S6. We determined N-terminal methionine removal (ATP synthase subunit e, Figure S5), N-terminal truncation, mitochondrion transit peptide cleavage (Cytochrome c oxidase subunit 4 isoform 1, mitochondrial, Cox4i1, Figure S4), N-terminal acetylation, phosphorylation (cell cycle exit and neuronal differentiation protein 1, Cend1, Figure S5), and myristoylation on the N-terminal glycine residue (Brain acid soluble protein 1, Basp1, Figure S4) on those proteoforms. The Basp1 proteoform has a mass shift of 79.160 Da on the first ten amino acid residues, which matches with the methionine removal and myristoylation on N-terminal glycine [210.198 Da–131.040 Da = 79.159 Da]. The PTM information matches well with that in the UniProt database (https://www.uniprot.org/uniprotkb/Q91XV3/entry). The N-myristoylation is required for Basp1 as a transcriptional corepressor to remove histone modifications H3K9ac and H3K4me3.45 We identified over 60 Cend1 proteoforms (Supporting Information II), and the majority of them have N-terminal acetylation. We also detected various mass shifts (e.g., 78–81 Da, 122 Da, 159–161, 238, and 478 Da) on the Cend1 proteoforms. Those mass shifts could be explained as single phosphorylation (80 Da), multiple phosphorylation (i.e., 160, 240, and 480 Da), or a combination of phosphorylation and acetylation (i.e., 122 Da). Cend1 has multiple phosphorylation sites according to the UniProt database and PhosphoSitePlus database (https://www.phosphosite.org). Cend1 is a neuronal protein highly expressed in the postnatal mouse brain and modulates cell cycle exit and neuronal differentiation.46 However, the functions of specific phosphorylated Cend1 proteoforms are still not known. Extracted ion electropherograms of some of the example proteoforms are shown in Figures S7 and S8.
Figure 2.
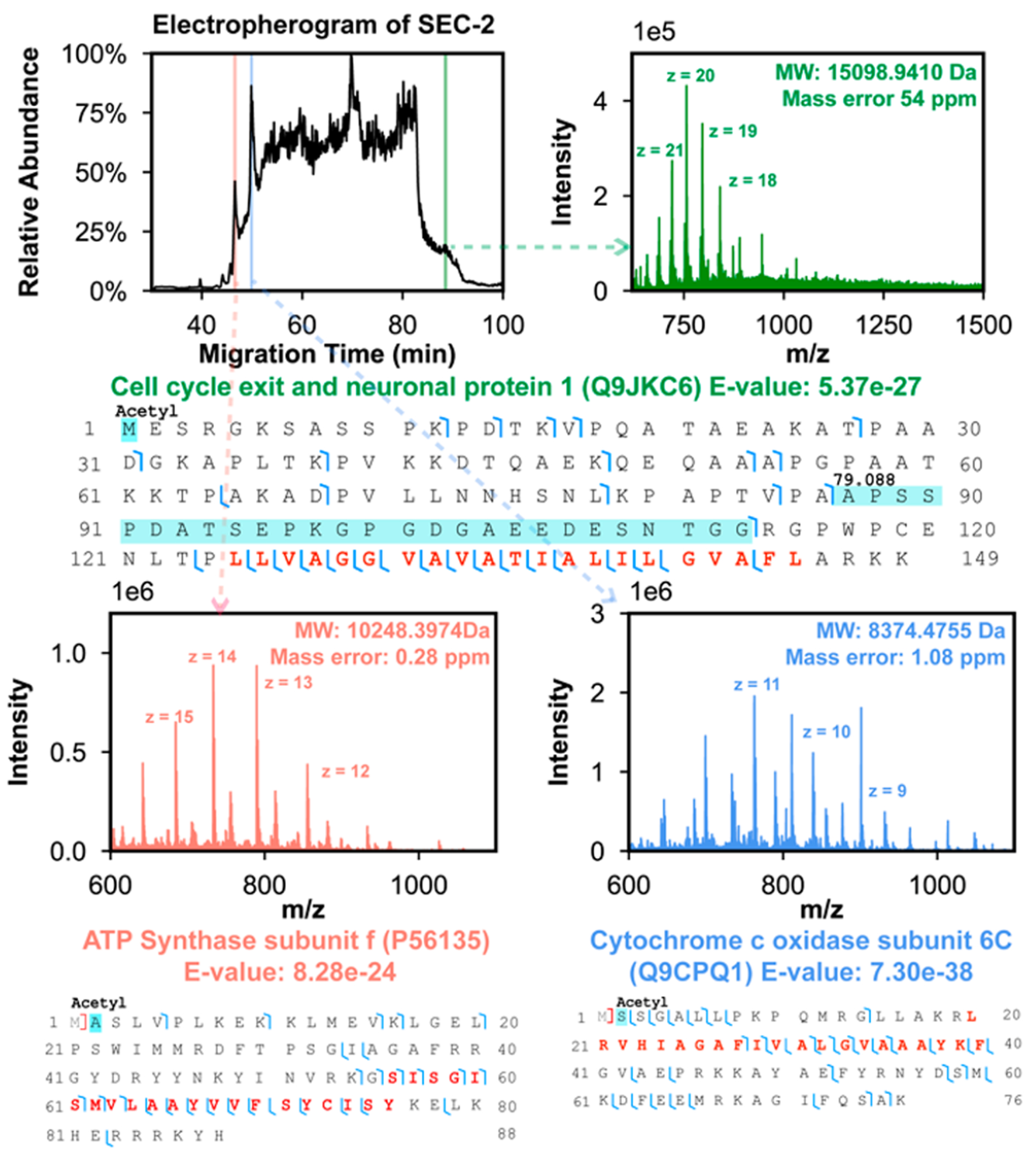
Mouse brain IMP data of SEC fraction 2 by CZE-MS/MS. The electropherogram of SEC fraction 2 and mass spectra of three IMP proteoforms identified at three different migration times are presented. The proteoform sequences and fragmentation patterns of the three IMP proteoforms are also shown. The TMD of proteoforms are marked in red in the proteoform sequences. The detected mass shifts and PTMs are labeled.
