Abstract
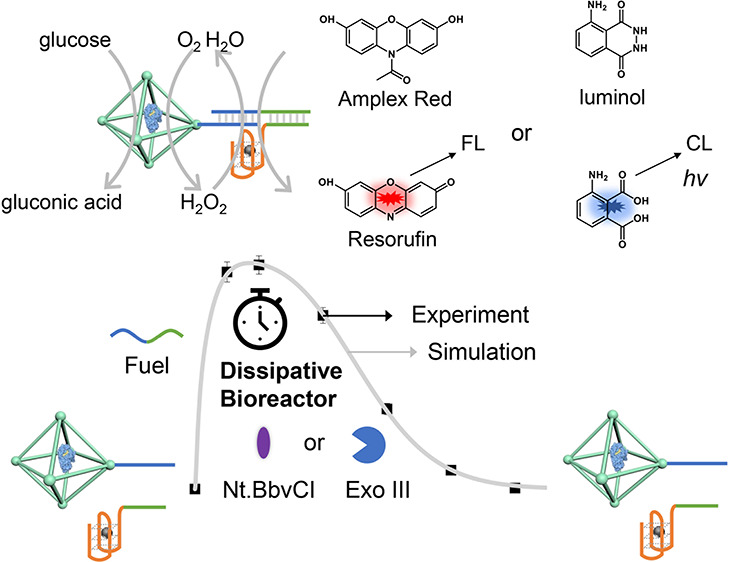
Glucose oxidase-loaded ZIF-90 metal–organic framework nanoparticles conjugated to hemin-G-quadruplexes act as functional bioreactor hybrids operating transient dissipative biocatalytic cascaded transformations consisting of the glucose-driven H2O2-mediated oxidation of Amplex-Red to resorufin or the glucose-driven generation of chemiluminescence by the H2O2-mediated oxidation of luminol. One system involves the fueled activation of a reaction module leading to the temporal formation and depletion of the bioreactor conjugate operating the nickase-guided transient biocatalytic cascades. The second system demonstrates the fueled activation of a reaction module yielding a bioreactor conjugate operating the exonuclease III-dictated transient operation of the two biocatalytic cascades. The temporal operations of the bioreactor circuits are accompanied by kinetic models and computational simulations enabling us to predict the dynamic behavior of the systems subjected to different auxiliary conditions.
Keywords: DNA, Chemiluminescence, Dissipative circuit, DNAzyme, Biocatalytic cascade, Strand displacement
Emulating native transient dissipative circuits by synthetic networks attracts substantial research efforts within the rapidly developing area of systems chemistry.1−3 Particularly the information encoded in the base sequence of nucleic acids provides a versatile means to dynamically reconfigure the biopolymer by a rich “tool-box” of triggers into diverse structural motives.4 Indeed, diverse dynamic reversible reconfiguration processes of nucleic acids, such as strand displacement,5,6 formation and dissociation of G-quadruplex,7,8 stabilization/destabilization of duplex nucleic acids by metal ions,9−11 light-stimulated stabilization and separation of nucleic acid duplexes,12,13 and enzymatic manipulation of the oligonucleotide biopolymer,14−19 were used to develop DNA switches20,21 and machines,22,23 logic gates,24 programmed DNA nanostructures,25−27 and switchable DNA-based materials.28−30 Moreover, the integration of control units within the switchable reconfigurable DNA circuits provides a means to develop transiently operating dissipative nucleic acid-based networks.31−33 Different triggers controlling the temporal transient operation of the circuits were introduced, including the application of enzymes, such as endonuclease,34 nickase,35 or ligase,36,37 as catalytic units regulating the formation and temporal depletion of DNA structures, the application of DNAzymes,38 the ion-induced formation and dissociation of G-quadruplexes,39 the ribonuclease (RNase)-stimulated control over transcription machineries,40 or the application of light-signals as temporal regulating stimuli of DNA circuits.38,41 Identification of useful applications of transient dissipative DNA circuits is, however, still challenging. Several applications of transient nucleic acid-based circuits were demonstrated, including the temporal aggregation/deaggregation of metal nanoparticles or semiconductor quantum dots and control over their optical properties of the systems,42,43 the temporal formation and separation of DNA nanostructures, such as microtubules or wires,44 and the conjugation of the DNA circuits to enzymes, driving transient biocatalytic cascades.45
Development of catalytic nanoparticles,46−48 such as metal,49−52 metal oxide,53−58 metal–organic framework nanoparticles,59−63 or carbon-based nanoparticles,64−66 “nanozymes”, attracts substantial research efforts in catalysis and materials science. Also, integration of biocatalysts and catalytic nanoparticles into nanoparticle frameworks generated hybrid composites acting as bioreactor systems driving catalytic cascades.67 Tethering of aptamer strands to nanozymes led to aptananozymes revealing the binding and catalytic functions mimicking native enzymes68 and to cell-targeting catalytic nanoparticles for therapeutic applications.69 Moreover, catalytic nanoparticles conjugated to catalytic nucleic acids, DNAzymes, acted as hybrids operating as bioreactor systems driving catalytic or biocatalytic cascades.70 Diverse applications of nucleic acid-modified nanozymes were reported for sensing,71−73 imaging,74,75 and therapeutic uses.69,76−78 Particularly, the loading of metal–organic framework nanoparticles with nanoparticle catalysts,79 biocatalyst,80 or drugs81 and capping the loaded nanocarriers with stimuli-responsive nucleic acid gates82 for controlled release of the loads were demonstrated.
Accordingly, the conjugation of transiently operating nucleic acid gates to nanozymes could introduce dynamic dimensions to nucleic acid/nanozyme hybrids. Here, we report on the assembly of supramolecular bioreactor systems consisting of nucleic acid-modified glucose oxidase (GOx)-loaded ZIF-90 metal–organic framework nanoparticles (NMOFs) and hemin-G-quadruplex constituents demonstrating transient-peroxidase like activities in the presence of the nickase, Nt.BbvCI, or exonuclease III, Exo III, as control units. The ZIF-90 nanoparticles were selected due to the ease to integrate GOx into the ZIF framework and due to the surface carboxaldehyde functionalities of ZIF-90 that allow the covalent conjugation of the nucleic acids to the scaffold.70
Transient Nickase-Driven Biocatalytic Cascades Using GOx-Loaded/Hemin-G-Quadruplex-Conjugated ZIF-90 as Functional Frameworks
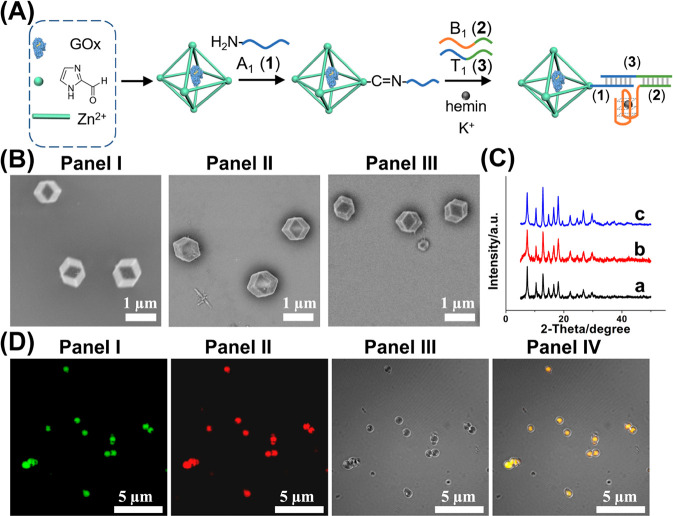
Figure 1 depicts the synthesis and characterization of the (1)+(2)/(3) supramolecular GOx-loaded ZIF-90 NMOFs hybrid as the functional conjugate for the nickase-activated, transient bioreactor system. Glucose oxidase (GOx)-loaded ZIF-90 NMOFs were prepared according to the literature70 by reacting imidazole-2-carboxaldehyde with Zn2+-ion, in the presence of GOx, Figure 1A. The carboxaldehyde functions associated with the NMOFs were functionalized with amino-nucleic acid tethers A1, (1), that provide the anchoring sites for the assembly of the GOx-loaded ZIF-90 NMOFs/G-quadruplex conjugate. Reacting the (1)-functionalized NMOFs with the strand T1, (3), and the G-rich strand B1, (2), in the presence of K+-ion yielded the GOx-loaded ZIF-90 NMOFs/G-quadruplex hybrid, and the subsequent treatment of the assembly with hemin yielded the GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex bioreactor hybrid system. Figure 1B depicts the SEM images of bare ZIF-90 NMOFs, the GOx-loaded ZIF-90 NMOFs, and the (1)+(2)/(3)-modified GOx-loaded ZIF-90 NMOFs. Identical dodecahedral crystalline NMOFs are observed, indicating that the functionalization of the NMOFs with the biomaterials does not affect the crystalline structure of the particles. This is further supported by identical XRD patterns of the NMOFs in the different states of modification of the NMOFs (Figure 1C). The loading of the NMOFs with GOx corresponded to 82 μg/mg NMOFs, and the loading of the tether (1) on the NMOFs and of the G-quadruplexes on the NMOFs both corresponded to 10 nmol/mg (for experimental details evaluating the loading of the components on the NMOFs see Supporting Information, Figures S1–S3). Confocal microscopy imaging experiments, Figure 1D, further supported the construction of the (1)+(2)/(3) supramolecular GOx-loaded ZIF-90 NMOFs/G-quadruplex conjugate. The loading of the hybrid with FITC (fluorescein isothiocyanate isomer I)-modified GOx demonstrated green fluorescent (λ = 420 nm) particles, panel I, consistent with the GOx-loaded NMOFs. Subjecting the particles to Zn(II) protoporphyrin IX, Zn(II)PPIX, resulted in the characteristic red fluorescence, λ = 590 nm, of Zn(II)PPIX bound to G-quadruplexes,83 panel II. The bright-field image of the particles is depicted in panel III, and the overlay of the images is displayed in panel IV, revealing a yellow fluorescence of the combined constituents associated with the hybrid NMOFs. The successful construction of (1)+(2)/(3) supramolecular GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex bioreactor was further characterized by the efficient biocatalytic cascades of glucose-driven H2O2-channeled oxidation of Amplex-Red or generation of chemiluminescence, Figure S4. (We note that the added K+-ion is essential to assemble the (1)+(2)/(3) supramolecular GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex bioreactor; see Figure S4B, panels I and II, curve c.)
