Abstract
Fungal pathogens cause life-threatening diseases in humans, and the increasing prevalence of these diseases emphasizes the need for new targets for therapeutic intervention. Nutrient acquisition during infection is a promising target, and recent studies highlight the contributions of endomembrane trafficking, mitochondria, and vacuoles in the sensing and acquisition of heme by fungi. These studies have been facilitated by genetically encoded biosensors and other tools to quantitate heme in subcellular compartments and to investigate the dynamics of trafficking in living cells. In particular, the applications of biosensors in fungi have been extended beyond the detection of metabolites, cofactors, pH, and redox status to include the detection of heme. Here, we focus on studies that make use of biosensors to examine mechanisms of heme uptake and degradation, with guidance from the model fungus Saccharomyces cerevisiae and an emphasis on the pathogenic fungi Candida albicans and Cryptococcus neoformans that threaten human health. These studies emphasize a role for endocytosis in heme uptake, and highlight membrane contact sites involving mitochondria, the endoplasmic reticulum and vacuoles as mediators of intracellular iron and heme trafficking.
Keywords: Iron acquisition, Genetically-encoded sensor, Endocytosis, CFEM proteins, Nutritional immunity
1. Introduction
Metal ions and heme are essential cofactors for numerous proteins and play key roles in multiple cellular processes (Galaris et al., 2019). In the context of microbial pathogenesis, the acquisition of metal ions, especially iron and copper, is critical for the ability of microbes to cause disease in vertebrate hosts (Monteith and Skaar, 2021). This is true for fungal pathogens of humans including Aspergillus fumigatus, Candida albicans, and Cryptococcus neoformans, and recent reviews provide excellent overviews of the mechanisms of iron homeostasis in these fungi (Bairwa et al., 2017; Gerwien et al., 2018; Horianopoulos and Kronstad, 2019; Roy and Kornitzer, 2019; Kornitzer and Roy, 2020; Labbé et al., 2020; Stanford and Voigt, 2020; Jung et al., 2021; Misslinger et al., 2021). During disease, fungal pathogens acquire iron from iron-containing proteins and, importantly, from heme that accounts for the bulk of the iron quota in vertebrate hosts (Donegan et al., 2019). Importantly, an understanding of mechanisms of iron and heme acquisition may lead to new therapeutic approaches to combat fungal diseases. Novel approaches and new targets are critically needed because of the devastating impact of fungal pathogens in immunocompromised individuals, the emergence of new pathogens, and the limited arsenal of antifungal drugs (Fisher et al., 2020).
The analysis of heme use by fungi has benefited recently by the application of genetically encoded sensors, which are powerful tools to monitor and quantify molecules of biological interest including metabolites or environmental signals in real time and in living cells. A number of excellent reviews provide detailed descriptions of genetically encoded sensors that generally consist of a protein that senses a specific signal (e.g., a small molecule, a protein-protein interaction, or an enzyme activity) coupled to a protein that provides a fluorescent or bioluminescent readout (Greenwald et al., 2018; Lin et al., 2019; Qiu et al., 2019; Terai et al., 2019; Kostyuk et al., 2020; Marsafari et al., 2020; Zhou et al., 2020; Kim et al., 2021; Nasu et al., 2021). There are numerous examples of applications of genetically encoded biosensors in fungal research. For example, Van Genechten et al., (2021) recently reviewed the applications of fluorescent proteins and imaging tools for the detection of amino acids, glucose, glutathione, lipids, oxygen, pH changes, redox homeostasis, and signaling activities (e.g., kinase activation). Approaches that are particularly relevant for fungal pathogens include monitoring intracellular pH, e.g., in response to antifungal drugs (Liu and Köhler, 2016; Tournu et al., 2017), monitoring oxygen levels during infection (Eichhof and Ernst, 2016), detecting signaling activities (Demuyser et al., 2018), and assessing redox status during colonization of host tissue (Mentges et al., 2015; Huang et al., 2017).
In this review, we focus on recent studies of heme acquisition and trafficking in fungi, with an emphasis on the use of the genetically encoded sensors and genetic approaches to provide insights into intracellular trafficking (Hanna et al., 2016, 2017, 2018; Sweeny et al., 2018; Bairwa et al., 2020; Martinez-Guzman et al., 2020; Weissman et al., 2021). We also highlight recent studies that reinforce an appreciation of mitochondria and vacuoles as hubs of iron and heme sensing, and that showcase the role of the endomembrane system in trafficking. We conclude with perspectives on areas for future emphasis with a focus on mitochondrial functions.
2. Heme acquisition and trafficking in fungal pathogens
Heme is an iron-containing prosthetic group that serves as a cofactor and a signaling molecule for crucial cellular activities such as iron homeostasis, gas sensing, electron transfer, cell cycle progression and proliferation, mitophagy, apoptosis and the response to oxidative stress (Donegan et al., 2019; Chambers et al., 2021; Gallio et al., 2021). Studies in the last 15 years are starting to reveal how fungi obtain iron from heme, the most abundant iron source in mammalian hosts (Ganz and Nemeth, 2015; Donegan et al., 2019). Much of our understanding of heme acquisition in fungal pathogens comes from studies with C. albicans, a commensal yeast normally found in the oral, gastrointestinal, and genital tracts but with the potential to cause life threatening systemic infections in immunocompromised patients (Kuznets et al., 2014; Kumamoto et al., 2020; Lopes and Lionkis, 2022). Like other pathogens, C. albicans must acquire essential nutrients and micronutrients from the host, but free iron is extremely scarce due to nutritional immunity. Therefore, heme, which is mostly bound to hemoglobin, represents an alternative iron source in the host. Indeed, many gram-positive bacteria such as Bacillus anthracis and Staphylococcus aureus, and gram-negative bacteria such as Serratia marcescens have independently acquired hemophores to hijack the host’s iron from heme (Caza and Kronstad, 2013; Contreras et al., 2014). Similar strategies have been identified in C. albicans including a heme-iron uptake system in addition to a high-affinity, reductive iron uptake system, and transporters for siderophores (Kornitzer and Roy, 2020).
The initial analyses of C. albicans uptake mechanisms for heme and hemoglobin identified several subunits of the vacuolar ATPase (VMA genes) and components of the ESCRT (endosomal sorting complex required for transport) and HOPS (homotypic fusion and vacuole protein) sorting complexes involved in endocytosis and targeting vesicles to the vacuole (Weissman et al., 2008) (Fig. 1A). The approach to identify these endomembrane trafficking functions involved expressing the heme-binding protein Rbt5 (described below) in Saccharomyces cerevisiae and using Synthetic Genetic Analysis to identify yeast mutants with defective growth on hemoglobin (Weissman et al., 2008). Subsequent testing of mutants with defects in the homologous genes in C. albicans confirmed roles for the vacuolar ATPase, the HOPS protein Vps41, proteins of ESCRT complexes, a type I myosin (Myo5), the Sla2 actin-binding protein involved in endocytosis, and a number of transporters. The emerging model is that endocytosis internalizes extracellular components, such as heme, into cytoplasmic vesicles. The cargo-loaded vesicles then fuse with early endosomes where they are sorted by ESCRT machinery into late endosomes or multivesicular bodies.
Fig. 1 -. Overview of heme acquisition systems in pathogenic and model fungi. (A) Candida albicans, (B) Schizosaccharomyes pombe and (C) Cryptococcus neoformans.
