Abstract
mecA, the gene that mediates methicillin resistance, and its accompanying mec locus DNA, insert near the gyrA gene in Staphylococcus aureus. To investigate whether there is a similar relationship between mecA and gyrA in coagulase-negative staphylococci (CNS), mecA- and gyrA-specific DNA fragments were used to probe methicillin-resistant isolates of Staphylococcus epidermidis (MRSE) (n = 11) and Staphylococcus haemolyticus (MRSH) (n = 11). The gyrA probe hybridized to the same SmaI DNA fragment as the mecA probe in all strains tested. However, since the size of the SmaI fragments containing mecA and gyrA varied from 73 to 600 kb, the distance between the two genes was determined more precisely. Cloned mecA or gyrA fragments plus vector sequences each containing a SmaI site were introduced into the chromosome of three isolates each of MRSE and methicillin-resistant S. aureus (MRSA), and the sizes of the generated SmaI fragments were determined by pulsed-field gel electrophoresis. The distance between gyrA and mecA was found to be between 38 and 42 kb in both MRSE and MRSA, and the two genes were in the same relative orientation in all strains. Restriction fragment length polymorphism (RFLP) patterns around the gyrA gene in CNS were identical, but species specific, for all 10 MRSE and 10 MRSH isolates examined. In contrast, 8 of 11 methicillin-susceptible S. epidermidis isolates and 7 of 7 methicillin-susceptible S. haemolyticus isolates had different gyrA RFLP patterns. These data show that mecA is site and orientation specific, relative to gyrA, in both MRSE and MRSA. In addition, the local environment around gyrA in methicillin-resistant CNS, in contrast to methicillin-susceptible isolates, is similar, suggesting clonality or the requirement for specific DNA sequences with which the mec complex must interact for chromosomal integration to occur.
Penicillin-binding proteins (PBPs), enzymes that cross-link peptidoglycan in the bacterial cell wall, are the targets of β-lactam antibiotics. One of the ways in which staphylococci have become resistant to β-lactam antibiotics is through the acquisition of mecA, a gene which is found in the chromosome and which encodes a PBP (PBP2A) with reduced affinity for these antibiotics (12, 33). PBP2A catalyzes all required cell wall cross-linking functions when the β-lactam-susceptible PBPs are bound and inactivated by antibiotics (18). The mecA gene is contained within a larger fragment of DNA, typically 32 to 60 kb in size and of unknown origin, known as the mec locus or mec complex (7). mec-associated DNA is not found in methicillin-susceptible staphylococci and, therefore, is assumed to be exogenously acquired. Furthermore, the mecA genes and much of the mec complexes appear to be identical among all of the methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant coagulase-negative staphylococci (CNS) examined (4, 34). Even though the ultimate origin of the mec locus is unknown, several investigators have postulated that S. aureus acquired methicillin resistance from CNS. One piece of evidence supporting this view is that a single Staphylococcus haemolyticus-specific insertion sequence (IS1272) was found within the mec region of some isolates of S. aureus (5). Additionally, Wu and colleagues have identified a mecA homolog within Staphylococcus sciuri that has a predicted 80% amino acid identity with PBP2A (35). Interestingly, this mecA homolog does not confer resistance to methicillin on S. sciuri, suggesting that mecA may have evolved from a PBP that is not associated with β-lactam resistance.
Sequence analysis of S. aureus mec DNA found the integration of the mec complex to be site specific (14, 15). The insertion site (intM) mapped between spaA (protein A) and gyrA (the A subunit of DNA gyrase) on the SmaI DNA fragment G of the NCTC 8325 chromosome. However, the precise location of the mec complex on the chromosome of CNS has not been identified. Since the gyrA gene region is highly conserved among eubacteria (20), we investigated the relationship between gyrA and mecA among methicillin-resistant CNS and compared it to the same relationship in MRSA. Similar chromosomal locations for mec among both S. aureus and different CNS species would suggest a common mode of acquisition and insertion.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are shown in Table 1. We identified isolates as staphylococci by their Gram stain appearance and catalase production. Species identification was performed by coagulation of rabbit plasma (Difco, Detroit, Mich.) and with Staph-Ident strips (Analytab Products, Plainview, N.Y.). The staphylococci examined represent a diverse collection of clinical isolates with unique pulsed-field gel electrophoresis (PFGE) patterns collected from the United States and Canada over a period from the early 1970s to 1987.
TABLE 1.
