Abstract
Background
Medulloblastoma is the most common malignant pediatric brain tumor, and leptomeningeal dissemination (LMD) of medulloblastoma both portends a poorer prognosis at diagnosis and is incurable at recurrence. The biological mechanisms underlying LMD are unclear. The Abelson (ABL) tyrosine kinase family members, ABL1 and ABL2, have been implicated in cancer cell migration, invasion, adhesion, metastasis, and chemotherapy resistance, and are upstream mediators of the oncogene c-MYC in fibroblasts and lung cancer cells. However, their role in medulloblastoma has not yet been explored. The purpose of this work was to elucidate the role of ABL1/2 in medulloblastoma LMD.
Methods
ABL1 and ABL2 mRNA expression of patient specimens was analyzed. shRNA knockdowns of ABL1/2 and pharmacologic inhibition of ABL1/2 were used for in vitro and in vivo analyses of medulloblastoma LMD. RNA sequencing of ABL1/2 genetic knockdown versus scrambled control medulloblastoma was completed.
Results
ABL1/2 mRNA is highly expressed in human medulloblastoma and pharmacologic inhibition of ABL kinases resulted in cytotoxicity. Knockdown of ABL1/2 resulted in decreased adhesion of medulloblastoma cells to the extracellular matrix protein, vitronectin (P = .0013), and significantly decreased tumor burden in a mouse model of medulloblastoma LMD with improved overall survival (P = .0044). Furthermore, both pharmacologic inhibition of ABL1/2 and ABL1/2 knockdown resulted in decreased expression of c-MYC, identifying a putative signaling pathway, and genes/pathways related to oncogenesis and neurodevelopment were differentially expressed between ABL1/2 knockdown and control medulloblastoma cells.
Conclusions
ABL1 and ABL2 have potential roles in medulloblastoma LMD upstream of c-MYC expression.
Keywords: ABL1, ABL2, c-MYC, medulloblastoma, metastases
Key Points.
ABL1 and ABL2 are highly expressed in all subgroups of medulloblastoma.
Pharmacological and genetic knockdown of ABL1/2 results in decreased c-MYC expression.
ABL1/2 genetic knockdown decreased leptomeningeal dissemination and increased survival in mice.
Importance of the Study.
We show for the first time that ABL1 and ABL2 mRNA expression is associated with medulloblastoma dissemination and poor overall survival. We found that pharmacologic inhibition of ABL1 and ABL2 decreased medulloblastoma viability as well as expression of c-MYC in a time- and dose-dependent manner. Genetic knockdown of ABL1/2 resulted in decreased c-MYC expression, decreased adhesion to the extracellular matrix protein vitronectin, and decreased tumor burden (resulting in increased survival) in a rodent model of medulloblastoma leptomeningeal dissemination. Furthermore, mRNA sequencing of ABL1/2 knockdown medulloblastoma cells showed enrichment in the hallmark pathway EPITHELIAL_MESENCHYMAL_TRANSITION, further evidence of the importance of ABL1 and ABL2 as putative drivers of medulloblastoma dissemination. ABL1 and ABL2 are putative therapeutic targets for the prevention and treatment of medulloblastoma dissemination.
Brain cancer has now replaced leukemia as both the most common pediatric cancer and the leading cause of pediatric cancer-related deaths in the United States.1 Medulloblastoma is the most common pediatric brain cancer and is characterized by 4 distinct molecular subgroups: WNT, SHH, group 3, and group 4.2 In certain subgroups such as group 3 and group 4, up to 40% of patients have leptomeningeal dissemination (LMD) at diagnosis. The presence of LMD at diagnosis is an independent risk factor for poor overall survival (OS).2 The drivers of LMD are poorly understood, and the lack of actionable targets is a major barrier to the development of effective therapies in treating this disease.
The Abelson (ABL) family of tyrosine kinases, including ABL1 and ABL2, are novel targets of interest in uncovering the biology of medulloblastoma LMD. In 1982, ABL1 was recognized as a key player in the oncogenic fusion protein, BCR-ABL1, which drives most cases of chronic myeloid leukemia (CML).3 The broader family of ABL tyrosine kinases has since been implicated in a variety of cellular processes relevant to cancer (eg, proliferation, motility, adhesion, and polarity).3,4 The ABL kinase inhibitor imatinib (Gleevec) revolutionized treatment in CML, and newer generations of ABL kinase inhibitors have displayed activity against other forms of leukemia and solid tumors.5,6
To the best of our knowledge, the role of ABL tyrosine kinases in medulloblastoma and/or LMD prior to this study was unknown, outside of a few studies observing ABL1 expression in medulloblastoma.7,8 Through this work, we report that ABL1 and ABL2 mRNA are highly expressed in medulloblastoma, that pharmacologic inhibition of ABL kinases decreases medulloblastoma cell viability, that ABL kinases help regulate protein expression of the oncogenic transcription factor c-MYC, that ABL1/2 contribute to leptomeningeal tumor burden and survival in a mouse model of medulloblastoma and adhesion to the extracellular matrix protein, vitronectin, and that expression of genes related to neurodevelopment and oncogenesis differs significantly between ABL1/2-depleted and control cells. Additionally, we describe RNA sequencing analysis of ABL1/2-genetically depleted cells demonstrating enrichment in the epithelial-mesenchymal transition (EMT) pathway, further reinforcing the importance of ABL1 and ABL2 in the oncogenesis of medulloblastoma. Together, these data provide insight into the roles of ABL1 and ABL2 as putative drivers of medulloblastoma LMD.
Materials and Methods
mRNA Analysis of Patient Specimens
mRNA expression of ABL1 and ABL2 was analyzed using publicly available datasets9 as previously described.10,11
Cell Culture
The human medulloblastoma cell lines D458 and D556 were obtained from the laboratory of Dr. Darell Bigner at the Preston Robert Tisch Brain Tumor Center (Durham, NC), and D283 was purchased from the American Type Culture Collection (Manassas, VA). All of these cell lines have amplification of MYC.12 D283 was derived from peritoneal metastases from a ventriculoperitoneal shunt and contains an extra copy of chromosome 11.13 D556 has a 1q gain and isochromosome 17q.14 All are P53 wild type.15 Together, all likely cluster within Group 3.10
Assessment of Cell Viability/Proliferation
Cells were plated in 96-well plates at a seeding density of 25 000 cells/well and incubated for 24 h at 37°C. ABL kinase inhibitors or cisplatin were added at various concentrations to all wells and incubated for 18 hours. Dosing and treatment duration was selected from previous publications of asciminib, GNF5, and nilotinib in solid tumors.16,17 CellTiter-Glo assays were performed according to the manufacturer’s protocol (Promega) and in at least duplicate. All final concentrations of DMSO (the solvent for all ABL kinase inhibitors) were < 0.5%.
