ABSTRACT
Although immune checkpoint inhibitor (ICI) therapy has dramatically improved outcome for metastatic melanoma patients, many patients do not benefit. Since adverse events may be severe, biomarkers for resistance would be valuable, especially in the adjuvant setting. We performed high-plex digital spatial profiling (DSP) using the NanoString GeoMx® on 53 pre-treatment specimens from ICI-treated metastatic melanoma cases. We interrogated 77 targets simultaneously in four molecular compartments defined by S100B for tumor, CD68 for macrophages, CD45 for leukocytes, and nonimmune stromal cells defined as regions negative for all three compartment markers but positive for SYTO 13. For DSP validation, we confirmed the results obtained for some immune markers, such as CD8, CD4, CD20, CD68, CD45, and PD-L1, by quantitative immunofluorescence (QIF). In the univariable analysis, 38 variables were associated with outcome, 14 of which remained significant after multivariable adjustment. Among them, CD95 was further validated using multiplex immunofluorescence in the Discovery immunotherapy (ITX) Cohort and an independent validation cohort with similar characteristics, showing an association between high levels of CD95 and shorter progression-free survival. We found that CD95 in stroma was associated with resistance to ICI. With further validation, this biomarker could have value to select patients that will not benefit from immunotherapy.
KEYWORDS: Biomarkers, DSP, immunotherapy, melanoma
Introduction
Immune checkpoint inhibitors (ICIs) have changed practice in treatment for advanced or metastatic melanoma, achieving 60% response with a combination of the anti-PD-1 (programmed cell death 1) and anti-CTLA4 (cytotoxic T-lymphocyte associated protein 4) drugs nivolumab and ipilimumab, respectively1. However, there is no broadly accepted method for the selection of patients who benefit from this therapy. In fact, many patients do not benefit, and some suffer adverse events2–5. In the advanced setting, these events can largely be managed, but the ICIs have also been approved in the adjuvant setting where many patients are cured by surgery alone. We suggest that biomarkers of resistance are especially important in the adjuvant setting to help clinicians decide which patients may be spared ICI therapy.
Towards the goal of finding biomarkers of resistance to ICIs, we used a new spatially informed discovery method. Digital Spatial Profiling (DSP) using the GeoMx® system (NanoString Technologies) allows quantitative spatially resolved measurements of multiple proteins on a single formalin-fixed, paraffin-embedded (FFPE) tissue section6. We previously published a study where we explored the predictive value of a 44-marker panel of immune markers in a cohort of immunotherapy-treated metastatic melanoma patients by DSP7. Here, we used an independent, newly collected cohort to validate the previous findings and to further characterize the tumor microenvironment with newly validated, larger antibody cocktails.
Materials and methods
Patient cohort and tissue microarray construction
Tissue specimens were prepared in a tissue microarray (TMA) format as previously described8. In this study, we evaluated pre-treatment specimens from two independent immunotherapy-treated cohorts collected from Yale Pathology archives: the Discovery ITX Cohort and the Validation ITX Cohort. The Validation ITX Cohort was previously described7,9, being patients with unresectable stage III-IV melanoma treated with anti-PD-1 alone or with anti-CTLA-4. The Discovery ITX Cohort was built in 2020 and includes pretreatment samples from 53 metastatic melanoma patients treated with immune checkpoint inhibitors (pembrolizumab, nivolumab, or ipilimumab plus nivolumab) from 2013 to 19. RECIST 1.1 was used to classify best overall response as complete response (CR), partial response (PR), stable disease (SD), or progressive disease (PD), and to determine progression-free survival (PFS)10. Representative tumor areas were obtained from formalin-fixed, paraffin-embedded (FFPE) specimens, and 0.6 mm cores were arrayed in a recipient block in TMA format. Three additional cores from placenta were inserted in the TMA as a control. For antibody validation, a Melanoma Index TMA was used, which consists of tumor tissue from 30 untreated melanoma patients from a historical cohort and control cores including placenta, tonsil, and melanoma cell lines (Yugen8, MEL624 WT, MEL624 B7-H1, MEL1335). Additionally, a non-ITX treated Historic Control Cohort of 131 untreated melanoma patients was used as the control group to be compared to the ITX Cohorts. The clinicopathological characteristics of the patients of the three cohorts obtained from clinical records and pathology reports are included in Supplementary Table S1. Tissues were collected with written-informed or waiver consent from patients under the approved Yale Human Investigation Committee protocol #9505008219 and conducted in accordance with the Declaration of Helsinki.
Digital spatial profiling
Our lab’s use of protein-based DSP has been previously described7,11. Briefly, FFPE tissue slides were deparaffinized and subjected to antigen retrieval procedures. Afterwards, we incubated them overnight with a cocktail of 77 unique photocleavable oligonucleotide-labeled primary antibodies (Supplementary Table S2). Additionally, tissue was incubated with primary antibodies against specific targets and conjugated with fluorophores to define the cellular compartments or Areas of Interest (AOI) on each core: S100B for “tumor”, CD68 for “macrophages”, and CD45 for “leukocytes”. SYTO 13 was used for counterstain. Once slides were loaded on the GeoMx® DSP instrument, a scanned fluorescent image was used to create adjusted compartments for each core or Region of Interest (ROI), with 650 µm of diameter. Next, each ROI was segmented in four molecular compartments based fluorescent signal: tumor (S100B+), macrophage (CD68+), leukocyte (CD45+), and nonimmune stromal cells (SYTO13+/S100B-/CD45-/CD68-) (Figure 1A-C). Figure 1D corresponds to a hematoxylin & eosin (H&E) staining of serial section, showing the same spot as in Figure 1A–C. Upon UV light exposure, oligos were released from S100B+, CD68+, CD45+, and “SYTO13+ only” regions, sequentially, for each ROI, and collected through microcapillary aspiration and transferred into microwell plates. Then, hybridization to 4-color, 6-spot optical barcodes and digital counting in the nCounter® System (NanoString Technologies) was conducted. Digital counts from barcodes corresponding to protein probes were first normalized with internal spike-in controls to account for system variation and then normalized to S6 and histone H3 housekeeping markers.
Figure 1.
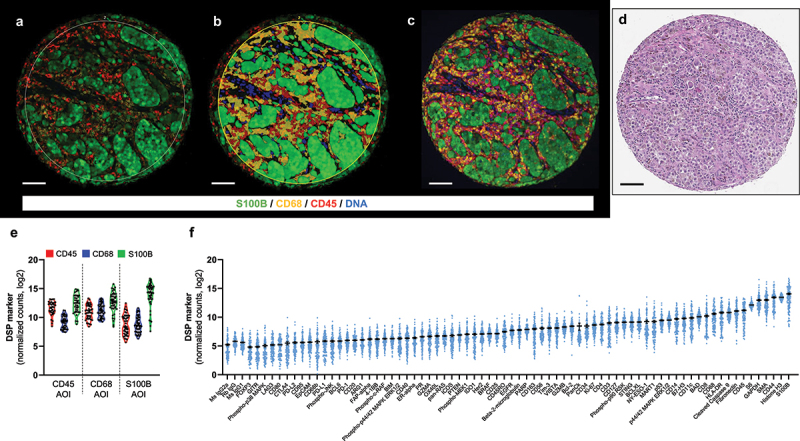
Compartments generated by DSP and an overview of target counts. representative TMA melanoma spot showing the fluorescence image (A) and the compartmentalized image created by molecular compartmentalization (fluorescence colocalization) (B) using GeoMx® DSP. C) Fluorescence image of same spot from serial section using AQUA platform. D) H&E staining of the same spot scanned on Aperio AT2 (Leica). E) Violin plot including DSP counts for CD45, CD68, and S100B on the corresponding compartments/AOIs. F) Mean DSP counts ± SEM in all compartments for each DSP marker. Scale bar, 100 µm.