Figure 1.
(A) Schematic synthesis of the (1)+(2)/(3) supramolecular GOx-loaded ZIF-90 NMOFs/hemin-G-Quadruplex bioreactor particles. (B) SEM images corresponding to panel I, bare ZIF-90 NMOFs; panel II, GOx-loaded ZIF-90 NMOFs; panel III, (1)+(2)/(3) supramolecular GOx-loaded ZIF-90 NMOFs. (C) XRD spectra corresponding to (a) bare ZIF-90 NMOFs; (b) GOx-loaded ZIF-90 NMOFs; (c) (1)+(2)/(3) modified GOx-loaded ZIF-90 NMOFs. (D) Confocal microscopy images corresponding to (1)+(2)/(3) supramolecular nanoparticles consisting of FITC-modified-GOx-loaded NMOFs and Zn(II)-protoporphyrin IX (Zn(II)PPIX)-modified G-quadruplex units: panel I, fluorescence confocal microscopy image of the NMOFs using the FITC fluorescence channel, λ = 420 nm; panel II, fluorescence confocal microscopy image of the NMOFs using the Zn(II)PPIX/G-quadruplex fluorescence channel, λ = 590 nm; panel III, bright-field microscopy image; panel IV, overlay of panels I–III.
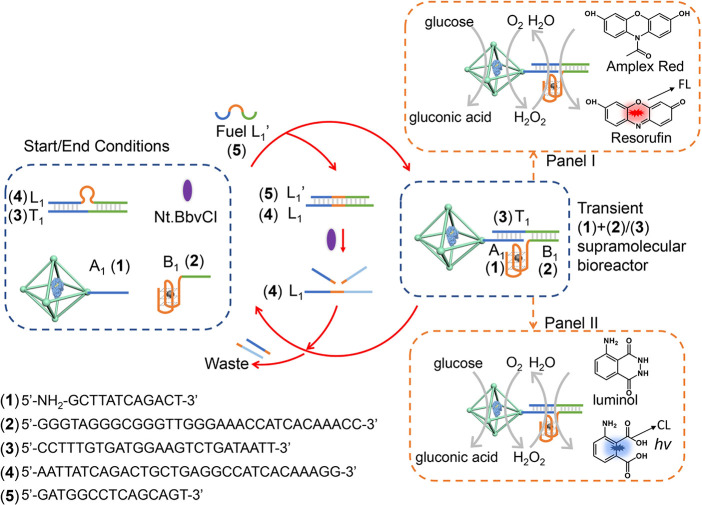
Figure 2 depicts schematically the fuel-driven nickase-modulated transient operation of the (1)+(2)/(3) GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex bioreactor system. The initial reaction module consists of the A1, (1)-functionalized GOx-loaded ZIF-90 NMOFs, the hemin-G-quadruplex, B1, (2) constituent, the duplex composed of L1/T1, (4)/(3), and the nicking enzyme Nt.BbvCI. Subjecting the reaction module to the fuel strand L1′, (5) results in the displacement of the duplex L1/T1, (4)/(3) to yield the duplex L1/L1′, (4)/(5) and free T1, (3). The released T1, (3) self-assembles the (1)+(2)/(3) GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex bioreactor hybrid system that drives two different biocatalytic cascades. One biocatalytic cascade, panel I, involves the aerobic oxidation of glucose, yielding gluconic acid and H2O2 and the subsequent hemin-G-quadruplex-catalyzed oxidation of Amplex-Red by H2O2 to form the fluorescent resorufin. The second biocatalytic cascade, driven by the GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex bioreactor, is displayed in Figure 2, panel II and involves the aerobic oxidation of glucose to gluconic acid and H2O2 and the subsequent hemin-G-quadruplex-catalyzed oxidation of luminol by the H2O2 to generate chemiluminescence, λ = 425 nm.84 The resulting duplex L1/L1′, (4)/(5), generated upon the fueled operation of the reaction module and the formation of the hybrid NMOFs bioreactor, is engineered to include in the sequence of L1′, (5) the nicking site to be cleaved by Nt.BbvCI. Cleavage of the L1′, (5) leads to fragmented “waste” products that are separated from the duplex, leading to the separation of L1, (4). The separated strand L1, (4) displaces T1, (3) from the (1)+(2)/(3) hybrid bioreactor structure, resulting in the separation of two bioreactor constituents, the blockage of the two biocatalytic cascades displayed in panels I and II, and the regeneration of the parent inactive reaction module. That is, the L1′, (5) triggered activation of the reaction module leads to the temporal activation of the two biocatalytic cascades that reveal a guided transient operation and depletion mechanism to regenerate the parent inactive state.
Figure 2.
Schematic reaction module for the fueled transient operation of the (1)+(2)/(3) GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex bioreactor system leading to the temporal biocatalytic cascades consisting of the following: panel I, GOx/hemin-G-quadruplex catalyzed oxidation of Amplex-Red to fluorescent resorufin; panel II, GOx/hemin-G-quadruplex catalyzed generation of chemiluminescence through the catalyzed H2O2 oxidation of luminol.