Western Blotting
Cells were seeded in 6-well plates at a density of 1.0 × 106 cells/well in 4 mL appropriate culture medium approximately 24 hours prior to treatment and protein harvest. Drugs were added at the proper concentration either directly or by resuspension in a small amount of fresh culture medium (~200 μL) to appropriate wells. After 1, 6, 24, or 48 hours, cell pellets were harvested and resuspended in 1 mL ice-cold PBS (pretreated with 1X sodium orthovanadate at 1 mM). Cells were lysed in Cell Lysis Buffer (Cell Signaling Technology) with protease/phosphatase inhibitor cocktail, with 1X PMSF (1 mM), and frozen down overnight at −80° C. Cell debris were then removed by microcentrifugation, and protein supernatant was extracted and resuspended in a 3:4 ratio with NuPAGE LDS Sample Buffer.
Protein assays were performed using either the EZQ Protein Quantitation Kit (Invitrogen) or reagents for the Bradford protein assay. Alternatively, a REVERT Total Protein Stain was used to normalize loading volumes. Equal amounts of protein were loaded into NuPAGE gels and separated by SDS-PAGE electrophoresis, transferred onto nitrocellulose membranes and blocked in SuperBlock Blocking Buffer (Thermo Fisher), and probed with primary antibodies. Antibodies included c-ABL (ABL1) (BD Pharmingen #554148), ABL2 (Abcam #134134), c-MYC (Cell Signaling #5605), and GAPDH (Cell Signaling #2118). Membranes were then probed with the appropriate secondary antibody, saturated with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher), and imaged on an Odyssey Fc imaging machine (LI-COR Biosciences). Membranes were washed with ~5 mL TBST 3× for at least 10 min between all steps. Blot images were analyzed in Image Studio Lite software. Western blots were performed in at least duplicate.
Vitronectin Cell Binding Assay
A total of 15 000 cells in 100 uL were pipetted into each well of a Greiner CELLSTAR 96-well plate (655180). The plate was coated beforehand at a concentration of 0.5 µg/cm2 using an appropriate dilution of vitronectin solution (Advanced Biomatrix #5051) in PBS that was left to set at room temperature for 1 hour before being rinsed off with distilled water. The cells were kept overnight on a rocker at room temperature until they were fixed and stained with the Richard-Allan Scientific Differential Quik Stain kit after 24 hours according to the kit’s protocol. FiJi (NIH) was used to count cells. Experiments were performed in quadruplicate.
In Vivo Experiments
Animal studies were approved by and in accordance with standards set by the Duke University Institutional Animal Care and Use Committee. Nude female mice were obtained from the Duke University Division of Laboratory Animal Resources. Mice were anesthetized by isoflurane (2.5% flow rate). Using the lambda point as a reference point (x = 0.2, z = 1.0), a burr hole was created by microdrill. The injecting needle (30 Gy) was inserted through the burr hole to a depth of y = 3.0. A 5 μL suspension of 200 000 D283 AAn tdTomato or D283 SCn tdTomato tumor cells in 6% methylcellulose was infused at 2.5 μL/min. The cranial opening was sealed by bone wax, and the wound was closed by surgical glue.
Formalin-fixed paraffin-embedded slides were used for immunohistochemistry (IHC) using the DISCOVERY ULTRA automated staining platform. The tissue sections were pretreated (epitope retrieval) with Roche cell conditioning solution CC1 (Roche catalog #: 950-124) for 56 minutes, incubated with rabbit monoclonal Ki67 (Abcam catalog #: ab16667), and diluted 1:200 with Discovery Ab Diluent (catalog #: 760-108) for 60 minutes at 36°C. ABL1 antibody (Abcam catalog # 15130) diluted 1:1,600 and ABL2 (ThermoFisher catalog # 17693) diluted 1:200 were also used. Rabbit IgG, substituted for the primary antibody, was used as the negative control. After binding of the primary antibody, Roche anti-rabbit HQ (catalog #: 760-4815) was applied and incubated for 12 minutes, followed by 12 minutes of incubation with anti-HQ HRP (catalog #: 760-4820) for antigen detection. The IHC reaction was visualized with DAB chromogen and counterstained with hematoxylin.
Photos of each specimen were taken in triplicate. QuPath18 was used to automate cell counting and calculate percentile positive cells in a given region of interest.
Generating shRNA Knockdowns of ABL1/2
To generate double knockdowns of ABL1/2, we used methods as previously described.19 shRNA sequences targeting ABL1 were (GGTGTATGAGCTGCTAGAGAA), targeting ABL2 were (CCTTATCTCACCCACTCTGAA), and for scrambled shRNA were (GGTGTATGGGCTACTATAGAA); all were lentivirally packaged. Briefly, D283 cells were plated and transfected with packaging and corresponding DNAs using FuGENE6 reagent (Promega). Virus-containing culture supernatants were harvested and filtered 24 and 48 hours following transfection and added to cell cultures in the presence of polybrene. Cells were then selected by FACS using their GFP reporter. These cells were then transduced with a luciferase-Tomato reporter for bioluminescence studies.
RNA Sequencing and Analysis
The Sequencing and Genomic Technologies Core Facility at Duke University performed RNA sequencing. The Sequencing and Genomic Technologies Core Facility extracted total RNA from cell pellets using the RNeasy Plus Mini Kit from Qiagen (catalog #: 74134). mRNA was enriched from total RNA and reverse-transcribed into cDNA to build sequencing libraries using the KAPA mRNA HyperPrep Kit from Roche (KK8581). Libraries were pooled to equimolar concentrations and sequenced on the NovaSeq 6000 S2 flow cell to produce 50 bp paired-end reads. Triplicates were run of both 283 AAn and 283 SCn, each in 2 lanes.
Sample fastq file quality metrics were assessed via FastQC v0.11.9 and MultiQC v1.11. Paired-end reads were aligned to the GRCh38.p14 human reference genome via STAR v2.7.2b with default parameter settings. Post-alignment quality metrics were assessed via the log file output of STAR. Quantification and generation of the raw counts' matrix were performed by featureCounts v1.6.3. The raw counts' matrix was then normalized, and differential expression was calculated via DESeq2 R package v1.36.0. Differential expression calculation was performed within RStudio 2022.07.0 Build 548 running R version 4.2.1. Gene set enrichment analysis (GSEA) was performed with the fgsea R package v1.23.4. Three samples of D283 AAn and D283 SCn were analyzed, and the mean was used for differential expression and GSEA.