Strains, plasmids, and probes used
| Strain, plasmid, or probe | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| MRSE | ||
| SE42NR | Boston, Mass., 1980; Novr Rifr Ems | This work |
| SE43NR | Canada, early 1970s; Novr Rifr Ems | This work |
| SE22NR | Richmond, Va., 1979; Novr Rifr Ems | This work |
| MRSA | ||
| 450M | 8325-4 transformed with mec region; Ems | 23 |
| 27216 | Boston, Mass., 1980; Ems | This work |
| Col | United Kingdom, 1961; Ems | 23 |
| MRSH Y176 | Richmond, Va., 1989 | 3 |
| S. aureus RN4220 | Restriction-deficient 8325-4; accepts E. coli DNA | 17 |
| Plasmids | ||
| pE194(Ts) | Temperature-sensitive gram-positive vector; confers Emr | 25 |
| pROJ6448(Ts) | pE194(Ts) with a 700-bp AluI fragment containing the nick site from pC221 cloned into the ClaI site | 25 |
| pC221 | Mobilizeable staphylococcal plasmid; confers Cmr | 24 |
| pGO1 | Conjugative staphylococcal plasmid; confers Gmr | 21 |
| pGO164 | 1.2-kb XbaI-BglII intragenic fragment from mecA cloned into pUC19 | This work |
| pGO567 | pGO164 with pROJ6448(Ts) cloned into the PstI site | This work |
| pGO592 | 7.5-kb SalI fragment containing gyrA (isolated from an SE43NR λ library) cloned into pUC19; clone contains gyrA (2.6 kb) plus 2.2 kb of downstream DNA and 2.7 kb of upstream DNA | This work |
| pGO597 | pGO592 with pROJ6448(Ts) cloned into the PstI site | This work |
| pGO557 | 735-bp PCR fragment located downstream from gyrA in S. aureus cloned into the HincII site of pUC19 | This work |
| pGO634 | pGO557 with pE194(Ts) cloned into the PstI site | This work |
| Probes | ||
| pGO533 | 450-bp PCR fragment from gyrA of SE43NR cloned into the SmaI site of pUC19 | This work |
| pGO546 | 450-bp PCR fragment from gyrA of Y176 cloned into the SmaI site of pUC19 | This work |
| mecI | 439-bp PCR fragment including all of mecI from SE42NR | This work |
| dnaA-dnaN | 723-bp PCR fragment including the 3′ end of dnaA and the 5′ end of dnaN from 450M | 1 |
| IS1272 | 1.2-kb EcoRI fragment isolated from pGO198 | 3 |
Nov, novobiocin; Rif, rifampin; Em, erythromycin; Cm, chloramphenicol; Gm, gentamicin.
PCR amplification and sequencing.
Two oligonucleotide primers complementary to the nucleotide sequence of the gyrA gene (29) of S. aureus were used to generate 450-bp PCR fragments from a methicillin-resistant Staphylococcus epidermidis (MRSE) (SE43NR) isolate and a methicillin-resistant S. haemolyticus (MRSH) (Y176) isolate for use as DNA probes. The two primers synthesized (Oligos Etc., Newtown, Conn.) were (5′)GGGTAAATATCAAAATCATCATGG(3′) and (5′)GCAGTTGGAAATCAGGACC(3′). The primers amplified the sequence encoding amino acids corresponding to nucleotides 76 through 221 of S. aureus gyrA. The PCR products were subsequently cloned into pUC19 (36). Sequence analysis was performed with the Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems Inc., Foster City, Calif.) on an Applied Biosystems Inc. 373A sequencer, and both DNA strands were sequenced.
Additional primers used and gene sequences amplified are as follows: (5′)GAAATGGAATTAATATAATG(3′) and (5′)GACTTGATTGTTTCCTC(3′) for the complete mecI gene; (5′)GGGCGTGATCATACGACCG(3′) and (5′)CCTGATGTAATTAATGTCTGG(3′) for sequences including the 3′ end of dnaA and the 5′ end of dnaN; and (5′)CCTAAGCGTAAAGAAGATTCAC(3′) and (5′)GATGGTGGCACCACATAC(3′) for the sequences downstream of gyrA in S. aureus.
Generation of λ library.
Unsheared genomic DNA was isolated from SE43NR by a variation of the Marmur technique as described previously (10). Genomic DNA was partially digested with Sau3A, and the fragments were separated by sucrose density gradient centrifugation. Fractions with DNA fragments from 9 to 23 kb long were isolated and subsequently ligated to Lambda DASH II (Stratagene, La Jolla, Calif.) digested with BamHI. Packaging of Lambda DASH II and infection of Escherichia coli XL-1 Blue (P2) (Stratagene) was performed according to the manufacturer’s recommendations. Plaques containing gyrA were isolated after transfer of the denatured phage DNA to nylon membranes (Boehringer Mannheim, Indianapolis, Ind.) and hybridization with gyrA-specific probes. DNA from the positive plaques was isolated and mapped by standard methods (27).
DNA isolation and manipulation.
Recombinant plasmids were generated in E. coli DH5α (23) with either pUC18 or pUC19 as the vector (36). The temperature-sensitive gram-positive plasmid pROJ6448(Ts) was subsequently ligated onto each clone (25). E. coli plasmid DNA was isolated by the boiling method of Holmes and Quigley (16). S. aureus and S. epidermidis plasmid DNA was prepared by the cetyltrimethylammonium bromide lysis method of Townsend et al. (32). Electroporation of recombinant plasmids containing pROJ6448(Ts) into S. aureus RN4220 was performed by the method of Schenk and Laddiga (28). Transduction was utilized to move recombinant plasmids between strains of S. aureus by using phage 80α according to previously described methods (8).
DNA hybridization.