Multiplex immune panels for DSP reproducibility evaluation
Two multiplex panels were performed on the Discovery ITX Cohort to validate DSP observations using an orthogonal method (QIF): panel A (CD8/CD20/CD4) and panel B (CD45/CD68/PD-L1). Panel A is a multiplexed immunofluorescence (IF) protocol for cytotoxic T CD8+, helper T CD4+, and B CD20+ cells that were previously published12. Briefly, tissue sections were subjected to deparaffinization, antigen retrieval, and blocking protocol mentioned above. The following antibody cocktail of primary target antibodies was incubated overnight: CD4 (rabbit IgG, clone SP35, Spring Bioscience), CD8 (mouse IgG1, clone C8/144B, Dako), CD20 (mouse IgG2a, clone L26, Dako). Sequential incubations of isotype-specific horseradish peroxidase (HRP)-conjugated secondary antibody with fluorescent reagents were performed: anti-rabbit Envision (K4009, Dako) with biotinylated tyramide/Streptavidin-AlexaFluor750 conjugate (Akoya), anti-mouse IgG1 antibody (ThermoFisher) with Cy3 plus-tyramide (Akoya), anti-mouse IgG2a antibody (Abcam) with Cy5-tyramide (Akoya). For panel B, which was adjusted from Liu et al.13, after conducting similar procedures as for panel A until primary antibody incubation, the antibody cocktail of primary antibodies used was as follows: PD-L1 (rabbit IgG, clone E1L3N, Cell Signaling Technology), CD68 (mouse IgG3, clone PG-M1, Dako), and CD45 (mouse IgG1, clone 2B11+PD7/26, Dako). HRP-conjugated secondary antibodies and fluorophores for this panel were as follows: anti-mouse IgG3 antibody (Abcam) with biotinylated tyramide/Streptavidin-AlexaFluor750 conjugate (Akoya), anti-rabbit Envision (K4009, Dako) with Cy5-tyramide (Akoya), and anti-mouse IgG1 Cy3 plus-tyramide (Akoya).
For both panels, incubations with 1 mM benzoic hydrazide and 0.15% hydrogen peroxide were performed to quench residual HRP activity. Finally, anti-S100B (Rabbit, clone 1706 R, Novus Biologicals) and goat anti-rabbit Alexa Fluor 488 (Invitrogen) were used to identify melanoma cells, counterstained with 4’,6-diamidino-2-phenylindole (DAPI) to visualize nuclei and mounted with ProLong Gold Antifade (Invitrogen), in both panels.
To validate the NanoString DSP technology, we performed QIF experiments using the AQUA method8,14 on the discovery cohort for well-characterized markers in the context of immunotherapy in melanoma using different immunohistochemistry approaches9,12,15–18. Both DSP and AQUA platforms create molecular compartments based on positive immunofluorescence signal with a region of interest to measure selected targets. Visually, for most of the cores, the compartments generated by both methods were comparable, as shown in Figure 1A–C. Regression of QIF scores and DSP counts for lymphoid markers in three compartments between the two assays for two independent blocks are included in Supplementary Fig. S2A. Whereas CD8 cytotoxic T cell marker (r = 0.48 (block 1)/0.54 (block 3); P < 0.01–0.0001) and CD4 helper T cell marker (r = 0.37/0.53; P < 0.05–0.001) showed high concordance between both methods when measured in near serial sections, there was a low degree of agreement between QIF and DSP platforms for CD20 B cell marker. These results support similar findings obtained by our group on an independent ITX-treated melanoma cohort9,12, as the cell frequency directly impacts on accurate measurements based on cell density. Moreover, CD8 but not CD4 nor CD20 was associated with better outcome by QIF (Supplementary Fig. S2B-D), validating previous analysis on an independent cohort of melanoma patients treated with immune checkpoint inhibitors from Yale (Validation ITX Cohort)7,9. Similarly, CD8 in CD68+CD45+SYTO13 compartment by DSP showed a similar trend as by QIF (HR: 0.86, 95% CI = 0.39–1.89) but did not reach statistical significance, whereas survival analysis for CD4 and CD20 by DSP was confirmed by QIF analysis. Counting CD68 in stroma compartment (r = 0.60 (block 1)/0.64 (block 3); P < 0.0001) had a higher concordance compared with tumor measurements (r = 0.36 (block 1)/0.46 (block 3); P < 0.01–0.001), which can be attributed to a prominent presence of macrophages in the stroma in comparison to the tumor (Supplementary Fig. S3A). Both PD-L1 and CD45 were highly correlated between QIF and DSP, regardless of the evaluated compartment (Supplementary Fig. S3A). In terms of outcome benefit, high levels of PD-L1 were associated with longer PFS compared with patients expressing low PD-L1 protein by QIF (Supplementary Fig. S3D). By DSP, high levels of PD-L1 showed longer PFS when measured in CD68+CD45 compartment (HR: 0.86, 95% CI = 0.41–1.80), but it was not statistically significant. As previously observed7,9, neither CD45 nor CD68 were associated with outcome (Supplementary Fig. S3B-C), which was correlated with DSP analyses.
CD95 antibody validation and multiplex CD95 panel
We tested three anti-CD95 monoclonal antibodies: C18C12 (CST), 13/Fas (BD Biosciences), EPR5700 (Abcam). First, we optimized the titer for all the clones using the standardization array Melanoma Index array at five concentrations covering two orders of magnitude in serial sections. A titration curve was plotted using the average scores of the highest 10% (“signal”) and lowest 10% (“noise”) of the patient tumor cores, including the signal-to-noise ratio (Supplementary Fig. S4H). The optimal antibody concentration selected was the one that had the highest dynamic range of signal with the highest signal-to-noise ratio. For antibody validation, a test cell array from a previous study19 was used, containing a variety of tumor cell lines.