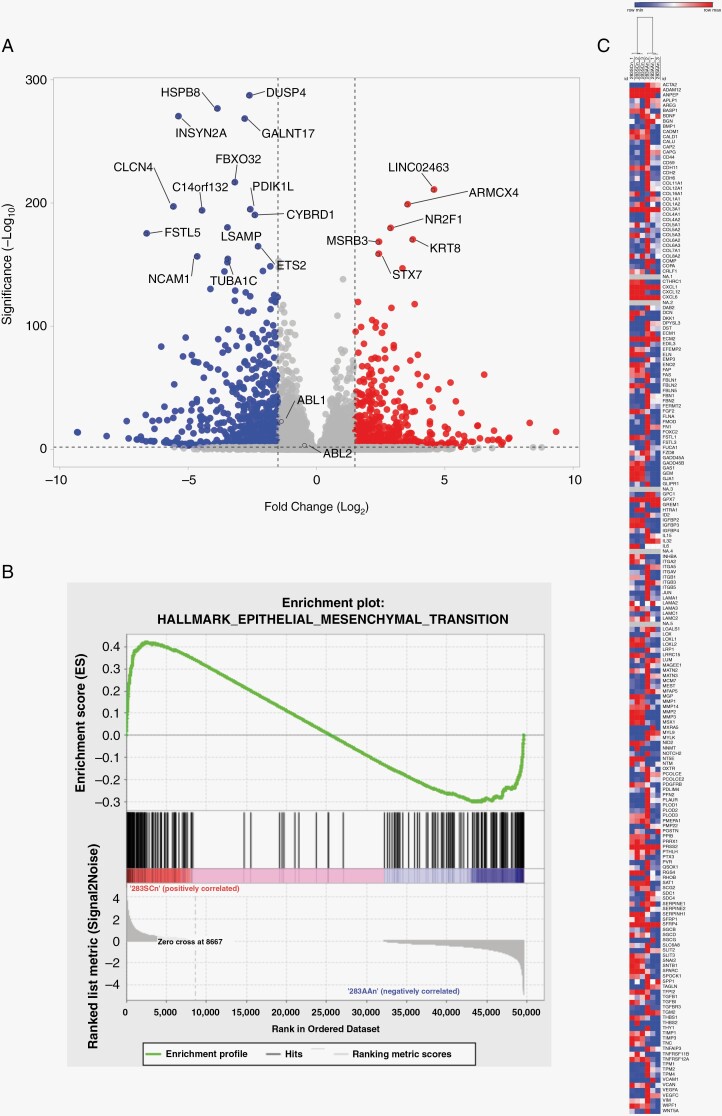
The volcano plot featured in Figure 5A was made using VolcaNoseR (https://huygens.science.uva.nl/VolcaNoseR/),20 the GSEA figure was created using GSEA v4.3.2 (Broad), and the heatmap was created using Morpheus (https://software.broadinstitute.org/morpheus/).
Figure 5.
Differential expression via RNA sequencing of D283 SCn and D283 AAn cells. (A) Volcano plot of differentially expressed genes in ABL1/2 knockdown cells (D283 AAn) and scrambled controls (D283 SCn). Blue represents decreased expression in ABL1/2 knockdown cells while red represents increased expression. Both ABL1 and ABL2 transcripts were significantly reduced in the double knockdown cells. ABL2 log2fold −0.4248, adjusted P = 7.74 × 10−4; ABL1 log2fold −1.32144, adjusted P = 1.66 10-22. (B) Gene set enrichment analysis enrichment plot of the single significant hallmark pathway: EPITHELIAL_MESENCHYMAL_TRANSITION. (C) Heatmap of the three D283 SCn samples, 2 columns each, and the 3 D283 AAn samples, 2 columns each. Hierarchical clustering demonstrates clustering into D283 SCn (left) and D283 AAn (right).
Statistical Analysis
GraphPad Prism v6.05 (La Jolla, California) was used for all statistical analyses. P values ≤ .05 were considered statistically significant. The Benjamini and Hochberg method was used to correct for multiple testing for the differential expression and GSEA analyses and to generate an adjusted statistically significant cutoff of P ≤ .05. The Wilcoxon test was used to compare animal survival.
For analysis of the data in Figure 1, M0 versus M + comparisons were performed using a non-paired student’s t-test. For analysis of the data presented in Supplementary Figure 1, the Kaplan–Meier estimator was used as previously described.11 The R packages “survival” (v3.2-10) and “survminer” (v0.4.9) were used, and the log-rank (Mantel-Cox) test was employed to compare subgroup survival. For analysis of the data presented in Figure 4, the mean values of each specimen were used in a non-paired student’s t-test of D283 AAn versus D283 SCn groups.
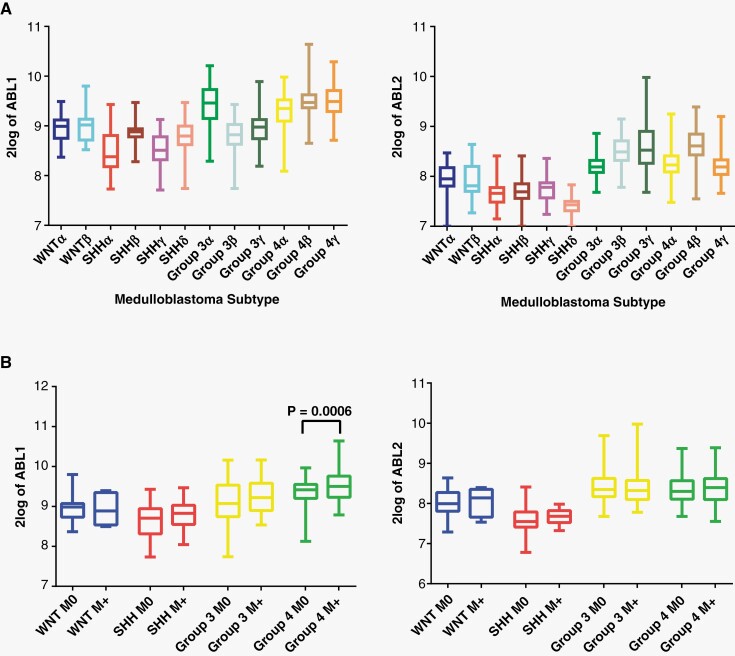
Figure 1.
ABL1/2 mRNA is expressed in medulloblastoma. (A) Box plot (mean and SD) of ABL1 and ABL2 mRNA expression by medulloblastoma subtype. (B) Box plot of ABL1 and ABL2 mRNA expression by medulloblastoma subgroup and metastatic status demonstrating generally increased mean ABL1 and ABL2 expression in M + patients (with metastasis), with significant elevation in group 4 expression of ABL1.