In summary, for the first time, we developed a CZE-MS/MS technique for TDP of IMPs, and by employing SEC-CZE-MS/MS, we achieved the identification of hundreds of IMP proteoforms from mouse brains. We expect that coupling SEC fractionation with CZE-MS/MS will be a useful tool for large-scale TDP of IMPs. The current limitation of the technique is the identification of large IMP proteoforms due to the limited mass resolution of the mass spectrometer used in this study. We believe that coupling our SEC-CZE separations to a high-end Orbitrap,47 FT ion cyclotron resonance (ICR),48 or time-of-flight (TOF)49 mass spectrometers will improve the characterization of large IMP proteoforms substantially.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Yuan Wang’s group at the Department of Animal Science, Michigan State University for kindly providing the mouse samples for our experiment. We also thank Prof. Julian Whitelegge at University of California, Los Angeles for all the valuable discussions. The work was funded by the National Cancer Institute (NCI) through the grant R01CA247863 (to LS) and the National Science Foundation through grant MCB-2034631 (to PKL). We thank the support from the National Institute of General Medical Sciences (NIGMS) through grants R01GM125991 and R01GM118470. We also thank the support from the National Science Foundation (CAREER Award, grant DBI1846913).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.3c02346.
Experimental details; the electropherogram and MS/MS spectra of single-shot CZE-MS/MS; mass distribution of the identified proteoforms for CZE-MS/MS and SEC-CZE-MS/MS; overlap of identified proteoforms between CZE-MS/MS and SEC-CZE-MS/MS; the electropherogram and mass spectra of SEC fraction 1, fraction 3, fraction 4; the extracted ion electropherograms of selected proteoforms in SEC fraction 3 and fraction 4 (PDF)
Lists of identified proteoforms (XLSX)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.analchem.3c02346
The authors declare no competing financial interest.
Contributor Information
Qianjie Wang, Department of Chemistry, Michigan State University, East Lansing, Michigan 48824, United States; Department of Biochemistry and Molecular Biology and Plant Resilience Institute, Michigan State University, East Lansing, Michigan 48824, United States.
Tian Xu, Department of Chemistry, Michigan State University, East Lansing, Michigan 48824, United States.
Fei Fang, Department of Chemistry, Michigan State University, East Lansing, Michigan 48824, United States.
Qianyi Wang, Department of Chemistry, Michigan State University, East Lansing, Michigan 48824, United States.
Peter Lundquist, Department of Biochemistry and Molecular Biology and Plant Resilience Institute, Michigan State University, East Lansing, Michigan 48824, United States.
Liangliang Sun, Department of Chemistry, Michigan State University, East Lansing, Michigan 48824, United States.