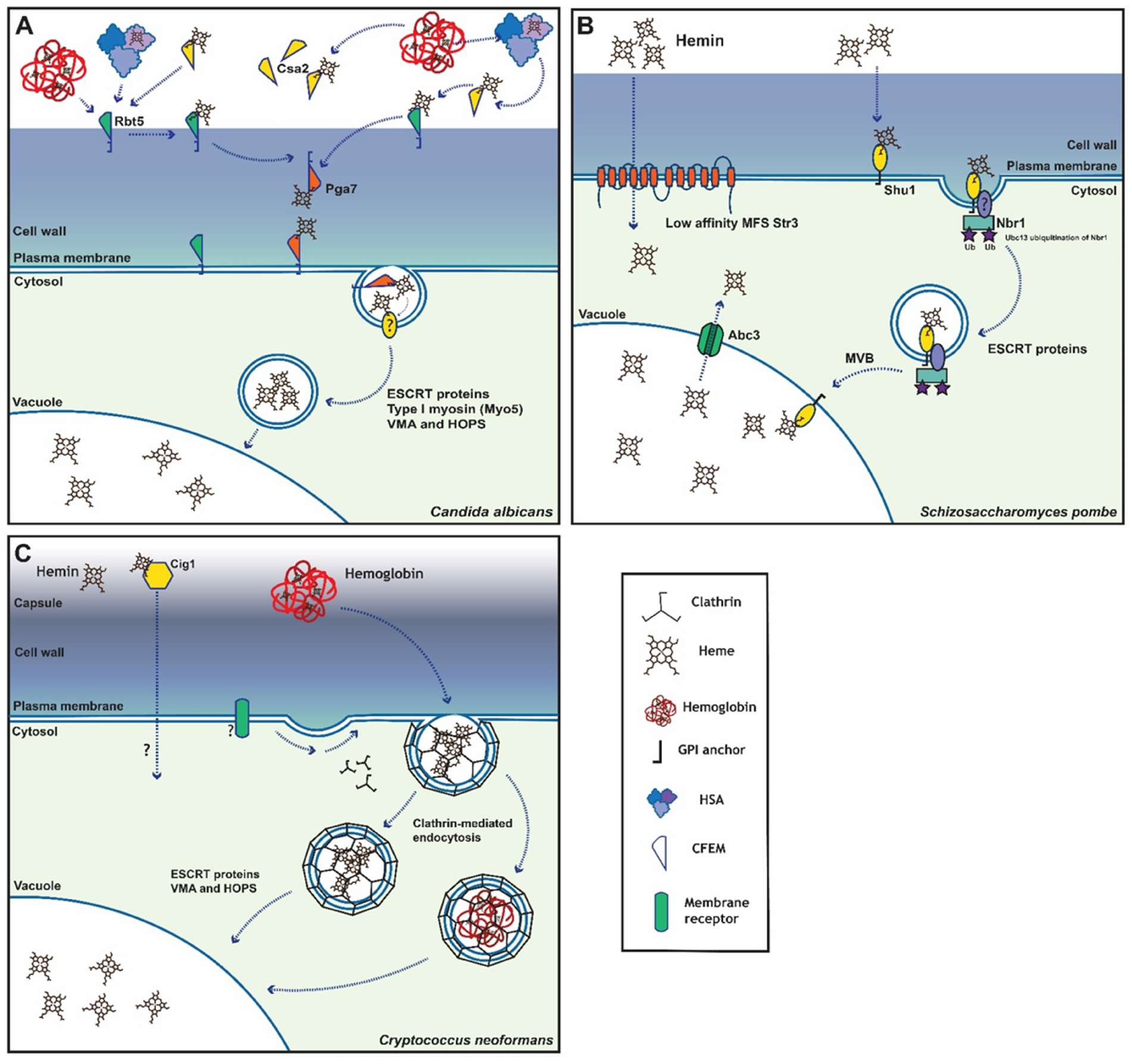
As described in the main text, the system in C. albicans consists of CFEM proteins (i.e., Rbt5, Csa2 and Pga7) that form a relay network to access heme (e.g., from hemoglobin). Subsequent internalization involves endocytic trafficking and participation of the ESCRT system for delivery to the vacuole. For S. pombe, the Shu1 protein binds heme and translocates from the plasma membrane to the vacuolar membrane. The ESCRT system and the Nbr1 protein also participate in internalization, while the Abc3 transporter is thought to mobilize heme from the vacuole to the cytoplasm. A lower affinity heme uptake system also exists in S. pombe and requires the Str3 transporter of the major facilitator superfamily (MFS). The system for heme use in C. neoformans involves the mannoprotein Cig1 and the participation of the machinery for endocytosis as well as the ESCRT system.
The mechanisms of acquisition of heme from extracellular hemoglobin have been revealed by elegant studies from the Kornitzer group (Weissman and Kornitzer, 2004; Kuznets et al., 2014; Nasser et al., 2016; Roy and Kornitzer, 2019; Kornitzer and Roy, 2020; Pinksy et al., 2020). Specifically, a relay network of the CFEM (Common in Fungal Extracellular Membrane) proteins Csa2, Rbt5 and Pga7 mediates the process of heme binding for delivery to the endocytic pathway (Weissman and Kornitzer, 2004; Kuznets et al., 2014; Nasser et al., 2016). The CFEM proteins have different locations: Csa2 is exported into the medium, Rbt5 is mainly localized to the cell wall via a GPI anchor, and Pga7 is GPI-anchored to the plasma membrane. These proteins act as hemophores to bind heme (e.g., from hemoglobin), and extracellular Csa2 and Rbt5 are thought to act in a relay to shuttle heme across the cell wall to Pga7 for eventual endocytosis (Fig. 1A). It is not yet clear, however, how the hemophores are connected to the endocytic machinery and whether a plasma membrane receptor is involved in directing heme to the endocytic/ESCRT pathway (Kornitzer and Roy, 2020). In addition to hemoglobin, other host proteins may also serve as sources of heme including human serum albumin (HSA) (Pinsky et al., 2020). Incubation with HSA stimulated the use of heme and hemoglobin by C. albicans, and the influence of HSA was dependent on the CFEM hemophore system. Interestingly, the drugs naproxen and salicylic acid that bind HSA interfere with heme use by C. albicans.
The importance of the hemophore relay system for the virulence of C. albicans was assessed in a mouse model of systemic candidiasis (Kuznets et al., 2014). In particular, a mutant lacking Pga7 is attenuated in virulence compared to the wild type strain or the mutant containing a reintroduced copy of the wild type PGA7 gene. Earlier work did not reveal a virulence contribution of Rbt5 in the mouse model, and this may reflect a smaller contribution of Rbt5 to heme and hemoglobin use compared to Pga7 (Kuznets et al., 2014; Braun et al., 2000). An ortholog of Rbt5 also functions in the uptake of hemoglobin in strains of Paracoccidioides, a dimorphic fungal pathogen that causes paracoccidioidomycosis in Latin America (Bailão et al., 2014; de Souza et al., 2020, 2021; Seki Kioshima et al., 2021). Reduced Rbt5 expression in Paracoccidioides by an antisense RNA approach led to lower survival of the fungus in macrophages and in the spleens of infected mice (Bailão et al., 2014).
Similar to the system in C. albicans, the saprophytic yeast Schizosaccharomyces pombe uses a GPI-anchored cell surface protein Shu1 and the vacuolar transporter Abc3 to acquire heme (Fig. 1B). Shu1 has 4 Cys residues that are reminiscent of the canonical 8-Cys containing CFEM motif and was found to interact with Nbr1, a receptor for ESCRT-dependent endosomal microautophagy, in the presence of heme (Mourer et al., 2017). These results suggest that the Shu1-Nbr1 heteroprotein complex could be recognized by the ESCRT machinery leading to endocytosis. Additionally, S. pombe has another lower affinity heme uptake system dependent on the Str3 protein, and this system appears to function independently from the higher affinity Shu1-Abc3 uptake system. Str3 is a cell surface transmembrane protein belonging to the major facilitator superfamily of transporters (MFS transporters) (Normant et al., 2018; Labbé et al., 2020).
Heme use has also been studied in the fungal pathogen C. neoformans that causes life threatening meningoencephalitis in immunocompromised individuals (Cadieux et al., 2013; Bairwa et al., 2019, 2020). This pathogen has a global health impact in that cryptococcal disease accounts for ~15% of deaths in the HIV/AIDS population (Shroufi et al., 2021). The fungus has multiple acquisition mechanisms to obtain iron including a high-affinity, reductive uptake system, transporters for siderophores, and the ability to use heme (Kronstad et al., 2013, Bairwa et al., 2019; Horianopoulos and Kronstad, 2019). As in C. albicans, heme/iron uptake by C. neoformans involves extracellular binding by a putative hemophore, followed by endocytosis and trafficking via vesicles and ESCRT and HOPS machinery to the vacuole (Fig. 1C). Four candidate CFEM proteins have been identified in C. neoformans but these appear to be dispensable for heme acquisition (G. Hu, unpublished results; Bairwa et al., 2019). Instead, a secreted mannoprotein Cig1 may be a hemophore because it possesses weak heme-binding activity and is required for optimal growth of the fungal cells at physiological pH (Cadieux et al., 2013). Notably, Cig1 does not share sequence similarity to the CFEM hemophores of C. albicans. Genetic screens revealed that C. neoformans takes up heme by clathrin‐mediated endocytosis (CME) and targets it to the vacuole via endosomes and ESCRT and HOPS complexes. Components of CME, such as the Chc1, Las17, Rvs161, and Rvs167 proteins, are critical in uptake of heme by influencing the endocytosis and intracellular trafficking of heme and hemoglobin, as revealed by both heme uptake assays and heme sensor measurements described below (Bairwa et al., 2019, 2020). The ESCRT machinery generally targets monoubiquitinated membrane proteins for internalization and trafficking to the vacuole (Hurley and Emr, 2006), which are the major sites of iron storage and use in cells. It is not yet known whether the heme-Cig1 binding complex, and/or other heme-binding hemophores are ubiquinated, or if any specific receptors are involved, and targeted to ESCRT machinery in C. neoformans. The ESCRT proteins Vps23 (ESCRT-I), Vps22 (ESCRT-II), and Snf7 (ESCRT-III) are involved in the use of heme iron, likely by influencing the intracellular trafficking of heme to the vacuole (Hu et al., 2013; Hu et al., 2015). Furthermore, the contribution of the ESCRT machinery for heme occurs at least, in part, via activation of the Rim101 pH signaling pathway leading to expression of Cig1 (Hu et al., 2015). Loss of Cig1 alone does not cause a virulence defect in a mouse model of cryptococcosis, in comparison with the attenuation that occurs upon loss of ferroxidase Cfo1 of the high affinity uptake system (Jung et al., 2008, 2009). However, deletion of the CIG1 gene in the background of a strain lacking Cfo1 further attenuated virulence thus revealing a contribution of the protein (Cadieux et al., 2013).