Next, we performed a multiplexed IF protocol for CD95, CD68, and CD45 markers on discovery and validation ITX melanoma cohorts, using two-fold redundancy per cohort. Briefly, tissue sections were subjected to deparaffinization, antigen retrieval, and blocking protocol mentioned above12. Primary antibodies against CD45 (mouse IgG1, clone 2B11+PD7/26, Dako) and CD95 (rabbit, clone EPR5700, Abcam) were incubated overnight. Afterwards, two isotype-specific HRP-conjugated secondary antibodies and fluorescent reagents were incubated with the tissue: anti-rabbit Envision (K4009, Dako) with Cy5-tyramide (Akoya), and anti-mouse IgG1 antibody with Cy3 plus-tyramide (Akoya). A benzoic hydrazide solution was used to block endogenous HRP. Then, anti-CD68 (mouse IgG3, clone PG-M1, Dako) antibody incubation for 1 hour at room temperature was performed, followed by anti-mouse IgG3 antibody with biotinylated tyramide/Streptavidin-AlexaFluor750 incubation. Lastly, melanoma cells were detected with anti-S100B and goat anti-rabbit AlexaFluor488 antibodies. DAPI was used for counterstain.
Fluorescent measurement and scoring
Quantitative immunofluorescence (QIF) of each target was performed using the Automated Quantitative Analysis (AQUATM) software v3 (Navigate BioPharma Inc.) as previously described20. Briefly, the QIF scores were obtained by dividing the target compartment pixel intensities by the area of a specific compartment such as all cells (DAPI+), tumor (S100B+), stroma (DAPI+ nuclei minus tumor), macrophages (CD68+), or leukocytes (CD45+). Additionally, QIF scores were normalized by exposure time and bit depth at which the images were captured. Besides visual evaluation, cases with staining artifacts or less than 3% tumor were excluded from the analysis.
Hematoxylin & eosin (H&E) staining of the Discovery ITX Cohort
After removing paraffin and rehydration steps, the slide was stained with hematoxylin (Dako) for 5 minutes, followed by eosin Y for 60 seconds, and underwent sequential incubations on alcohol and xylene for dehydration. A brightfield image was digitized at 20× using the ScanScope AT2 platform (Leica Biosystems, Wetzlar, Germany).
Gene expression data and protein enrichment analyses
RNAseq data from public databases were used to analyze the expression of FAS in different tumors (TCGA dataset; www.cbioportal.com) and tumor cell lines (CCLE (Cancer Cell Line Encyclopedia) dataset, Broad Institute). In addition, the relationship between FAS and immune infiltrates in melanoma was studied by Tumor Immune Estimation Resource (TIMER 2.0, http://timer.cistrome.org/).
Statistical analysis
The software used for statistical purposes was GraphPadTM PrismⓇ v7.0 software for Windows (GraphPad Software, Inc., La Jolla, CA), JMP Pro software (version Pro 13, SAS Institute Inc., Cary, NC), and RStudio version 1.4.1717. Pearson’s correlation coefficient (r) was used to assess the agreement between two blocks for 71 targets analyzed by DSP in CD68+, CD45+, or S100B+ compartments or AOIs on Yale Melanoma Discovery ITX Cohort, as well as QIF scores and DSP counts from near serial sections for immune markers. Overall survival (OS) and PFS curves were constructed using Kaplan–Meier analysis, and statistical significance was determined using the log-rank test. Multivariable Cox proportional hazards models included age, sex, stage, ECOG status and specimen category as covariates21–23. For statistical analysis, the average AQUA scores from two available cores of each case were used, for both DSP and QIF experiments. All statistical tests were two-sided, and significance was represented as (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001.
Results
DSP standardization to QIF and validation
We generated 106 ROIs from 53 melanoma cases, each represented by two TMA cores in two master blocks of the Discovery ITX Cohort. ROIs were compartmentalized in four molecular compartments (tumor = S100B+; macrophages = CD68+; leukocytes = CD45+ without CD68+ cells as they were included in the macrophagic compartment; SYTO 13+/S100B-/CD68-/CD45- = nonimmune stromal cells, like fibroblasts) and 77 protein markers (including controls) were separately measured. It is important to mention that due to the heterogenicity of S100B levels melanoma cells and the fact that some melanomas are CD68+, the classification of tumor AOIs was based on not only S100B positivity but also morphology and previous QIF experiments using secondary antibody amplification system for S100B. For this analysis, we only selected the compartments with ≥20 nuclei for accurate target measurement in the tumor compartment (n = 50), in CD68 compartment (n = 43), in CD45 compartment (n = 37), and in nonimmune stromal cell compartment (n = 12), resulting in 142 AOIs. To analyze the effect of all cells in the stroma on treatment response, we created two aggregate stromal compartments for each target: one defined as the sum of DSP counts in leukocytes and macrophages, and a second one defined as the sum of DSP counts in leukocytes, macrophages and nonimmune stromal cells (CD68-/CD45-/SYTO13+), such as fibroblasts.
To evaluate the reproducibility of DSP and to determine the correlation of target count measurements between nonadjacent tumor areas, we compared counts of DSP markers in S100B, CD68, and CD45 compartments from each of the two independent cores from the 53 specimens, collected in separate DSP runs (Supplementary Fig. S1). For the tumor (S100B) compartment, almost all the markers analyzed in the tumor compartment, including immune targets, had high correlation with r > 0.4 and P < 0.05 for most of them (Supplementary Fig. S1C). This could be explained by the size of the tumor area that some immune cells infiltrated in the tumor were included in the tumor compartment due to either variability of S100B expression and limitations of fluorescent anti-S100B primary antibody to detect low levels of the protein or contamination from adjacent immune cells CD68+ and CD45+. For CD68+ compartment, markers related to monocytic lineage such as CD163, CD40, CD11c, and CD14, and other immune markers were highly correlated between blocks. In CD45+ compartment, CD8 (r = 0.91; P < 0.0001), Tim-3 (r = 0.89; P < 0.0001), CD45RO (r = 0.85; P < 0.0001), or GZMA (r = 0.83; P < 0.0001) were highly correlated between blocks, showing tissue heterogeneity might not be relevant for these markers in this cohort. Additionally, these markers were expressed at higher levels than the negative controls (Figure 1F). Then, we studied the DSP counts of the markers used to create the compartments by IF (S100B, CD68, CD45) in each respective compartment (Figure 1E), and the selected marker in the corresponding compartment was higher than the rest. There was a spillover of S100B counts in both CD68 and CD45 compartments, most likely due to segmentation issues related to 10 µm resolution of the UV light mirrors and higher abundance of this marker compared to CD68 or CD45.
Identifying new biomarkers for immunotherapy in melanoma
We assessed the association between the expression of 71 targets measured by DSP in four compartments and outcome in pretreatment specimens of 53 melanoma patients who received immunotherapy. Two nonadjacent TMA cores per each patient were collected in separate runs from two independent TMA master blocks.