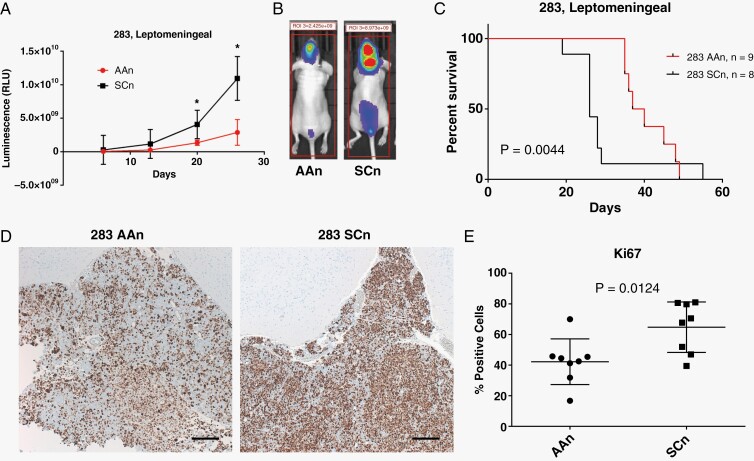
Figure 4.
Knockdown of ABL1/2 decreases tumor burden in a mouse model of medulloblastoma with leptomeningeal dissemination. (A) Animals harboring ABL1/2 knockdown medulloblastoma cells (283 AAn, n = 8) demonstrated significantly less tumor spread as shown by whole-body bioluminescence compared to scrambled controls (283 SCn, n = 9) (mean and SD displayed, *=P < .05). (B) Representative bioluminescence profiles at day 20-post tumor implantation for ABL1/2 knockdown-harboring animals (283AAn) compared to scrambled controls (283 SCn). Bioluminescence images from every animal at every timepoint can be found in Supplementary Figure 3. (C) Animals harboring ABL1/2 double knockdown had significantly longer overall survival than animals harboring scrambled controls (P = .0044). (D,E) Histological analysis of tumor specimens reveals significantly lower proliferation index (Ki67) in 283 AAn compared to 283 SCn (P = .0124). Representative images (D) of tumor specimens found in the leptomeningeal compartment on the brain’s surface stained with Ki67 (brown) demonstrate less Ki67 positivity in the 283 AAn group (quantification in (E)). Additional examples of histological tumor location in the ventricle and the surface of the brain can be found in Supplementary Figure 4. All images were taken at 5× magnification. Scale bar = 200 µm. The mean values of each specimen were used in a non-paired student’s t-test of D283 AAn versus D283 SCn groups.
Results
Relationship Between ABL Kinase Expression and Presence of Metastasis or Survival Outcomes in Medulloblastoma
In a cohort of 763 patients with medulloblastoma,9 ABL1 and ABL2 mRNA expression levels were highest in group 3 and group 4 subtypes (Figure 1A). In particular, high levels of ABL1 corresponded to the subtypes with highest percentage of metastases at diagnosis including group 3α, group 3γ, group 4α, group 4β, and group 4γ.9 Patients with metastatic disease at diagnosis (M+) generally had higher mRNA expression of ABL1 and ABL2 in their tumors compared to patients without metastatic disease at diagnosis (M0), with ABL1 expression reaching significance for group 4 patients (Figure 1B). Dividing the cohort into high and low ABL1/2 expression groups, tumors with high ABL1 expression showed a nonsignificant trend for worse overall survival (OS) in the SHH molecular subgroup (Supplementary Figure 1A), whereas tumors with high ABL2 expression showed a nonsignificant trend for worse overall survival in the SHH and group 3 molecular subgroups (Supplementary Figure1B).
Inhibition of ABL Kinases Results in Medulloblastoma Cytotoxicity and Decreases c-MYC Expression
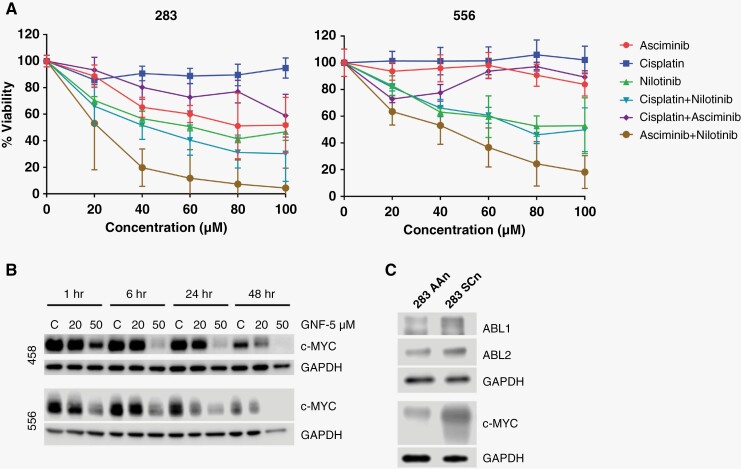
Next, we assessed the in vitro effects of treatment with the allosteric, specific ABL kinase asciminib (ABL001) as well as the multi-tyrosine kinase inhibitor nilotinib,21 and cisplatin, a genotoxic chemotherapy agent that is part of the backbone of standard of care adjuvant therapy for medulloblastoma. Two patient-derived medulloblastoma cell lines (D283 and D556) were explored. Using a standard viability assay (CellTiter-Glo, Promega), we found that ABL kinase inhibitors overall decreased cell viability more potently than cisplatin at micromolar concentrations over the course of 18 hours (Figure 2A) and that the combination of asciminib plus nilotinib was more potent than either agent alone. Interestingly, asciminib and nilotinib have been shown to co-bind single molecules of the BCR-ABL1 fusion protein, and asciminib plus nilotinib cotreatment has been shown to inhibit CML colony growth in vitro, suppress emergence of resistance point mutations in vitro, and decrease CML xenograft tumor growth in vivo more markedly than either agent alone.22,23
Figure 2.
Inhibition/inactivation of ABL1 and ABL2 results in medulloblastoma cell death and decreased expression of c-MYC. (A) Dose–response curves of D283 and D556 medulloblastoma cells treated for 18 hours with cisplatin, a mainstay alkylating agent for the adjuvant treatment of medulloblastoma; the ABL allosteric inhibitor, asciminib; and the nonselective tyrosine kinase inhibitor, nilotinib. The combination of nilotinib and asciminib was the most lethal. (B) Representative Western blot demonstrating the time- and dose-dependent decrease in expression of c-MYC when treated with the ABL allosteric inhibitor GNF5. (C) Western blot of ABL1/2 knockdown cells (283 AAn) demonstrating decreased c-MYC expression compared to scrambled controls (283 SCn).