Additional studies in C. neoformans also revealed contributions to heme trafficking for endosomal trafficking proteins including the noncatalytic subunit of flippase (P4-ATPase) Cdc50, the Sec1/Munc18 (SM) protein Vps45, and two Vam6/Vps39/TRAP1-domain proteins, Vps3 and Vam6/Vps39 (Caza et al., 2018; Hu et al., 2017, 2021). Cdc50 functions as a chaperone to facilitate the exit of flippases and other cargo from the ER. In C. neoformans, Cdc50 contributes to heme uptake and internal trafficking of iron‐containing molecules (e.g., to the vacuole, mitochondria, and ER), and iron processing, possibly by interaction with flippases (Hu et al., 2017). SM proteins such as Vps45 regulate vesicle trafficking and fusion by interacting with SNARE (soluble N-ethylmaleimide-sensitive attachment protein receptor) proteins that are components of the fusion machinery. In C. neoformans, Vps45 co-localizes with mitochondria, and is required for mitochondrial functions, calcium homeostasis, and resistance to reactive oxygen species (Caza et al., 2018). Vps45 also plays a significant role in the uptake of exogenous iron/heme and intracellular sorting of the iron permease Cft1 (Caza et al., 2018). Vam6/Vps39/TRAP1-domain proteins, Vps3 and Vam6, are essential components, respectively, of CORVET and HOPS complexes of the endosomal pathway, mediating the fusion events between endosomes and the vacuole (Bröcker et al., 2012). In C. neoformans, the Vam6/Vps39 protein in particular supports fungal growth on heme and influences the trafficking and expression of iron uptake proteins (Hu et al., 2021). Moreover, Vam6 is a key component of the vacuole and mitochondrial patch (vCLAMP) contact site that may participate in vesicle transport between the endomembrane system and mitochondria in fungi (Elbaz-Alon et al., 2014; Hönscher et al., 2014). A role for Vam6 supports the possibility that mitochondria contribute to the iron acquisition from heme via a connection with the vacuole. Notably, functions involved in uptake and cellular trafficking of heme iron in C. neoformans are all required for virulence in a mouse model of cryptococcosis, including Cig1, Las17, ESCRT complex components, Vps45, Cdc50, Vam6, and Vps3, highlighting the importance of heme uptake and intracellular trafficking in fungal pathogenesis (Cadieux et al., 2013, Bairwa et al., 2017, 2019; Caza et al., 2018; Hu et al., 2013, 2015, 2017, 2021).
The importance of heme as an iron source has also been explored in Aspergillus fumigatus. This filamentous fungus is acquired via inhalation of airborne spores (conidia) and can cause invasive aspergillosis, particularly in immunocompromised individuals (Cadena et al., 2021). This disease is often fatal due to challenges in diagnosis and the limited options for antifungal therapy. There is a wealth of information on mechanisms of iron sensing, acquisition and homeostasis in A. fumigatus (Blatzer and Latgé, 2017; Misslinger et al., 2021). In particular, the contributions of siderophores to iron acquisition and storage, and the role of reductive iron assimilation have been studied in detail. In the context of virulence, the analysis of a mutant lacking SidA, the L-ornithine-N5-monooxygenase, revealed that this enzyme is essential for virulence in a neutropenic mouse model of aspergillosis (Schrettl et al., 2004). In contrast, a mutant lacking the iron permease, FtrA, for high affinity, reductive iron uptake, was still able to cause disease. The availability of a sidA/ftrA double deletion mutant provided an opportunity to assess the ability of A. fumigatus to use host-related iron sources. Specifically, the mutant was unable to grow on blood agar or hemoglobin, hemin, holotransferrin or ferritin (Schrettl et al., 2004). These results led to the conclusion that A. fumigatus lacks mechanisms to acquire iron from these sources. Consistent with this interpretation, single, double or triple deletion mutants lacking the three genes encoding candidate CFEM hemophores in A. fumigatus did not reveal contributions to heme uptake or virulence (Vaknin et al., 2014).
More recently, Michels et al. (2022) revisited the possibility that A. fumigatus exploits heme as an iron source during infection. Lung hemorrhage, a feature of invasive aspergillosis, was examined and found to correlate with increased lung hemoglobin during the first three days of infection; a correlation with increased iron in heme was also detected. Heme uptake was assessed in fungal cultures by examining fluorescence in cells exposed to tin (IV)-protoporphyrin, a non-iron heme analog. Increased fluorescence was consistent with uptake, as was the detection of increased iron associated with heme in the cytoplasm of cells grown on heme as the sole iron source. Importantly, administration of heme during infection increased the severity of disease and increased fungal burden in the lungs (Michels et al., 2022). Pre-culture of germinated conidia with heme but not tin (IV)-protoporphyrin also resulted in increased lung damage and fungal burden upon infection. Taken together, these results suggest that A. fumigatus can use heme during infection, although questions remain about the mechanisms of acquisition, including the relationship with iron acquisition via siderophore production and potential influences of heme on the host during aspergillosis.
3. Development and application of a genetically encoded heme sensor in S. cerevisiae
Genetically encoded biosensors have been deployed in several organisms to investigate mechanisms involved in heme signaling, dynamics, trafficking, and allocation to different subcellular compartments (Hanna et al., 2016; Gallio et al., 2021; Leung et al., 2021). These biosensors are supporting efforts to understand heme availability in cell compartments given that heme can be stably bound to proteins or present in a labile or exchangeable form to enable transfer to heme-binding proteins. The responsiveness of the heme sensors must therefore be considered in the context of the relative affinities and concentrations of the sensor versus other heme-binding proteins. A detailed discussion of these considerations has been presented by Gallio et al. (2021). Of importance to fungal cells, a ratiometric biosensor (HS1) was engineered by the Reddi laboratory for the analysis of labile or exchangeable heme in S. cerevisiae (Hanna et al., 2016). The yeast HS1 heme biosensor functions similar to the mammalian heme probe (CISDY) in which the fluorescence resonance energy transfer (FRET) effect of fluorescent molecules is modulated by the heme binding activities of sensory modules (Song et al., 2015; Hanna et al., 2016). Specifically, the HS1 sensor consists of a bacterial heme binding moiety cytochrome b562 (cyt b562) inserted within the coding sequence of an EGFP protein that is N-terminally linked to the red fluorescent protein Katushka 2 (mKATE2) (Arnesano et al., 2000; Takeda et al., 2003; Hanna et al., 2016) (Fig. 2A). Heme binding to cyt b562 enables resonance energy transfer from eGFP to cyt b562 quenching almost all of the fluorescent signal while not influencing the fluorescence of mKATE2. Thus, the sensor allows the ratiometric analysis of the intracellular labile heme pools independent of sensor concentration (Arpino et al., 2012; Hanna et al., 2016; Bairwa et al., 2020) (Fig. 2B). The read out of heme binding to the sensor is therefore measured as a decrease in the GFP signal relative to the mKATE2 fluorescence.
Fig. 2 -. Deployment of a genetically encoded heme biosensor in Cryptococcus neoformans.