Genomic DNA was extracted from staphylococcal isolates by a variation of the Marmur technique as previously described (10). Genomic digests were then transferred by alkaline capillary transfer to Zeta-Probe (Bio-Rad, Hercules, Calif.) nylon membranes ([α-32P]dCTP-labeled probes) or nylon membranes purchased from Boehringer Mannheim (digoxigenin-labeled probes) according to the method of Sambrook et al. (27). Southern hybridization was performed with DNA probes that were either labeled with 32P (Bethesda Research Laboratories, Inc., Gaithersburg, Md.) by nick translation or with digoxigenin-11-dUTP (Boehringer Mannheim) by random primed labeling.
PFGE.
Genomic DNA was prepared for PFGE by previously described methods (6). If large fragments were to be visualized, the following parameters were used: 6 V/cm; initial pulse time, 1 s; final pulse time, 30 s for 22 h at 14°C. If small fragments (<200 kb) were to be visualized, the parameters were changed to 6 V/cm; initial pulse time, 0.5 s; final pulse time, 12 s for 24 h at 14°C. PFGE size standards were purchased from New England Biolabs (Beverly, Mass.). Band sizes were estimated with an IS1000 digital imaging system (Alpha Innotech Corp., San Leandro, Calif.).
Conjugative mobilization and allelic replacement.
pROJ6448(Ts)-containing clones were electroporated into RN4220, which contained both pGO1 (conjugative; gentamicin resistant [Gmr]) and pC221 (mobilizeable; chloramphenicol resistant [Cmr]) to create the donor. pROJ6448(Ts) (erythromycin resistant [Emr]) contains the nick site from pC221 cloned into the unique ClaI site. Therefore pROJ6448(Ts)-containing plasmids can be mobilized by pGO1 if mobA and mobB from pC221 are supplied in trans, as previously described (25, 27). S. epidermidis isolates, chosen for their susceptibility to erythromycin, were selected for resistance to both rifampin and novobiocin by serial passage in the antibiotics. The mating procedure was performed by the syringe method as described previously (18). Transconjugants were selected on phenol red mannitol agar (or MRY agar) (Difco) supplemented with 1% yeast extract (Difco), 10 μg of rifampin/ml, 1 μg of novobiocin/ml, and 10 μg of erythromycin/ml. Phenol red mannitol agar was used to differentiate between S. epidermidis and S. aureus. S. aureus ferments mannitol, which creates a yellow colony, while S. epidermidis colonies remain red. Transconjugants were confirmed by restriction endonuclease analysis of plasmid DNA. Transfer frequencies were determined by dividing the number of transconjugants by the number of donor cells. Staphylococcal strains harboring plasmids containing pROJ6448(Ts) were cured by three rounds of growth at the nonpermissive temperature (43°C) in order to detect homologous recombination of the plasmid into either gyrA or mecA.
Construction of strains.
RN4220 donor strains containing pGO1, pC221, and pGO567 or pGO597 were mated into SE42NR, SE43NR, and SE22NR. Mobilization of recombinant plasmids into S. epidermidis strains occurred at a frequency of 10−8 per donor cell as previously described (31). The recombinant plasmids pGO567 and pGO634 were transferred from RN4220 into S. aureus strains by transduction with phage 80α. The presence of undeleted recombinant plasmids was confirmed by agarose gel electrophoresis of plasmid DNA. All appropriate strains were subsequently cured of pGO634, pGO597, or pGO567 and analyzed by PFGE to determine whether homologous recombination had occurred within the appropriate SmaI band (the band containing both gyrA and mecA). Homologous recombination of the recombinant plasmid into the appropriate gene was confirmed by PCR or Southern analysis. Strains containing pGO567, pGO634, or pGO597 within the appropriate gene were named by first listing the strain and then adding “/mecA::pGO567”, “/gyrA::pGO597”, or “/gyrA::pGO634”.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nucleotide sequences presented are AF005934 for the 450-bp fragment of the gyrA gene of S. epidermidis (SE43NR) and AF005935 for the 450-bp fragment of the gyrA gene of S. haemolyticus (Y176).
RESULTS
Nucleotide sequence comparisons of gyrA genes.
The nucleotide sequences of the gyrA genes obtained from S. epidermidis (SE43NR) and S. haemolyticus (Y176) compared to the sequence of S. aureus gyrA are presented in Fig. 1. The three nucleotide sequences are highly similar: the S. epidermidis and the S. haemolyticus gyrA nucleotide sequences each show 83% identity to the nucleotide sequence of gyrA of S. aureus. The sequence determined for S. epidermidis is identical, in its region of overlap, to that published by Sreedharan et al. (29) and extends the known sequence of S. epidermidis gyrA from nucleotides 1 to 642. The encoded proteins are also highly similar, with the S. haemolyticus GyrA showing 90% identity and the S. epidermidis GyrA showing 93% identity to the amino acid sequence of S. aureus GyrA.
FIG. 1.
Comparison of the gyrA DNA sequences from nucleotides 246 to 642 in S. aureus, S. epidermidis, (S. epi), and S. haemolyticus (S. haem). Nucleotides in S. epidermidis and S. haemolyticus which are dissimilar to those in S. aureus are shown in bold.
RFLPs among CNS.