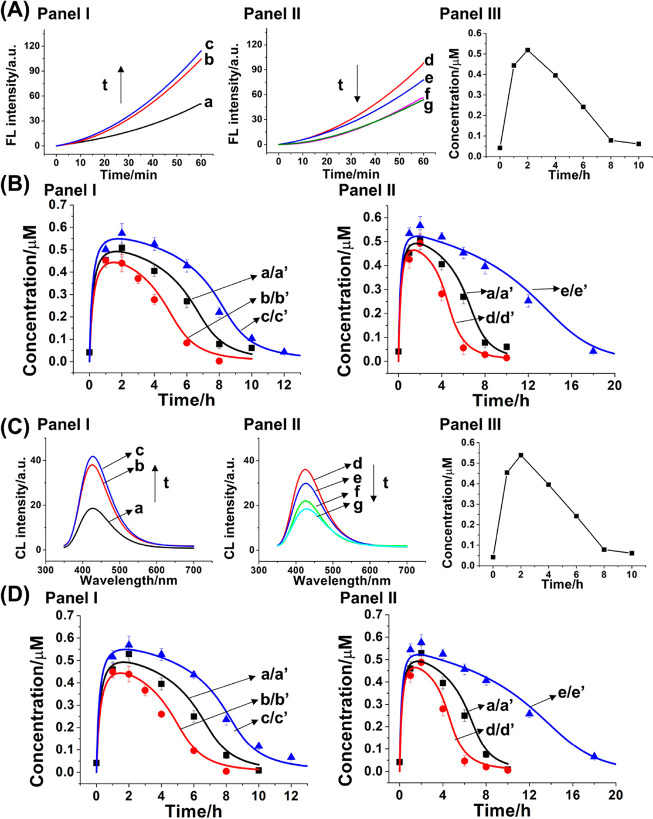
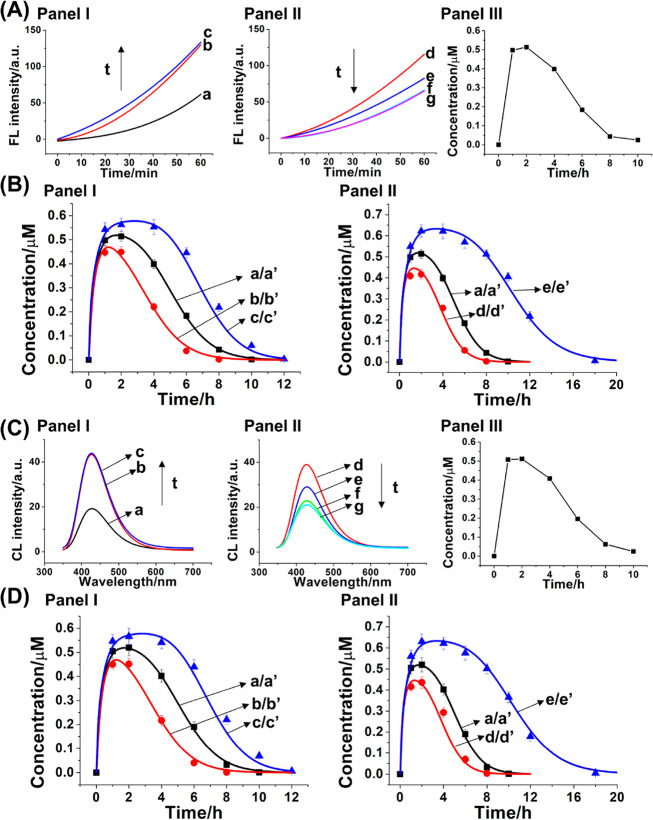
Panels I and II of Figure 3A display the time-dependent fluorescence changes of resorufin generated at time intervals of the L1′-triggered operation of the reaction module (L1′ = 3 μM and Nt.BbvCI = 0.046 μM). The rates of resorufin formation increase for a time interval of 2 h and then decrease for a depletion time interval for 10 h leading to regeneration of the parent system. Using a calibration curve relating the fluorescence intensities of resorufin to the concentrations of (1)+(2)/(3) supramolecular structure (Figure S6, Supporting Information), the transient concentrations of (1)+(2)/(3) supramolecular bioreactor, corresponding to the transient catalytic formation of resorufin, were calculated, and these are displayed in Figure 3A, panel III. A transient behavior of the catalytic rates generating resorufin is, indeed, observed. The transient supramolecular catalytic bioreactor generating resorufin is anticipated to be controlled by the concentration of the fuel L1′ triggering the reaction module and by the concentration of the nicking enzyme Nt.BbvCI. Qualitatively, increasing the concentration of the fuel strand L1′ is anticipated to enhance and enrich the content of the transiently formed resorufin whereas elevating the content of the Nt.BbvCI is expected to decrease the peak content of the catalytically generated resorufin and to shorten the depletion time-interval of the temporal process. The efficiency of the GOx-loaded ZIF-90/hemin-G-quadruplex biocatalytic cascade is controlled by the loading of GOx in the NMOFs and the resulting H2O2 generated by the GOx-catalyzed aerobic oxidation of glucose. The results are displayed in Figure S5 and accompanied discussion. As the loading of GOx increases, the biocatalytic cascade is enhanced, yet the background signal of the separated GOx-loaded ZIF-90/hemin-G-quadruplex constituents is intensified.
Figure 3.
(A) Time-dependent fluorescence changes of resorufin generated by samples withdrawn, at time intervals, from the dynamic reaction module depicted in panel I of Figure 2: panel I, samples withdrawn at (a) t = 0 h, (b) t = 1 h, (c) t = 2 h; panel II, samples withdrawn at (d) t = 4 h, (e) t = 6 h, (f) t = 8 h, (g) t = 10 h; panel III, temporal, transient concentration changes of the (1)+(2)/(3) supramolecular structure generating the fluorescent resorufin, upon operation of the dynamic reaction module shown in Figure 2, panel I. The experimental conditions operating the dynamic process shown in panels I–III are A1-(1), 1 μM; B1-(2), 1 μM; T1/L1-(3)/(4), 2 μM; L1′-(5), 3 μM; Nt.BbvCI, 0.046 μM. (B) Probing the effects of the concentrations of the fuel strand L1′ (panel I) and of the Nt.BbvCI (panel II) on the temporal transient concentrations of the (1)+(2)/(3) supramolecular structure, generating fluorescent resorufin according to Figure 2, panel I. Part B, panel I: (a) Dotted points correspond to experimental data recorded at the conditions specified in (A); (a′) solid curves correspond to computationally simulated results using the kinetic model formulated in Figure S8, Supporting Information. (b′, c′) Computationally simulated transient concentrations of (1)+(2)/(3) supramolecular structure, generating fluorescent resorufin, at auxiliary conditions L1′ = 2 and 4 μM, respectively. (b, c) Dotted data correspond to experimentally validated results in the presence of L1′ = 2 and 4 μM, respectively. Part B, panel II: (a/a′) is the same condition with (a/a′) in panel I. (d′, e′) Computationally simulated transient concentrations of (1)+(2)/(3) supramolecular structure, generating fluorescent resorufin, at auxiliary conditions Nt.BbvCI = 0.069 and 0.023 μM, respectively. (d, e) Dotted data correspond to experimentally validated results in the presence of Nt.BbvCI = 0.069 and 0.023 μM, respectively. (C) Time-dependent chemiluminescence changes of luminol generated by samples withdrawn at time intervals from the dynamic reaction module depicted in panel II, Figure 2: panel I, samples withdrawn at (a) t = 0 h, (b) t = 1 h, (c) t = 2 h; panel II, samples withdrawn at (d) t = 4 h, (e) t = 6 h, (f) t = 8 h, (g) t = 10 h; panel III, temporal, transient concentration changes of the (1)+(2)/(3) supramolecular structure generating chemiluminescence upon operation of the dynamic reaction module shown in Figure 2, panel II. Experimental conditions operating the dynamic process shown in panels I–III are A1-(1), 1 μM; B1-(2), 1 μM; T1/L1-(3)/(4), 2 μM; L1′-(5), 3 μM; Nt.BbvCI, 0.046 μM. (D) Probing the effects of the concentrations of the fuel strand L1′ (panel I) and the concentrations of the Nt.BbvCI (panel II) on the temporal transient generation of the (1)+(2)/(3) supramolecular structure and generation of chemiluminescence according to Figure 2, panel II. Part D, panel I: (a′) Dotted points correspond to experimental data recorded at the conditions specified in (C); (a′) solid curves correspond to computationally simulated result using the kinetic model formulated in Figure S8, Supporting Information. (b′, c′) Computationally simulated transient concentrations of (1)+(2)/(3) supramolecular structure generating chemiluminescence at auxiliary conditions L1′ = 2 and 4 μM, respectively. (b, c) Dotted data correspond to experimentally validated results in the presence of L1′ = 2 and 4 μM, respectively. Part D, panel II: (a/a′) is the same condition with (a/a′) in panel I. (d′, e′) Computationally simulated transient concentrations of (1)+(2)/(3) supramolecular structure generating chemiluminescence at auxiliary conditions Nt.BbvCI = 0.069 and 0.023 μM, respectively. (d, e) Dotted data correspond to experimentally validated results in the presence of Nt.BbvCI = 0.069 and 0.023 μM, respectively.
As an attempt to understand the effect of these auxiliary parameters on the transient process, we formulated a kinetic model that follows the transient process, Figure S8. This model was adopted to simulate the experimental results (dotted transient points a) in Figure 3B, panel I, with a computationally fitted transient (solid transient curve a′), generated by the set of rate constants comprising the model, that are summarized in Table S1 (for a detailed description of the simulation process see Figure S8). This set of rate-constants has a value provided that it can predict the transient behavior of the supramolecular bioreactor at other auxiliary conditions. Accordingly, the transient behavior of the formation and depletion of (1)+(2)/(3) supramolecular bioreactor generating fluorescent resorufin was predicted at different auxiliary concentrations of L1′ (curves b′ and c′, Figure 3B, panel I) and the nicking enzyme concentrations (curves d′ and e′, Figure 3B, panel II). The predicted results were experimentally validated, curves b and c in Figure 3B, panel I and curves d and e in Figure 3B, panel II, respectively. Very good fit between the experimental results and the computationally predicted transients is demonstrated, supporting the kinetic model.