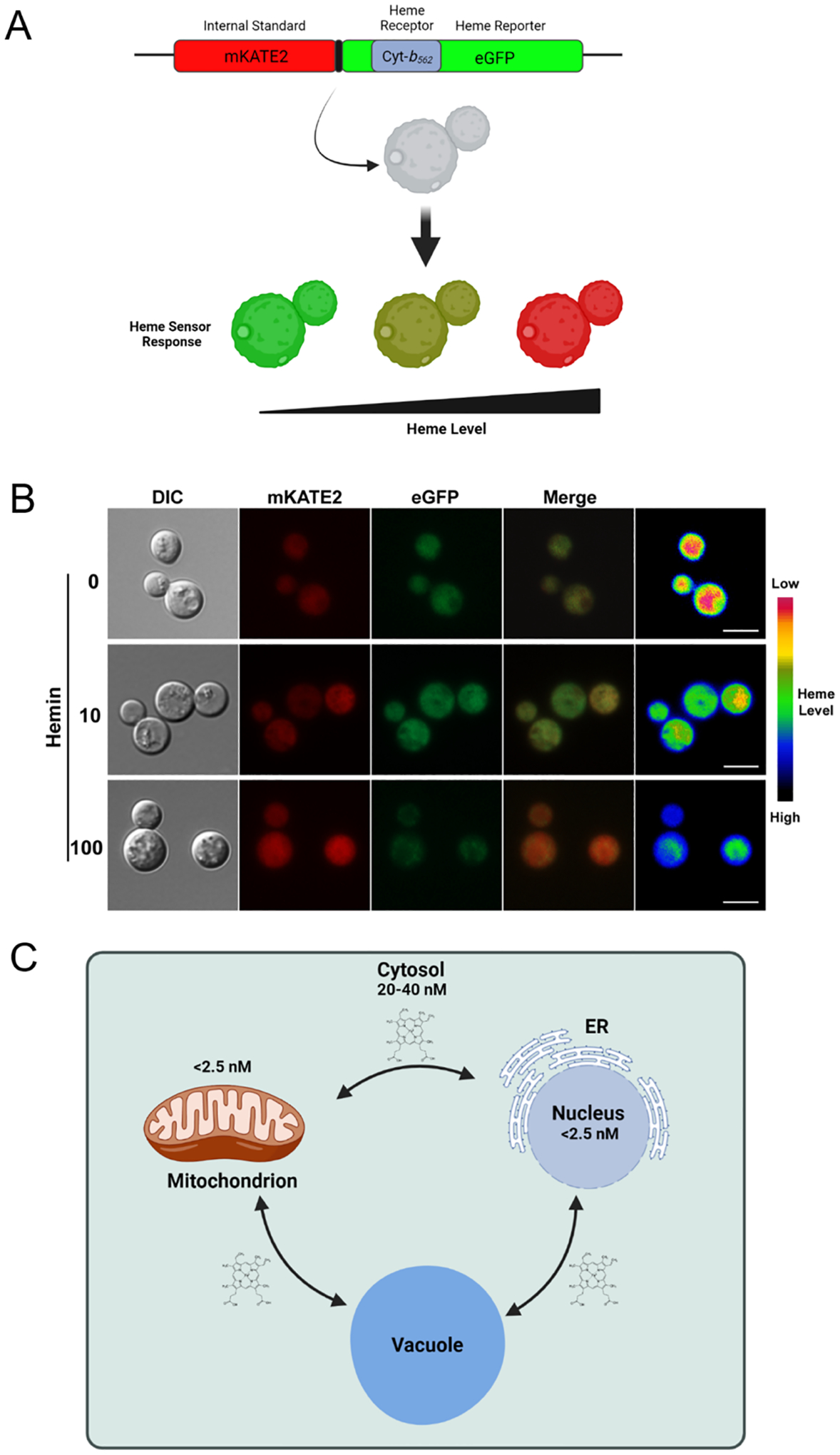
(A) Diagram of sensor composition including the heme binding domain of Cyt-b562 within eGFP and the mKATE2 protein to allow ratiometric measurements (Hanna et al., 2016). Heme binding quenches the GFP signal but does not influence mKATE2 thus resulting in a shift in the ratio of fluorescence from the two proteins. (B) Example of the evaluation of the labile heme pool in C. neoformans by microscopy. The images are from a replicate of an experiment presented by Bairwa et al. (2020). Scale bar = 5 μm. (C) Diagram of cellular compartments and labile heme concentrations estimated by Hanna et al. (2016) for S. cerevisiae.
Different versions of the biosensor have been tuned to different binding affinities and expression levels to detect a range of concentrations of heme in specific subcellular compartments without interfering with cellular physiology or heme homeostasis. For example, an HS1 variant (HS1-M7A) detected a labile heme level in the cytosol of 20 to 40 nM, whereas the levels detected in the nucleus or mitochondria remained ~4-fold lower (2.5 nM) than the cytosol (Hanna et al., 2016) (Fig. 2C). Utilizing the same probe, it was found that heme levels could vary from different subcellular compartments in response to signaling molecules such as nitric oxide (NO). In particular, it was observed that the nitric oxide donor NOC-7 induced rapid changes of labile heme in the cytosol and nucleus, with increases of two and 100-fold, respectively, possibly through the labilization of hemoproteins by weakening the affinity of bound heme to promote dissociation. Notably, a mutant screen revealed that deletion of the gene encoding the heme binding protein glyceraldehyde phosphate dehydrogenase (GAPDH) (or inhibition of heme binding to this protein) altered cytosolic labile heme levels and the influence of heme on the activity of the heme-dependent transcription factor Hap1 (Hanna et al., 2016; Sweeny et al., 2018).
To further explore the potential role(s) of labile heme, experiments with the heme sensor revealed that S. cerevisiae cells treated with succinyl acetone (SA), an inhibitor of heme synthesis, exhibited greater reduction of labile heme levels than total heme suggesting that yeast cells preferentially mobilize labile heme when experiencing cellular heme deficiency (Hanna et al., 2018). The same study revealed that stress induced by lead (Pb2+) negatively affects total cellular heme by disrupting heme biosynthesis. Targeting of heme biosensors to specific subcellular compartments in S. cerevisiae also helped elucidate not only the levels of heme but also mechanisms of heme mobilization after synthesis at the mitochondrial inner membrane (Hanna et al., 2016; Martinez-Guzman et al., 2020). For example, the use of in vivo pulse-chase assays using the sensors in combination with succinyl acetone (SA) revealed higher heme trafficking rates towards the nucleus than to the cytosol or the mitochondrial matrix suggesting the participation of heme as an interorganellar signal. The analysis of nuclear-mitochondrial heme mobility in different yeast mutants also revealed the contribution of various mitochondria-associated proteins to heme trafficking. For instance, the GTPases Gem1 and Mgm1, regulators of the endoplasmic reticulum-mitochondria encounter structure (ERMES) and mitochondrial fusion, respectively, both positively regulate heme transport towards the nucleus. This was in contrast to the negative regulation observed for Dnm1, a mitochondrial fission-related GTPase. Overall, the versatility and scope of heme biosensors to understand the impact of stress on heme homeostasis and the mechanisms involved in heme uptake and trafficking make of these molecular probes valuable tools for investigating heme trafficking in medically important fungal pathogens, as described in the next section.
4. Application of a genetically encoded heme sensor in the fungal pathogens Cryptococcus neoformans and Candida albicans
As outlined earlier, the mechanisms for the uptake and homeostasis of heme-iron have been examined in the fungal pathogen C. neoformans (Jung et al., 2008, 2009; Kronstad et al., 2013; Hu et al., 2013; Caza and Kronstad, 2013; Cadieux et al., 2013; Saikia et al., 2014; Hu et al., 2015; Bairwa et al., 2017, 2019). The generation and application of a codon optimized version (CnHS) of the yeast heme sensor (HS1-M7A) developed by Hanna et al. (2016) enabled further analysis of the intracellular heme dynamics in C. neoformans including the impact of oxidative stress, mitochondrial and vacuolar functions on heme homeostasis (Bairwa et al., 2020). The use of the CnHS heme probe also allowed further examination and validation of the role of clathrin-mediated endocytosis (CME) in the heme uptake process (Bairwa et al., 2019). For example, disruption of the CME machinery through chemical inhibition using chlorpromazine or deletion of the endocytic components Chc1 or Las7 induced a reduced response of the heme sensor to exogenous heme, thus supporting the role of the CME in heme internalization. The CnHS heme sensor was also used to characterize other mechanisms involved in the uptake and intracellular trafficking of heme-iron, such as the hemophore Cig1 and the vesicular trafficking regulator Vps45 (Bairwa et al., 2020; Cadieux et al., 2013; Caza et al., 2018). Importantly, the heme sensor also detected alterations in the labile heme pool when vacuolar or mitochondrial functions were compromised. For example, inhibition of vacuolar acidification or the activity of electron transfer chain complexes resulted in drastic differences of probe ratiometric values compared to the read outs obtained for the untreated fungal cells. In addition, expression of CnHS in a wild-type strain revealed a gradual increase for the ratiometric values of the probe after cells were phagocytosed by murine alveolar macrophages. The increase in the GFP signal relative to mKATE2 fluorescence suggests that C. neoformans may experience a restriction of its intracellular labile heme pool. This result suggests limited availability of heme within the host and/or cellular adaptation to heme homeostasis during the intracellular proliferation in phagocytic cells (Bairwa et al., 2020). Overall, the utility of the heme biosensor validated previous observations on heme-iron homeostasis in C. neoformans and, importantly, expanded the molecular toolkit to more thoroughly dissect heme uptake and processing mechanisms.
