Abstract
In this study, we present the structures of human urea transporters UT-A and UT-B to characterize them at molecular level and to detail the mechanism of UT-B inhibition by its selective inhibitor, UTBinh-14. High-resolution structures of both transporters establish the structural basis for the inhibitor’s selectivity to UT-B, and the identification of multiple binding sites for the inhibitor will aid with the development of drug lead molecules targeting both transporters. Our study also discovers phospholipids associating with the urea transporters by combining structural observations, native MS, and lipidomics analysis. These insights improve our understanding of urea transporter function at a molecular level and provide a blueprint for a structure-guided design of therapeutics targeting these transporters.
Crystal and cryo-EM structures of human urea transporters UT-A and UT-B enable structure-based design of selective inhibitors.
INTRODUCTION
Urea is a key molecule in the excretion of nitrogenous waste from complex organisms and plays a secondary role in a number of physiological processes, most notably the maintenance of osmotic balance at both cellular and broader physiological levels (1–5). Urea is primarily excreted in the kidney, where it passes the membrane filter in the glomerulus of the nephron then reabsorbed in the renal tubules. The second step of reabsorption is an important process in regulating the urine volume, hence blood pressure, as this facilitates reabsorption of water by osmotic gradient (6). While the initial excretion occurs through diffusion across the membrane, the subsequent reabsorption is mediated by urea transporters UT-A (HsUT-A; SLC14A2) and UT-B (HsUT-B; SLC14A1) which are differentially expressed in specific renal tubule compartments (2, 3, 7). This physiological role has made HsUT-A and HsUT-B targets for the development of a new class of salt-sparing diuretics, and a number of inhibitors selective to either member have been described (8–15). These inhibitors have been proven useful for not only the drug discovery programs but also uncovering other roles of urea transport in new disease areas. For example, cell volume dysregulation by HsUT-B–facilitated urea transport was assessed with a selective inhibitor UTBinh-14 in neuroinflammation and in erythrocyte deformation (4, 16, 17), further adding to the profile of urea channel inhibitors as potential therapeutics for neurodegenerative diseases (4, 18).
Crystal structures of bovine UT-B (BtUT-B) and Desulfovibrio vulgaris urea transporter (DvUT) have shown urea transporters to be homotrimers with a pore in each subunit (19, 20). In these structures, 10 transmembrane (TM) helices of each subunit are arranged in a pseudo-C2-symmetry and allow urea transport in a channel-like mechanism. While these structures reveal the structural basis for substrate recognition and urea transport, information on other structural aspects of HsUT-A and HsUT-B, such as insights regarding interactions with small-molecule inhibitors and their selectivity, are not available.
In this study, we present high-resolution structures of human UT-A and UT-B determined with protein crystallography and cryo–electron microscopy (cryo-EM). While our structures show human urea transporters to have a similar overall architecture to BtUT-B and DvUT, we also observe substantial biophysical differences such as surface charges, with implications on their physiological roles. We also determined HsUT-B in complex with UTBinh-14 inhibitor, which shows a different binding location and mode to those predicted in docking studies using BtUT-B, and further characterized the biophysical properties of the inhibitor binding to HsUT-B. Our structures also reveal key residue differences between HsUT-A and HsUT-B at the binding site of UTBinh-14, establishing the structural basis for inhibitor selectivity. Last, we present phospholipids closely associating with HsUT-A and HsUT-B with our cryo-EM structure and mass spectrometry (MS), revealing common features with structurally and functionally homologous ammonia transporters.
RESULTS AND DISCUSSION
Structures of human UT-A and UT-B reveal similar mechanism of urea transport
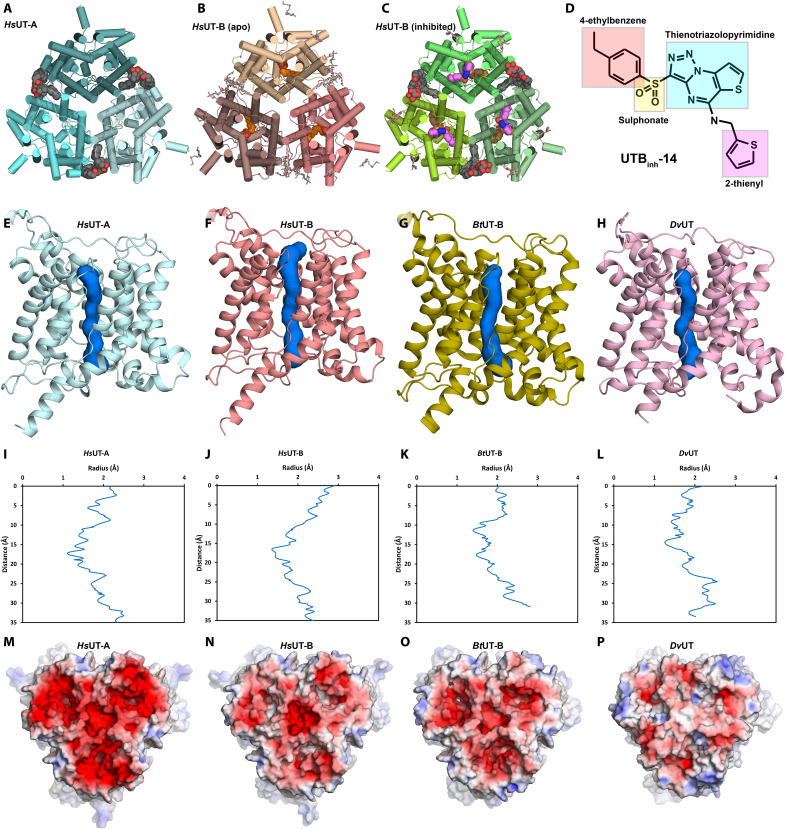
We aimed to determine structures of both HsUT-A and HsUT-B to understand the functional differences between the two paralogs and to enable structure-guided design for selective inhibitors. Full-length HsUT-B, while readily expressed in human embryonic kidney 293F cells (Thermo Fisher Scientific), failed to produce crystals with robust diffraction. Therefore, we engineered the construct for minimal truncation mimicking the limited proteolysis of BtUT-B. Because a previous study has shown that glycosylation-knockout mutation N211I in HsUT-B1 has no discernible effects on plasma membrane expression and urea uptake in Xenopus oocytes (21), we also removed N-linked glycosylation by site-directed mutagenesis. Together, these construct modifications yielded well-diffracting crystals, which resulted in a 2.4-Å x-ray structure (Fig. 1B). As the expression level for HsUT-A was too low for crystallography, we opted for its structure determination by cryo-EM instead. The use of a graphene grid allowed us to overcome its inherent preferred orientation issues and low yield, eventually leading to an electrostatic potential (ESP) map of 2.9 Å nominal resolution (Fig. 1A and fig. S1D).
Fig. 1. Overall structures of HsUT-A and HsUT-B are similar to each other and to their orthologs.
(A to C) Overall structure models of HsUT-A determined with cryo-EM (A), apo-HsUT-B determined with crystallography (B), and inhibitor-bound HsUT-B determined with cryo-EM (C). (D) Structure of UTBinh-14 used in this study. (E to H) Urea channel pores of HsUT-A (E), HsUT-B (F), BtUT-B (G), and DvUT (H). (I to L) Pore radius plots of HsUT-A (I), HsUT-B (J), BtUT-B (K), and DvUT (L). (M to P) Electrostatic surface representations (−5 kT/e in red to 5 kT/e in blue) of the extracellular side of HsUT-A (M), HsUT-B (N), BtUT-B (O), and DvUT (P).