REFERENCES
- (1).Kar U; Simonian M; Whitelegge JP Expert Rev. Proteomics 2017, 14, 715–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wu CC; Yates JR Nat. Biotechnol 2003, 21, 262–267. [DOI] [PubMed] [Google Scholar]
- (3).Sinitcyn P; Richards AL; Weatheritt RJ; Brademan DR; Marx H; Shishkova E; Meyer JG; Hebert AS; Westphall MS; Blencowe BJ; Cox J; Coon JJ Nat. Biotechnol 2023, DOI: 10.1038/s41587-023-01714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Fagerberg L; Jonasson K; von Heijne G; Uhlén M; Berglund L Proteomics 2010, 10, 1141–1149. [DOI] [PubMed] [Google Scholar]
- (5).Uhlén M; Fagerberg L; Hallström BM; Lindskog C; Oksvold P; Mardinoglu A; Sivertsson Å; Kampf C; Sjöstedt E; Asplund A; Olsson I; Edlund K; Lundberg E; Navani S; Szigyarto CA-K; Odeberg J; Djureinovic D; Takanen JO; Hober S; Alm T; Edqvist P-H; Berling H; Tegel H; Mulder J; Rockberg J; Nilsson P; Schwenk JM; Hamsten M; von Feilitzen K; Forsberg M; Persson L; Johansson F; Zwahlen M; von Heijne G; Nielsen J; Pontén F Science 2015, 347, 1260419. [DOI] [PubMed] [Google Scholar]
- (6).Chu H; Zhao Q; Liu J; Yang K; Wang Y; Liu J; Zhang K; Zhao B; He H; Zheng Y; Zhong S; Liang Z; Zhang L; Zhang Y Anal. Chem 2022, 94, 758–767. [DOI] [PubMed] [Google Scholar]
- (7).Zhao Q; Fang F; Shan Y; Sui Z; Zhao B; Liang Z; Zhang L; Zhang Y Anal. Chem 2017, 89, 5179–5185. [DOI] [PubMed] [Google Scholar]
- (8).Fischer F; Wolters D; Rögner M; Poetsch A Mol. Cell. Proteomics 2006, 5, 444–453. [DOI] [PubMed] [Google Scholar]
- (9).Lee HC; Carroll A; Crossett B; Connolly A; Batarseh A; Djordjevic MA Front. Plant Sci 2020, 11, 595726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Chen EI; McClatchy D; Park SK; Yates JR Anal. Chem 2008, 80, 8694–8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Smith LM; Kelleher NL Nat. Methods 2013, 10, 186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Smith LM; Kelleher NL Science 2018, 359, 1106–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Costa HA; Leitner MG; Sos ML; Mavrantoni A; Rychkova A; Johnson JR; Newton BW; Yee M-C; De La Vega FM; Ford JM; Krogan NJ; Shokat KM; Oliver D; Halaszovich CR; Bustamante CD Proc. Natl. Acad. Sci. U S A 2015, 112, 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yang X; Coulombe-Huntington J; Kang S; Sheynkman GM; Hao T; Richardson A; Sun S; Yang F; Shen YA; Murray RR; Spirohn K; Begg BE; Duran-Frigola M; MacWilliams A; Pevzner SJ; Zhong Q; Wanamaker SA; Tam S; Ghamsari L; Sahni N; Yi S; Rodriguez MD; Balcha D; Tan G; Costanzo M; Andrews B; Boone C; Zhou XJ; Salehi-Ashtiani K; Charloteaux B; Chen AA; Calderwood MA; Aloy P; Roth FP; Hill DE; Iakoucheva LM; Xia Y; Vidal M Cell 2016, 164, 805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jenuwein T; Allis CD Science 2001, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- (16).Kerner J; Lee K; Tandler B; Hoppel CL Biochim. Biophys. Acta 2012, 1818, 1520–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kerner J; Lee K; Hoppel CL Free Radic. Res 2011, 45, 16–28. [DOI] [PubMed] [Google Scholar]
- (18).Keener JE; Zhang G; Marty MT Anal. Chem 2021, 93, 583–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Barrera NP; Isaacson SC; Zhou M; Bavro VN; Welch A; Schaedler TA; Seeger MA; Miguel RN; Korkhov VM; van Veen HW; Venter H; Walmsley AR; Tate CG; Robinson CV Nat. Methods 2009, 6, 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yen H-Y; Liko I; Song W; Kapoor P; Almeida F; Toporowska J; Gherbi K; Hopper JTS; Charlton SJ; Politis A; Sansom MSP; Jazayeri A; Robinson CV Nat. Chem 2022, 14, 1375–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Keener JE; Zambrano DE; Zhang G; Zak CK; Reid DJ; Deodhar BS; Pemberton JE; Prell JS; Marty MT J. Am. Chem. Soc 2019, 141, 1054–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Susa AC; Lippens JL; Xia Z; Loo JA; Campuzano IDG; Williams ER J. Am. Soc. Mass Spectrom 2018, 29, 203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Harvey SR; O’Neale C; Schey KL; Wysocki VH Anal. Chem 2022, 94, 1515–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ro SY; Schachner LF; Koo CW; Purohit R; Remis JP; Kenney GE; Liauw BW; Thomas PM; Patrie SM; Kelleher NL; Rosenzweig AC Nat. Commun 2019, 10, 2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Fantin SM; Parson KF; Niu S; Liu J; Polasky DA; Dixit SM; Ferguson-Miller SM; Ruotolo BT Anal. Chem 2019, 91, 15469–15476. [DOI] [PubMed] [Google Scholar]