We next sought to investigate potential mechanisms behind the role of ABL kinases in medulloblastoma progression. c-MYC is an oncogene that promotes medulloblastoma cell proliferation, and elevated c-MYC expression is an independent risk factor for poor prognosis in medulloblastoma.24 The molecular mechanisms that regulate c-MYC in medulloblastoma are largely unknown. ABL kinases have been identified as key regulators of c-MYC expression in both fibroblasts involving PDGFR/c-Src signaling cascades, and in lung cancer cells.25,26 To explore the relationship of ABL kinases and c-MYC in medulloblastoma, we treated the medulloblastoma cell lines D458 and D556 with GNF5, a highly selective allosteric ABL kinase inhibitor similar to asciminib, for 1, 6, 24, or 48 hours. We found that GNF5 specifically decreased expression of c-MYC in a time- and concentration-dependent manner (Figure 2B). We then used an shRNA approach to genetically inactivate ABL1 and ABL2 in D283 cells and found a corresponding decrease in c-MYC expression in ABL1/2 knockdown cells compared to control (Figure 2C). Furthermore, using mRNA data, we found a significant positive correlation between ABL2 and MYC in SHH (r = 0.1513, P = .023), Group 3 (r = 0.3274, P < .001), and group 4 (r = 0.2305, P < .001) and a significant negative correlation between ABL1 and MYC in group 3 (r = −0.3021, P = .0002) (Supplementary Figure 2). Given that both the pharmacologic inhibition of ABL kinases and genetic inactivation of ABL1/2 resulted in decreased c-MYC expression, while mRNA levels of ABL2 correlated positively with MYC, we hypothesize that ABL2 and likely ABL1 are upstream mediators of c-MYC expression in medulloblastoma.
Genetic Inactivation of ABL1/2 Decreases Adhesion in Medulloblastoma
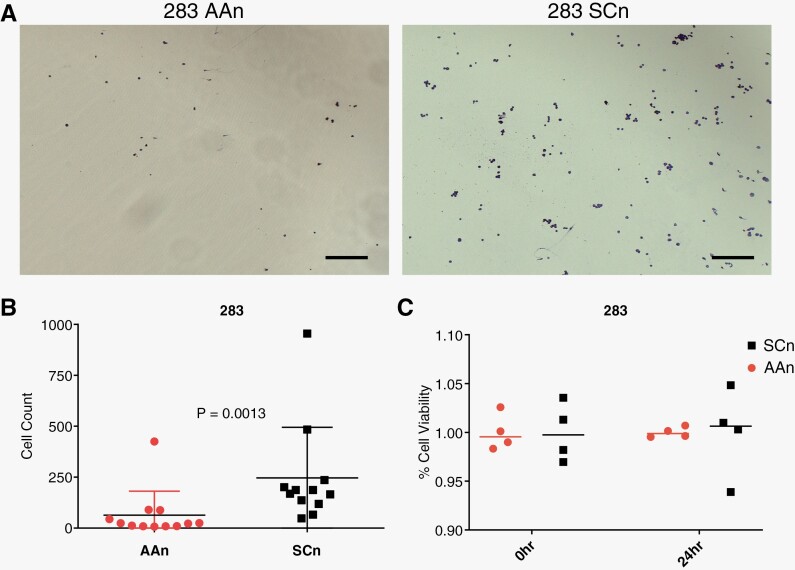
Following our exploration of ABL kinases’ roles in medulloblastoma cell viability and c-MYC signaling, we explored potential roles for ABL kinases in a process critical to LMD: tumor cell adhesion. ABL1 and ABL2 are critical in the regulation of cadherin-mediated cellular adhesion via Rho GTPases27 and E-cadherin is concentrated at the arachnoid cell surface,28 a key component of the leptomeningeal extracellular matrix (ECM). Engagement of tumor cell cadherins and leptomeningeal cadherins results in tight junctions and adhesion.4 Since medulloblastoma has been shown to express a protein related to E-cadherin called neural (N)-cadherin,29 we explored the role of ABL kinases in binding to vitronectin, another key component of the leptomeningeal ECM and brain pericytes30,31 that mediates cell adhesion, in tandem with cadherins. High levels of vitronectin have been found to be expressed in the meninges of the brain and spinal cord31 and vitronectin is considered the ”master controller” of the ECM.32 We observed significantly decreased binding to vitronectin in the ABL1/2 knockdown cells compared to control (Figure 3A, B). Notably, this decreased binding could not be explained by a difference in cell viability between control and knockdown groups (Figure 5C).
Figure 3.
Genetic knockdown of ABL1/2 decreases binding to vitronectin. (A) Representative images taken at 5× and (B) quantification of vitronectin binding assays for 283 AAn and 283 SCn cells at 24 hours, demonstrating a statistically significant decrease in binding capacity to vitronectin in the ABL1/2 knockdown cells (AAn). (C) Quantification of cell viability for 283 AAn and 283 SCn cells demonstrates no significant decrease in viability at 24 hours, indicating decreased vitronectin binding and not decreased viability. Scale bar = 200 µm.
Genetic Inactivation of ABL1/2 Decreases Tumor Burden and Increases Overall Survival in a Mouse Model of Medulloblastoma LMD
Our next aim was to investigate how our in vitro findings of ABL kinases’ roles in medulloblastoma cell viability and adhesion might translate to an improvement in survival. To do so, we assessed the effects of an ABL1/2 genetic knockdown in a mouse model of medulloblastoma with LMD. Animals harboring ABL1/2 double knockdown medulloblastoma cells developed significantly less LMD and smaller tumor volumes compared to controls, as measured by whole-body bioluminescence (Figure 4A, B). This decrease in leptomeningeal tumor burden corresponded with significantly increased OS (Figure 5C). The decrease in tumor spread and resultant increased OS was associated with significantly decreased proliferative index (Ki67) in the ABL1/2 knockdown group (Figure 4D,E). Histologically, tumor was confirmed on the surface of the brain in the leptomeningeal compartment and in the intraventricular space (Supplementary Figure 4). Staining of both ABL1 and ABL2 was found predominantly in the cytoplasm (Supplementary Figure 5), consistent with previous literature.4
Differential Gene Expression of ABL1/2-Depleted D283 Cells
Finally, given our data demonstrating ABL1/2-dependent differences in the capacity for cell adhesion and presumably migration/invasion at the protein level, as well as medulloblastoma survival in vitro and in vivo at the cell and organism level, we asked whether ABL1/2 dependency might also be observed in the expression of different genes at the transcriptomic level. We thus performed RNA sequencing and analyzed differential expression patterns between D283 SCn (scrambled) shRNA control cells and D283 AAn cells (harboring a double knockdown of ABL1/2, n = 3 for each group). Using an adjusted statistically significant cutoff of P ≤ .05, there were 6,022 differentially expressed genes between the control and genetically manipulated groups including both ABL1 (log2FoldChange −1.32, adjusted P = 1.66 × 10−22) and ABL2 (log2FoldChange −0.42, adjusted P = 7.75 × 10−4) (Figure 5A). The most significantly upregulated genes among the ABL1/2 knockdown cells included LINC02463, ARMCX4, NR2F1, KRT8, MSRB3, and STX7, while the most significantly downregulated genes included DUSP4, HSPB8, INSYN2A, GALNT17, and FBXO32 (Figure 5A, Supplementary Table 1). In GSEA, the pathway HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION was significantly different between the two cell types (adjusted P = .03962, Supplementary Table 2, Figure 5B). Notably, unsupervised hierarchical clustering suggested a similar gene expression profile for the 3 replicates for each respective condition (Figure 5C).