The overall structures of HsUT-A and HsUT-B are similar to each other and to BtUT-B and DvUT (19, 20), showing the trimeric arrangement with 10 TM helices and two pore (P) helices per subunit (Fig. 1, A and B). HsUT-A’s trimeric arrangement was an unexpected finding as our construct was equivalent to full-length UT-A1 isoform, which is a gene duplication of UT monomers (fig. S3). Closer observation of HsUT-A’s map indicated that this sample was largely that of its N-terminal half (equivalent to HsUT-A3) as most of its side chains matched with it (fig. S2, A, B, D, and E). However, of the 88 differing residues examined, there were a few notable exceptions such as A401/P863 where the map feature matched better to the C-terminal residues (fig. S2 F). Supporting the observation that both C- and N-terminal halves of HsUT-A are present, MS of its tryptic digests showed the presence of both N-terminal and C-terminal peptides. Despite extensive data processing efforts, including static classification and particle heterogeneity analysis tools, it was not possible to separate the two species or to isolate maps with pseudo-heterooligomeric arrangement. As the majority of residues in the map are in line with the N-terminal half of HsUT-A (i.e., HsUT-A3), we built the model for its N-terminal sequences and based our structural analyses primarily on this model. For the 17 residues where the map was ambiguous or favoring C-terminal half, we left them without building side chains to make clear that the model may not properly represent the map for these regions.
In both structures, each subunit hosts a channel-like pore for urea transport and the channels are in the open state with approximately 2 to 3 Å in diameter at constriction formed by V69, T177, L292, and F342 (Fig. 1, I and J). This is similar to BtUT-B and DvUT, which also feature open pores of 2.5 and 2 Å at constriction (Fig. 1, K and L). This indicates that HsUT-A3 and HsUT-B in our structures are in similar urea-conducting states.
HsUT-A3 shows significantly more negative surface charge on its extracellular side compared to other urea transporters (Fig. 1M). Whereas the extracellular surfaces of HsUT-B and BtUT-B are only mildly negatively charged and DvUT close to net neutral (Fig. 1, N to P), nearly the entire extracellular surface of HsUT-A shows electrostatic charges of less than −5 kT/e with Adaptive Poisson-Boltzmann Solver (APBS) electrostatics. HsUT-A’s extracellular surface becomes less negatively charged with lower pH values, being similar to HsUT-B’s when the pH is 5.0 (fig. S4). The urinary filtrate becomes significantly acidic in the inner medullary collecting duct (IMCD) where UT-A1 and UT-A3 are expressed with nominal pH as low as 5.5 (22, 23), and local pH at the luminal surface is likely even lower as urine pH control is an active process (24). Hence, the surface charge of HsUT-A3 in a physiological condition may vary depending on the IMCD luminal pH, which in turn is influenced by factors such as vasopressin. As negatively charged extracellular surface is a feature common to structurally related transporters of amines and urea (fig. S7), this surface charge may be an important feature of these proteins to attract their ligands.
UTBinh-14 primarily inhibits UT-B by blocking its extracellular pore entry
UTBinh-14 is a highly selective inhibitor for UT-B, with over a thousand-fold selectivity over UT-A (15). Owing to such properties, it has been used as a chemical probe to study physiological roles of UT-B and is an attractive lead molecule for a drug discovery program (4). Therefore, our effort concentrated on obtaining the structure of HsUT-B in complex with UTBinh-14 compound.
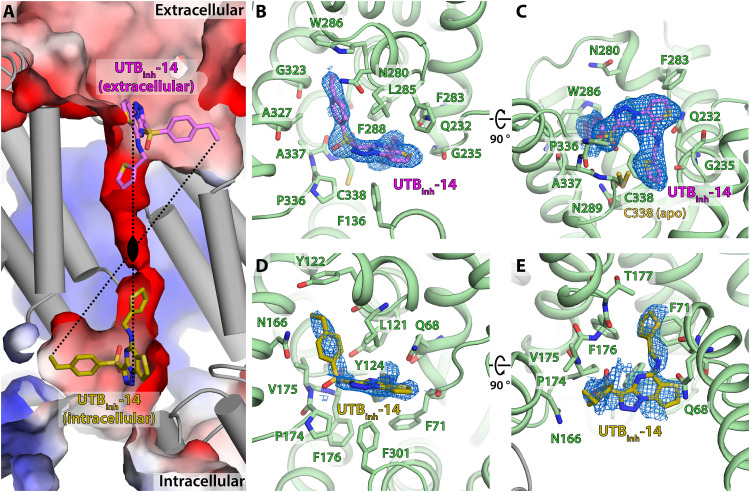
We were not successful in determining the structure of HsUT-B in the inhibitor-bound state with crystallography despite repeated attempts. Hypothesizing that this could be due to either the combination of low solubility and low affinity of UTBinh-14 or its low stability, we changed our structure determination technique to cryo-EM. This resulted in a refined ESP map with 2.6 Å nominal resolution, showing little conformational change (Fig. 1C) and revealing UTBinh-14 blocking the channel on the extracellular side (Fig. 2, B and C). Such a location is different from the prediction by docking studies, where it was thought to block UT-B from the intracellular side (25). In the crystal structure of apo-HsUT-B, this site is occupied by a polyethylene glycol (PEG) molecule instead (fig. S5, A and B), so the high concentration of PEG400 in the crystallizing solution might have competed for this inhibitor-binding site and prevented us from obtaining UTBinh-14–bound structures with crystallography.
Fig. 2. UTBinh-14 inhibits HsUT-B at both extracellular and intracellular sides of its urea channel pore.
(A) Transsectional view of HsUT-B channel pore showing the locations of UTBinh-14. Black oval denotes the center of C2 pseudosymmetry axis of the inhibitor binding modes. (B) Extracellular view of the primary UTBinh-14 binding mode and (C) its transsectional view. (D) Intracellular view of the secondary UTBinh-14 binding mode and (E) its transsectional view.
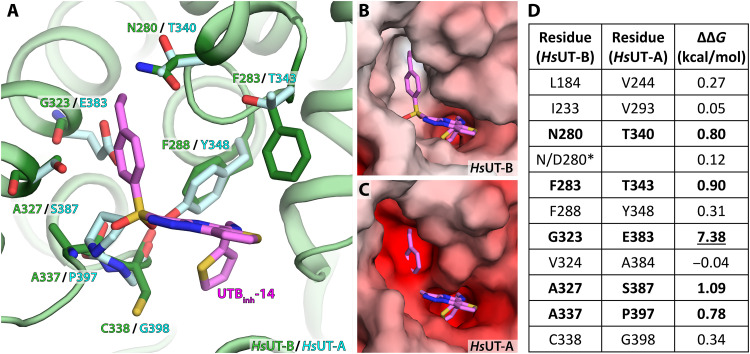
UTBinh-14 fits well into this extracellular pocket, with its 2-thienyl group directly blocking the pore entry. The inhibitor forms hydrophilic interactions with the side chain of Q232 and the backbone carbonyl of A337 of HsUT-B, and its 4-ethylbenzene group sits in a hydrophobic pocket formed by W286, F288, G323, and A327 (Fig. 2, B and C). Superimposing the HsUT-A3 structure suggests that UTBinh-14 will not fit into its equivalent site due to its unfavorable environment (Fig. 3A): HsUT-A3’s binding pocket is significantly more negatively charged than that of HsUT-B, in particular for the 4-ethylbenzene binding region (Fig. 3, B and C). In silico binding free-energy calculations confirm this observation: Mutation of G323 in HsUT-B (equivalent to E383 in HsUT-A) to glutamic acid results in positive binding free-energy loss of 7.38 kcal/mol (from −14.32 to −6.94 kcal/mol), and A327S mutation results in loss of 1.09 kcal/mol (Fig. 3D). In addition, the 4-ethylbenzene group is in close proximity to N/D280 (~6 Å) (Fig. 2, B and C), whose variation is responsible for the Kidd blood group system (26). While mutating this residue from asparagine to aspartic acid does not change UTBinh-14’s binding free energy in our in silico calculations (0.12 kcal/mol), mutation to threonine (HsUT-A equivalent) did lead to a minor loss of 0.8 kcal/mol, suggesting its involvement in the inhibitor binding. Therefore, it is possible for a hypothetical inhibitor based on UTBinh-14 to show selectivity toward certain Kidd blood group with consequences for its potential as a drug candidate.