In the log-rank and univariable unadjusted analysis using three exploratory cut points (median, top tertile, top quartile), we found 38 variables (a target in a compartment) associated with PFS and/or OS (Tables 1, 2). Figure 2 depicts DSP counts of representative markers in CD45, CD68 and S100B AOI and Kaplan–Meier curves for ERK 1/2, CD95 and HLA-DR. In the multivariable analysis, 14 variables remained statistically significantly associated with beneficial outcome or resistance to therapy (Tables 1, 2). In the tumor compartment, high levels of CD95 and S100B were associated with resistance to immunotherapy and with shorter PFS (HR: 2.98, P = 0.025; and HR: 3.29, P = 0.038, respectively) (Table 1). In contrast, high levels of Ki-67 statistically predicted longer PFS (HR: 0.033, P = 0.033). For the CD68 compartment, relevant immune markers, such as PD-1 (HR: 6.86, P = 0.001) and HLA-DR (HR: 0.09, P = 0.004) were significantly associated with resistance to therapy and poor survival, respectively. Only OX40L measured in the CD45 compartment was significantly associated with shorter PFS (HR: 6.51, P = 0.0025) and OS (HR: 5.83, P = 0.005) (Tables 1, 2). When the 71 targets were interrogated in the combined stromal compartment (CD45+/CD68+/SYTO13- or CD45+/CD68+/SYTO13+), none of them reached statistical significance after multivariable analysis, although additional markers were found relevant by log-rank test, like CD27, BIM, LAG3, and CD80. When we analyzed PD-L1 in our Discovery ITX Cohort, it was associated with better outcome when analyzed in the stroma by QIF (Supplementary Fig. S3D) but not by DSP, although the trend was similar (CD68 AOI by DSP = cutoff point: tertile; HR (95% IC): 0.54 (0.24–1.22)), as previously shown by our group on an independent ITX melanoma cohort, the Validation ITX Cohort7.
Table 1.
Markers significantly associated with PFS benefit under immune checkpoint inhibitors. HR = Hazard ratio; 95% CI = 95% confidence interval.
| Markers associated with PFS benefit | ||||||||
|---|---|---|---|---|---|---|---|---|
| Compartment | Marker | Cut-off point |
Log-rank HR (95% CI) |
P | Univariable HR (95% CI) |
P | Multivariable HR (95% CI) |
P |
| Tumor | CD95 | Median | 2.98 (1.46–6.09) | .0022 | 3.16 (1.46–6.86) | .0024 | 2.98 (1.11–8.01) | .025 |
| S100B | Median | 2.05 (1.01–4.17) | 0.044 | 2.12 (1.00–4.46) | 0.044 | 2.08 (.80–5.39) | 0.12 | |
| S100B | Top quartile | 2.19 (.91–5.27) | 0.031 | 2.24 (1.06–4.74) | 0.044 | 3.29 (1.04–1.37) | 0.038 | |
| Ki67 | Top tertile | .34 (.16–.70) | 0.018 | .33 (.13–.86) | 0.012 | .31 (.10–.99) | 0.033 | |
| ERa | Top tertile | 1.99 (.92–4.32) | 0.050 | 2.01 (.99–4.09) | 0.060 | 1.82 (.64–5.16) | 0.26 | |
| CD68 | PD-1 | Median | 2.62 (1.18–5.83) | .016 | 2.65 (1.17–6.01) | .017 | 2.93 (1.16–7.43) | .018 |
| PD-1 | Top tertile | 2.69 (1.10–6.62) | 0.0095 | 2.73 (1.24–6.03) | 0.015 | 5.09 (1.78–14.57) | 0.002 | |
| PD-1 | Top quartile | 2.98 (1.02–8.69) | 0.0054 | 4.52 (1.78–11.47) | 0.0024 | 6.86 (2.21–21.3) | 0.001 | |
| CD95 | Median | 2.79 (1.22–6.34) | 0.0069 | 3.00 (1.31–6.89) | 0.0086 | 1.96 (.76–5.08) | 0.15 | |
| CD95 | Top tertile | 2.59 (1.03–6.56) | 0.011 | 2.88 (1.24–6.71) | 0.017 | 2.84 (.92–8.81) | 0.065 | |
| CD95 | Top quartile | 3.64 (1.15–11.50) | 0.0006 | 4.52 (1.78–11.5) | 0.002 | 5.20 (1.46–18.5) | 0.011 | |
| EpCAM | Top quartile | 3.17 (1.13–8.93) | 0.0026 | 3.27 (1.45–7.37) | 0.007 | 2.99 (1.07–8.38) | 0.034 | |
| P-ERK1/2 | Top quartile | 2.23 (.84–5.94) | 0.046 | 2.28 (.99–5.22) | 0.063 | 2.06 (.73–5.81) | 0.2 | |
| BCLXL | Top tertile | .38 (.17–.85) | 0.045 | .38 (.14–1.02) | 0.036 | .33 (.10–1.06) | 0.044 | |
| ERK1/2 | Median | .44 (.20–.96) | 0.039 | .43 (.19–.98) | 0.040 | .39 (.16–1.00) | 0.046 | |
| Pan-CK | Top tertile | 2.77 (1.12–6.84) | 0.0054 | 3.25 (1.36–7.76) | 0.0083 | 2.42 (.86–6.86) | 0.096 | |
| CD45 | SMA | Median | 2.59 (1.09–6.14) | .026 | 2.74 (1.09–6.91) | .028 | 2.65 (0.75–9.41) | .12 |
| LAG3 | Median | 2.85 (1.17–6.94) | 0.012 | 3.08 (1.23–7.68) | 0.014 | 1.82 (.55–6.04) | 0.32 | |
| p53 | Median | .41 (.17–.96) | 0.040 | .38 (.15–.99) | 0.039 | .35 (.12–1.04) | 0.049 | |
| ERa | Median | 2.30 (.97–5.48) | 0.044 | 2.48 (1.00–6.17) | 0.048 | 2.11 (.62–7.22) | 0.21 | |
| OX40L | Top tertile | 2.68 (.95–7.55) | 0.018 | 2.82 (1.15–6.91) | 0.028 | 6.51 (1.92–22.07) | 0.0025 | |
| PARP | Top quartile | .27 (.10–.71) | 0.057 | .27 (.06–1.15) | 0.036 | .39 (.06–2.52) | 0.3 | |
| CD45+CD68 | BIM | Top tertile | 2.33 (0.90–6.08) | .048 | 1.59 (0.73–3.45) | .3 | 0.93 (0.38–2.31) | .9 |
| BIM | Top quartile | 2.44 (0.94–6.34) | .015 | 1.74 (0.77–3.92) | .4 | 0.90 (0.35–2.34) | .8 | |
| PD-1 | Median | 2.15 (1.00–4.60) | .036 | 1.77 (0.83–3.74) | .14 | 1.48 (0.66–3.33) | .3 | |
| CD27 | Top quartile | 2.14 (0.86–5.35) | .046 | 1.59 (0.72–3.53) | .3 | 1.54 (0.60–3.94) | .4 | |
| CD45+CD68+SYTO13 | CD20 | Top quartile | 2.22 (0.88–5.60) | .034 | 2.74 (1.09–6.91) | .3 | 0.94 (0.38–2.32) | .9 |
| S100B | Median | 2.62 (1.23–5.57) | .010 | 2.02 (0.94–4.33) | .066 | 2.58 (0.94–7.13) | .054 | |
| LAG3 | Top tertile | 2.09 (0.87–4.99) | .045 | 1.75 (0.80–3.79) | .2 | 1.55 (0.62–3.90) | .4 | |
| CD80 | Top tertile | 2.51 (1.04–6.04) | .011 | 0.38 (0.15–0.99) | .065 | 1.78 (0.77–4.14) | .2 | |
| S100B | Top tertile | 2.11 (0.91–4.89) | .044 | 2.48 (1.00–6.17) | .15 | 1.60 (0.64–4.00) | .3 | |
| P-ERK1/2 | Top tertile | 2.19 (0.94–5.12) | .033 | 2.82 (1.15–6.91) | .12 | 1.08 (0.46–2.53) | .9 | |
Bold values represent the Values equal or below 0.05.