Similarly, Figure 3C, panels I and II, depicts the temporal chemiluminescence spectra generated by samples withdrawn at time intervals from the transient operating bioreactor system according to Figure 2, panel II. The L1′-triggered activation of the reaction module yields in the first 2 h of operating the bioreactor system reaction samples where the resulting chemiluminescence is temporally intensified, and afterward the bioreactor samples withdrawn from the reaction system show a continuous temporal decrease in the chemiluminescence, reaching the parent chemiluminescence intensities after a time interval of ∼10 h. These results are consistent with the temporal L1′-triggered buildup of the (1)+(2)/(3) supramolecular GOx-loaded NMOFs/hemin-G-quadruplex catalytic bioreactor system that undergoes transient depletion by the nickase-induced separation of the bioreactor conjugate, leading to the recovery of the original reaction module. The transient concentration changes of (1)+(2)/(3) supramolecular bioreactor generating chemiluminescence are displayed in Figure 3C, panel III. As before, decreasing the concentration of the fuel strand L1′ that activates the reaction module results in lower temporally generated yields of the bioreactor conjugate, leading to lower chemiluminescence intensities, Figure 3D, panel I. Also, increasing the concentration of the nicking enzyme results in lower yields of chemiluminescence generated by the bioreactor system and enhanced depletion rates of the transiently operated bioreactor intermediate, Figure 3D, panel II.
Transient Exonuclease III-Driven Biocatalytic Cascades Using GOx-Loaded/Hemin-G-Quadruplex-Conjugated ZIF-90 as Functional Frameworks
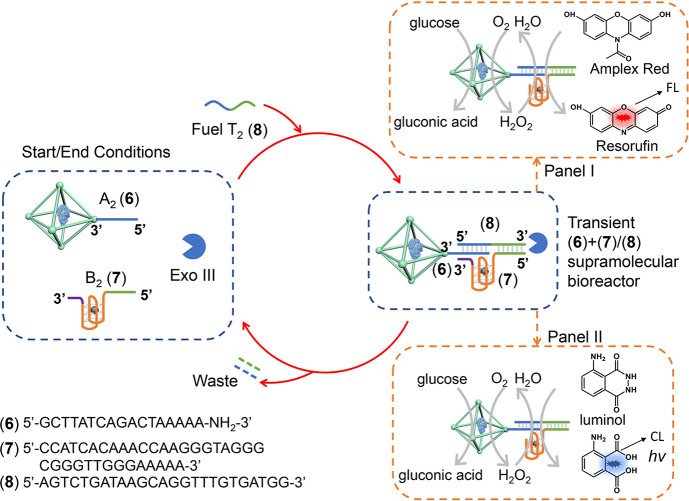
An alternative transient GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex reaction module leading to the fuel-triggered and transient operation of the bioreactor catalyzing the biocatalytic oxidation of Amplex-Red to resorufin or the biocatalytic generation of chemiluminescence is depicted in Figure 4. In this system, exonuclease III, Exo III, acts as the enzyme controlling the dissipative, transient operation of the bioreactor. The reaction module consists of the GOx-loaded ZIF-90 NMOFs functionalized with the nucleic acid A2, (6) (loading of GOx 82 μg/mg NMOFs; loading of A2 10 nmol/mg NMOFs), and the hemin-G-quadruplex B2, (7). The enzyme Exo III is also included in the reaction module. Subjecting the reaction module to the fuel strand T2, (8), results in the assembly of the (6)+(7)/(8) supramolecular bioreactor complex consisting of the (8)-bridged GOx-loaded ZIF-90 NMOFs and hemin-G-quadruplex constituents. The supramolecular complex operates two different biocatalytic cascades. One biocatalytic cascade, panel I, involves the aerobic GOx-catalyzed oxidation of glucose to yield gluconic acid and H2O2 and the subsequent hemin-G-quadruplex-catalyzed oxidation of Amplex-Red to resorufin. The second biocatalytic cascade driven by the (6)+(7)/(8) supramolecular GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex bioreactor involves the aerobic GOx-loaded ZIF-90 NMOFs-catalyzed oxidation of glucose to gluconic acid and H2O2 and the subsequent hemin-G-quadruplex catalyzed oxidation of luminol by H2O2 and the generation of chemiluminescence, panel II. The Exo III included in the system selectively degrades the 3′-ended fuel strand T2, leading to the separation of the supramolecular complex regenerating the rest of the reaction module and producing base fragments T2 as waste products. The separation of the supramolecular complex by Exo III leads to the temporal activation of the biocatalytic cascades shown in panels I and II and to the recovery of the parent reaction module. That is, the T2-triggered activation of the reaction module leads to the temporal and transient formation of the (6)+(7)/(8) supramolecular complex consisting of the GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex that guides the temporal biocatalytic cascades shown in Figure 4, panels I and II.
Figure 4.
Schematic reaction module for the fueled transient operation of the Exo III-guided (6)+(7)/(8) GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex bioreactor system leading to the temporal biocatalytic cascades consisting of the following: panel I, GOx/hemin-G-quadruplex catalyzed oxidation of Amplex-Red to fluorescent resorufin; panel II, GOx/hemin-G-quadruplex catalyzed generation of chemiluminescence through the catalyzed H2O2 oxidation of luminol.
It should be noted that in order to operate the Exo III driven transient generation of the bioreactor, the supramolecular GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex complex had to be engineered to retain an intact, Exo III-resistant structure, to the extent that only the T2 that forms the duplex with B2 is being digested by Exo III. We find that it is essential to tether to the G-quadruplex B2 a single strand 3′-terminated tether; otherwise, the G-quadruplex is also degraded by Exo III.
Panels I and II of Figure 5A depict the time-dependent fluorescence changes of resorufin generated upon subjecting the reaction module shown in Figure 4 to the fuel strand T2 and withdrawing at time interval samples driving the glucose-initiated H2O2-mediated oxidation of Amplex Red to resorufin. The withdrawn samples reveal a temporal initial increase in the oxidation rates of Amplex-Red for a time interval of ∼2 h, and afterward, the withdrawn sample shows a continuous temporal decline in the bioreactor catalyzed oxidation rates, which are fully blocked after 8 h, reaching the parent background rates of Amplex-Red oxidation to resorufin by the parent reaction module. Readdition of the fuel strand T2 to the system reactivated the transient operation of the reaction module, following the oxidation of Amplex-Red to form resorufin. These results are consistent with the T2-fueled/Exo III-guided operation of the reaction module shown in Figure 4. The bioreactor Exo III-driven, T2-triggered, transient formation of (6)+(7)/(8) supramolecular bioreactor generating fluorescent resorufin is controlled by the concentrations of T2, Figure 5B, panel I, and Exo III, Figure 5B, panel II. As the concentrations of T2 increases, the peak rates of temporal resorufin formation are higher, and as the concentration of Exo III increases, the peak rates of catalyzed formation of resorufin are lower and the depletion of the catalytic rates generating resorufin is enhanced. Figure S11 formulates the kinetic model corresponding to the transient operation of the T2/Exo III driven (6)+(7)/(8) supramolecular GOx-loaded NMOFs/hemin-G-quadruplex bioreactor system. Figure 5B, panel I, curve a′, depicts the fitted, computationally simulated, temporal concentrations of the catalytic (6)+(7)/(8) supramolecular structure generating resorufin by the T2/Exo III-driven bioreactor system, overlapping the experiment transient curve a, using the kinetic models. The derived rate constants following the kinetic model are summarized in Table S2. Using this set of rate constants, the predicted curves corresponding to temporal transient concentrations of the catalytic bioreactor system at different concentrations of T2 and Exo III were computed, curves b′ and c′, Figure 5B, panel I, and d′ and e′, Figure 5B, panel II, and the computational results are experimentally validated, curves b and c, Figure 5B, panel I, and curves d and e, Figure 5B, panel II. Indeed, the experimental results fit well to the predicted transient systems. Moreover, the T2/Exo III-driven activation of the GOx-loaded ZIF-90 NMOFs/hemin-G-quadruplex bioreactor, Figure 4, panel II, was applied to stimulate the temporally catalyzed generation of chemiluminescence. Panels I and II of Figure 5C depict the temporal chemiluminescence spectra generated by samples withdrawn at time intervals from the T2/Exo III-activated reaction module. The triggered reaction module reveals intensified chemiluminescence spectra for a time interval of 2 h and, subsequently, temporal depletion and blockage of the chemiluminescence after 8 h. Figure 5C, panel III shows the temporal, transient concentrations of (6)+(7)/(8) supramolecular structure generating chemiluminescence upon the T2/Exo III-triggered activation of the bioreactor system in the presence of 3 μM T2 and 0.4 μM Exo III. Figure 5D depicts the experimental and simulated temporal, transient formation of the (6)+(7)/(8) supramolecular bioreactor leading to chemiluminescence upon activation of the reaction module at different concentrations of T2 and Exo III. As the concentration of T2 increases, the temporal peak chemiluminescence intensity is higher, and as the concentration of Exo III is elevated, the peak chemiluminescence intensity is lower and the dissipative depletion of the bioreactor driven process is enhanced.