Fig. 3. Structural basis for the selectivity of UTBinh-14 to UT-B.
(A) Overlay of aligned HsUT-A residues (cyan) against HsUT-B residues (green) involved in UTBinh-14 binding. (B) Electrostatic surface charge representation of HsUT-B at the UTBinh-14 binding pocket. (C) Electrostatic surface charge representation of HsUT-A at the UTBinh-14 binding pocket. (D) Table of relative binding free-energy difference calculations for each residue mutation (from HsUT-B to HsUT-A).
Simple superimposition of HsUT-A3 and inhibitor-bound HsUT-B structure models suggests that the sulfonate ester group of the compound would sterically clash with HsUT-A’s P397 (equivalent to A337 in HsUT-B) (Fig. 3A). This sulfonate group at first appears to be important for the selectivity of UTBinh-14, as compounds featuring such moiety generally favor UT-B over UT-A (15, 27), and even the one with highest UT-A affinity is only 13 times more potent for UT-A than for UT-B (13). However, in silico binding free-energy calculations suggest that this may not be a major influence on the compound selectivity, with an energy loss of only 0.78 kcal/mol (Fig. 3D). On closer examination, this is due to A337’s side chain facing away from the sulfonate group in HsUT-B structure, hence mutation to proline making little difference to the compound binding (in contrast, side chain of HsUT-A’s P397 faces toward the binding pocket). Therefore, while the sulfonate group may be a contributing factor in UTBinh-14’s selectivity, it may not be as significant as its 4-ethylbenzene group.
An interesting feature of the binding mode of UTBinh-14 is its 2-thienyl group at the channel pore entry (Fig. 2C). This group is in close proximity to C338 with its sulfur atom facing toward the residue, and this has forced the side chain of C338 to rotate away (with respect to the apo state) from the pore entry due to steric clash. If the side chain did not rotate away from the inhibitor, its thiol group would have been within 2.3 Å of the 2-thienyl group’s sulfur atom. Therefore, it is possible that the inhibitor could form a covalent bond with UT-B at C338 if the 2-thienyl group was replaced with a thiol group. Therefore, our structure of inhibitor-bound HsUT-B suggests the possibility to generate irreversible covalent and selective inhibitors for UT-B.
UTBinh-14 and β-OG can bind to UT-B on its intracellular side
Our cryo-EM map of UTBinh-14–bound HsUT-B shows a small molecule feature on the intracellular side of the channel pore (Fig. 2, D and E). While its ESP feature is not as defined as that of UTBinh-14 on the extracellular side, it is still consistent with the shape of the inhibitor and not with any other potential molecules [e.g., β-dodecyl maltopyranoside (β-DDM)]. Real-space refinement with Phenix estimates its occupancy at 0.38, and the incomplete occupancy by UTBinh-14 suggests low binding affinity at this site compared to the extracellular one. While a published docking experiment predicted UTBinh-14 to also bind to the intracellular side (25), it is significantly different from our experimentally determined binding mode in that its 2-thienyl group is not directed toward the channel pore.
UTBinh-14’s binding mode on the intracellular side closely resembles that of the primary binding site on the extracellular side (Fig. 2A). Its 2-thienyl group is directed toward the channel pore, sterically blocking it, and its 4-ethylphenyl moiety is located in an adjacent hydrophobic pocket bounded by L116, Y122, and V175 (Fig. 2, D and E). These lead to UTBinh-14’s intracellular binding mode effectively being a rotational symmetry equivalent to its extracellular one, with the rotational axis located at the center of the pore (Fig. 2A). Such similarity of UTBinh-14’s extracellular and intracellular binding modes is not unexpected given that urea transporters feature a C2 pseudosymmetry around the same axis in the pore center. This also suggests that the binding of UTBinh-14 is driven by the sterics of the binding pocket (i.e., its shape) in addition to specific interactions between its functional groups and the protein.
While UTBinh-14 can bind to both intracellular and extracellular sides of UT-B, it is likely that its primary in vivo mode of inhibition is extracellular given our observations. First, drug molecules generally have higher bioavailability for extracellular surfaces than intracellular components. UTBinh-14’s apparent higher affinity (as inferred by higher occupancy) toward UT-B’s extracellular side would compound the bioavailability difference to saturate the extracellular binding pocket at far lower applied doses than it would the intracellular pocket in cell-based experiments. In addition, there is very little sequence difference between HsUT-B and HsUT-A in this intracellular pocket, and in silico mutation experiments of the few divergent residues (L116A and Y122H) showed less than 0.5 kcal/mol change in binding free energies (0.36 and 0.28 kcal/mol, respectively). Given that UTBinh-14 is a highly selective inhibitor for UT-B, such lack of difference in the intracellular pocket between UT-A and UT-B indicates that this is unlikely to be the main binding pocket for UTBinh-14 in a physiological setting.
Another ligand for the intracellular side of HsUT-B in our structures is n-octyl-β-d-glucopyranoside (β-OG), which is present in the crystal structure of apo-HsUT-B (fig. S5, C and D) since this short-chain detergent was used for crystallization experiments to improve crystal packing and diffraction but not for the EM studies. The glucoside group of β-OG forms hydrophilic interactions with several residues, including Q68, L364, and V367. On the other hand, its hydrophobic chain, which partially blocks the channel pore, is located in a mildly hydrophilic pocket formed by L121, Y122, N165, and V175. Such binding mode of β-OG closely matches that of UTBinh-14: β-OG’s hydrophilic glucoside positioned at the same site as the UTBinh-14’s thieno[2,3-e][1,2,3]triazolo[1,5-a]pyrimidine (TTP) group, which is also hydrophilic, and its hydrophobic tail at the same site as the UTBinh-14’s 4-ethylbenzene group (which is hydrophobic as well).
While the intracellular UTBinh-14 and β-OG in our HsUT-B structures are likely artifacts of nonphysiological levels of each compound in a highly purified environment, they still provide useful clues about the potential binding poses of small molecules for de novo design of therapeutic inhibitors targeting this side of urea transporters. This can be particularly useful for UT-A inhibitor designs because (i) the extracellular side of UT-A1 is less accessible by drug molecules than the intracellular side in physiological settings since it faces the lumen of IMCD (3, 6), and (ii) structural similarities between UT-A and UT-B allow extrapolation of UT-B’s interactions with UTBinh-14 and β-OG to UT-A inhibitor designs.