Table 2.
Markers significantly associated with OS benefit under immune checkpoint inhibitors. HR = Hazard ratio; 95% CI = 95% confidence interval.
| Markers associated with OS benefit | ||||||||
|---|---|---|---|---|---|---|---|---|
| Compartment | Marker | Cut-off point |
Log-rank HR (95% CI) |
P | Univariable HR (95% CI) |
P | Multivariable HR (95% CI) |
P |
| CD68 | ERK1/2 | Median | 0.39 (0.15–0.98) | .043 | 0.36 (0.13–1.01) | .042 | 0.24 (0.07–0.77) | .012 |
| ERK1/2 | Top tertile | .32 (.12–.82) | 0.052 | .31 (.09–1.08) | 0.039 | .14 (.03–.74) | 0.0079 | |
| HLA-DR | Median | .26 (.10–.67) | 0.0048 | .22 (.07–.70) | 0.0045 | .09 (.01–.55) | 0.0035 | |
| HLA-DR | Top tertile | .30 (.12–.76) | 0.039 | .29 (.08–1.01) | 0.028 | .22 (.05–1.02) | 0.037 | |
| HLA-DR | Top quartile | .12 (.04–.30) | 0.010 | .12 (.02–.89) | 0.010 | .11 (.01–1.18) | 0.019 | |
| CD45 | PD-1 | Median | 2.95 (1.10–7.94) | .033 | 3.01 (1.04–8.73) | .034 | 1.93 (0.49–7.54) | .34 |
| LAG3 | Median | 3.00 (1.11–8.08) | 0.032 | 3.02 (1.05–8.73) | 0.032 | 3.02 (.65–13.94) | 0.14 | |
| MART1 | Median | 2.75 (1.03–7.38) | 0.047 | 2.82 (.97–8.19) | 0.047 | 1.77 (.50–6.28) | 0.37 | |
| OX40L | Top tertile | 3.07 (.98–9.59) | 0.016 | 3.28 1.18–9.11) | 0.025 | 5.83 (1.71–19.84) | 0.0046 | |
| BCL6 | Top quartile | 2.61 (.74–9.16) | 0.049 | 2.75 (.96–7.85) | 0.072 | 3.20 (.88–11.7) | 0.087 | |
| CD45+CD68 | BIM | Top quartile | 2.70 (0.96–7.61) | .020 | 1.99 (0.81–4.87) | .14 | 1.18 (0.40–3.53) | .8 |
Bold values represent the Values equal or below 0.05.
Figure 2.
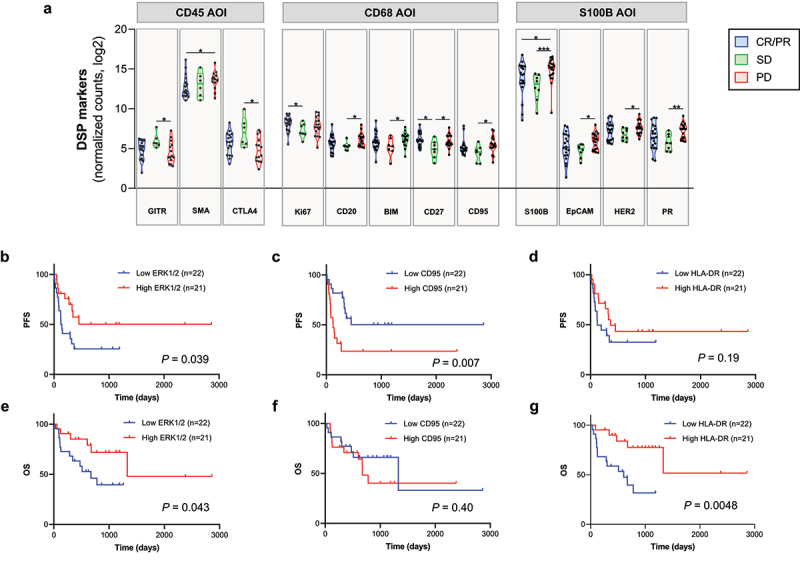
Candidate predictive markers to immunotherapy in the discovery cohort by DSP. A) Violin plot showing RECIST groups (CR/PR: complete response/partial response, SD: stable disease, PD: progression disease) for specific DSP markers in the corresponding compartments (CD45, CD68, S100B). *: P < 0.05; **: P < 0.01; ***: P < 0.001. Kaplan–Meier curves showing progression-free survival (PFS) (B–D) and overall survival (OS) (E–G) for DSP markers in CD68 compartment using the median as cutoff point. Survival analysis by log-rank (Mantel–Cox) test was performed and results were included in table 1 and 2.
CD95 as a candidate biomarker for resistance to immunotherapy
Based on the results obtained with DSP technology, the next step was to confirm the observations by an orthogonal method such as standard multiplexed immunofluorescence method first on the Discovery ITX Cohort and then on a second independent cohort, the Validation ITX Cohort. Among all the potential candidates that were associated with PFS benefit after multivariable analysis, we decided to pursue CD95 (also known as APO1 or Fas), which is a cell surface receptor member of the TNF-α (Tumor Necrosis Factor) family that, upon Fas Ligand (FasL) binding, induces extrinsic apoptosis24. We first validated our antibody as previously described25. Briefly, after finding the optimal concentration of the three monoclonal anti-CD95 antibodies commercially available (Supplementary Fig. S4H), we chose clone EPR5700 based on specific membranous signal (Supplementary Fig. S4B-G), comparison between the other antibodies (Supplementary Fig. S4J, K) and orthogonal validation by comparing protein and mRNA on multiple cancer cell lines (Supplementary Fig. S4I). After multiplex IF (Figure 3A–J), we observed that CD95 expression was higher in the tumor compartment than in the stroma compartment, and non-responder groups (“PD”) expressed statistically significantly higher amount of CD95 protein compared to the rest of RECIST groups (“CR/PR” and/or “SD”) in all compartments (Supplementary Fig. S5A). As observed by DSP, high levels of CD95 in CD68 compartment were associated with shorter PFS (Figure 3L). More importantly, when we explored CD95 in our independent Validation ITX Cohort, we confirmed that low levels of CD95 were associated with longer PFS (Figure 3M). Similar to the Discovery ITX Cohort, the levels of CD95 protein in the Validation ITX Cohort were slightly higher in the tumor compartment with respect to the stroma compartment, without differences between RECIST groups (Supplementary Fig. S5B). To determine if CD95 was indicative or associated with resistance to ICIs, we needed to determine if CD95 was a prognostic factor in metastatic melanoma-treated patients with standard of care therapy prior to immunotherapy. For that purpose, we applied the same multiplex QIF protocol on an immunotherapy non-treated cohort of melanoma patients from the Yale archives, named as Historic Control Cohort. Whereas CD95 was higher in tumor than in stroma as in the immunotherapy-treated cohorts (Supplementary Fig. S5C), we found opposite results regarding survival analysis: patients that expressed higher levels of CD95 in the stroma showed longer OS than patients expressing lower levels based (Figure 3N). Furthermore, we confirmed this result using RNA-seq data from TCGA (Figure 3O). FAS mRNA distribution in multiple cancer types is shown in Supplementary Fig. S5D. Interestingly, FAS mRNA expression was positively correlated with multiple immune infiltrates, such as neutrophils, T cell CD8+ cells, T cell CD4+ memory, B cells, monocytes, macrophages M1, NK cells, and T regulatory cells, and inversely correlated with T cell CD4+ T helper type 1, and T-like NK cell abundance (Supplementary Fig. S6).