All 22 S. epidermidis and 18 S. haemolyticus isolates were unique, as assessed from both differing geographic origins and unrelated SmaI fragment patterns. The results of Southern blot hybridization with either pGO533 (S. epidermidis) or pGO456 (S. haemolyticus) gyrA DNA probes are presented in Table 2. All 11 MRSE isolates examined had identical 12-kb ClaI, 10-kb EcoRI, and 4-kb HindIII fragments that hybridized with the gyrA probe. In contrast, there were several restriction fragment length polymorphisms (RFLPs) seen among methicillin-susceptible S. epidermidis (MSSE) isolates. Only 3 of 11 isolates had RFLPs identical to that seen among MRSE. Ten of the 11 MRSH isolates hybridizing with the gyrA probe had identical 2-kb ClaI fragments, while all 11 isolates hybridized with an approximately 20-kb EcoRI fragment and a 2-kb HindIII fragment. Again, in contrast to the methicillin-resistant isolates, there were several RFLPs seen among methicillin-susceptible S. haemolyticus (MSSH); none was the same as the MRSH pattern when all three restriction enzymes were compared. While MRSE and MRSH had identical RFLPs within each species, none of the RFLP patterns was the same between species.
TABLE 2.
Hybridization of CNS isolates with gyrA probes
| Isolate (no.) | Restriction enzyme fragment
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
ClaIa
|
EcoRI
|
HindIII
|
SmaIb
|
|||||
| Size (kb) | No. | Size (kb) | No. | Size (kb) | No. | Size (kb) | No. | |
| MRSE (11) | 12 | 11 | 10 | 11 | 4 | 11 | 600 | 4 |
| 280 | 1 | |||||||
| 130 | 1 | |||||||
| 115 | 2 | |||||||
| 100 | 2 | |||||||
| 92 | 1 | |||||||
| MSSE (11)c | 15 | 3 | 15 | 1 | 12 + 6 | 1 | ||
| 12 | 5 | 12 | 4 | 8 | 1 | |||
| 7 | 1 | 10 | 6 | 6 | 1 | |||
| 5 | 1 | |||||||
| 2.5 | 1 | 4 | 6 | |||||
| 2 | 1 | |||||||
| MRSH (11) | 2 | 10 | 20 | 11 | 2 | 11 | 218 | 1 |
| 194 | 1 | |||||||
| 0.5 | 1 | 177 | 1 | |||||
| 130 | 1 | |||||||
| 125 | 1 | |||||||
| 97 | 3 | |||||||
| 93 | 3 | |||||||
| MSSH (7)d | 6 | 2 | 20 | 4 | 2 | 5 | ||
| 5 | 2 | 20 + 9 | 1 | |||||
| 4.8 | 2 | 12 | 1 | 4 | 1 | |||
| 4 | 1 | 9 | 1 | |||||
MRSH and MSSH isolates were probed with pGO546 (gyrA). MRSE and MSSE isolates were probed with pGO533 (gyrA).
MRSH isolates were probed with both pGO164 (mecA) and pGO546. MRSE isolates were probed with both pGO164 and pGO533.
Only 10 of 11 MSSE isolates were digested with ClaI and probed with gyrA.
Only six of seven MSSH isolates were digested with HindIII and probed with gyrA.
Chromosomal location of gyrA in relation to mecA.
Both the gyrA probe and the mecA probe hybridized on the same SmaI fragment in each of 25 isolates tested (3 MRSA, 11 MRSH, and 11 MRSE isolates). The size of the SmaI fragment to which the gyrA and mecA probes hybridized ranged from 73 to 600 kb (Table 2).
Introduction of plasmids into CNS.
Even though DNA hybridization localized gyrA and mecA to the same SmaI fragment in all isolates examined, we could say nothing about the distance between the genes because of the variable sizes of the fragments (Table 2). We sought to estimate the intergenic mecA-gyrA distances in CNS by introducing new SmaI sites into or near each of these genes and measuring the sizes of the resulting fragments by PFGE. Three erythromycin-susceptible MRSE isolates (SE42NR, SE43NR, and SE22NR) were used as recipients for mobilizeable plasmids from S. aureus donors. Plasmids for chromosomal integration were created as follows. An intragenic 1.2-kb mecA fragment was ligated into pUC19 (pGO164), and the mobilization plasmid [pROJ6448(Ts)] was added to create pGO567 (Table 1). Similarly, a 7.5-kb fragment was obtained from a SE43NR λ library that contained the 3′ end of gyrA and downstream sequences. It was similarly ligated into pUC19, and the mobilization plasmid was added to produce pGO597 (Table 1). Each of these plasmids, containing a unique SmaI site, could be introduced into each of the three MRSE recipients at low frequency (5 to 10 colonies per plate) by conjugative mobilization. However, attempts to introduce these plasmids into two erythromycin-susceptible MRSH isolates were unsuccessful. Growing cells at the nonpermissive temperature for pROJ6448(Ts) replication allowed detection of cells containing mec and gyr sequences integrated into the chromosome by homologous recombination. Integration was confirmed by PFGE as shown in Fig. 2. The same mecA plasmid described above was used for three MRSA isolates (Col, 450M, and 27216), but a different gyrA vector (pGO634), containing a 735-bp PCR fragment 3′ to gyrA, was constructed by referring to published sequences in the database (29).