Cryo-EM and MS identify phospholipids associating with UT-A and UT-B
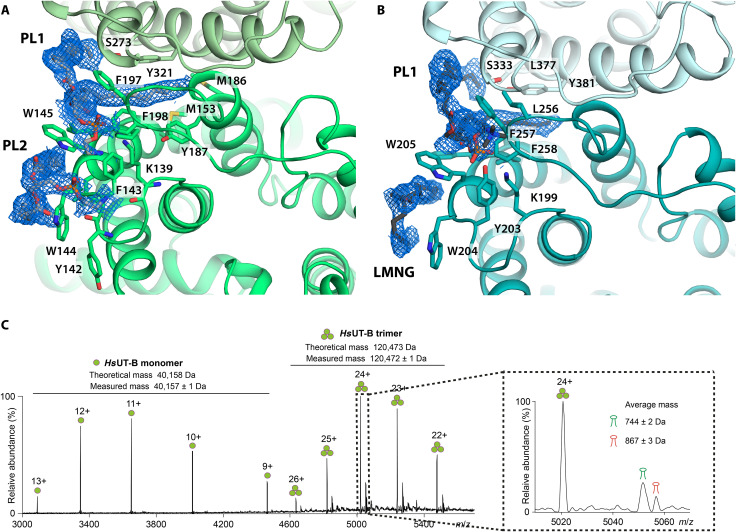
Cryo-EM maps of both HsUT-A and UTBinh-14–bound HsUT-B structures show multiple features consistent with phospholipids. These features are located on the extracellular side of the TM surface, including one between subunits (Fig. 4, A and B). For each of the lipids at the subunit interface, one of the phospholipid tails extends far into a hydrophobic cleft formed at the interface of the subunits, bounded by L274 and Y321 of one subunit and M153, M186, and Y187 of the other subunit in the case of HsUT-B (Fig. 4A). HsUT-A and HsUT-B appear to have similar lipid tail groups, with the lipid features in both HsUT-A and HsUT-B showing at least 16 carbon chains. For both proteins, the ends of the lipid tail groups are positioned close to the large internal cavities at the center of the trimers. Since the internal cavity of urea transporters is generally hypothesized to be filled with phospholipids (28), it is possible that this tail-binding cleft serves as a gate for lipids to move in and out of the cavity.
Fig. 4. Cryo-EM structures of HsUT-A and HsUT-B show phospholipids occupying clefts at the subunit interfaces.
(A) Extracellular view of phospholipids in inhibitor-bound HsUT-B structure. PE lipids are modeled in. Light gray, phospholipids. (B) Extracellular view of phospholipids in HsUT-A structure. Dark gray, phospholipid and LMNG at equivalent lipid site to HsUT-B’s. (C) Native MS spectrum of HsUT-B. Apo-HsUT-B and two series of lipid-bound peaks are observed with average masses of 744 ± 2 Da and 867 ± 3 Da, respectively (insert).
The map features for the phospholipid head groups are either very weak or nonexistent, especially for HsUT-B’s (Fig. 4A), making their modeling inaccurate. Therefore, we could not determine the identity of the lipids from our structural data alone. We subsequently performed native MS analysis of HsUT-B, where two adducts peaks were observed (Fig. 4C). In combination with lipidomics analysis, we identified these two adducts as phosphatidylethanolamines (PEs) and phosphatidylinositols (PIs) (fig. S6, A and B). Unlike PE, PI is only a minor component of mammalian plasma membrane, with the small amount rapidly phosphorylated to produce phosphatidylinositides for signaling, and even less is present on the outer side of the membrane bilayer (29, 30). Therefore, the enrichment of PI suggests either specific binding to UT-B or its sequestration to cytoplasmic organelles such as the Golgi complex, which have significantly higher levels of PI (31).
The lipid binding modes in our structures are similar to that observed in a crystal structure of an Escherichia coli ammonia transporter EcAmtB (fig. S6, C and D) (32), which is structurally and evolutionarily related to urea transporters. EcAmtB’s annular lipids were identified as phosphatidylglycerols (PGs) and cardiolipin, whose head groups are net negatively charged like the PI lipids present in UT-B. Given the structural similarities, PIs may have a stabilizing influence on human urea transporters as PGs do on EcAmtB, for example, by maintaining its homotrimeric state (33).
Of note, we do not observe lipid- or detergent-like feature in the central cavities of urea transporters despite hypotheses that they would be filled with lipids (19, 20). Both crystal maps and cryo-EM reconstructions at subatomic resolutions of <4 Å commonly show “belts” of well-associating detergents and/or phospholipids around the TM surfaces of membrane proteins, such as those of EcAmtB [Protein Data Bank (PDB) ID: 4NH2] (34), a lipid scramblase TMEM16F (6QP6) (35), a motor protein prestin (7LGU) (36), and a ligand-gated calcium channel TRPA1 (6PQQ) (37). Our urea transporter structures also show a number of lipid and detergent-like features around their TM surfaces, some of which are modeled; however, such features are lacking in their internal cavities, with only non–lipid-like disordered features seen at low contour levels. It also appears that these cavities would not be sterically favorable for phospholipids, as insufficient openings exist to accommodate the lipid head groups on either side of the bilayer for all urea transporters, including HsUT-A and HsUT-B. Nevertheless, it is still possible for them to be filled with lipids and/or have functional roles, as was the case with ATG9A whose central cavity is thought to aid in its lipid scramblase activity and in maintaining osmotic gradient across the membrane (38). Supporting this speculation is the structures of a functionally related (but structurally distinct) bacterial urea channel UreI in Helicobacter pylori (39, 40), which feature lipid-like densities in their internal cavities. As this cavity is a consistent feature of urea transporters not present in their structural and functional homologs such as ammonia transporters EcAmtB, HsRhCG, and HpUreI, as well as it will be interesting to study any physiological roles it may have.
In conclusion, both UT-A and UT-B are major targets for the development of a new class of diuretics, and their structural characterizations in our study will be valuable inputs for the drug discovery programs. We have identified the extracellular pocket as the primary binding site for the inhibitor UTBinh-14 with our structures, rationalized its selectivity to HsUT-B over HsUT-A, and provided specific ways to further improve the inhibitor design. In addition, our discovery of a second UTBinh-14 binding site on the intracellular side and the presence of β-OG in the apo-HsUT-B structure can be used to design new inhibitors targeting this site, with potentials not only for HsUT-B but also for HsUT-A, which features nearly identical pocket.
Our human urea transporter structures and native MS also provide the basis for studying their close interactions with phospholipids. The identification of PI lipids associating with HsUT-B is particularly interesting, and this suggests potential physiological roles such as protein stabilization, localization via lipid raft formation, or regulation by sequestration to PI-rich organelles. The last possibility is particularly compelling, since HsUT-A1 and HsUT-A3 are already known to be regulated in this way upon vasopressin-triggered phosphorylation (41, 42). While we could not directly test such hypothesis with lipidomics MS on purified HsUT-A due to its expression level issues in this study, this presents an exciting area of further investigation to understand the molecular physiology of urea transporters. The positioning of the lipid tails near the trimers’ central cavities is another interesting feature, and it remains to be investigated whether this cavity harbors lipids or has other physiological roles instead.
Last, our HsUT-B structure allows mapping of exonic variants responsible for the wide range of Kidd blood group antigens (fig. S8). In addition to the major N/D280 variations for Jk(a) and Jk(b) blood types, there are a number of Jk(null) phenotypes caused by a number of point mutations (26, 43, 44). Mapping these variants on the HsUT-B structure provides characterization of blood group antigens at molecular level, and this can also aid drug lead optimization programs by avoiding or using pockets near such variants. Our structures of human urea transporters therefore improve both the understanding of their function and the development of new diuretics targeting urea transport.
MATERIALS AND METHODS
Molecular biology, virus production, and protein expression
HsUT-B with N-terminal truncation to aid crystallization and N211A mutation for deglycosylation (SLC14A1Δ30N211A) and full-length HsUT-A were cloned into a pHTBV vector with C-terminal twin-Strep, 10-His, and green fluorescent protein tags. Baculoviruses for these constructs were generated following the standard protocol outlined in (45). E. coli DH10Bac cells were transformed with plasmids containing the target genes. Baculoviral DNA extracted from the cells was used to transfect Sf9 cells (Thermo Fisher Scientific) grown in Sf-900TM II media supplemented with 2% fetal bovine serum (Thermo Fisher Scientific) and incubated on an orbital shaker for 70 hours at 27°C. Produced baculovirus particles were harvested by centrifugation at 900g for 10 min and collecting the supernatants, and these were further amplified with Sf9 cells.