Genetically encoded heme sensors have also been employed with C. albicans to further understand mechanisms of heme and hemoglobin use. A recent study demonstrated the ability of the fungus to use heme both as an iron source and directly, and examined the role of the heme oxygenase Hmx1 in iron acquisition from heme and hemoglobin (Weissman et al., 2021). Hmx1 appears to play a minor role in iron acquisition from these sources, but likely has a more important role in preventing heme toxicity. The use of cells encoding the heme sensor revealed that loss of Hmx1 function results in a higher labile heme concentration in the cytosol in response to heme or hemoglobin in the medium, compared to the wild type strain. As discussed above, the uptake of heme from hemoglobin and from human serum albumin carrying heme is dependent on the CFEM hemophore relay to deliver heme to the vacuole (Pinsky et al., 2020; Weissman et al., 2021). The appearance of heme in the cytoplasm therefore suggests the presence of a vacuolar transporter (e.g., similar to Abc3 in S. pombe). The study also revealed that heme may be taken up by another mechanism that is energy dependent and separate from the CFEM hemophore cascade (Weissman et al., 2021). Overall, the studies with C. albicans further demonstrate the utility of the genetically encoded heme sensor in understanding internalization dynamics and identifying uptake and processing functions.
5. Connections between iron and heme trafficking, endomembrane systems and organelles.
The connections between endocytosis and iron/heme acquisition identified for fungal pathogens (C. neoformans and C. albicans) are consistent with earlier studies in S. cerevisiae. In this model yeast, endocytic processes influence the activities of two iron uptake systems in the plasma membrane, a reductive, a high-affinity system involving the multicopper oxidase Fet3 and the iron permease Ftr1, and a siderophore transport system involving the Arn1-4 transporters (Li and Kaplan, 2004; Philpott, 2006; Li et al., 2010; Rutherford et al., 2001). The role of endocytosis can be illustrated with the siderophore uptake system in which the Arn1-4 proteins internalize iron bound to siderophores, and fluctuations in the level of the siderophore ferrichrome influence recycling of the Arn1 transporter from endosomes to the plasma membrane (Kim et al., 2002). Sorting of Arn1 in the Golgi to endosomal compartment was found in cells in the absence of ferrichrome, and addition of the siderophore at a low concentration (0.1 μM) resulted in relocalization of Arn1 to the plasma membrane. Interestingly, Arn1 at the plasma membrane undergoes endosome-to-plasma membrane recycling at an intermediate ferrichrome concentration (10 μM) (Kim et al., 2002). Together, these studies establish a paradigm for the connections between iron/heme trafficking and the endomembrane system including the functions of the ESCRT complexes and the vacuole in fungi (Xu et al., 2014; Gerwien et al., 2018; Rizzollo et al., 2021).
Mitochondria represent a key nexus in efforts to understand heme homeostasis because many mitochondrial proteins such as those involved in respiration bind iron ions, heme, or iron–sulfur clusters, and the organelle contains part of the machinery for heme biosynthesis (Paul et al., 2017). Additionally, mitochondrial activities such as the generation of iron-sulfur clusters and heme biosynthesis regulate iron homeostasis via iron or heme-responsive transcription factors. For example, activation of S. cerevisiae master iron regulators Aft1- or Aft2-dependent iron regulon requires mitochondrial iron-sulfur protein biogenesis (Babcock et al., 1997; Rutherford et al., 2005). Additionally, and as mentioned above, the Hap1 transcription factor is activated by heme (Zhang and Guarente, 1995). Similarly, the transcription factors Cir1 and HapX in C. neoformans are iron-binding proteins and regulate genes involved in mitochondrial iron metabolism including iron-sulfur cluster biosynthesis (Jung et al., 2006; Jung et al., 2010; Do et al., 2020). Additional connections between iron/heme sensing and the endomembrane system are also emerging. For example, Xue and colleagues (2017) reported that the protein complex endoplasmic reticulum (ER) mitochondria encounter structure (ERMES) mediated iron regulation via the transcription factor Aft1. The expression of the iron regulon controlled by the Aft1 was activated in the absence of ERMES components, resulting in accumulation of iron in the yeast cells, perhaps due to an influence on mitochondrial proteins important for regulation of the iron–sulfur cluster biosynthesis (Xue et al., 2017). ERMES functions in the exchange of materials between the ER and mitochondria, and is required for iron homeostasis in S. cerevisiae (Xue et al., 2017). It is possible that heme is delivered to the vacuole, mitochondria and ER via ERMES complexes in other fungi. A similar mechanism may be in place in C. neoformans because inhibition of mitochondrial respiration causes a decrease in the cytosolic labile heme pool (Bairwa et al., 2020). Overall, these findings support emerging insights into the roles of endomembrane trafficking systems and mitochondria in mediating iron and heme uptake.
6. Conclusions and perspectives
Genetically-encoded sensors are providing new insights into the mechanisms of heme uptake and homeostasis in pathogenic and non-pathogenic fungi. These mechanisms include hemophore dependent and independent heme acquisition systems, and a role for endosomal trafficking in heme acquisition and processing. In fact, endosomal trafficking mediated by ESCRT machinery and the involvement of vacuole and mitochondrial functions are common features in heme uptake in C. albicans, C. neoformans, and S. pombe. These studies suggest that the heme sensor and additional genetic approaches could further clarify aspects of heme uptake and homeostasis in other pathogenic fungi such as A. fumigatus and Paracoccioides sp. In general, the mechanisms by which fungal (and other) pathogens extract heme from host sources need to be more thoroughly examined (Tolosano et al., 2010; Kornitzer and Roy, 2020; Pinsky et al., 2020). This information will be useful in further understanding fungal pathogenesis and, potentially, for therapeutic intervention by exploiting heme uptake systems, e.g., with non-iron metalloprotophyrins (Bairwa et al., 2019).
Despite the accumulating information on heme acquisition in fungi, there are several areas where further investigation is needed. These include more detailed characterization of the different contributions of the CFEM proteins and hemophores in heme acquisition in C. albicans and C. neoformans. In both species, there is evidence for additional heme uptake mechanisms, and there may be other hemophores for C. neoformans. Additionally, functions that act upstream (e.g., receptors and transporters) of the endocytic machinery remain to be identified in the pathogenic fungi. More mechanistic insights are also needed to understand the roles of downstream components such as endomembrane proteins and proteins at membrane contact sites that shuttle heme inside the cell. In this regard, the role of GAPDH found in S. cerevisiae needs to also be examined in the pathogens (Sweeny et al., 2018). Finally, the recent work on the Hmx1 heme oxygenase in C. albicans provides valuable insights into heme degradation or recycling, and more work is needed to build a clear picture of intracellular heme processing (Weissman et al., 2021).
The role of mitochondria in heme homeostasis in fungi deserves particular emphasis for future work. That is, additional connections between endosomal trafficking and mitochondrial functions are likely to be relevant to iron and heme transport and homeostasis. Mitochondria are vital for fungi to adapt to environmental change or to cause disease, and the majority of mitochondrial proteins are encoded by nuclear genes, translated in cytosol or at the outer mitochondrial membrane, and subsequently imported (Becker and Wagner, 2018; Béthune et al., 2019; Pfanner et al., 2019; Vardi-Oknin and Arava, 2019; Müntjes et al., 2021). In this regard, we raise the speculative possibility that iron and heme homeostasis influences mitochondrial functions via regulation of the endosomal transport of mRNA to mitochondria (Schatton and Rugarli, 2018; Tsuboi et al., 2020). mRNA localization and subsequent local translation enables spatiotemporal gene expression, which allows fungi to finely adjust cellular physiology in response to intracellular and extracellular signals (Das et al., 2021; Niessing et al., 2018). There are detailed studies on mRNA localization in fungi including S. cerevisiae and the plant pathogen Ustilago maydis that provide a foundation for studying the impact of iron/heme homeostasis on mRNA localization and mitochondrial function (Vollmeister et al., 2012; Haag et al., 2015, 2017; Salogiannis and Reck-Peterson, 2017; Béthune et al., 2019; Müller et al., 2019; Das et al., 2021). In this context, powerful methods are available to study RNA localization including localized RNA recording, APEX-seq, APEX-RIP, proximity-specific ribosome profiling, fluorescence in situ hybridization (FISH) and live cell imaging (Williams et al., 2014; Chen et al., 2015; Kaewsapsak et al., 2017; Tutucci et al., 2018; Padrón et al., 2019; Medina-Munoz et al., 2020). Finally, we also note that iron homeostasis is known to involve regulation of mRNA stability and this area should be investigated in the context of iron and heme pools in different cellular compartments of fungal pathogens (Perea-García et al., 2022; Romero et al., 2022).