- (26).Donnelly DP; Rawlins CM; DeHart CJ; Fornelli L; Schachner LF; Lin Z; Lippens JL; Aluri KC; Sarin R; Chen B; Lantz C; Jung W; Johnson KR; Koller A; Wolff JJ; Campuzano IDG; Auclair JR; Ivanov AR; Whitelegge JP; Paša-Tolić L; Chamot-Rooke J; Danis PO; Smith LM; Tsybin YO; Loo JA; Ge Y; Kelleher NL; Agar JN Nat. Methods 2019, 16, 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Catherman AD; Li M; Tran JC; Durbin KR; Compton PD; Early BP; Thomas PM; Kelleher NL Anal. Chem 2013, 85, 1880–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Catherman AD; Durbin KR; Ahlf DR; Early BP; Fellers RT; Tran JC; Thomas PM; Kelleher NL Mol. Cell. Proteomics 2013, 12, 3465–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Whitelegge JP; Zhang H; Aguilera R; Taylor RM; Cramer WA Mol. Cell. Proteomics 2002, 1, 816–827. [DOI] [PubMed] [Google Scholar]
- (30).Brown KA; Tucholski T; Alpert AJ; Eken C; Wesemann L; Kyrvasilis A; Jin S; Ge Y Anal. Chem 2020, 92, 15726–15735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Han X; Wang Y; Aslanian A; Fonslow B; Graczyk B; Davis TN; Yates JRI J. Proteome Res 2014, 13, 6078–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Chen D; McCool EN; Yang Z; Shen X; Lubeckyj RA; Xu T; Wang Q; Sun L Mass Spectrom. Rev 2023, 42, 617–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).McCool EN; Xu T; Chen W; Beller NC; Nolan SM; Hummon AB; Liu X; Sun L Sci. Adv 2022, 8, eabq6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Johnson KR; Gao Y; Greguš M; Ivanov AR Anal. Chem 2022, 94, 14358–14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Josić D; Zeilinger K; Reutter W; Böttcher A; Schmitz G G. J. Chromatogr. A 1990, 516, 89–98. [DOI] [PubMed] [Google Scholar]
- (36).Nelson BC; Malik S; Seeley SK; Uden PC Chromatographia 1999, 49, 28–34. [Google Scholar]
- (37).Cheng J; Morin GB; Chen DDY ELECTROPHORESIS 2020, 41, 370–378. [DOI] [PubMed] [Google Scholar]
- (38).Cheng J; Chen DDY ELECTROPHORESIS 2018, 39, 1216–1221. [DOI] [PubMed] [Google Scholar]
- (39).Lu A; Wiśniewski JR; Mann MJ Proteome Res. 2009, 8, 2418–2425. [DOI] [PubMed] [Google Scholar]
- (40).Zhao Q; Sun L; Liang Y; Wu Q; Yuan H; Liang Z; Zhang L; Zhang Y Talanta 2012, 88, 567–572. [DOI] [PubMed] [Google Scholar]
- (41).Kou Q; Xun L; Liu X Bioinformatics 2016, 32, 3495–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Krogh A; Larsson B; von Heijne G; Sonnhammer ELL J. Mol. Biol 2001, 305, 567–580. [DOI] [PubMed] [Google Scholar]
- (43).Doucette AA; Vieira DB; Orton DJ; Wall MJ J. Proteome Res. 2014, 13, 6001–6012. [DOI] [PubMed] [Google Scholar]
- (44).Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Pérez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yilmaz S; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Ternent T; Brazma A; Vizcaíno JA Nucleic Acids Res. 2019, 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Moorhouse AJ; Loats AE; Medler KF; Roberts SGE iScience 2022, 25, 104796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Segklia K; Stamatakis A; Stylianopoulou F; Lavdas AA; Matsas R Front. Cell. Neurosci 2019, 12, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Wang C; Liang Y; Zhao B; Liang Z; Zhang L; Zhang Y Anal. Chem 2022. 94, 6172–6179. [DOI] [PubMed] [Google Scholar]
- (48).Tucholski T; Knott S; Chen B; Pistono P; Lin Z; Ge Y Anal. Chem 2019, 91, 3835–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Fornelli L; Parra J; Hartmer R; Stoermer C; Lubeck M; Tsybin YO Anal. Bioanal. Chem 2013, 405, 8505–8514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