Each liter of Expi293F cell culture (Thermo Fisher Scientific) in Freestyle 293 Expression Medium (Thermo Fisher Scientific) was infected with 30 ml of P3 baculovirus-containing supernatant in the presence of 5 mM sodium butyrate. Cells were grown in an orbital shaker for 45 hours at 37°C and 8% CO2 before being harvested by centrifugation at 900g for 10 min, washed with phosphate-buffered saline, then centrifuged again. The cell-washed cell pellets were flash-frozen with liquid nitrogen (LN2), then stored at −80°C until needed.
Protein purification
For the purification of apo-HsUT-B, the following protocol was used. Whole-cell pellets expressing the target construct were resuspended to a total volume of 50 ml per 15 g of cell pellet with buffer A [20 mM Hepes (pH 7.5) and 150 mM NaCl] supplemented with 0.7% n-dodecyl beta-maltoside [(β-DDM) Generon] and 0.07% cholesteryl hemisuccinate (CHS; Generon). The cells were solubilized at 4°C for 1 hour with gentle rotation. Cell debris was pelleted by centrifugation at 45,000g for 1 hour. The clarified lysate was added to 0.5-ml bed volume of Strep-Tactin Superflow (IBA) per 100 ml of lysate and allowed to bind at 4°C for 1 hour. The resin was collected on a gravity-flow column and washed with buffer B (buffer A with 0.02% β-DDM and 0.002% CHS), then with buffer B supplemented with 2 mM adenosine 5′-triphosphate and 5 mM MgCl2. Protein was eluted with 10 CV of buffer B containing 5 mM d-desthiobiotin followed by tag cleavage by tobacco etch virus protease overnight and reverse purification. The samples were subjected to size exclusion chromatography with a Superose 6 Increase 10/300 column (GE Healthcare) pre-equilibrated with buffer C (buffer A with 0.7% β-OG). Peak fractions were pooled, concentrated to 100 μM and immediately used for crystallization experiments.
For the purification of UTBinh-14–bound HsUT-B, all buffers were supplemented with 10 μM UTBinh-14 by adding 100 mM dimethyl sulfoxide stocks. Buffer B was used instead of buffer C for size exclusion chromatography, and the peak fractions were concentrated to 100 μM.
For the purification of HsUT-A, same protocol as HsUT-B was used, except 0.07% lauryl maltose neopentyl glycol (LMNG) replacing β-DDM at solubilization step, and 0.005% LMNG and 0.00025% CHS being used instead of other detergents in the rest of buffers. Peak fractions from size exclusion chromatography were concentrated to 1 μM for graphene-coated grid sample and 20 μM for Quantifoil grid sample.
Crystallization of apo-HsUT-B
Apo-HsUT-B was crystallized by sitting-drop vapor diffusion method. Protein (1.9 mg/ml) was mixed using the Mosquito crystallization robot (TTP Labtech) with equal volume of precipitant containing 27% (v/v) PEG400, 0.1 M glycine (pH 9.5), and 0.05 M sodium chloride, in total volumes of 100, 150, or 200 nl. Successfully grown crystals were harvested using cryo-loops of appropriate size (Mitegen) and flash-frozen in liquid nitrogen. Diffraction data were collected at I24 Microfocus Beamline at Diamond Light Source (DLS, Harwell Science and Innovation Campus, Didcot, UK). Wavelength of x-ray radiation at the synchrotron was adjusted to 0.9686 Å. Raw x-ray diffraction data were analyzed using 3dii option of Xia2 (46). Details of the dataset collection parameters are available in Table 1.
Table 1. Data table for x-ray crystallography dataset of apo-HsUT-B.
RMSD, root mean square deviation.
| Property | Apo-HsUT-B |
|---|---|
| Data collection and processing | |
| Wavelength (Å) | 0.9686 |
| Resolution range (Å) | 56.16–2.40 (2.46–2.40) |
| Space group | P63 |
| Unit cell (Å) | 112.331 112.331 87.94 90 90 120 |
| Total reflections | 480,757 (28,936) |
| Unique reflections | 24,811 (1834) |
| Multiplicity | 19.4 |
| Completeness (%) | 100 (100) |
| Mean I/sigma(I) | 1.49 |
| Wilson B factor | 50.8 |
| R merge | 0.217 (3.143) |
| R meas | 0.229 (3.358) |
| CC1/2 | 0.997 (0.460) |
| Refinement | |
| Resolution range (Å) | 47.33–2.40 (2.48–2.40) |
| R work | 0.1754 (0.2826) |
| R free | 0.2251 (0.3307) |
| Number of atoms | |
| All nonhydrogen atoms | 2974 |
| Macromolecules | 2702 |
| Ligands | 201 |
| Solvent | 71 |
| Number of protein residues | 356 |
| RMSD | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.046 |
| Ramachandran plot | |
| Favored (%) | 97.46 |
| Allowed (%) | 2.54 |
| Outliers (%) | 0.00 |
| Validation | |
| Molprobity score | 1.22 |
| Rotamer outliers (%) | 0.00 |
| Clash score | 3.24 |
| B factor | |
| Average (Å2) | 61.0 |
| Macromolecules (Å2) | 59.27 |
| Ligands (Å2) | 92.03 |
| Solvent (Å2) | 58.71 |
| Number of TLS groups | 1 |
| PDB ID | 6QD5 |
Model building and refinement of apo-HsUT-B diffraction dataset
The structure was determined by molecular replacement with Phaser (47) using the BtUT-B structure model (PDB ID: 4EZC) used as the search model. Nonconserved residues of the search model were cropped using Chainsaw (48) and the output was used as template for manual refinement in Coot (49). The models were subjected to cycles of manual refinements in Coot and Phenix real-space refine (50). Geometries of the models were verified with MolProbity function (51) in Phenix.
Cryo-EM sample preparation and data collection
HsUT-B at 100 μM was frozen on Quantifoil Au R1.2/1.3 300-mesh grids glow discharged on a high setting for 30 s, with plunge-freezing performed on a Vitrobot Mark IV (Thermo Fisher Scientific) set to 100% humidity and 4°C. HsUT-A at 1 μM was frozen on Quantifoil Au R0.6/1 300-mesh grids with a graphene monolayer support using a protocol similar to Han et al. (52) and Naydenova et al. (53). Graphene grids were glow discharged on a low setting for 5 s, with plunge-freezing performed on Vitrobot Mark IV set to 100% humidity and 4°C. HsUT-A at 10 μM was frozen on Quantifoil Au R1.2/1.3 300-mesh grids glow discharged on a high setting for 30 s, with pluge-freezing performed on a Vitrobot Mark IV set to 100% humidity and 4°C.
The cryo-EM datasets were collected on a Titan Krios (Thermo Fisher Scientific) operating at 300 keV at Oxford Particle Imaging Centre (Oxford, UK), where dose-fractionated micrographs were collected on a K2 detector with beam tilt collection mode on Serial EM. Details for each collected dataset are available in Table 2.