Figure 5.
(A) Time-dependent fluorescence changes of resorufin generated by samples withdrawn, at time intervals, from the dynamic reaction module depicted in panel I of Figure 4: panel I, samples withdrawn at (a) t = 0 h, (b) t = 1 h, (c) t = 2 h; panel II, samples withdrawn at (d) t = 4 h, (e) t = 6 h, (f) t = 8 h, (g) t = 10 h; panel III, temporal, transient concentration changes of the (6)+(7)/(8) supramolecular structure generating the fluorescent resorufin, upon operation of the dynamic reaction module shown in Figure 4, panel I. The experimental conditions operating the dynamic process shown in panels I–III are A2-(6), 1 μM; B2-(7), 1 μM; T2-(8), 3 μM; and Exo III, 0.4 μM. (B) Probing the effects of the concentrations of the fuel strand T2 (panel I) and of the Exo III (panel II) on the temporal transient generation of the (6)+(7)/(8) supramolecular structure, generating fluorescent resorufin according to Figure 4, panel I. Part B, panel I: (a) Dotted points correspond to experimental data recorded at the conditions specified in (A); (a′) solid curves correspond to computationally simulated results using the kinetic model formulated in Figure S11, Supporting Information. (b′, c′) Computationally simulated transient concentrations of (6)+(7)/(8) supramolecular structure, generating fluorescent resorufin, at auxiliary conditions T2 = 2 and 4 μM, respectively; (b) and (c) Dotted data correspond to experimentally validated results in the presence of T2 = 2 and 4 μM, respectively. Part B, panel II: (a/a′) is the same condition with (a/a′) in panel I. (d′, e′) Computationally simulated transient concentrations of (6)+(7)/(8) supramolecular structure, generating fluorescent resorufin, at auxiliary conditions Exo III = 0.6 and 0.2 μM, respectively. (d, e) Dotted data correspond to experimentally validated results in the presence of Exo III = 0.6 and 0.2 μM, respectively. (C) Time-dependent chemiluminescence changes of luminol generated by samples withdrawn at time intervals from the dynamic reaction module depicted in panel II of Figure 4: panel I, samples withdrawn at (a) t = 0 h, (b) t = 1 h, (c) t = 2 h; panel II, samples withdrawn at (d) t = 4 h, (e) t = 6 h, (f) t = 8 h, (g) t = 10 h; panel III, temporal, transient concentration changes of the (6)+(7)/(8) supramolecular structure generating chemiluminescence upon operation of the dynamic reaction module shown in Figure 4, panel II. Experimental conditions operating the dynamic process shown in panels I–III are A2-(6), 1 μM; B2-(7), 1 μM; T2-(8), 3 μM; Exo III, 0.4 μM. (D) Probing the effects of the concentrations of the fuel strand T2 (panel I) and the concentrations of the Exo III (panel II) on the temporal transient generation of the (6)+(7)/(8) supramolecular structure and generation of chemiluminescence according to Figure 4, panel II. Part D, panel I: (a) Dotted points correspond to experimental data recorded at the conditions specified in (C); (a′) solid curves correspond to computationally simulated result using the kinetic model formulated in Figure S11, Supporting Information. (b′, c′) Computationally simulated transient concentrations of (6)+(7)/(8) supramolecular structure generating chemiluminescence at auxiliary conditions T2 = 2 and 4 μM, respectively. (b, c) Dotted data correspond to experimentally validated results in the presence of T2 = 2 and 4 μM, respectively. Part D, panel II: (a/a′) is the same condition with (a/a′) in panel I. (d′, e′) Computationally simulated transient concentrations of (6)+(7)/(8) supramolecular structure generating chemiluminescence at auxiliary conditions Exo III = 0.6 and 0.2 μM, respectively. (d, e) Dotted data correspond to experimentally validated results in the presence of Exo III = 0.6 and 0.2 μM, respectively.
Conclusion
The study introduced nucleic acid-modified/GOx-loaded ZIF-90 conjugates as functional assemblies guiding transient biocatalytic cascades. Such frameworks could find important nanomedical applications for dose-controlled, temporal release of therapeutic agents. For example, the GOx-stimulated activation of the hemin-G-quadruplex peroxidase-mimicking DNAzyme yields reactive oxygen species (ROS) that might act for temporal chemodynamic treatment of cancer cells.69 Alternatively, the triggered temporal unlocking of the NMOFs could be used for the dose-controlled release of drugs, e.g., insulin from the NMOFs carriers.85 Moreover, the present study introduced the fueled strand displacement principle and coupled enzyme driven transformation as control motives of the dynamic process. Other triggering stimuli for temporal operation of the reaction modules such as miRNA/RNase,86 redox-triggered aptamer-ligand complexes,87 or light,38 may be envisaged.
Acknowledgments
The research is supported by the Israel Science Foundation (Project 2049/20).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.3c02542.
Experimental section; calibration curves for determination of the loading of GOx and DNA strands A1 or A2 involved in the bioreactor; efficient biocatalytic cascades performed by (1)+(2)/(3) supramolecular structure; effect of loading amount of GOx on the transient biocatalytic cascades; calibration curve for determination of the concentration of (1)+(2)/(3) supramolecular structure; kinetic model and simulation details of Nt.BbvCI-guided dissipative bioreactor system; calibration curve for determination of the concentration of (6)+(7)/(8) supramolecular structure; kinetic model and simulation details of Exo III-guided dissipative bioreactor system (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Ashkenasy G.; Hermans T. M.; Otto S.; Taylor A. F. Systems chemistry. Chem. Soc. Rev. 2017, 46 (9), 2543–2554. 10.1039/C7CS00117G. [DOI] [PubMed] [Google Scholar]
- Ludlow R. F.; Otto S. Systems chemistry. Chem. Soc. Rev. 2008, 37 (1), 101–108. 10.1039/B611921M. [DOI] [PubMed] [Google Scholar]
- Mattia E.; Otto S. Supramolecular systems chemistry. Nat. Nanotechnol. 2015, 10 (2), 111–119. 10.1038/nnano.2014.337. [DOI] [PubMed] [Google Scholar]
- Wang F.; Lu C.-H.; Willner I. From Cascaded Catalytic Nucleic Acids to Enzyme–DNA Nanostructures: Controlling Reactivity, Sensing, Logic Operations, and Assembly of Complex Structures. Chem. Rev. 2014, 114 (5), 2881–2941. 10.1021/cr400354z. [DOI] [PubMed] [Google Scholar]
- Simmel F. C.; Yurke B.; Singh H. R. Principles and Applications of Nucleic Acid Strand Displacement Reactions. Chem. Rev. 2019, 119 (10), 6326–6369. 10.1021/acs.chemrev.8b00580. [DOI] [PubMed] [Google Scholar]
- Zhang D. Y.; Turberfield A. J.; Yurke B.; Winfree E. Engineering Entropy-Driven Reactions and Networks Catalyzed by DNA. Science 2007, 318 (5853), 1121–1125. 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- Dong J.; O’Hagan M. P.; Willner I. Switchable and dynamic G-quadruplexes and their applications. Chem. Soc. Rev. 2022, 51 (17), 7631–7661. 10.1039/D2CS00317A. [DOI] [PubMed] [Google Scholar]