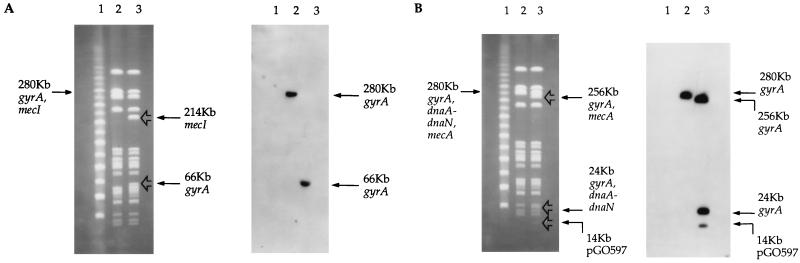
FIG. 2.
PFGE (left) and corresponding Southern blots (right) of SE43NR (A and B), SE43NR with a SmaI-containing plasmid integrated into mecA (SE43NR/mecA::pGO567) (A), and SE43NR with a SmaI-containing plasmid integrated near gyrA (SE43NR/gyrA::pGO597) (B). (A) Gel probed with gyrA PCR fragment. Lanes: 1, PFGE ladder; 2, SE43NR; 3, SE43NR/mecA::pGO567. The arrows indicate bands that hybridized with gyrA in both SE43NR (280-kb) and SE43NR/mecA::pGO567 (66 kb). A 214-kb fragment hybridized with the mecI probe in SE43NR/mecA::pGO567, while mecI hybridized to the 280-kb fragment in SE43NR (see Results) (data not shown). (B) Gel probed with the entire plasmid pGO592, which includes pUC19 as well as 7.5 kb of SE43NR sequences (gyrA plus 3′ and 5′ sequences). Lanes: 1, PFGE ladder; 2, SE43NR; 3, SE43NR/gyrA::pGO597. The arrows indicate bands that hybridized with pGO592 (gyrA) in both SE43NR (280 kb) and SE43NR/gyrA::pGO597 (256, 24, and 14 kb). Due to insertion and duplication of the entire 7.5-kb gyrA chromosomal fragment in the chromosome of SE43NR, both new SmaI fragments (256 and 24 kb) hybridized with pGO592. Additionally, a percentage of the population of SE43NR/gyrA::pGO597 contained uncured copies of pGO597 (pGO592 plus the pROJ6448 (Ts) staphylococcal replicon) that hybridized to pGO592 (14 kb). A probe for sequences 5′ to gyrA (dnaA-dnaN) hybridized with the 24-kb fragment in SE43NR/gyrA::pGO597, while mecA hybridized with the 256-kb fragment. Both mecA and dnaA-dnaN hybridized to the 280-kb fragment in SE43NR (see Results) (data not shown).
Determination of the distance between mecA and terminal SmaI sites.
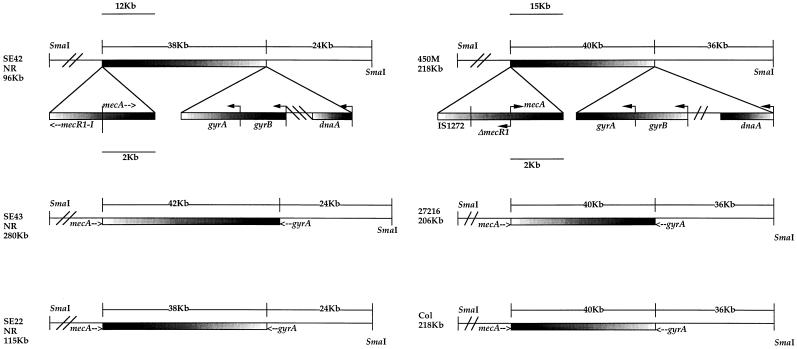
The distance between mecA and the terminal SmaI sites was estimated by comparison to a PFGE size standard. The wild-type SmaI fragments that contained both mecA and gyrA were 96 kb for SE42NR, 115 kb for SE22NR, and 280 kb for SE43NR. However, when a new SmaI site was introduced into mecA by homologous recombination, a band of similar size was found in each isolate after digestion with SmaI. This new SmaI band was estimated to be 62 kb in SE42NR, 66 kb in SE43NR, and 62 kb in SE22NR. When a new SmaI site was introduced into mecA in S. aureus 450M, Col, and 27216, the wild-type SmaI fragments that contained both mecA and gyrA (218 kb for 450M and Col and 206 kb for 27216) were cleaved into two bands by SmaI (142 and 76 kb for 450M and Col and 130 and 76 kb for 27216). Southern blot analysis with gyrA as a probe revealed that gyrA was present on the 62-kb SmaI fragment of SE42NR/mecA::pGO567 and SE22NR/mecA::pGO567, the 66-kb fragment of SE43NR/mecA::pGO567, and the 76-kb fragment of 450M/mecA::pGO567, Col/mecA::pGO567, and 27216/mecA::pGO567.