Discussion
Many patients with medulloblastoma experience LMD at recurrence and/or exhibit significant toxicities related to adjuvant therapy.33 Current standard of care is chemotherapy (cisplatin, vincristine, cyclophosphamide) plus craniospinal irradiation. There is a paramount need for more efficacious, less-toxic therapies for medulloblastoma LMD. Here, we present ABL family kinases as novel therapeutic targets of interest in leptomeningeal disseminated medulloblastoma. Targeted therapy against these proteins, which are highly implicated in cell proliferation, adhesion, and migration, could present a paradigm shift from cytotoxic alkylating chemotherapy and ionizing radiation.
Within cohorts containing hundreds of patients, we show that ABL1 and ABL2 mRNA is most highly expressed in group 3 and 4 medulloblastoma, and that this expression trends toward differentiating patients with metastatic disease compared to those without. We also show a trend that high ABL1/2 expression is associated with worse survival outcomes in select subgroups of medulloblastoma.
We corroborate these data with in vitro studies, showing that patient-derived medulloblastoma cell lines are sensitive to pharmacologic inhibition of ABL kinases and that this ABL-dependent cell viability may also depend on c-MYC signaling. Although the doses of small molecules used in this work were relatively high, they were within doses previously used in solid tumors.17 Furthermore, because both asciminib and GNF5 are specific allosteric ABL1/2 inhibitors,21 it is very unlikely any results observed were due to off-target effects. To this point, double genetic knockdown of ABL1/2 decreases c-MYC expression by Western blot analysis. These results build off prior work that suggests possible direct interactions between ABL kinases and c-MYC in non-cancer25 and other solid tumors.26 Data from Furstoss et al.25 suggest that inhibiting c-ABL with imatinib reduces transcriptional expression of c-MYC, and experiments with mutated c-ABL suggest that c-ABL lies downstream of c-Src, implying a more direct rather than indirect effect of ABL1/2 on c-MYC levels. Data from Gu et al. suggest that ABL-mediated activation of TAZ and β-catenin, which both target MYC, is required for metastasis of NSCLC.26
Genetic knockdown of ABL1/2 also decreases the adhesion capacity of medulloblastoma cells to vitronectin, possibly through reliance on the regulation of cadherins as well as other molecules such as integrins. In vivo, a mouse model of medulloblastoma demonstrated decreased tumor burden with less metastatic disease and increased overall survival upon knockdown of ABL1/2. It is likely that this decreased tumor burden was due to both decreased c-MYC-mediated cell proliferation and adhesion to the ECM. Further studies will explore the exact mechanistic underpinnings of which proteins are involved in ABL-mediated regulation of adhesion and invasion/migration in medulloblastoma, working off other evidence that ABL2 gene silencing decreases migration/invasion in cancer cell lines34 and that higher vitronectin expression/binding correlates with metastasis and worse clinical prognosis.35–37 Further work should also identify how ABL1/2 knockdown may mitigate LMD by decreasing cell proliferation (as evidenced by decreased Ki67 staining in our samples), given the crucial role of c-MYC in regulating this process as well. Given that ABL1/2 knockdown did not decrease cell viability in vitro, it is likely that ABL1/2 knockdown primarily decreases LMD by first disrupting existing interactions of ABL1/2 with matrix proteins like vitronectin and integrins as suggested by our group and observed by others.38,39 Furthermore, given the known independent relationships of ABL1/2 and medulloblastoma metastasis to critical signal transducers like PDGF receptors (specifically, PDGFRB), it is possible that the phenotypic effects may be explained by a signaling cascade that involves ABL1/2 acting downstream of PDGFRB in cancer cells.25,40
Finally, we show that ABL1/2 knockdown medulloblastoma cells display a significant number of differentially expressed genes compared to controls. The most significantly upregulated genes included: LINC0246, a lncRNA whose SNPs (single nucleotide polymorphisms) have been associated with lymphoma; ARMCX4, mutations of which have been associated with the malignant childhood brain tumor atypical teratoid rhabdoid tumor; KRT8, whose expression has been suggested as a prognostic marker for many cancer types; MSRB3, whose expression may confer stress resistance in cancer; and STX7, which encodes a vesicle transport protein implicated in breast cancer cell invasion (see Supplementary Table 3 for references). Downregulated genes included: DUSP4, which encodes a phosphatase that targets the kinases ERK1, ERK2, and JNK (among others), which are often dys-/upregulated in tumors and play crucial roles in cell growth and survival41,42; HSPB8, which encodes a heat shock protein that likely regulates cell proliferation, apoptosis, and carcinogenesis; GALNT17, mutations of which have been associated with cerebellar dysfunction and astrocytoma growth; and FBXO32, an F-box protein that has been shown to target c-MYC for degradation and has, when dysregulated, been implicated in cancer stemness (see Supplementary Table 3 for references). Taken together, these results provide novel insight into genes lying at the intersection of neurodevelopment and oncogenesis, and whose dysregulation in the positive or negative direction may couple with ABL1 and ABL2 expression to drive medulloblastoma tumorigenesis and LMD (see Supplementary Table 3). Interestingly, GSEA revealed enrichment in the hallmark pathway43 EPITHELIAL_MESENCHYMAL_TRANSITION. This is striking as the EMT, whereby epithelial cells lose their polarity to acquire mesenchymal cell features (eg, increased motility), is presumed to be a dynamic process critical for tumor progression and metastasis, and ABL kinases have been directly implicated in this plasticity.44,45
Future research will be directed at unraveling molecular pathways downstream of ABL1 and ABL2 involved in tumor proliferation, adhesion, and invasion, and will determine how intra- and intertumoral heterogeneity may affect how these cascades present throughout different tumors. Given our data showing that ABL inhibition decreases c-MYC expression, further research should also determine if ABL kinase inhibitors are effective in all medulloblastoma subgroups or preferentially in the high-c-MYC subgroup Group 310 and the relative contribution of ABL1 and ABL2 in MYC regulation.