- Lu C.-H.; Qi X.-J.; Orbach R.; Yang H.-H.; Mironi-Harpaz I.; Seliktar D.; Willner I. Switchable Catalytic Acrylamide Hydrogels Cross-Linked by Hemin/G-Quadruplexes. Nano Lett. 2013, 13 (3), 1298–1302. 10.1021/nl400078g. [DOI] [PubMed] [Google Scholar]
- Freeman R.; Finder T.; Willner I. Multiplexed Analysis of Hg2+ and Ag+ Ions by Nucleic Acid Functionalized CdSe/ZnS Quantum Dots and Their Use for Logic Gate Operations. Angew. Chem., Int. Ed. 2009, 48 (42), 7818–7821. 10.1002/anie.200902395. [DOI] [PubMed] [Google Scholar]
- Miyake Y.; Togashi H.; Tashiro M.; Yamaguchi H.; Oda S.; Kudo M.; Tanaka Y.; Kondo Y.; Sawa R.; Fujimoto T.; Machinami T.; Ono A. MercuryII-Mediated Formation of Thymine–HgII–Thymine Base Pairs in DNA Duplexes. J. Am. Chem. Soc. 2006, 128 (7), 2172–2173. 10.1021/ja056354d. [DOI] [PubMed] [Google Scholar]
- Ono A.; Cao S.; Togashi H.; Tashiro M.; Fujimoto T.; Machinami T.; Oda S.; Miyake Y.; Okamoto I.; Tanaka Y. Specific interactions between silver(i) ions and cytosine–cytosine pairs in DNA duplexes. Chem. Commun. 2008, (39), 4825–4827. 10.1039/b808686a. [DOI] [PubMed] [Google Scholar]
- Kamiya Y.; Asanuma H. Light-Driven DNA Nanomachine with a Photoresponsive Molecular Engine. Acc. Chem. Res. 2014, 47 (6), 1663–1672. 10.1021/ar400308f. [DOI] [PubMed] [Google Scholar]
- O’Hagan M. P.; Duan Z.; Huang F.; Laps S.; Dong J.; Xia F.; Willner I. Photocleavable Ortho-Nitrobenzyl-Protected DNA Architectures and Their Applications. Chem. Rev. 2023, 123 (10), 6839–6887. 10.1021/acs.chemrev.3c00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K.; Li W.; Nie Z.; Huang Y.; Liu Z.; Nie L.; Yao S. Enzyme-Regulated Activation of DNAzyme: A Novel Strategy for a Label-Free Colorimetric DNA Ligase Assay and Ligase-Based Biosensing. Chem. – Eur. J. 2012, 18 (13), 3992–3999. 10.1002/chem.201102290. [DOI] [PubMed] [Google Scholar]
- Holland P. M.; Abramson R. D.; Watson R.; Gelfand D. H. Detection of specific polymerase chain reaction product by utilizing the 5′----3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 1991, 88 (16), 7276–7280. 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.-M.; Zhang X.-B.; Kong R.-M.; Yang B.; Tan W. A Ligation-Triggered DNAzyme Cascade for Amplified Fluorescence Detection of Biological Small Molecules with Zero-Background Signal. J. Am. Chem. Soc. 2011, 133 (30), 11686–11691. 10.1021/ja203693b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol C. D.; Kuo C.-F.; Thayer M. M.; Cunningham R. P.; Tainer J. A. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature 1995, 374 (6520), 381–386. 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- Weizmann Y.; Cheglakov Z.; Willner I. A Fok I/DNA Machine that Duplicates its Analyte Gene Sequence. J. Am. Chem. Soc. 2008, 130 (51), 17224–17225. 10.1021/ja806222e. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Wang Z.; Liu Y.; Yang J.; Zhang X.; Li Y.; Pan L.; Ke Y.; Yan H. Nicking-Assisted Reactant Recycle To Implement Entropy-Driven DNA Circuit. J. Am. Chem. Soc. 2019, 141 (43), 17189–17197. 10.1021/jacs.9b07521. [DOI] [PubMed] [Google Scholar]
- Harroun S. G.; Prévost-Tremblay C.; Lauzon D.; Desrosiers A.; Wang X.; Pedro L.; Vallée-Bélisle A. Programmable DNA switches and their applications. Nanoscale 2018, 10 (10), 4607–4641. 10.1039/C7NR07348H. [DOI] [PubMed] [Google Scholar]
- Wang F.; Liu X.; Willner I. DNA Switches: From Principles to Applications. Angew. Chem., Int. Ed. 2015, 54 (4), 1098–1129. 10.1002/anie.201404652. [DOI] [PubMed] [Google Scholar]
- Bath J.; Turberfield A. J. DNA nanomachines. Nat. Nanotechnol. 2007, 2 (5), 275–284. 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- Teller C.; Willner I. Functional nucleic acid nanostructures and DNA machines. Curr. Opin. Biotechnol. 2010, 21 (4), 376–391. 10.1016/j.copbio.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Fan D.; Wang J.; Wang E.; Dong S. Propelling DNA Computing with Materials′ Power: Recent Advancements in Innovative DNA Logic Computing Systems and Smart Bio-Applications. Adv. Sci. 2020, 7 (24), 2001766. 10.1002/advs.202001766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F.; Zhang F.; Liu Y.; Yan H. DNA Origami: Scaffolds for Creating Higher Order Structures. Chem. Rev. 2017, 117 (20), 12584–12640. 10.1021/acs.chemrev.6b00825. [DOI] [PubMed] [Google Scholar]
- Pinheiro A. V.; Han D.; Shih W. M.; Yan H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011, 6 (12), 763–772. 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Li Z.; Willner I. Dynamic Reconfigurable DNA Nanostructures, Networks and Materials. Angew. Chem., Int. Ed. 2023, 62 (18), e202215332. 10.1002/anie.202215332. [DOI] [PubMed] [Google Scholar]
- Kahn J. S.; Hu Y.; Willner I. Stimuli-Responsive DNA-Based Hydrogels: From Basic Principles to Applications. Acc. Chem. Res. 2017, 50 (4), 680–690. 10.1021/acs.accounts.6b00542. [DOI] [PubMed] [Google Scholar]
- Vázquez-González M.; Willner I. Stimuli-Responsive Biomolecule-Based Hydrogels and Their Applications. Angew. Chem., Int. Ed. 2020, 59 (36), 15342–15377. 10.1002/anie.201907670. [DOI] [PubMed] [Google Scholar]
- Vázquez-González M.; Willner I. DNA-Responsive SiO2 Nanoparticles, Metal–Organic Frameworks, and Microcapsules for Controlled Drug Release. Langmuir 2018, 34 (49), 14692–14710. 10.1021/acs.langmuir.8b00478. [DOI] [PubMed] [Google Scholar]
- Amano S.; Borsley S.; Leigh D. A.; Sun Z. Chemical engines: driving systems away from equilibrium through catalyst reaction cycles. Nat. Nanotechnol. 2021, 16 (10), 1057–1067. 10.1038/s41565-021-00975-4. [DOI] [PubMed] [Google Scholar]
- Li Z.; Wang J.; Willner I. Transient Out-of-Equilibrium Nucleic Acid-Based Dissipative Networks and Their Applications. Adv. Funct. Mater. 2022, 32 (37), 2200799. 10.1002/adfm.202200799. [DOI] [Google Scholar]
- Liu Q.; Li H.; Yu B.; Meng Z.; Zhang X.; Li J.; Zheng L. DNA-Based Dissipative Assembly toward Nanoarchitectonics. Adv. Funct. Mater. 2022, 32 (37), 2201196. 10.1002/adfm.202201196. [DOI] [Google Scholar]
- Deng J.; Walther A. Autonomous DNA nanostructures instructed by hierarchically concatenated chemical reaction networks. Nat. Commun. 2021, 12 (1), 5132. 10.1038/s41467-021-25450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.; Ouyang Y.; Wang J.; Willner I. Dissipative Gated and Cascaded DNA Networks. J. Am. Chem. Soc. 2021, 143 (13), 5071–5079. 10.1021/jacs.1c00486. [DOI] [PubMed] [Google Scholar]