In order to orient mecA in relation to gyrA, probes for DNA 5′ to mecA were used. This was the mecI repressor gene for the three MRSE isolates and a copy of IS1272 for the three MRSA isolates. Since the new SmaI site was introduced on the 3′ end of mecA, if mecI or IS1272 hybridized with the same new SmaI fragment as gyrA, then gyrA was upstream of mecA. If mecI or IS1272 hybridized with the fragment opposite from the one hybridized by gyrA, then gyrA was downstream of mecA. In all isolates gyrA was found downstream (3′) of mecA (data not shown).
Determination of the distance between gyrA and terminal SmaI sites.
Integration of pGO597, containing the S. epidermidis gyrA gene and flanking sequences, into the chromosome of the three MRSE isolates introduced a new SmaI site that generated a fragment of 24 kb in each isolate. Integration of pGO634, containing the S. aureus gyrA intragenic fragment, into the three MRSA isolates generated a new 36-kb SmaI fragment. The chromosomal map distance between mecA and gyrA was calculated by subtracting the size of the small gyrA SmaI fragment (24 to 36 kb) from the size of the chromosomal SmaI fragment 3′ to mecA (62 to 76 kb), shown above to contain gyrA. As with mecA, the orientation of gyrA relative to mecA was determined by using a probe for a gene 5′ to gyrA (dnaA-dnaN), since the new SmaI site was introduced 3′ of gyrA. In each case, dnaA-dnaN hybridized to the small (24- to 36-kb) SmaI fragment while mecA hybridized to the larger fragment. This oriented the direction of transcription of the gyr operon away from the nearest natural SmaI site toward mecA. Southern blot analysis of both SE43NR/mecA::pGO567 and SE43NR/gyrA::pGO597 is shown in Fig. 2. Maps of the distances between the mecA and gyrA genes for the isolates studied are shown in Fig. 3.
FIG. 3.
Chromosomal map distance between mecA and gyrA in strains studied. The distances below the strain names represent the sizes of the wild-type SmaI fragments containing both mecA and gyrA. The distances between the two genes and the terminal SmaI sites, as indicated above the linear maps, was determined by the addition of new SmaI restriction sites at the 3′ end of mecA (nucleotide 1904 of 2,144) in both MRSE and MRSA, 2.2 kb downstream of gyrA in MRSE and 735 bp downstream of gyrA in MRSA. The triangular schematics below SE43NR and 450M represent the organization of the mec and gyr regions within the strains studied.
DISCUSSION
The amount of DNA within mec varies among MRSA isolates (9, 15, 30). The mec region was initially identified in isogenic S. aureus strains, one of which was susceptible while the other was the same strain made methicillin resistant by transformation with DNA from a clinical methicillin-resistant isolate (7). In those experiments, the methicillin-resistant transformant contained approximately 32 kb of DNA not found in the susceptible parent. Subsequent mapping and sequencing experiments performed by Hiramatsu et al. have identified clinical isolates with from 32 to 63 kb of DNA flanking mecA that were not found in methicillin-susceptible isolates (15). Mapping and sequence data from a number of investigators have shown features common to S. aureus mec regions (3, 4, 9, 30). First, 3′ to mecA is a region that varies in length, containing a variable number of 40-bp repeated sequences and an open reading frame always flanked by a copy of IS431 (26). Second, on the other side of IS431, farther downstream of mecA, investigators have found a number of integrated plasmids and resistance genes that are usually flanked by another copy of IS431 (30). Presumably, the terminus of mec DNA 3′ to mecA is close to the second copy of IS431 and contains sequences that are repeated in inverted order at the other terminus (14). Therefore, the DNA 3′ to mecA in most S. aureus isolates varies from 10 to 20 kb in length, depending upon the size of the IS431-flanked integrated elements, but has been reported to be as little as 5 or as much as 30 kb (15).
DNA 5′ to mecA is much more extensive. The two-gene regulatory operon, mecR1-mecI, is immediately 5′ to the mecA promoter-operator in most isolates, but in some isolates these sequences are partially deleted and replaced by a truncated copy of IS1272 (5). The DNA 5′ to mecI or IS1272 has been completely sequenced in one Japanese isolate, N315, and is said to contain an open reading frame with homology to a site-specific recombinase gene (15).
There is less known about mec DNA in CNS, but DNA hybridization and PCR amplification in both MRSE and MRSH suggest that the mec sequences 3′ to mecA are essentially the same (2, 4). In addition, the DNA at least 4 kb 5′ to mecA in MRSE is the same, as shown by hybridization, as that in MRSA (4). Other elements, such as Tn554 and pUB110, have been identified in the mec DNA of both MRSE (4, 34) and MRSA (9, 30) as well. The DNA 5′ to mecA in MRSH seems to be arranged somewhat differently, with only 20 bp of mecR1 remaining before the insertion of unidentified DNA (4). Thus, there is sufficient evidence to conclude that substantial portions of mec are homologous among these staphylococci and, therefore, that all three species acquired the DNA from a common source.
We found the chromosomal distances from the gyrA to the mecA genes to be very similar among all of the six staphylococci (three MRSA and three MRSE isolates) that we mapped, and therefore, we documented site specificity for mec insertion in two different staphylococcal species. It has been reported that there is a putative mec insertion site common to all S. aureus isolates and that the mec termini contain inverted repeats, suggesting that, in MRSA, mec is or has been a site-specific mobile element (14, 15). However, there are no target site duplications flanking mec in MRSA (15). The putative S. haemolyticus insertion site is reported to be different from that of S. aureus (14), and none has been reported for S. epidermidis.