Highlights.
Heme is an abundant iron source for fungi that infect vertebrate hosts
Proteins at the cell surface play important roles in iron acquisition from heme
Internalization of heme depends on endocytosis in model and pathogenic fungi
Genetically-encoded sensors support the analysis of heme uptake and trafficking
More work is needed to understand heme processing and movement between organelles
Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT and Future Planning 2021M3A9I4021431 and 2022R1F1A1065306 (to WHJ), and by grants from the Canadian Institutes of Health Research (MOP-13234 and PJT-166043) and the National Institute of Allergy and Infectious Diseases (RO1AI053721) (to JWK). Additional support was obtained from CREATE training award for the Plant Response to Eliminate Critical Threats (PRoTECT) program (https://www.protect-ubc.com) from NSERC. The authors apologize to those researchers whose work we were unable to cite due to space limitations. JWK is a Burroughs Wellcome Fund Scholar in Molecular Pathogenic Mycology, and a fellow of the CIFAR program: Fungal Kingdom, Threats & Opportunities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
The authors declare that no competing interests exist.
REFERENCES
- Arnesano F, Banci L, Bertini I, Ciofi-Baffoni S, Woodyear TL, Johnson CM, Barker PD 2000. Structural consequences of b- to c-type heme conversion in oxidized Escherichia coli cytochrome b562. Biochem 39, 1499–1514. [DOI] [PubMed] [Google Scholar]
- Arpino JA, Czapinska H, Piasecka A, Edwards WR, Barker P, Gajda MJ, Bochtler M, Jones DD 2012. Structural basis for efficient chromophore communication and energy transfer in a constructed didomain protein scaffold. J. Am Chem Soc, 134, 13632–13640. [DOI] [PubMed] [Google Scholar]
- Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J 1997. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Bailão EF, Parente JA, Pigosso LL, de Castro KP, Fonseca FL, Silva-Bailão MG, Báo SN, Bailão AM, Rodrigues ML, Hernandez O, McEwen JG, Soares CM 2014. Hemoglobin uptake by Paracoccidioides spp. is receptor-mediated. PLoS Negl. Trop. Dis 8, e2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairwa G, Jung WH, Kronstad JW 2017. Iron acquisition in fungal pathogens of humans. Metallomics 9, 215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairwa G, Sánchez-León E, Do E, Jung WH, Kronstad JW 2020. A cytoplasmic heme sensor illuminates the impacts of mitochondrial and vacuolar functions and oxidative stress on heme-iron iomeostasis in Cryptococcus neoformans. mBio 11, e00986–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairwa G, Caza M, Horianopoulos L, Hu G, Kronstad J 2019. Role of clathrin-mediated endocytosis in the use of heme and hemoglobin by the fungal pathogen Cryptococcus neoformans. Cell. Microbiol 21, e12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Wagner R 2018. Mitochondrial outer membrane channels: Emerging diversity in transport processes. BioEssays 40, e1800013. [DOI] [PubMed] [Google Scholar]
- Béthune J, Jansen RP, Feldbrügge M, Zarnack K 2019. Membrane-associated RNA-binding proteins orchestrate organelle-coupled translation. Trends Cell biol 29, 178–188. [DOI] [PubMed] [Google Scholar]
- Blatzer M, Latgé JP 2017. Metal-homeostasis in the pathobiology of the opportunistic human fungal pathogen Aspergillus fumigatus. Curr. Op. Microbiol 40, 152–159. [DOI] [PubMed] [Google Scholar]
- Braun BR, Head WS, Wang MX, Johnson AD 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröcker C, Kuhlee A, Gatsogiannis C, Balderhaar HJ, Hönscher C, Engelbrecht-Vandré S, Ungermann C, Raunser S 2012. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. USA 109, 1991–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena J, Thompson GR III, Patterson TF 2021. Aspergillosis: Epidemiology, Diagnosis, and Treatment. Infect Dis Clin North Am, 35, 415–434. [DOI] [PubMed] [Google Scholar]
- Cadieux B, Lian T, Hu G, Wang J, Biondo C, Teti G, Liu V, Murphy ME, Creagh AL, Kronstad JW 2013. The Mannoprotein Cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans. J. Infect. Dis 207, 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caza M, Kronstad JW 2013. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Frontiers Cell. Inf. Microbiol 3, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caza M, Hu G, Nielson ED, Cho M, Jung WH, Kronstad JW 2018. The Sec1/Munc18 (SM) protein Vps45 is involved in iron uptake, mitochondrial function and virulence in the pathogenic fungus Cryptococcus neoformans. PLoS Path 14, e1007220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers IG, Willoughby MM, Hamza I, Reddi AR 2021. One ring to bring them all and in the darkness bind them: The trafficking of heme without deliverers. Biochim. Biophys. Acta 1868, 118881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, & Zhuang X 2015. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras H, Chim N, Credali A, Goulding CW 2014. Heme uptake in bacterial pathogens. Curr. Op. Chem. Biol 19, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Vera M, Gandin V, Singer RH, Tutucci E 2021. Intracellular mRNA transport and localized translation. Nat. Revs. Mol. Cell Biol 22, 483–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza AF, de Paula MS, Lima RM, Silva MG, de Curcio JS, Pereira M, de Almeida Soares CM 2020. The “Little Iron Waltz”: The ternary response of Paracoccidioides spp. to iron deprivation. J. Fungi (Basel), 6, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza AF, Tomazett MV, Freitas E Silva KS, de Curcio JS, Pereira CA, Baeza LC, Paccez JD, Gonçales RA, Rodrigues F, Pereira M, de Almeida Soares CM 2021. Interacting with hemoglobin: Paracoccidioides spp. recruits hsp30 on its cell surface for enhanced ability to use this iron source. J. Fungi (Basel), 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuyser L, Van Genechten W, Mizuno H, Colombo S, Van Dijck P 2018. Introducing fluorescence resonance energy transfer-based biosensors for the analysis of cAMP-PKA signalling in the fungal pathogen Candida glabrata. Cell. Microbiol 20, e12863. [DOI] [PubMed] [Google Scholar]
- Do E, Cho YJ, Kim D, Kronstad JW, Jung WH 2020. A transcriptional regulatory Map of iron homeostasis reveals a new control circuit for capsule formation in Cryptococcus neoformans. Genetics 215, 1171–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donegan RK, Moore CM, Hanna DA, Reddi AR 2019. Handling heme: The mechanisms underlying the movement of heme within and between cells. Free Rad. Biol. Med 133, 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhof I, Ernst JF 2016. Oxygen-independent FbFP: Fluorescent sentinel and oxygen sensor component in Saccharomyces cerevisiae and Candida albicans. Fun. Genet. Biol 92, 14–25. [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M 2014. A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell, 30, 95–102. [DOI] [PubMed] [Google Scholar]
- Fisher MC, Gurr SJ, Cuomo CA, Blehert DS, Jin H, Stukenbrock EH, Stajich JE, Kahmann R, Boone C, Denning DW, Gow N, Klein BS, Kronstad JW, Sheppard DC, Taylor JW, Wright GD, Heitman J, Casadevall A, Cowen LE 2020. Threats posed by the fungal kingdom to humans, wildlife, and agriculture. mBio 11, e00449–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaris D, Barbouti A, Pantopoulos K 2019. Iron homeostasis and oxidative stress: An intimate relationship. Biochim. Biophys. Acta 1866, 118535. [DOI] [PubMed] [Google Scholar]
- Gallio AE, Fung SS, Cammack-Najera A, Hudson AJ, Raven EL 2021. Understanding the logistics for the distribution of heme in cells. J. Amer. Chem. Soc 1, 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T, Nemeth E 2015. Iron homeostasis in host defence and inflammation, Nat. Rev. Immunol 15, 500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwien F, Skrahina V, Kasper L, Hube B, Brunke S 2018. Metals in fungal virulence. FEMS Microbiol. Rev 42, fux050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald EC, Mehta S, Zhang J 2018. Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem. Rev 118, 11707–11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag C, Pohlmann T, Feldbrügge M 2017. The ESCRT regulator Did2 maintains the balance between long-distance endosomal transport and endocytic trafficking. PLoS Genet 13, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag C, Steuten B, Feldbrügge M 2015. Membrane-coupled mRNA trafficking in fungi. Ann. Rev. Microbiol 69, 265–281. [DOI] [PubMed] [Google Scholar]