- Deng J.; Liu W.; Sun M.; Walther A. Dissipative Organization of DNA Oligomers for Transient Catalytic Function. Angew. Chem., Int. Ed. 2022, 61 (10), e202113477. 10.1002/anie.202113477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen L.; Walther A. Programmable dynamic steady states in ATP-driven nonequilibrium DNA systems. Sci. Adv. 2019, 5 (7), eaaw0590. 10.1126/sciadv.aaw0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Li Z.; Zhou Z.; Ouyang Y.; Zhang J.; Ma X.; Tian H.; Willner I. DNAzyme- and light-induced dissipative and gated DNA networks. Chem. Sci. 2021, 12 (33), 11204–11212. 10.1039/D1SC02091A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J.; Ouyang Y.; Wang J.; O’Hagan M. P.; Willner I. Assembly of Dynamic Gated and Cascaded Transient DNAzyme Networks. ACS Nano 2022, 16 (4), 6153–6164. 10.1021/acsnano.1c11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Wang J.; Willner I. Autoinhibited transient, gated, and cascaded dynamic transcription of RNAs. Sci. Adv. 2022, 8 (33), eabq5947. 10.1126/sciadv.abq5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J.; Bezold D.; Jessen H. J.; Walther A. Multiple Light Control Mechanisms in ATP-Fueled Non-equilibrium DNA Systems. Angew. Chem., Int. Ed. 2020, 59 (29), 12084–12092. 10.1002/anie.202003102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.; Xuan M.; Huo S.; Fan J.; Chakraborty G.; Wang Y.; Zhao H.; Herrmann A.; Zheng L. Four-Dimensional Deoxyribonucleic Acid–Gold Nanoparticle Assemblies. Angew. Chem., Int. Ed. 2020, 59 (39), 17250–17255. 10.1002/anie.202007616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y.; Zhang P.; Manis-Levy H.; Paltiel Y.; Willner I. Transient Dissipative Optical Properties of Aggregated Au Nanoparticles, CdSe/ZnS Quantum Dots, and Supramolecular Nucleic Acid-Stabilized Ag Nanoclusters. J. Am. Chem. Soc. 2021, 143 (42), 17622–17632. 10.1021/jacs.1c07895. [DOI] [PubMed] [Google Scholar]
- Rizzuto F. J.; Platnich C. M.; Luo X.; Shen Y.; Dore M. D.; Lachance-Brais C.; Guarné A.; Cosa G.; Sleiman H. F. A dissipative pathway for the structural evolution of DNA fibres. Nat. Chem. 2021, 13 (9), 843–849. 10.1038/s41557-021-00751-w. [DOI] [PubMed] [Google Scholar]
- Ouyang Y.; Zhang P.; Willner I. Dissipative biocatalytic cascades and gated transient biocatalytic cascades driven by nucleic acid networks. Sci. Adv. 2022, 8 (18), eabn3534. 10.1126/sciadv.abn3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Ren J.; Qu X. Nanozymes: Classification, Catalytic Mechanisms, Activity Regulation, and Applications. Chem. Rev. 2019, 119 (6), 4357–4412. 10.1021/acs.chemrev.8b00672. [DOI] [PubMed] [Google Scholar]
- Liang M.; Yan X. Nanozymes: From New Concepts, Mechanisms, and Standards to Applications. Acc. Chem. Res. 2019, 52 (8), 2190–2200. 10.1021/acs.accounts.9b00140. [DOI] [PubMed] [Google Scholar]
- Ragg R.; Tahir M. N.; Tremel W. Solids Go Bio: Inorganic Nanoparticles as Enzyme Mimics. Eur. J. Inorg. Chem. 2016, 2016 (13–14), 1906–1915. 10.1002/ejic.201501237. [DOI] [Google Scholar]
- Jin L.; Meng Z.; Zhang Y.; Cai S.; Zhang Z.; Li C.; Shang L.; Shen Y. Ultrasmall Pt Nanoclusters as Robust Peroxidase Mimics for Colorimetric Detection of Glucose in Human Serum. ACS Appl. Mater. Interfaces 2017, 9 (11), 10027–10033. 10.1021/acsami.7b01616. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Wang C.; Cai N.; Long S.; Yu F. Negatively charged gold nanoparticles as an intrinsic peroxidase mimic and their applications in the oxidation of dopamine. J. Mater. Sci. 2014, 49 (20), 7143–7150. 10.1007/s10853-014-8422-x. [DOI] [Google Scholar]
- Sun Z.; Zhang N.; Si Y.; Li S.; Wen J.; Zhu X.; Wang H. High-throughput colorimetric assays for mercury(ii) in blood and wastewater based on the mercury-stimulated catalytic activity of small silver nanoparticles in a temperature-switchable gelatin matrix. Chem. Commun. 2014, 50 (65), 9196–9199. 10.1039/C4CC03851G. [DOI] [PubMed] [Google Scholar]
- Wu Y.-S.; Huang F.-F.; Lin Y.-W. Fluorescent Detection of Lead in Environmental Water and Urine Samples Using Enzyme Mimics of Catechin-Synthesized Au Nanoparticles. ACS Appl. Mater. Interfaces 2013, 5 (4), 1503–1509. 10.1021/am3030454. [DOI] [PubMed] [Google Scholar]
- André R.; Natálio F.; Humanes M.; Leppin J.; Heinze K.; Wever R.; Schröder H.-C.; Müller W. E. G.; Tremel W. V2O5 Nanowires with an Intrinsic Peroxidase-Like Activity. Adv. Funct. Mater. 2011, 21 (3), 501–509. 10.1002/adfm.201001302. [DOI] [Google Scholar]
- Gao L.; Zhuang J.; Nie L.; Zhang J.; Zhang Y.; Gu N.; Wang T.; Feng J.; Yang D.; Perrett S.; Yan X. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat. Nanotechnol. 2007, 2 (9), 577–583. 10.1038/nnano.2007.260. [DOI] [PubMed] [Google Scholar]
- Natalio F.; André R.; Hartog A. F.; Stoll B.; Jochum K. P.; Wever R.; Tremel W. Vanadium pentoxide nanoparticles mimic vanadium haloperoxidases and thwart biofilm formation. Nat. Nanotechnol. 2012, 7 (8), 530–535. 10.1038/nnano.2012.91. [DOI] [PubMed] [Google Scholar]
- Pirmohamed T.; Dowding J. M.; Singh S.; Wasserman B.; Heckert E.; Karakoti A. S.; King J. E. S.; Seal S.; Self W. T. Nanoceria exhibit redox state-dependent catalase mimetic activity. Chem. Commun. 2010, 46 (16), 2736–2738. 10.1039/b922024k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragg R.; Natalio F.; Tahir M. N.; Janssen H.; Kashyap A.; Strand D.; Strand S.; Tremel W. Molybdenum Trioxide Nanoparticles with Intrinsic Sulfite Oxidase Activity. ACS Nano 2014, 8 (5), 5182–5189. 10.1021/nn501235j. [DOI] [PubMed] [Google Scholar]
- Wang L.; Min Y.; Xu D.; Yu F.; Zhou W.; Cuschieri A. Membrane lipid peroxidation by the peroxidase-like activity of magnetite nanoparticles. Chem. Commun. 2014, 50 (76), 11147–11150. 10.1039/C4CC03082F. [DOI] [PubMed] [Google Scholar]
- Abdelhamid H. N.; Mahmoud G. A.-E.; Sharmouk W. A cerium-based MOFzyme with multi-enzyme-like activity for the disruption and inhibition of fungal recolonization. J. Mater. Chem. B 2020, 8 (33), 7548–7556. 10.1039/D0TB00894J. [DOI] [PubMed] [Google Scholar]
- Chen W.-H.; Vázquez-González M.; Kozell A.; Cecconello A.; Willner I. Cu2+-Modified Metal–Organic Framework Nanoparticles: A Peroxidase-Mimicking Nanoenzyme. Small 2018, 14 (5), 1703149. 10.1002/smll.201703149. [DOI] [PubMed] [Google Scholar]
- Li M.; Chen J.; Wu W.; Fang Y.; Dong S. Oxidase-like MOF-818 Nanozyme with High Specificity for Catalysis of Catechol Oxidation. J. Am. Chem. Soc. 2020, 142 (36), 15569–15574. 10.1021/jacs.0c07273. [DOI] [PubMed] [Google Scholar]
- Yuan A.; Lu Y.; Zhang X.; Chen Q.; Huang Y. Two-dimensional iron MOF nanosheet as a highly efficient nanozyme for glucose biosensing. J. Mater. Chem. B 2020, 8 (40), 9295–9303. 10.1039/D0TB01598A. [DOI] [PubMed] [Google Scholar]
- Zheng L.; Wang F.; Jiang C.; Ye S.; Tong J.; Dramou P.; He H. Recent progress in the construction and applications of metal-organic frameworks and covalent-organic frameworks-based nanozymes. Coord. Chem. Rev. 2022, 471, 214760. 10.1016/j.ccr.2022.214760. [DOI] [Google Scholar]