One would expect that, since there is variability in the distances between mecA and the mec terminus 3′ to mecA, we would have found diversity in the distances between mecA and gyrA. However, in our isolates we found the distance between the two genes to be highly conserved. This may be due to the fact that we chose isolates that were susceptible to erythromycin and, thus, were less likely to be multiresistant. Therefore, these isolates may not contain integrated plasmids or resistance elements within 3′ mec DNA. On the basis of our data on the distance from gyrA to mecA in MRSE, the orientation of gyrA 3′ to mecA, and the reported lengths of mec DNA 3′ to mecA in MRSA, we can estimate the distance from gyrA to the 3′ mec terminus at 18 to 32 kb, but it could be as little as 8 or as much as 37 kb, depending on the amount of mec DNA 3′ to mecA. It should be relatively straightforward to locate the S. epidermidis mec insertion site in S. epidermidis genomic libraries or genome sequences. Since our data show that S. epidermidis and, probably, S. haemolyticus have the same relative chromosomal insertion site specificity as S. aureus, identification of specific insertion site sequences in different staphylococcal species may help define the mechanism of acquisition and insertion of mec.
Site-specific insertion of mec DNA in staphylococci may be due to the presence of target insertion sites for a phage or mobile element. However, our RFLP data show that there is a lack of polymorphism around the gyrA genes among MRSE and MRSH isolates, in contrast to extensive RFLP heterogeneity in these areas among methicillin-susceptible isolates. This apparent gyrA RFLP homogeneity in the context of extensive genome diversity, as determined by PFGE, suggests that mec may have entered the chromosome by homologous recombination, bringing a portion of the gyr locus or intervening DNA from a donor organism. The availability of genome sequences from a variety of related organisms may also help resolve this question.
Finally, it has been extremely difficult to introduce plasmid DNA into random clinical S. epidermidis isolates. Techniques such as electroporation, protoplast fusion, protoplast transformation, and nonspecific mobilization have been successful for only a few isolates (2, 11, 13, 19, 22). However, we have shown that conjugative mobilization can be used to introduce recombinant plasmids into virtually any S. epidermidis isolate and that allelic replacement mutagenesis can be successfully performed as a result. The only limitation that we faced was the requirement that the recipient be erythromycin susceptible. This requirement may have been the cause of our failure to introduce plasmids into S. haemolyticus, since we could find only two isolates in our extensive S. haemolyticus collection that were susceptible to erythromycin. However, this problem can be easily overcome by construction of mobilizeable plasmids containing a variety of resistance markers. Systems for conjugative mobilization of plasmids into CNS should expand studies on the genetic basis for pathogenesis in these species.
ACKNOWLEDGMENTS
These studies were supported in part by Public Health Service grant R37 AI35705 from the National Institute of Allergy and Infectious Diseases and by a grant from Bristol-Myers Squibb.
REFERENCES
- 1.Alonso J C, Fisher L M. Nucleotide sequence of the recF gene cluster from Staphylococcus aureus and complementation analysis in Bacillus subtilis recF mutants. Mol Gen Genet. 1995;246:680–686. doi: 10.1007/BF00290713. [DOI] [PubMed] [Google Scholar]
- 2.Archer, G. L. Unpublished data.
- 3.Archer G L, Niemeyer D M, Thanassi J A, Pucci M J. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archer G L, Niemeyer D M. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 1994;10:343–347. doi: 10.1016/0966-842x(94)90608-4. [DOI] [PubMed] [Google Scholar]
- 5.Archer G L, Thanassi J A, Niemeyer D M, Pucci M J. Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1996;40:924–929. doi: 10.1128/aac.40.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannerman T L, Hancock G A, Tenover F C, Miller J M. Pulsed-field gel electrophoresis as a replacement for phage typing of Staphylococcus aureus. J Clin Microbiol. 1995;33:551–555. doi: 10.1128/jcm.33.3.551-555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck W D, Berger-Bachi B, Kayser F H. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J Bacteriol. 1986;165:373–378. doi: 10.1128/jb.165.2.373-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Climo M W, Sharma V K, Archer G L. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J Bacteriol. 1996;178:4975–4983. doi: 10.1128/jb.178.16.4975-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubin D T, Chikramane S G, Inglis B, Matthews P R, Stewart P R. Physical mapping of the mec region of an Australian methicillin-resistant staphylococcus lineage and a closely related American strain. J Gen Microbiol. 1992;138:169–180. doi: 10.1099/00221287-138-1-169. [DOI] [PubMed] [Google Scholar]
- 10.Galetto D W, Johnston J L, Archer G L. Molecular epidemiology of trimethoprim resistance among coagulase-negative staphylococci. Antimicrob Agents Chemother. 1987;31:1683–1688. doi: 10.1128/aac.31.11.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gotz F, Schumacher B. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol Lett. 1987;40:285–288. [Google Scholar]