In conclusion, we demonstrate the putative association of ABL1 and ABL2 as upstream regulators of c-MYC in medulloblastoma. Inactivation of ABL1 and ABL2 resulted in decreased LMD in mice and improved OS. Future research will uncover the more detailed biology of ABL kinases’ roles in medulloblastoma and hopefully lead to targeted therapies that have the potential to improve overall survival and quality of life for thousands of patients with this incurable disease.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Peter Pytel for assistance with interpretation of the histological specimens. The authors thank other lab members for technical assistance, the staff of the Duke Preston Robert Tisch Brain Tumor Center BioRepository for slide preparation of mouse specimens, the Duke Pathology Research Immunohistology Laboratory for immunostaining of mouse specimens, and Vaibhav Jain for assistance in uploading the RNA sequencing data.
Contributor Information
Jill K Jones, Harvard/MIT MD-PhD Program, Boston, MA, USA; Department of Neurosurgery, Duke University, Durham, NC, USA.
Hengshan Zhang, Department of Neurosurgery, Duke University, Durham, NC, USA.
Anne-Marie Lyne, Institut Curie, PSL Research University, Paris, France; Inserm, U900, Paris, France; MINES ParisTech, CBI – Centre for Computational Biology, PL Research University, Paris, France.
Florence M G Cavalli, Institut Curie, PSL Research University, Paris, France; Inserm, U900, Paris, France; MINES ParisTech, CBI – Centre for Computational Biology, PL Research University, Paris, France.
Wafa E Hassen, Department of Neurosurgery, Duke University, Durham, NC, USA.
Kevin Stevenson, Duke University Molecular Physiology Institute, Durham, NC, USA.
Reb Kornahrens, Department of Neurosurgery, Duke University, Durham, NC, USA.
Yuanfan Yang, Department of Neurosurgery, Duke University, Durham, NC, USA; Department of Neurosurgery, The University of Alabama at Birmingham, Birmingham, AL, USA.
Sean Li, Department of Neurosurgery, Duke University, Durham, NC, USA; Case Western University School of Medicine, Cleveland, OH, USA.
Samuel Dell, Department of Neurosurgery, Duke University, Durham, NC, USA; Division of Hematologic Malignancies and Cellular Therapy, Duke Cancer Institute.
Zachary J Reitman, Department of Radiation Oncology, Duke University School of Medicine, Durham, NC, USA.
James E Herndon, II, Department of Biostatistics and Bioinformatics, Duke University School of Medicine, Durham, NC, USA.
Jacob Hoj, Department of Pharmacology and Cancer Biology, Duke University, Durham, NC, USA.
Ann Marie Pendergast, Department of Pharmacology and Cancer Biology, Duke University, Durham, NC, USA.
Eric M Thompson, Department of Neurosurgery, Duke University, Durham, NC, USA; Preston Robert Tisch Brain Tumor Center, Duke University, Durham, NC, USA; Department of Neurosurgery, The University of Chicago, Chicago, IL, USA.
Funding
This work was supported by NIH (R03 NS114629-01), NIH (P50 CA190991), NIH (P30 CA014236), and the Chetna and Meena Trust (all to EMT); and by NIH (K08 CA256045), the Pediatric Brain Tumor Foundation, and the St. Baldrick’s Foundation (all to ZJR); and by NIH 1R01CA246133 (to AMP).
Conflict of interest statement
EMT is a scientific advisor to OncoHeroes Biosciences. AMP is a consultant and advisory board member for The Pew Charitable Trusts. The other authors declare no potential competing interests.
Authorship statement
EMT designed the project. JKJ, HZ, WH, SD, and EMT contributed to experimental design. JKJ and EMT drafted the manuscript. JKJ, HZ, RK, YY, SL, and SD performed experiments. AL and FC provided and analyzed patient mRNA data. JH and AMP contributed to the experimental design and provided insights and advice throughout the project. KS and EMT designed and analyzed cell RNA sequencing. JEH designed and analyzed the statistics. ZR helped with analysis of RNA sequencing data. All authors reviewed and revised the manuscript.
Data Availability
All relevant data are available from the authors upon request. RNA sequencing data has been uploaded to the Gene Expression Omnibus (GSE224691, https://www.ncbi.nlm.nih.gov/geo/).
References
- 1. Curtin SC, Minino AM, Anderson RN.. Declines in cancer death rates among children and adolescents in the United States, 1999-2014. NCHS Data Brief. 2016(257):1–8. [PubMed] [Google Scholar]
- 2. Thompson EM, Hielscher T, Bouffet E, et al. Prognostic value of medulloblastoma extent of resection after accounting for molecular subgroup: A retrospective integrated clinical and molecular analysis. Lancet Oncol. 2016;17(4):484–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Klein A, van Kessel AG, Grosveld G, et al. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300(5894):765–767. [DOI] [PubMed] [Google Scholar]
- 4. Khatri A, Wang J, Pendergast AM.. Multifunctional Abl kinases in health and disease. J Cell Sci. 2016;129(1):9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hochhaus A, Larson RA, Guilhot F, et al. ; IRIS Investigators. Long-term outcomes of imatinib treatment for chronic myeloid leukemia. N Engl J Med. 2017;376(10):917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grobner SN, Worst BC, Weischenfeldt J, et al. ; ICGC PedBrain-Seq Project. The landscape of genomic alterations across childhood cancers. Nature. 2018;555(7696):321–327. [DOI] [PubMed] [Google Scholar]
- 7. Chen J, Kang Z, Li S, et al. Molecular profile reveals immune-associated markers of medulloblastoma for different subtypes. Front Immunol. 2022;13:911260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu S, Lin F, Chen Z, et al. Identification of a twelve-gene signature and establishment of a prognostic nomogram predicting overall survival for medulloblastoma. Front Genet. 2020;11:563882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cavalli FMG, Remke M, Rampasek L, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31(6):737–754.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thompson EM, Keir ST, Venkatraman T, et al. The role of angiogenesis in Group 3 medulloblastoma pathogenesis and survival. Neuro Oncol. 2017;19(9):1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li S, McLendon R, Sankey E, et al. CD155 is a putative therapeutic target in medulloblastoma. Clin Transl Oncol. 2022;25(3):696–705. [DOI] [PubMed] [Google Scholar]
- 12. Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41(4):465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friedman HS, Burger PC, Bigner SH, et al. Establishment and characterization of the human medulloblastoma cell line and transplantable xenograft D283 Med. J Neuropathol Exp Neurol. 1985;44(6):592–605. [DOI] [PubMed] [Google Scholar]