Table 2. Data table for cryo-EM data collection, refinement, and validation statistics of UTBinh-14–bound HsUT-B and apo-HsUT-A.
| HsUT-B (inhibited) | HsUT-A (graphene coated) | HsUT-A (normal/tilt) | |
|---|---|---|---|
| Data collection | |||
| Microscope | Titan Krios | Titan Krios | Titan Krios |
| Detector | K2 | K2 | K2 |
| Voltage (kV) | 300 | 300 | 300 |
| Magnification | 105,000 | 105,000 | 105,000 |
| Collection mode | Counting | Counting | Counting |
| Electron exposure (e/Å2) | 39.6 | 52.6 | 53.3/52.0 |
| Number of frames | 40 | 50 | 50 |
| Pixel size | 0.82 | 0.82 | 0.82 |
| Defocus range (μm; steps) | −0.8 to −2.6 (−0.2) | −1.0 to −2.6 (−0.2) | −1.0 to −2.6 (−0.2)/−1.6 to −1.8 (0.2) |
| Number of movies | 8221 | 4431 | 4237/7028 |
| Data processing | |||
| Initial number of particles | 2,288,882 | 723,119 | 1,581,566/2,527,272 |
| Number of particles after 2D classification | 284,410 | 180,511 | 137,735/81,266 |
| Symmetry | C3 | C3 | C3 |
| Number of particles used for 3D refinement | 258,739 | 180,448 | 218,993 |
| Map resolution [Å; Fourier Shell Correction (FSC) threshold = 0.143] | 2.6 | 2.9 | 3.0 |
| Resolution range (Å) | 2.4–22.1 | 2.6–5.6 | 2.7–32.3 |
| Map sharpening B factor (Å2) | 109 | 138 | 141 |
| Refinement | |||
| Model resolution(Å; FSC threshold = 0.5) | 2.7 | 3.1 | |
| Model composition | |||
| Nonhydrogen atoms | 9378 | 8487 | |
| Protein residues | 1065 | 1005 | |
| Ligands | 66 | 24 | |
| RMSD | |||
| Bond lengths (Å) | 0.012 | 0.009 | |
| Bond angles (°) | 1.156 | 0.852 | |
| Validation | |||
| Molprobity score | 1.72 | 1.64 | |
| Clash score | 7.08 | 7.97 | |
| Rotamer outliers | 0.35 | 0.98 | |
| Ramachandran plot | |||
| Favored (%) | 95.18 | 96.70 | |
| Allowed (%) | 4.82 | 2.97 | |
| Disallowed (%) | 0.00 | 0.33 | |
| EMDB code | EMD-16112 | EMD-16110 | EMD-16111 |
| PDB code | 8BLP | 8BLO | |
Cryo-EM data processing
Micrographs were imported to Cryosparc 2.14 and motion-corrected with its Patch Motion Correction function (54). After defocus estimation with its Patch CTF function, particles were picked initially with blob picking and extracted with 100 pixel box size. After iterative two-dimensional (2D) classifications, classes with rare particle orientations were selected, which were then subjected to Topaz training and extraction jobs (55). These particles were then cleaned up with duplicated particle removal function, followed by extraction to 100 pixel box size and 2D classifications. The polished particles were used for ab initio model reconstruction function on Cryosparc with two classes generated for further polishing. These were then used for two cycles of nonuniform refinement, per-particle motion correction, and CTF refinement, then the reconstructions with highest nominal resolutions on Fourier Shell Correlation curve and map quality on visual inspection were used for further analyses.
Model building and refinement of cryo-EM datasets
The HsUT-B structure model from crystallography was fitted to the ESP map of the UTBinh-14–bound HsUT-B. In the case of the HsUT-A dataset, the HsUT-B structure model’s sequence was replaced with HsUT-A’s using the Chainsaw function in CCP4i2 (48, 56), which was then fitted to its ESP map. The models were subjected to cycles of manual refinements in Coot and Phenix real-space refine (49, 50). Geometries of the models were verified with MolProbity function in Phenix (51). Model coordinates and restraints for the UTBinh-14 compound were generated using AceDRG (57). UTBinh-14 models were fitted to ESP features consistent with the compound, with its occupancy estimated with Phenix.
Relative binding free energy calculations
The model of UTBinh-14–bound HsUT-B structure was loaded to ICM-Pro (Molsoft) and converted to an ICM-compliant model with pH set to 7.5. Relative binding energy differences (ΔΔG) of residue mutation from HsUT-B to equivalent amino acids in HsUT-A were calculated with Try Mutations function of ICM-Pro (58).
Native MS
HsUT-B samples were buffer exchanged to 200 mM ammonium acetate with 2× CMC β-OG, loaded into a gold-coated needle, and introduced into a Q-Exactive UHMR mass spectrometer (Thermo Fisher Scientific). The following parameters were used: Capillary voltage was set to 1.2 kV, source fragmentation was set to 25 eV, injection flatapole was set to 5 V, inter flatapole was set to 4 V, bent flatapole was set to 2 V, resolution was set to 17500 at mass/charge ratio (m/z) 200, higher-energy C-trap dissociation (HCD) energy was set to 100 eV, and trapping gas pressure was set to 7.5.
Lipidomics analysis
The bound lipids of HsUT-B samples were extracted by chloroform-methanol (2:1, v/v) as described previously (59) and dried using a speedVac vacuum concentrator (Thermo Fisher Scientific). The samples was then dissolved in 80% methanol. For liquid chromatography–tandem MS (LC-MS/MS) analysis, lipids were separated on a C18 column (Acclaim PepMap 100, C18, 75 μm × 150 mm, 3 μm, Thermo Fisher Scientific) by a Dionex UltiMate 3000 RSLC Nano System connected to an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher Scientific). The mobile phase A is acetonitrile:H2O (60:40), 10 mM ammonium formate, and 0.1% formic acid, and mobile phase B is isopropanol:acetonitrile (90:10), 10 mM ammonium formate, and 0.1% formic acid. Lipids were separated with a gradient of 32 to 99% buffer B at a flow rate of 300 nl/min. The Orbitrap Eclipse was operated in negative mode. Spray voltage was set to −2.1 kV and ion transfer tube temperature was at 320°C. For data-dependent acquisition, full MS scan range was set to 200 to 2000 with a resolution of 120,000. Fragments were acquired using a normalized collision energy at 25/30/35. Phospholipid identifications were performed manually using Xcalibur 4.4.
In-gel digest MS
The identity of proteins in solution or in gel bands excised following SDS–polyacrylamide gel electrophoresis (PAGE) was confirmed by tryptic digestion and tandem MS (LC-MS/MS). SDS-PAGE gel bands were excised as 1 mm by 4 mm slices using a gel cutting tip (GeneCatcher, web Scientific) and stored in 10% MeOH at 4°C. Before digestion, the methanol solution was removed and replaced with 100% acetonitrile for 2 min. The solution was then removed and replaced with 100 μl of 100 mM NH4HCO3 (pH 8.0). One microliter of 1 M dithiothreitol was added and incubated at 56°C for 40 min. Four microliters of 1 M iodoacetamide was then added and the reaction incubated at ambient temperature in the dark for 20 min. A further 1 μl of 1 M dithiothrietol, 200 μl of 100 mM NH4HCO3, and 1 μl of trypsin solution (sequencing grade, Sigma-Aldrich; 1 mg/ml in 0.01 M HCl) was then added. Tryptic digestion proceeded at 37°C for 16 hours and was terminated by addition of 3 μl of formic acid. LC-MS/MS was performed using a Dionex U3000 Nano HPLC coupled to a Bruker Amazon ETD ion trap mass spectrometer. One microliter of tryptic digest was loaded on to a 200 μm inside diameter × 5 cm PS-DVB monolith column (PepSwift, Dionex Corp.) A linear gradient of 0% B to 15% B was developed over 5 min, followed by a second linear gradient from 15% B to 40% B over 2 min. The column was washed at 90% for 2 min and then equilibrated at 90% B for a further 6 min. Solvent A was 2% (v/v) acetonitrile, 0.1% formic acid in water, and solvent B was 80% acetonitrile and 0.1% formic acid. The flow rate was 2.5 μl/min. The mass spectrometer was operated in positive ion, standard enhanced mode with a scan rate of 8100 m/z per sec and a scan range of 250 to 1800 m/z. The trap accumulation time was 200 ms and the accumulation target was 200,000 counts. Data-dependent peptide fragmentation was performed in Auto MSMS mode. Compound extraction and peptide deconvolution was performed using the DA data analysis program (Bruker Daltonik). Database searching was performed using the Mascot 2.2.04 search algorithm (Matrix Science) with the following search parameters: Charge states +2, +3; MS tolerance 1.5 Da; MSMS tolerance 0.5 Da; UniProt_SwissProt database without taxonomic restrictions.