- 12.Hartman B J, Tomasz A. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heilman C, Gerke C, Perdreau-Remington F, Gotz F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect Immun. 1996;64:277–282. doi: 10.1128/iai.64.1.277-282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiramatsu K. Molecular evolution of MRSA. Microbiol Immunol. 1995;39:531–543. doi: 10.1111/j.1348-0421.1995.tb02239.x. [DOI] [PubMed] [Google Scholar]
- 15.Hiramatsu K, Kondo N, Ito T. Genetic basis for molecular epidemiology of MRSA. J Infect Chemother. 1996;2:117–129. doi: 10.3412/jsb.52.417. [DOI] [PubMed] [Google Scholar]
- 16.Holmes D S, Quigley M. A rapid method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 17.Kriesworth B N, Lofdahl S, Betley M J, O’Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 18.Labischinski H. Consequences of the interaction of β-lactam antibiotics with penicillin binding proteins from sensitive and resistant Staphylococcus aureus strains. Med Microbiol Immunol. 1992;181:241–265. doi: 10.1007/BF00198846. [DOI] [PubMed] [Google Scholar]
- 19.Mack D, Nedelmann M, Krokotsch A, Schwarzkopf A, Heesemann J, Laufs R. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect Immun. 1994;62:3244–3253. doi: 10.1128/iai.62.8.3244-3253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margerrison E E C, Hopewell R, Fisher L M. Nucleotide sequence of the Staphylococcus aureus gyrB-gyrA locus encoding the DNA gyrase A and B proteins. J Bacteriol. 1992;174:1596–1603. doi: 10.1128/jb.174.5.1596-1603.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morton T M, Eaton D M, Johnston L J, Archer G L. DNA sequence and units of transcription of the conjugative gene complex (trs) of Staphylococcus aureus plasmid pGO1. J Bacteriol. 1993;175:4436–4447. doi: 10.1128/jb.175.14.4436-4447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller E, Hübner J, Gutierrez N, Takeda S, Goldmann D A, Pier G B. Isolation and characterization of transposon mutants of Staphylococcus epidermidis deficient in capsular polysaccharide/adhesin and slime. Infect Immun. 1993;61:551–558. doi: 10.1128/iai.61.2.551-558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niemeyer D M, Pucci M J, Thanassi J A, Sharma V K, Archer G L. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J Bacteriol. 1996;178:5464–5471. doi: 10.1128/jb.178.18.5464-5471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Projan S J, Kornblum J, Moghazeh S L, Edelman I, Gennaro M L, Novick R P. Comparative sequence and functional analysis of pT181 and pC221 cognate plasmid replicons from Staphylococcus aureus. Mol Gen Genet. 1983;199:452–464. doi: 10.1007/BF00330758. [DOI] [PubMed] [Google Scholar]
- 25.Projan S J, Archer G L. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J Bacteriol. 1989;171:1841–1845. doi: 10.1128/jb.171.4.1841-1845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryffel C, Bucher R, Kayser F H, Berger-Bachi B. The Staphylococcus aureus mec determinant comprises an unusual cluster of direct repeats and codes for a gene product similar to Escherichia coli sn-glycerophosphoryl diester phosphodiesterase. J Bacteriol. 1991;173:7416–7422. doi: 10.1128/jb.173.23.7416-7422.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schenk S, Laddiga R A. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;94:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 29.Sreedharan S, Peterson L R, Fisher L M. Ciprofloxacin resistance in coagulase-positive and -negative staphylococci: role of mutations at serine 84 in the DNA gyrase A protein of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1991;35:2151–2154. doi: 10.1128/aac.35.10.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart P R, Dubin D T, Chikramane S G, Inglis B, Matthews P R, Poston S M. IS257 and small plasmid insertions in the mec region of the chromosome of Staphylococcus aureus. Plasmid. 1994;31:12–20. doi: 10.1006/plas.1994.1002. [DOI] [PubMed] [Google Scholar]
- 31.Thomas W D, Archer G L. Mobilization of recombinant plasmids from Staphylococcus aureus into coagulase-negative staphylococci species. Plasmid. 1992;27:164–168. doi: 10.1016/0147-619x(92)90017-5. [DOI] [PubMed] [Google Scholar]
- 32.Townsend S E, Ashdown N, Bolton S, Grubb W B. The use of cetyltrimethylammonium bromide for the rapid isolation from Staphylococcus aureus of relaxable and non-relaxable plasmid DNA suitable for in-vitro manipulation. Lett Appl Microbiol. 1985;1:87–94. [Google Scholar]
- 33.Ubukata K, Nonoguchi R, Matsuhashi M, Konno M. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J Bacteriol. 1989;171:2882–2885. doi: 10.1128/jb.171.5.2882-2885.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ubukata K, Nonoguchi R, Matsuhashi M, Song M D, Konno M. Restriction maps of the regions coding for methicillin and tobramycin resistances on chromosomal DNA in methicillin-resistant staphylococci. Antimicrob Agents Chemother. 1989;33:1624–1626. doi: 10.1128/aac.33.9.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S, Piscitelli C, de Lancastre H E, Tomasz A. Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb Drug Resist. 1996;4:435–441. doi: 10.1089/mdr.1996.2.435. [DOI] [PubMed] [Google Scholar]
- 36.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]