Figure 3.
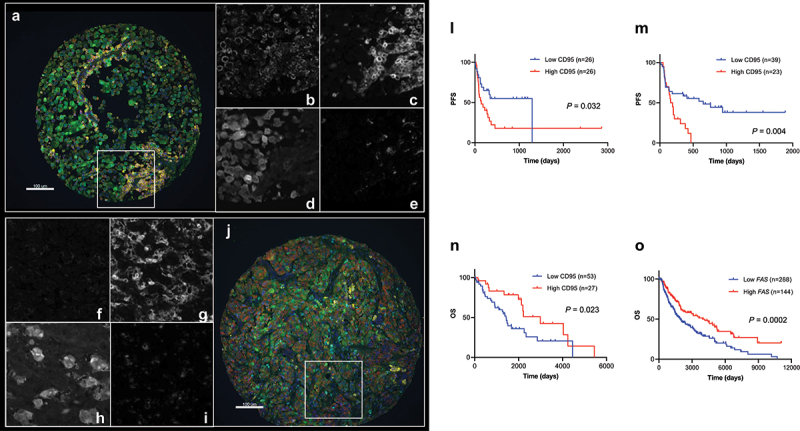
Validation of CD95 as an indicative immune marker for immunotherapy in melanoma identified by DSP. A, J) Two representative images of CD95 expression on melanoma tissue (CD95: red; S100B (melanocytes): green; CD45 (pan-leukocyte): yellow; CD68 (macrophages): cyan; nuclei: blue). (B–E) and (F–I) are regions from images A and J, respectively, showing CD95 (B, F), CD45 (C, G), S100B (D, H), and CD68 (E, I) in white. Kaplan–Meier survival curves showing: (L) PFS for CD95 protein quantified in CD68 compartment by QIF in discovery ITX cohort YTMA469 (cutoff point: median); (M) PFS for CD95 protein measured in stroma by QIF in validation ITX cohort YTMA376 (cutoff point: top tertile); (N) OS for CD95 protein quantified in stroma by QIF in non-ITX cohort YTMA192 (cutoff point: top tertile); (O) OS for FAS mRNA from TCGA dataset in non-ITX cohort (cutoff point: top tertile).
CD95 regulates the immunity at multiple levels, but it has also been described as a modulator of tumor activities24. When we divided our cohort in “high” and “low” p53 DSP counts using the median as a cutoff point, we observed statistically significant differences regarding CD95 DSP counts on S100B AOI (high p53: median = 30.25, 95% CI = 22.91–48.65; low p53: median = 15.72, 95% CI = 15.65–29.24; P = 0.020 by Mann–Whitney test) and CD45 AOI (high p53: median = 72.30, 95% CI = 56.71–128.8; low p53: median = 35.74, 95% CI = 35.38–74.58; P = 0.022 by Mann–Whitney test), confirming published results26.
Discussion
In this study, we used the novel GeoMx® DSP technology to identify novel candidate predictors of response or resistance to immunotherapy. Immune markers that are highly represented in the tissue, such as CD8, CD45 or CD68, were highly concordant between DSP and QIF, whereas CD20, which was present in the tissue at low proportion, showed poor correlations between methodologies. We found similar results previously when we compared Imaging Mass Cytometry (IMC) and DSP/QIF results for the same markers9.
PD-L1 is a controversial biomarker largely, since it is difficult to accurately measure. There are several studies comparing the performance of PD-L1 laboratory-developed test (LDT) assays as well as the four Food & Drug Administration (FDA)-approved PD-L1 immunohistochemistry assays linked to several therapies for multiple cancers, illustrating the necessity of harmonization between them at the expense of misclassifying patients for the corresponding treatment27–30. Moreover, whereas some studies showed association between PD-L1 expression and benefit upon ICI therapy, there are clearly responses in PD-L1 low or negative patients in both melanoma and lung cancer31,32. In our study, patients expressing high levels of PD-L1 in the stroma showed better outcome by QIF, confirming published data7.
In this study, we identified 38 variables associated with PFS and/or OS benefit in spatial context using GeoMx® DSP technology. On multivariable analysis, 14 of them remained statistically significant. One of the most relevant findings was the identification of high levels of CD95 in tumor and CD68 compartments as a predictor of resistance to immunotherapy in melanoma. Furthermore, this result was confirmed by an orthogonal method in the same cohort and validated by an orthogonal method in an independent ITX cohort (Figure 3, Supplementary Table S3). CD95 is ubiquitously expressed in normal tissues33, and it has a dynamic range of expression among melanoma and multiple tumors (Supplementary Fig. S5D). CD95 regulates extrinsic apoptosis upon binding its ligand CD95L. However, many pro-survival functions in tumor cells have been also described for this pathway, such as promoting metastasis, increasing cancer initiation and cancer stemness34,35. Furthermore, FAS mRNA expression has been associated with p53 mutational status in a melanoma cohort treated with ipilimumab26. In our study, we observed an association between p53 and CD95 protein levels by DSP. CD95 and its ligand are also expressed in immune cells. In macrophages, blocking CD95/CD95L interaction in vitro suppresses their activation, suggesting a role of this pathway in promoting chronic inflammation36, which may contribute to ICI resistance37,38. In our study, we observed an association between FAS expression and multiple immune populations in melanoma, such as cytotoxic CD8+ T cells and memory T cells in melanoma, using TIMER (Supplementary Fig. S6), supporting the relevance of CD95 role in immunotherapy response in our ITX cohorts.
By contrast, CD95 protein measured in CD68 compartment in the non-ITX cohort by QIF was a positive prognostic factor associated with OS benefit (Figure 3, Supplementary Table S3). Similar results were obtained for total FAS mRNA levels in an independent cohort from TCGA dataset. CD95 has been previously described as a prognostic factor in solid and hematologic cancers, both positively and negatively associated with disease progression33. During tumor progression, CD95 can be downregulated in order for the cancer cells to minimize the risk of undergoing apoptosis. However, high levels of CD95 (and CD95L) are associated with disease progression, suggesting that this receptor facilitates tumor growth and metastases.