Acknowledgments
We thank E. Maclean, L. Carrique, H. Duyvesteyn, and T. Walters at Oxford Particle Imaging Centre (Oxford, UK) for the assistance with electron microscopes. We also thank B. Marsden at Centre for Medicines Discovery (Oxford, UK) for assistance with cluster maintenance.
Funding: This research was carried out with funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement nos. 875510 (EUbOPEN) and 115766 (UltraDD). The JU receives support from the European Union’s Horizon 2020 research and innovation program and EFPIA and Ontario Institute for Cancer Research, Royal Institution for the Advancement of Learning, McGill University, Kungliga Tekniska Hoegskolan, and Diamond Light Source Ltd. This research was also funded in part by the Wellcome Trust (grant no. 106169/Z/14/Z). Oxford Particle Imaging Centre was funded by a Wellcome Trust JIF award (grant no. 060208/Z/00/Z) and is supported by equipment grants from WT (093305/Z/10/Z). H.T. and C.V.R. thank the support of the Wellcome Trust grant number 221795/Z/20/Z.
Author contributions: This project was designed by K.L.D. and G.C. G.C. and L.D. performed construct designs. N.A.B.-B., L.D., and D.W. performed molecular biology. G.M., S.M.M.M., M.S., G.C., and L.D. performed protein expression. G.C., M.S., and L.D. performed protein purification. G.C. and L.D. carried out protein crystallography and data processing. W.v.P. contributed graphene-coated EM grids used in this study. G.C. carried out cryo-EM sample preparation, data collection, and data processing. G.C., L.D., and A.S. refined structure models, which were proofread by G.C., A.S., and A.C.W.P. for internal validation. G.C. performed in silico experiments. H.T. performed MS experiments. R.C. performed gel-band MS. G.C. prepared the manuscript. G.C., K.L.D., H.T., C.V.R., J.-P.T. and W.v.P. revised the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The mammalian expression constructs can be provided by G.C. pending scientific review and a completed material transfer agreement. Requests for the constructs should be submitted to G.C. The coordinates and reflection files from the x-ray structure of apo-HsUT-B are deposited in the PDB under the entry 6DQ5. The cryo-EM map of HsUT-B in complex with antagonist is deposited in the Electron Microscopy Data Bank as entry EMD-16112 and the corresponding coordinates of the model are deposited in the PDB under entry 8BLP. The cryo-EM maps of HsUT-A have been deposited in the Electron Microscopy Data Bank as entries EMD-16110 (graphene coated) and EMD-16111 (tilted collection) and the corresponding model of HsUT-A (graphene coated) in the PDB as entry 8BLO.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Supplementary Text
References
REFERENCES AND NOTES
- 1.T. Titko, L. Perekhoda, I. Drapak, Y. Tsapko, Modern trends in diuretics development. Eur. J. Med. Chem. 208, 112855 (2020). [DOI] [PubMed] [Google Scholar]
- 2.G. Stewart, The emerging physiological roles of the SLC14A family of urea transporters. Br. J. Pharmacol. 164, 1780–1792 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.C. P. Smith, Mammalian urea transporters. Exp. Physiol. 94, 180–185 (2009). [DOI] [PubMed] [Google Scholar]
- 4.A. C. Jones, F. Pinki, G. S. Stewart, D. A. Costello, Inhibition of urea transporter (UT)-B modulates LPS-induced inflammatory responses in BV2 microglia and N2a neuroblastoma cells. Neurochem. Res. 46, 1322–1329 (2021). [DOI] [PubMed] [Google Scholar]
- 5.S. Azouzi, M. Gueroult, P. Ripoche, S. Genetet, Y. Colin Aronovicz, C. le van Kim, C. Etchebest, I. Mouro-Chanteloup, Energetic and molecular water permeation mechanisms of the human red blood cell urea transporter B. PLOS ONE 8, e82338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.J. D. Klein, M. A. Blount, J. M. Sands, Urea transport in the kidney. Compr. Physiol. 1, 699–729 (2011). [DOI] [PubMed] [Google Scholar]
- 7.J. M. Sands, M. A. Blount, Genes and proteins of urea transporters. Subcell. Biochem. 73, 45–63 (2014). [DOI] [PubMed] [Google Scholar]
- 8.S. A. Amin, S. Nandi, S. K. Kashaw, T. Jha, S. Gayen, A critical analysis of urea transporter B inhibitors: Molecular fingerprints, pharmacophore features for the development of next-generation diuretics. Mol. Divers. 26, 2549–2559 (2022). [DOI] [PubMed] [Google Scholar]
- 9.S. Wang, Y. Xu, Y. Zhao, S. Zhang, M. Li, X. Li, J. He, H. Zhou, Z. Ge, R. Li, B. Yang, N-(4-acetamidophenyl)-5-acetylfuran-2-carboxamide as a novel orally available diuretic that targets urea transporters with improved PD and PK properties. Eur. J. Med. Chem. 226, 113859 (2021). [DOI] [PubMed] [Google Scholar]
- 10.S. Nandi, S. Sanyal, S. A. Amin, S. K. Kashaw, T. Jha, S. Gayen, Urea transporter and its specific and nonspecific inhibitors: State of the art and pharmacological perspective. Eur. J. Pharmacol. 911, 174508 (2021). [DOI] [PubMed] [Google Scholar]
- 11.M. Li, Y. Zhao, S. Zhang, Y. Xu, S. Y. Wang, B. W. Li, J. H. Ran, R. T. Li, B. X. Yang, A thienopyridine, CB-20, exerts diuretic activity by inhibiting urea transporters. Acta Pharmacol. Sin. 41, 65–72 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Y. Zhao, M. Li, B. Li, S. Zhang, A. Su, Y. Xing, Z. Ge, R. Li, B. Yang, Discovery and optimization of thienopyridine derivatives as novel urea transporter inhibitors. Eur. J. Med. Chem. 172, 131–142 (2019). [DOI] [PubMed] [Google Scholar]
- 13.S. Lee, O. Cil, E. Diez-Cecilia, M. O. Anderson, A. S. Verkman, Nanomolar-potency 1,2,4-triazoloquinoxaline inhibitors of the kidney urea transporter UT-A1. J. Med. Chem. 61, 3209–3217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.F. Li, T. Lei, J. Zhu, W. Wang, Y. Sun, J. Chen, Z. Dong, H. Zhou, B. Yang, A novel small-molecule thienoquinolin urea transporter inhibitor acts as a potential diuretic. Kidney Int. 83, 1076–1086 (2013). [DOI] [PubMed] [Google Scholar]
- 15.M. O. Anderson, J. Zhang, Y. Liu, C. Yao, P. W. Phuan, A. S. Verkman, Nanomolar potency and metabolically stable inhibitors of kidney urea transporter UT-B. J. Med. Chem. 55, 5942–5950 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.B. Huang, H. Wang, D. Zhong, J. Meng, M. Li, B. Yang, J. Ran, Expression of urea transporter b in normal and injured brain. Front. Neuroanat. 15, 591726 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.X. Geng, T. Lei, H. Zhou, W. Yao, W. Xin, B. Yang, The knockout of urea transporter-B improves the hemorheological properties of erythrocyte. Clin. Hemorheol. Microcirc. 65, 249–257 (2017). [DOI] [PubMed] [Google Scholar]