- Hanna DA, Martinez-Guzman O, Reddi AR 2017. Heme gazing: Illuminating eukaryotic heme trafficking, dynamics, and signaling with fluorescent heme sensors. Biochemistry 56, 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna DA, Hu R, Kim H, Martinez-Guzman O, Torres MP, Reddi AR 2018. Heme bioavailability and signaling in response to stress in yeast cells. J. Biol. Chem 293, 12378–12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna DA, Harvey RM, Martinez-Guzman O, Yuan X, Chandrasekharan B, Raju G, Outten FW, Hamza I, Reddi AR 2016. Heme dynamics and trafficking factors revealed by genetically encoded fluorescent heme sensors. Proc. Natl. Acad. Sci. USA 113, 7539–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, Ungermann C 2014. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell 30, 86–94. [DOI] [PubMed] [Google Scholar]
- Horianopoulos LC, Kronstad JW 2019. Connecting iron regulation and mitochondrial function in Cryptococcus neoformans. Curr. Op. Microbiol 52, 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Bakkeren E, Caza M, Horianopoulos L, Sánchez-León E, Sorensen M, Jung W, Kronstad JW 2021. Vam6/Vps39/TRAP1-domain proteins influence vacuolar morphology, iron acquisition and virulence in Cryptococcus neoformans. Cell. Microbiol 23, e13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Caza M, Bakkeren E, Kretschmer M, Bairwa G, Reiner E, Kronstad J 2017. A P4-ATPase subunit of the Cdc50 family plays a role in iron acquisition and virulence in Cryptococcus neoformans. Cell. Microbiol 19, 10.1111/cmi.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Caza M, Cadieux B, Bakkeren E, Do E, Jung WH, Kronstad JW 2015. The endosomal sorting complex required for transport machinery influences haem uptake and capsule elaboration in Cryptococcus neoformans. Mol. Microbiol 96, 973–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Caza M, Cadieux B, Chan V, Liu V, Kronstad J 2013. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Inf. Immun 81, 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Caplan J, Sweigard JA, Czymmek KJ, Donofrio NM 2017. Optimization of the HyPer sensor for robust real-time detection of hydrogen peroxide in the rice blast fungus. Mol. Plant Pathol 18, 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Emr SD 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Ann. Rev. Biophys. Biomol. Struc 35, 277–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Saikia S, Hu G, Wang J, Fung CK, D’Souza C, White R, Kronstad JW 2010. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Path 6, e1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sánchez-León E, Kronstad JW 2021. Coordinated regulation of iron metabolism in Cryptococcus neoformans by GATA and CCAAT transcription factors: connections with virulence. Curr. Genet 67, 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, White R, Kronstad JW 2006. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol 4, e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, Lian T, Singh A, Kosman DJ, & Kronstad JW (2008). Iron source preference and regulation of iron uptake in Cryptococcus neoformans. PLoS Path 4, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Hu G, Kuo W, Kronstad JW 2009. Role of ferroxidases in iron uptake and virulence of Cryptococcus neoformans. Euk. Cell 8, 1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewsapsak P, Shechner DM, Mallard W, Rinn JL, Ting AY 2017. Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. eLife 6, e29224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ju J, Lee HN, Chun H, Seong J 2021. Genetically encoded biosensors based on fluorescent proteins. Sensors (Basel), 21, 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Yun CW, Philpott CC 2002. Ferrichrome induces endosome to plasma membrane cycling of the ferrichrome transporter, Arn1p, in Saccharomyces cerevisiae. EMBO J 21, 3632–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornitzer D, Roy U 2020. Pathways of heme utilization in fungi. Biochim. Biophys. Acta 1867, 118817. [DOI] [PubMed] [Google Scholar]
- Kostyuk AI, Panova AS, Kokova AD, Kotova DA, Maltsev DI, Podgorny OV, Belousov VV, Bilan DS 2020. In Vivo imaging with genetically encoded redox biosensors. Int. J. Mol. Sci 21, 8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad JW, Hu G, Jung WH 2013. An encapsulation of iron homeostasis and virulence in Cryptococcus neoformans. Trends Microbiol 21, 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA, Gresnigt MS, Hube B 2020. The gut, the bad and the harmless: Candida albicans as a commensal and opportunistic pathogen in the intestine. Curr. Op. Microbiol 56, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznets G, Vigonsky E, Weissman Z, Lalli D, Gildor T, Kauffman SJ, Turano P, Becker J, Lewinson O, Kornitzer D 2014. A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PLoS Path 10, e1004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé S, Mourer T, Brault A, Vahsen T 2020. Machinery for fungal heme acquisition. Curr. Genet 66, 703–711. [DOI] [PubMed] [Google Scholar]
- Leung GC, Fung SS, Gallio AE, Blore R, Alibhai D, Raven EL, Hudson AJ 2021. Unravelling the mechanisms controlling heme supply and demand. Proc. Natl. Acad. Sci. USA 118, e2104008118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kaplan J 2004. A mitochondrial-vacuolar signaling pathway in yeast that affects iron and copper metabolism. J. Biol. Chem 279, 33653–33661. [DOI] [PubMed] [Google Scholar]
- Li L, Murdock G, Bagley D, Jia X, Ward DM, Kaplan J 2010. Genetic dissection of a mitochondria-vacuole signaling pathway in yeast reveals a link between chronic oxidative stress and vacuolar iron transport. J. Biol. Chem 285, 10232–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Mehta S, Zhang J 2019. Genetically encoded fluorescent biosensors illuminate kinase signaling in cancer. J. Biol. Chem 294, 14814–14822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NN, Köhler JR 2016. Antagonism of fluconazole and a proton pump inhibitor against Candida albicans. Antimicrob. Agents Chemo 60, 1145–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JP, Lionakis MS 2022. Pathogenesis and virulence of Candida albicans. Virulence 13, 89–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsafari M, Ma J, Koffas M, Xu P 2020. Genetically-encoded biosensors for analyzing and controlling cellular process in yeast. Curr. Op. Biotechnol 64, 175–182. [DOI] [PubMed] [Google Scholar]
- Martinez-Guzman O, Willoughby MM, Saini A, Dietz JV, Bohovych I, Medlock AE, Khalimonchuk O, Reddi AR 2020. Mitochondrial-nuclear heme trafficking in budding yeast is regulated by GTPases that control mitochondrial dynamics and ER contact sites. J. Cell Sci 133, jcs237917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Munoz HC, Lapointe CP, Porter DF, Wickens M 2020. Records of RNA locations in living yeast revealed through covalent marks. Proc. Natl. Acad. Sci. USA 117, 23539–23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentges M, Bormann J 2015. Real-time imaging of hydrogen peroxide dynamics in vegetative and pathogenic hyphae of Fusarium graminearum. Sci. Reports, 5, 14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels K, Solomon AL, Scindia Y, Sordo Vieira L, Goddard Y, Whitten S, Vaulont S, Burdick MD, Atkinson C, Laubenbacher R, Mehrad B 2022. Aspergillus utilizes extracellular heme as an iron source during invasive pneumonia, driving infection severity. J. Inf. Dis 225, 1811–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misslinger M, Hortschansky P, Brakhage AA, Haas H 2021. Fungal iron homeostasis with a focus on Aspergillus fumigatus. Biochim. Biophys. Acta 1868, 118885. [DOI] [PubMed] [Google Scholar]
- Monteith AJ, Skaar EP 2021. The impact of metal availability on immune function during infection. Trends Endocrinol. Metab 32, 916–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourer T, Normant V, Labbé S 2017. Heme assimilation in Schizosaccharomyces pombe requires cell-surface-anchored protein Shu1 and vacuolar transporter Abc3. J. Biol. Chem 292, 4898–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Pohlmann T, Feldbrügge M 2019. Core components of endosomal mRNA transport are evolutionarily conserved in fungi. Fungal Genet. Biol 126, 12–16. [DOI] [PubMed] [Google Scholar]
- Müntjes K, Devan SK, Reichert AS, Feldbrügge M 2021. Linking transport and translation of mRNAs with endosomes and mitochondria. EMBO reports, 22, e52445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser L, Weissman Z, Pinsky M, Amartely H, Dvir H, Kornitzer D 2016. Structural basis of haem-iron acquisition by fungal pathogens. Nature Microbiol 1, 16156. [DOI] [PubMed] [Google Scholar]