- Vázquez-González M.; Liao W.-C.; Cazelles R.; Wang S.; Yu X.; Gutkin V.; Willner I. Mimicking Horseradish Peroxidase Functions Using Cu2+-Modified Carbon Nitride Nanoparticles or Cu2+-Modified Carbon Dots as Heterogeneous Catalysts. ACS Nano 2017, 11 (3), 3247–3253. 10.1021/acsnano.7b00352. [DOI] [PubMed] [Google Scholar]
- Wang H.; Liu C.; Liu Z.; Ren J.; Qu X. Specific Oxygenated Groups Enriched Graphene Quantum Dots as Highly Efficient Enzyme Mimics. Small 2018, 14 (13), 1703710. 10.1002/smll.201703710. [DOI] [PubMed] [Google Scholar]
- Wang S.; Cazelles R.; Liao W.-C.; Vázquez-González M.; Zoabi A.; Abu-Reziq R.; Willner I. Mimicking Horseradish Peroxidase and NADH Peroxidase by Heterogeneous Cu2+-Modified Graphene Oxide Nanoparticles. Nano Lett. 2017, 17 (3), 2043–2048. 10.1021/acs.nanolett.7b00093. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Zhang X.; Huang L.; Zhang Z.; Dong S. GOx@ZIF-8(NiPd) Nanoflower: An Artificial Enzyme System for Tandem Catalysis. Angew. Chem., Int. Ed. 2017, 56 (50), 16082–16085. 10.1002/anie.201710418. [DOI] [PubMed] [Google Scholar]
- Ouyang Y.; Biniuri Y.; Fadeev M.; Zhang P.; Carmieli R.; Vázquez-González M.; Willner I. Aptamer-Modified Cu2+-Functionalized C-Dots: Versatile Means to Improve Nanozyme Activities-“Aptananozymes”. J. Am. Chem. Soc. 2021, 143 (30), 11510–11519. 10.1021/jacs.1c03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y.; Fadeev M.; Zhang P.; Carmieli R.; Sohn Y. S.; Karmi O.; Qin Y.; Chen X.; Nechushtai R.; Willner I. Aptamer-Functionalized Ce4+-Ion-Modified C-Dots: Peroxidase Mimicking Aptananozymes for the Oxidation of Dopamine and Cytotoxic Effects toward Cancer Cells. ACS Appl. Mater. Interfaces 2022, 14 (50), 55365–55375. 10.1021/acsami.2c16199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.; Vázquez-González M.; O’Hagan M. P.; Ouyang Y.; Wang Z.; Willner I. Enzyme-Loaded Hemin/G-Quadruplex-Modified ZIF-90 Metal–Organic Framework Nanoparticles: Bioreactor Nanozymes for the Cascaded Oxidation of N-hydroxy-l-arginine and Sensing Applications. Small 2022, 18 (11), 2104420. 10.1002/smll.202104420. [DOI] [PubMed] [Google Scholar]
- Hun X.; Xie G.; Luo X. Scaling up an electrochemical signal with a catalytic hairpin assembly coupling nanocatalyst label for DNA detection. Chem. Commun. 2015, 51 (33), 7100–7103. 10.1039/C5CC00680E. [DOI] [PubMed] [Google Scholar]
- Jv Y.; Li B.; Cao R. Positively-charged gold nanoparticles as peroxidiase mimic and their application in hydrogen peroxide and glucose detection. Chem. Commun. 2010, 46 (42), 8017–8019. 10.1039/c0cc02698k. [DOI] [PubMed] [Google Scholar]
- Ouyang Y.; O’Hagan M. P.; Willner I. Functional catalytic nanoparticles (nanozymes) for sensing. Biosens. Bioelectron. 2022, 218, 114768. 10.1016/j.bios.2022.114768. [DOI] [PubMed] [Google Scholar]
- Fan K.; Cao C.; Pan Y.; Lu D.; Yang D.; Feng J.; Song L.; Liang M.; Yan X. Magnetoferritin nanoparticles for targeting and visualizing tumour tissues. Nat. Nanotechnol. 2012, 7 (7), 459–464. 10.1038/nnano.2012.90. [DOI] [PubMed] [Google Scholar]
- Ragg R.; Schilmann A. M.; Korschelt K.; Wieseotte C.; Kluenker M.; Viel M.; Völker L.; Preiß S.; Herzberger J.; Frey H.; Heinze K.; Blümler P.; Tahir M. N.; Natalio F.; Tremel W. Intrinsic superoxide dismutase activity of MnO nanoparticles enhances the magnetic resonance imaging contrast. J. Mater. Chem. B 2016, 4 (46), 7423–7428. 10.1039/C6TB02078J. [DOI] [PubMed] [Google Scholar]
- Hao C.; Qu A.; Xu L.; Sun M.; Zhang H.; Xu C.; Kuang H. Chiral Molecule-mediated Porous CuxO Nanoparticle Clusters with Antioxidation Activity for Ameliorating Parkinson’s Disease. J. Am. Chem. Soc. 2019, 141 (2), 1091–1099. 10.1021/jacs.8b11856. [DOI] [PubMed] [Google Scholar]
- Jiang D.; Ni D.; Rosenkrans Z. T.; Huang P.; Yan X.; Cai W. Nanozyme: new horizons for responsive biomedical applications. Chem. Soc. Rev. 2019, 48 (14), 3683–3704. 10.1039/C8CS00718G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Wang Z.; Li X.; Wang L.; Yin M.; Wang L.; Chen N.; Fan C.; Song H. Dietary Iron Oxide Nanoparticles Delay Aging and Ameliorate Neurodegeneration in Drosophila. Adv. Mater. 2016, 28 (7), 1387–1393. 10.1002/adma.201503893. [DOI] [PubMed] [Google Scholar]
- Ma Y.-C.; Zhu Y.-H.; Tang X.-F.; Hang L.-F.; Jiang W.; Li M.; Khan M. I.; You Y.-Z.; Wang Y.-C. Au nanoparticles with enzyme-mimicking activity-ornamented ZIF-8 for highly efficient photodynamic therapy. Biomater. Sci. 2019, 7 (7), 2740–2748. 10.1039/C9BM00333A. [DOI] [PubMed] [Google Scholar]
- Chen W.-H.; Vázquez-González M.; Zoabi A.; Abu-Reziq R.; Willner I. Biocatalytic cascades driven by enzymes encapsulated in metal–organic framework nanoparticles. Nat. Catal. 2018, 1 (9), 689–695. 10.1038/s41929-018-0117-2. [DOI] [Google Scholar]
- Chen W.-H.; Luo G.-F.; Vázquez-González M.; Cazelles R.; Sohn Y. S.; Nechushtai R.; Mandel Y.; Willner I. Glucose-Responsive Metal–Organic-Framework Nanoparticles Act as “Smart” Sense-and-Treat Carriers. ACS Nano 2018, 12 (8), 7538–7545. 10.1021/acsnano.8b03417. [DOI] [PubMed] [Google Scholar]
- Zhou Z.; Vázquez-González M.; Willner I. Stimuli-responsive metal–organic framework nanoparticles for controlled drug delivery and medical applications. Chem. Soc. Rev. 2021, 50 (7), 4541–4563. 10.1039/D0CS01030H. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Sharon E.; Freeman R.; Liu X.; Willner I. Fluorescence Detection of DNA, Adenosine-5′-Triphosphate (ATP), and Telomerase Activity by Zinc(II)-Protoporphyrin IX/G-Quadruplex Labels. Anal. Chem. 2012, 84 (11), 4789–4797. 10.1021/ac300348v. [DOI] [PubMed] [Google Scholar]
- Pavlov V.; Xiao Y.; Gill R.; Dishon A.; Kotler M.; Willner I. Amplified Chemiluminescence Surface Detection of DNA and Telomerase Activity Using Catalytic Nucleic Acid Labels. Anal. Chem. 2004, 76 (7), 2152–2156. 10.1021/ac035219l. [DOI] [PubMed] [Google Scholar]
- Chen W.-H.; Luo G.-F.; Sohn Y. S.; Nechushtai R.; Willner I. miRNA-Specific Unlocking of Drug-Loaded Metal–Organic Framework Nanoparticles: Targeted Cytotoxicity toward Cancer Cells. Small 2019, 15 (17), 1900935. 10.1002/smll.201900935. [DOI] [PubMed] [Google Scholar]
- Del Grosso E.; Ragazzon G.; Prins L. J.; Ricci F. Fuel-Responsive Allosteric DNA-Based Aptamers for the Transient Release of ATP and Cocaine. Angew. Chem., Int. Ed. 2019, 58 (17), 5582–5586. 10.1002/anie.201812885. [DOI] [PubMed] [Google Scholar]
- Biniuri Y.; Luo G.-F.; Fadeev M.; Wulf V.; Willner I. Redox-Switchable Binding Properties of the ATP-Aptamer. J. Am. Chem. Soc. 2019, 141 (39), 15567–15576. 10.1021/jacs.9b06256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.