- 14. Aldosari N, Wiltshire RN, Dutra A, et al. Comprehensive molecular cytogenetic investigation of chromosomal abnormalities in human medulloblastoma cell lines and xenograft. Neuro Oncol. 2002;4(2):75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ivanov DP, Coyle B, Walker DA, Grabowska AM.. In vitro models of medulloblastoma: Choosing the right tool for the job. J Biotechnol. 2016;236:10–25. [DOI] [PubMed] [Google Scholar]
- 16. Chahal KK, Li J, Kufareva I, et al. Nilotinib, an approved leukemia drug, inhibits smoothened signaling in Hedgehog-dependent medulloblastoma. PLoS One. 2019;14(9):e0214901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoj JP, Mayro B, Pendergast AM.. The ABL2 kinase regulates an HSF1-dependent transcriptional program required for lung adenocarcinoma brain metastasis. Proc Natl Acad Sci U S A. 2020;117(52):33486–33495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bankhead P, Loughrey MB, Fernandez JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoj JP, Mayro B, Pendergast AM.. A TAZ-AXL-ABL2 feed-forward signaling axis promotes lung adenocarcinoma brain metastasis. Cell Rep. 2019;29(11):3421–3434.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goedhart J, Luijsterburg MS.. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci Rep. 2020;10(1):20560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones JK, Thompson EM.. Allosteric inhibition of ABL kinases: Therapeutic potential in cancer. Mol Cancer Ther. 2020;19(9):1763–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eide CA, Zabriskie MS, Savage Stevens SL, et al. Combining the allosteric inhibitor asciminib with ponatinib suppresses emergence of and restores efficacy against highly resistant BCR-ABL1 mutants. Cancer Cell. 2019;36(4):431–443.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wylie AA, Schoepfer J, Jahnke W, et al. The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature. 2017;543(7647):733–737. [DOI] [PubMed] [Google Scholar]
- 24. Grotzer MA, Hogarty MD, Janss AJ, et al. MYC messenger RNA expression predicts survival outcome in childhood primitive neuroectodermal tumor/medulloblastoma. Clin Cancer Res. 2001;7(8):2425–2433. [PubMed] [Google Scholar]
- 25. Furstoss O, Dorey K, Simon V, et al. c-Abl is an effector of Src for growth factor-induced c-myc expression and DNA synthesis. EMBO J. 2002;21(4):514–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gu JJ, Rouse C, Xu X, et al. Inactivation of ABL kinases suppresses non-small cell lung cancer metastasis. JCI Insight. 2016;1(21):e89647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zandy NL, Playford M, Pendergast AM.. Abl tyrosine kinases regulate cell-cell adhesion through Rho GTPases. Proc Natl Acad Sci U S A. 2007;104(45):17686–17691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tohma Y, Yamashima T, Yamashita J.. Immunohistochemical localization of cell adhesion molecule epithelial cadherin in human arachnoid villi and meningiomas. Cancer Res. 1992;52(7):1981–1987. [PubMed] [Google Scholar]
- 29. Utsuki S, Oka H, Sato Y, et al. E, N-cadherins and beta-catenin expression in medulloblastoma and atypical teratoid/rhabdoid tumor. Neurol Med Chir (Tokyo). 2004;44(8):402–6; discussion 407. discussion 407. [DOI] [PubMed] [Google Scholar]
- 30. Jia C, Keasey MP, Malone HM, et al. Vitronectin from brain pericytes promotes adult forebrain neurogenesis by stimulating CNTF. Exp Neurol. 2019;312:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seiffert D, Iruela-Arispe ML, Sage EH, Loskutoff DJ.. Distribution of vitronectin mRNA during murine development. Dev Dyn. 1995;203(1):71–79. [DOI] [PubMed] [Google Scholar]
- 32. Ruzha Y, Ni J, Quan Z, Li H, Qing H.. Role of vitronectin and its receptors in neuronal function and neurodegenerative diseases. Int J Mol Sci . 2022;23(20):12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Packer RJ, Vezina G.. Management of and prognosis with medulloblastoma: therapy at a crossroads. Arch Neurol. 2008;65(11):1419–1424. [DOI] [PubMed] [Google Scholar]
- 34. Xing QT, Qu CM, Wang G.. Overexpression of Abl2 predicts poor prognosis in hepatocellular carcinomas and is associated with cancer cell migration and invasion. Onco Targets Ther. 2014;7:881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhong Wu X, Honke K, Long Zhang Y, Liang Zha X, Taniguchi N.. Lactosylsulfatide expression in hepatocellular carcinoma cells enhances cell adhesion to vitronectin and intrahepatic metastasis in nude mice. Int J Cancer. 2004;110(4):504–510. [DOI] [PubMed] [Google Scholar]
- 36. Heyman L, Leroy-Dudal J, Fernandes J, et al. Mesothelial vitronectin stimulates migration of ovarian cancer cells. Cell Biol Int. 2010;34(5):493–502. [DOI] [PubMed] [Google Scholar]
- 37. Shi K, Lan RL, Tao X, et al. Vitronectin significantly influences prognosis in osteosarcoma. Int J Clin Exp Pathol. 2015;8(9):11364–11371. [PMC free article] [PubMed] [Google Scholar]
- 38. Lewis JM, Schwartz MA.. Integrins regulate the association and phosphorylation of paxillin by c-Abl. J Biol Chem. 1998;273(23):14225–14230. [DOI] [PubMed] [Google Scholar]
- 39. Xu J, Millard M, Ren X, Cox OT, Erdreich-Epstein A.. c-Abl mediates endothelial apoptosis induced by inhibition of integrins alphavbeta3 and alphavbeta5 and by disruption of actin. Blood. 2010;115(13):2709–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gilbertson RJ, Clifford SC.. PDGFRB is overexpressed in metastatic medulloblastoma. Nat Genet. 2003;35(3):197–198. [DOI] [PubMed] [Google Scholar]
- 41. Samatar AA, Poulikakos PI.. Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat Rev Drug Discov. 2014;13(12):928–942. [DOI] [PubMed] [Google Scholar]
- 42. Wagner EF, Nebreda AR.. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9(8):537–549. [DOI] [PubMed] [Google Scholar]
- 43. Liberzon A, Birger C, Thorvaldsdottir H, et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dongre A, Weinberg RA.. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69–84. [DOI] [PubMed] [Google Scholar]
- 45. Sirvent A, Benistant C, Roche S.. Cytoplasmic signalling by the c-Abl tyrosine kinase in normal and cancer cells. Biol Cell. 2008;100(11):617–631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available from the authors upon request. RNA sequencing data has been uploaded to the Gene Expression Omnibus (GSE224691, https://www.ncbi.nlm.nih.gov/geo/).