- Nasu Y, Shen Y, Kramer L, Campbell RE 2021. Structure- and mechanism-guided design of single fluorescent protein-based biosensors. Nature Chem. Biol 17, 509–518. [DOI] [PubMed] [Google Scholar]
- Niessing D, Jansen RP, Pohlmann T, Feldbrügge M 2018. mRNA transport in fungal top models. Wiley Interdisc. Revs. RNA, 9, 10.1002/wrna.1453. [DOI] [PubMed] [Google Scholar]
- Normant V, Mourer T, Labbé S 2018. The major facilitator transporter Str3 is required for low-affinity heme acquisition in Schizosaccharomyces pombe. J. Biol. Chem 293, 6349–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrón A, Iwasaki S, Ingolia NT 2019. Proximity RNA labeling by APEX-Seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell 75, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BT, Manz DH, Torti FM, Torti SV 2017. Mitochondria and Iron: current questions. Expert Rev. Hematol 10, 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-García A, Puig S, Peñarrubia L 2022. The role of post-transcriptional modulators of metalloproteins in response to metal deficiencies. J. Exp. Botany 73, 1735–1750. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Warscheid B, Wiedemann N 2019. Mitochondrial proteins: from biogenesis to functional networks. Nature Rev. Mol. Cell Biol 20, 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott CC 2006. Iron uptake in fungi: a system for every source. Biochim. Biophys. Acta 1763, 636–645. [DOI] [PubMed] [Google Scholar]
- Pinsky M, Roy U, Moshe S, Weissman Z, Kornitzer D 2020. Human Serum Albumin facilitates heme-iron utilization by fungi. mBio, 11, e00607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Zhai H, Hou J 2019. Biosensors design in yeast and applications in metabolic engineering. FEMS Yeast Res 19, foz082. [DOI] [PubMed] [Google Scholar]
- Rizzollo F, More S, Vangheluwe P, Agostinis P 2021. The lysosome as a master regulator of iron metabolism. Trends Biochem Sci 46, 960–975. [DOI] [PubMed] [Google Scholar]
- Romero AM, García-Martínez J, Pérez-Ortín JE, Martínez-Pastor MT, Puig S 2022. Changes in mRNA stability play an important role in the adaptation of yeast cells to iron deprivation. Biochim. Biophys. Acta 1865, 194800. [DOI] [PubMed] [Google Scholar]
- Roy U, Kornitzer D 2019. Heme-iron acquisition in fungi. Curr. Op. Microbiol 52, 77–83. [DOI] [PubMed] [Google Scholar]
- Rutherford JC, Jaron S, Ray E, Brown PO, Winge DR 2001. A second iron-regulatory system in yeast independent of Aft1p. Proc. Natl. Acad. Sci. USA 98, 14322–14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford JC, Ojeda L, Balk J, Mühlenhoff U, Lill R, Winge DR 2005. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J. Biol. Chem 280, 10135–10140. [DOI] [PubMed] [Google Scholar]
- Saikia S, Oliveira D, Hu G, Kronstad J 2014. Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans. Infect. Imm 82, 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salogiannis J, Reck-Peterson SL 2017. Hitchhiking: A non-canonical mode of microtubule-based transport. Trends Cell Biol 27, 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatton D, Rugarli EI 2018. A concert of RNA-binding proteins coordinates mitochondrial function. Crit. Revs. Biochem Mol. Biol 53, 652–666. [DOI] [PubMed] [Google Scholar]
- Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, Arst HN Jr, Haynes K, Haas H 2004. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med 200, 1213–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Kioshima E, de Souza Bonfim de Mendonça P, de Melo Teixeira M, Grenier Capoci IR, Amaral A, Vilugron Rodrigues-Vendramini FA, Lauton Simões B, Rodrigues Abadio AK, Fernandes Matos L, Soares Felipe MS 2021. One century of study: What we learned about Paracoccidioides and how this pathogen contributed to advances in antifungal therapy. J. Fungi (Basel), 7, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroufi A, Chiller T, Jordan A, Denning DW, Harrison TS, Govender NP, Loyse A, Baptiste S, Rajasingham R, Boulware DR, Ribeiro I, Jarvis JN, Van Cutsem G 2021. Ending deaths from HIV-related cryptococcal meningitis by 2030. Lancet Infect. Dis 21, 16–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Yang M, Wegner SV, Zhao J, Zhu R, Wu Y, He C, Chen PR 2015. A genetically encoded FRET sensor for intracellular heme. ACS Chem. Biol 10, 1610–1615. [DOI] [PubMed] [Google Scholar]
- Stanford FA, Voigt K 2020. Iron assimilation during emerging infections caused by opportunistic fungi with emphasis on Mucorales and the development of antifungal resistance. Genes 11, 1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeny EA, Singh AB, Chakravarti R, Martinez-Guzman O, Saini A, Haque MM, Garee G, Dans PD, Hannibal L, Reddi AR, Stuehr DJ 2018. Glyceraldehyde-3-phosphate dehydrogenase is a chaperone that allocates labile heme in cells. J. Biol. Chem 293, 14557–14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Kamiya N, Nagamune T 2003. A novel protein-based heme sensor consisting of green fluorescent protein and apocytochrome b(562). Anal. Biochem 317, 116–119. [DOI] [PubMed] [Google Scholar]
- Terai K, Imanishi A, Li C, Matsuda M 2019. Two decades of genetically encoded biosensors based on Förster Resonance Energy Transfer. Cell Struct. Fun 44, 153–169. [DOI] [PubMed] [Google Scholar]
- Tolosano E, Fagoonee S, Morello N, Vinchi F, Fiorito V 2010. Heme scavenging and the other facets of hemopexin. Ant. Redox Signaling, 12, 305–320. [DOI] [PubMed] [Google Scholar]
- Tournu H, Luna-Tapia A, Peters BM, Palmer GE 2017. In Vivo indicators of cytoplasmic, vacuolar, and extracellular pH using pHluorin2 in Candida albicans. mSphere 2, e00276–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Leff J, Zid BM 2020. Post-transcriptional control of mitochondrial protein composition in changing environmental conditions. Biochem. Soc. Trans 48, 2565–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutucci E, Livingston NM, Singer RH, Wu B 2018. Imaging mRNA in vivo, from birth to death. Ann. Rev. Biophys 47, 85–106. [DOI] [PubMed] [Google Scholar]
- Vaknin Y, Shadkchan Y, Levdansky E, Morozov M, Romano J, Osherov N 2014. The three Aspergillus fumigatus CFEM-domain GPI-anchored proteins (CfmA-C) affect cell-wall stability but do not play a role in fungal virulence. Fun. Genet. Biol 63, 55–64. [DOI] [PubMed] [Google Scholar]
- Van Genechten W, Van Dijck P, Demuyser L 2021. Fluorescent toys ‘n’ tools lighting the way in fungal research. FEMS Microbiol. Rev 45, fuab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardi-Oknin D, Arava Y 2019. Characterization of factors involved in localized translation near mitochondria by ribosome-proximity labeling. Front. Cell Dev. Biol 7, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmeister E, Schipper K, Baumann S, Haag C, Pohlmann T, Stock J, Feldbrügge M 2012. Fungal development of the plant pathogen Ustilago maydis. FEMS Microbiol. Rev 36, 59–77. [DOI] [PubMed] [Google Scholar]
- Weissman Z, Kornitzer D 2004. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol 53, 1209–1220. [DOI] [PubMed] [Google Scholar]
- Weissman Z, Shemer R, Conibear E, Kornitzer D 2008. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol. Microbiol 69, 201–217. [DOI] [PubMed] [Google Scholar]
- Weissman Z, Pinsky M, Donegan RK, Reddi AR, Kornitzer D 2021. Using genetically encoded heme sensors to probe the mechanisms of heme uptake and homeostasis in Candida albicans. Cellular Microbiol 23, e13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS 2014. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346, 748–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Dong Y, Cheng X, Yu Q, Qian K, Mao J, Jia C, Ding X, Zhang B, Chen Y, Zhang B, Xing L, Li M 2014. Cellular iron homeostasis mediated by the Mrs4-Ccc1-Smf3 pathway is essential for mitochondrial function, morphogenesis and virulence in Candida albicans. Biochim. Biophys. Acta 1843, 629–639. [DOI] [PubMed] [Google Scholar]
- Xue Y, Schmollinger S, Attar N, Campos OA, Vogelauer M, Carey MF, Merchant SS, Kurdistani SK 2017. Endoplasmic reticulum-mitochondria junction is required for iron homeostasis. J. Biol. Chem 292, 13197–13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Guarente L 1995. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J 14, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Mehta S, Zhang J 2020. Genetically encodable fluorescent and bioluminescent biosensors light up signaling networks. Trends Biochem. Sci 45, 889–905. [DOI] [PMC free article] [PubMed] [Google Scholar]