Our study has a number of limitations. First, all the protein experiments were done on TMAs rather than whole tissue sections39. Despite its advantages as biomarker discovery tool, TMA is not used in clinical setting and is not the optimal format for immune cell evaluation. To compensate for this weakness, we used two nonadjacent TMA cores for each patient. Further, we argue that if a biomarker can be validated on the small area of a TMA spot, then it is likely to validate on whole tissue sections. Another limitation is the resolution of the GeoMx® DSP technology (10 µm) compared to well-characterized QIF technique using fluorescence microscopes (≤0.2 µm) and other high-plex methodologies40,41, which might account for reduced measurement accuracy in cells in close proximity in our study, such as S100B expressed in CD45 or CD68 compartments, and others7. Both AQUA and DSP use the molecular compartmentalization approach (instead of segmentation) for target measurement. This is considered a limitation by some investigators and we have shown that it can be a weakness when studying low-frequency events like rare immune cell populations12. Even though we validated our findings using a second independent cohort of pre-treatment specimens from ITX melanoma cohort, the analysis did not take into account the variety of immunotherapies that the patients received. Comparing additional samples from patients receiving different treatments provided by multiple institutions will be required to confirm our findings.
In conclusion, this study illustrates the potential of DSP technology to explore the tumor microenvironment, providing new predictive or indicative candidates for response and resistance to cancer therapy. Based on our findings, CD95 could be a potential biomarker for resistance to ICI blockade in melanoma, and with validation, could suggest alternative therapy or management for those patients likely to be refractory to immunotherapy.
Supplementary Material
Acknowledgments
This work was supported by the Yale SPORE in Skin Cancer P50 CA121974 (M. Bosenberg and H. Kluger, PIs) and by a sponsored research agreement from Navigate Biopharma (DLR). The authors also acknowledge the expert assistance of Lori Charette and the staff of the Yale Tissue Microarray Facility division of Yale Pathology Tissue Services for construction of the TMAs used in the study.
Funding Statement
This work was funded by a sponsored research agreement from Navigate Biopharma and in part by the Yale SPORE in Skin Cancer, P50 CA121974 (M. Bosenberg and H. Kluger, PIs).
Disclosure statement
SMM is a current employee at Boehringer Ingelheim Inc. PD is a current employee and stockholder at NanoString Technologies; PFW currently works at Verily Life Sciences, Alphabet Inc. (CA, USA); H.M. Kluger reports receiving commercial research grants from Merck, Bristol-Myers Squibb, and Apexigen, and is a consultant/advisory board member for Alexion, Corvus, Nektar, Biodesix, Genentech, Merck, Celldex, Pfizer, Iovance, Array Biopharma, Clinigen, Bristol-Myers Squibb, Instil Bio and Immunocore. D.L. Rimm declares that in the last two years he has served as a consultant to Astra Zeneca, Amgen, BMS, Cell Signaling Technology, Cepheid, Daiichi Sankyo, Danaher, GSK, Konica/Minolta, Merck, NanoString, Novartis, PAIGE.AI, Perkin Elmer/Akoya, and Roche/Ventana. No potential conflict of interest was reported by the author(s).
Authors’ contributions
SMM and DLR participated in the concept, design, and planning of the study. SMM and MM developed the methodology. SMM, SJ, PFW, and HMK constructed the discovery and validation ITX cohorts and conducted clinical data collection. SMM and MM acquired the data. SMM, AF, MM, PD, and RGM analyzed the data. SMM and DLR interpreted the results and drafted the manuscript. All authors critically reviewed, edited the manuscript, and approved the final draft for submission.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/2162402X.2023.2260618
References
- 1.Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, Linette GP, Meyer N, Giguere JK, Agarwala SS, et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med. 2015;372(21):2006–10. doi: 10.1056/NEJMoa1414428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti–PD-1 antibody in Cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob JJ, Cowey CL, Lao CD, Wagstaff J, Schadendorf D, Ferrucci PF, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377(14):1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber JS, D’Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, Hoeller C, Khushalani NI, Miller WH, Lao CD, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16(4):375–384. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- 5.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373(1):23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merritt CR, Ong GT, Church SE, Barker K, Danaher P, Geiss G, Hoang M, Jung J, Liang Y, McKay-Fleisch J, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol. 2020;38(5):586–599. doi: 10.1038/s41587-020-0472-9. [DOI] [PubMed] [Google Scholar]
- 7.Toki MI, Merritt CR, Wong PF, Smithy JW, Kluger HM, Syrigos KN, Ong GT, Warren SE, Beechem JM, Rimm DL.. High-plex predictive marker Discovery for melanoma immunotherapy–treated patients using Digital spatial profiling. Clin Cancer Res. 2019;25(18):5503–5512. doi: 10.1158/1078-0432.CCR-19-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camp RL, Charette LA, Rimm DL. Validation of tissue microarray technology in breast carcinoma. Lab Invest. 2000;80(12):1943–1949. doi: 10.1038/labinvest.3780204. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Morilla S, Villarroel-Espindola F, Wong PF, Toki MI, Aung TN, Pelekanou V, Bourke-Martin B, Schalper KA, Kluger HM, Rimm DL. Biomarker Discovery in patients with immunotherapy-treated melanoma with imaging mass cytometry. Clin Cancer Res. 2021;27(7):1987–1996. doi: 10.1158/1078-0432.CCR-20-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 11.Zugazagoitia J, Gupta S, Liu Y, Fuhrman K, Gettinger S, Herbst RS, Schalper KA, Rimm DL. Biomarkers associated with beneficial PD-1 checkpoint blockade in non–Small cell lung Cancer (NSCLC) identified using high-plex Digital spatial profiling. Clin Cancer Res. 2020;26(16):4360–4368. doi: 10.1158/1078-0432.CCR-20-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong PF, Wei W, Smithy JW, Acs B, Toki MI, Blenman KRM, Zelterman D, Kluger HM, Rimm DL. Multiplex Quantitative analysis of tumor-Infiltrating Lymphocytes and immunotherapy outcome in metastatic melanoma. Clin Cancer Res. 2019;25(8):2442–2449. doi: 10.1158/1078-0432.CCR-18-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Zugazagoitia J, Ahmed FS, Henick BS, Gettinger SN, Herbst RS, Schalper KA, Rimm DL. Immune cell PD-L1 colocalizes with macrophages and is associated with outcome in PD-1 pathway blockade therapy. Clin Cancer Res. 2020;26(4):970–977. doi: 10.1158/1078-0432.CCR-19-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumeister VM, Anagnostou V, Siddiqui S, England AM, Zarrella ER, Vassilakopoulou M, Parisi F, Kluger Y, Hicks DG, Rimm DL. Quantitative assessment of effect of preanalytic cold ischemic time on protein expression in breast cancer tissues. J Natl Cancer Inst. 2012;104(23):1815–1824. doi: 10.1093/jnci/djs438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards J, Wilmott JS, Madore J, Gide TN, Quek C, Tasker A, Ferguson A, Chen J, Hewavisenti R, Hersey P, et al. CD103+ tumor-resident CD8+ T cells are associated with improved survival in immunotherapy-naïve melanoma patients and expand significantly during anti–PD-1 treatment. Clin Cancer Res. 2018;24(13):3036–3045. doi: 10.1158/1078-0432.CCR-17-2257. [DOI] [PubMed] [Google Scholar]
- 17.Gide TN, Silva IP, Quek C, Ahmed T, Menzies AM, Carlino MS, Saw RPM, Thompson JF, Batten M, Long GV, et al. Close proximity of immune and tumor cells underlies response to anti-PD-1 based therapies in metastatic melanoma patients. Oncoimmunology. 2020;9(1):1659093. doi: 10.1080/2162402X.2019.1659093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halse H, Colebatch AJ, Petrone P, Henderson MA, Mills JK, Snow H, Westwood JA, Sandhu S, Raleigh JM, Behren A, et al. Multiplex immunohistochemistry accurately defines the immune context of metastatic melanoma. Scie Rep. 2018;8(1):11158. doi: 10.1038/s41598-018-28944-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toki MI, Cecchi F, Hembrough T, Syrigos KN, Rimm DL. Proof of the quantitative potential of immunofluorescence by mass spectrometry. Lab Invest. 2017;97(3):329–334. doi: 10.1038/labinvest.2016.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8(11):1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 21.Eton O, Legha SS, Moon TE, Buzaid AC, Papadopoulos NE, Plager C, Burgess AM, Bedikian AY, Ring S, Dong Q, et al. Prognostic factors for survival of patients treated systemically for disseminated melanoma. J Clin Oncol. 1998;16(3):1103–1111. doi: 10.1200/JCO.1998.16.3.1103. [DOI] [PubMed] [Google Scholar]
- 22.Manola J, Atkins M, Ibrahim J, Kirkwood J. Prognostic factors in metastatic melanoma: a pooled analysis of Eastern Cooperative Oncology group trials. J Clin Oncol. 2000;18(22):3782–3793. doi: 10.1200/JCO.2000.18.22.3782. [DOI] [PubMed] [Google Scholar]
- 23.Joosse A, Collette S, Suciu S, Nijsten T, Patel PM, Keilholz U, Eggermont AMM, Coebergh JWW, de Vries E. Sex is an independent prognostic indicator for survival and relapse/progression-free survival in metastasized stage III to IV melanoma: a pooled analysis of five European organisation for research and treatment of cancer randomized controlled trials. J Clin Oncol. 2013;31(18):2337–2346. doi: 10.1200/JCO.2012.44.5031. [DOI] [PubMed] [Google Scholar]
- 24.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116(2):205–219. doi: 10.1016/S0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 25.MacNeil T, Vathiotis IA, Martinez-Morilla S, Yaghoobi V, Zugazagoitia J, Liu Y, Rimm DL. Antibody validation for protein expression on tissue slides: a protocol for immunohistochemistry. Biotechniques. 2020;69(6):460–468. doi: 10.2144/btn-2020-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao W, Du N, Huang T, Guo J, Mo X, Yuan T, Chen Y, Ye T, Xu C, Wang W, et al. TP53 mutation as potential negative predictor for response of anti-CTLA-4 therapy in metastatic melanoma. EBioMedicine. 2018;32:119–124. doi: 10.1016/j.ebiom.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Morilla S, McGuire J, Gaule P, Moore L, Acs B, Cougot D, Gown AM, Yaziji H, Wang W-L, Cartun RW, et al. Quantitative assessment of PD-L1 as an analyte in immunohistochemistry diagnostic assays using a standardized cell line tissue microarray. Lab Invest. 2019;100(1):4–15. doi: 10.1038/s41374-019-0295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sompuram SR, Torlakovic EE, ‘t Hart, THart NA K, Bogen SA, ‘t Hart NA. Quantitative comparison of PD-L1 IHC assays against NIST standard reference material 1934. Mod Pathol. 2021;34(12):2242. doi: 10.1038/s41379-021-00898-4. [DOI] [PubMed] [Google Scholar]
- 29.Rimm DL, Han G, Taube JM, Yi ES, Bridge JA, Flieder DB, Homer R, West WW, Wu H, Roden AC, et al. A prospective, multi-institutional, pathologist-based assessment of 4 immunohistochemistry assays for PD-L1 expression in non–small cell lung Cancer. JAMA Oncol. 2017;3(8):1051–1058. doi: 10.1001/jamaoncol.2017.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirsch FR, McElhinny A, Stanforth D, Ranger-Moore J, Jansson M, Kulangara K, Richardson W, Towne P, Hanks D, Vennapusa B, et al. PD-L1 immunohistochemistry assays for lung Cancer: results from phase 1 of the blueprint PD-L1 IHC assay comparison project. J Thorac Oncol. 2017;12(2):208–222. doi: 10.1016/j.jtho.2016.11.2228. [DOI] [PubMed] [Google Scholar]
- 31.Daud AI, Wolchok JD, Robert C, Hwu WJ, Weber JS, Ribas A, Hodi FS, Joshua AM, Kefford R, Hersey P, et al. Programmed death-ligand 1 expression and response to the anti–programmed death 1 antibody pembrolizumab in melanoma. J Clin Oncol. 2016;34(34):4102–4109. doi: 10.1200/JCO.2016.67.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doroshow DB, Sanmamed MF, Hastings K, Politi K, Rimm DL, Chen L, Melero I, Schalper KA, Herbst RS. Immunotherapy in non–small cell lung Cancer: facts and hopes. Clin Cancer Res. 2019;25(15):4592–4602. doi: 10.1158/1078-0432.CCR-18-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peter ME, Hadji A, Murmann AE, Brockway S, Putzbach W, Pattanayak A, Ceppi P. The role of CD95 and CD95 ligand in cancer. Cell Death Differ. 2015;22(4):549–559. doi: 10.1038/cdd.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards DM, Merz C, Gieffers C, Krendyukov A. CD95L and anti-tumor immune response: Current understanding and New evidence. Cancer Manag Res. 2021;13:2477–2482. doi: 10.2147/CMAR.S297499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teodorczyk M, Kleber S, Wollny D, Sefrin JP, Aykut B, Mateos A, Herhaus P, Sancho-Martinez I, Hill O, Gieffers C, et al. CD95 promotes metastatic spread via Sck in pancreatic ductal adenocarcinoma. Cell Death Differ. 2015;22(7):1192–1202. doi: 10.1038/cdd.2014.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma Y, Liu H, Tu-Rapp H, Thiesen HJ, Ibrahim SM, Cole SM, Pope RM. Fas ligation on macrophages enhances IL-1R1–toll-like receptor 4 signaling and promotes chronic inflammation. Nat Immunol. 2004;5(4):380–387. doi: 10.1038/ni1054. [DOI] [PubMed] [Google Scholar]
- 37.Wang D, DuBois RN. Immunosuppression associated with chronic inflammation in the tumor microenvironment. Carcinogenesis. 2015;36(10):1085–1093. doi: 10.1093/carcin/bgv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang X, Wang J, Lu D, Xu X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Signal Transduct Target Ther. 2021;6(1):75. doi: 10.1038/s41392-021-00484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogino S, Galon J, Fuchs CS, Dranoff G. Cancer immunology–analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8:711–719. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schüffler PJ, Grolimund D, Buhmann JM, Brandt S, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 41.Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20(4):436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