- 18.R. R. Handley, S. J. Reid, R. Brauning, P. Maclean, E. R. Mears, I. Fourie, S. Patassini, G. J. S. Cooper, S. R. Rudiger, C. McLaughlan, P. J. Verma, J. F. Gusella, M. MacDonald, H. J. Waldvogel, C. S. Bawden, R. L. M. Faull, R. G. Snell, Brain urea increase is an early Huntington's disease pathogenic event observed in a prodromal transgenic sheep model and HD cases. Proc. Natl. Acad. Sci. U.S.A. 114, E11293–E11302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.E. J. Levin, Y. Cao, G. Enkavi, M. Quick, Y. Pan, E. Tajkhorshid, M. Zhou, Structure and permeation mechanism of a mammalian urea transporter. Proc. Natl. Acad. Sci. U.S.A. 109, 11194–11199 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.E. J. Levin, M. Quick, M. Zhou, Crystal structure of a bacterial homologue of the kidney urea transporter. Nature 462, 757–761 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.N. Lucien, F. Sidoux-Walter, N. Roudier, P. Ripoche, M. Huet, M. M. Trinh-Trang-Tan, J. P. Cartron, P. Bailly, Antigenic and functional properties of the human red blood cell urea transporter hUT-B1*. J. Biol. Chem. 277, 34101–34108 (2002). [DOI] [PubMed] [Google Scholar]
- 22.K. Ishibashi, S. Sasaki, N. Yoshiyama, T. Shiigai, J. Takeuchi, Generation of pH gradient across the rabbit collecting duct segments perfused in vitro. Kidney Int. 31, 930–936 (1987). [DOI] [PubMed] [Google Scholar]
- 23.H. H. Bengele, J. H. Schwartz, E. R. McNamara, E. A. Alexander, Chronic metabolic acidosis augments acidification along the inner medullary collecting duct. Am. J. Physiol. 250, F690–F694 (1986). [DOI] [PubMed] [Google Scholar]
- 24.R. C. Stanton, D. C. Boxer, J. L. Seifter, Expression of Na(+)-H+ exchange and ATP-dependent proton extrusion in growing rat IMCD cells. Am. J. Phys. 258, C416–C420 (1990). [DOI] [PubMed] [Google Scholar]
- 25.A. S. Verkman, C. Esteva-Font, O. Cil, M. O. Anderson, F. Li, M. Li, T. Lei, H. Ren, B. Yang, Small-molecule inhibitors of urea transporters. Subcell. Biochem. 73, 165–177 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.S. Lawicki, R. B. Covin, A. A. Powers, The Kidd (JK) blood group system. Transfus. Med. Rev. 31, 165–172 (2017). [DOI] [PubMed] [Google Scholar]
- 27.M. H. Levin, R. de la Fuente, A. S. Verkman, Urearetics: A small molecule screen yields nanomolar potency inhibitors of urea transporter UT-B. FASEB J. 21, 551–563 (2007). [DOI] [PubMed] [Google Scholar]
- 28.E. J. Levin, M. Zhou, Structure of urea transporters. Subcell. Biochem. 73, 65–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. P. Zewe, A. M. Miller, S. Sangappa, R. C. Wills, B. D. Goulden, G. R. V. Hammond, Probing the subcellular distribution of phosphatidylinositol reveals a surprising lack at the plasma membrane. J. Cell Biol. 219, e201906127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. G. Pemberton, Y. J. Kim, J. Humpolickova, A. Eisenreichova, N. Sengupta, D. J. Toth, E. Boura, T. Balla, Defining the subcellular distribution and metabolic channeling of phosphatidylinositol. J. Cell Biol. 219, e201906130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D. Casares, P. V. Escribá, C. A. Rosselló, Membrane lipid composition: Effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. Int. J. Mol. Sci. 20, 2167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.A. Laganowsky, E. Reading, T. M. Allison, M. B. Ulmschneider, M. T. Degiacomi, A. J. Baldwin, C. V. Robinson, Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.K. Gupta, J. A. C. Donlan, J. T. S. Hopper, P. Uzdavinys, M. Landreh, W. B. Struwe, D. Drew, A. J. Baldwin, P. J. Stansfeld, C. V. Robinson, The role of interfacial lipids in stabilizing membrane protein oligomers. Nature 541, 421–424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.S. Khademi, J. O'Connell III, J. Remis, Y. Robles-Colmenares, L. J. W. Miercke, R. M. Stroud, Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.35 Å. Science 305, 1587–1594 (2004). [DOI] [PubMed] [Google Scholar]
- 35.C. Alvadia, N. K. Lim, V. Clerico Mosina, G. T. Oostergetel, R. Dutzler, C. Paulino, Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F. eLife 8, e44365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J. Ge, J. Elferich, S. Dehghani-Ghahnaviyeh, Z. Zhao, M. Meadows, H. von Gersdorff, E. Tajkhorshid, E. Gouaux, Molecular mechanism of prestin electromotive signal amplification. Cell 184, 4669–4679.e13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Y. Suo, Z. Wang, L. Zubcevic, A. L. Hsu, Q. He, M. J. Borgnia, R. R. Ji, S. Y. Lee, Structural insights into electrophile irritant sensing by the human TRPA1 channel. Neuron 105, 882–894.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.S. Maeda, H. Yamamoto, L. N. Kinch, C. M. Garza, S. Takahashi, C. Otomo, N. V. Grishin, S. Forli, N. Mizushima, T. Otomo, Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat. Struct. Mol. Biol. 27, 1194–1201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Y. Cui, K. Zhou, D. Strugatsky, Y. Wen, G. Sachs, Z. H. Zhou, K. Munson, pH-dependent gating mechanism of the Helicobacter pyloriurea channel revealed by cryo-EM. Sci. Adv. 5, eaav8423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D. Strugatsky, R. McNulty, K. Munson, C. K. Chen, S. M. Soltis, G. Sachs, H. Luecke, Structure of the proton-gated urea channel from the gastric pathogen Helicobacter pylori. Nature 493, 255–258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.M. A. Blount, J. D. Klein, C. F. Martin, D. Tchapyjnikov, J. M. Sands, Forskolin stimulates phosphorylation and membrane accumulation of UT-A3. Am. J. Physiol. Renal Physiol. 293, F1308–F1313 (2007). [DOI] [PubMed] [Google Scholar]
- 42.C. Zhang, J. M. Sands, J. D. Klein, Vasopressin rapidly increases phosphorylation of UT-A1 urea transporter in rat IMCDs through PKA. Am. J. Physiol. Renal Physiol. 282, F85–F90 (2002). [DOI] [PubMed] [Google Scholar]
- 43.J. R. Hamilton, Kidd blood group system: A review. Immunohematology 31, 29–35 (2015). [PubMed] [Google Scholar]
- 44.T. Onodera, K. Sasaki, H. Tsuneyama, K. Isa, K. Ogasawara, M. Satake, K. Tadokoro, M. Uchikawa, JK null alleles identified from Japanese individuals with Jk(a−b−) phenotype. Vox Sang. 106, 382–384 (2014). [DOI] [PubMed] [Google Scholar]
- 45.P. Mahajan et al., in Structural Genomics: General Applications, Y. W. Chen, C.-P. B. Yiu, Eds. (Springer US, 2021), pp. 95–115. [Google Scholar]
- 46.G. Winter, xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 43, 186–190 (2010). [Google Scholar]
- 47.A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, R. J. Read, Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.N. Stein, CHAINSAW: A program for mutating pdb files used as templates in molecular replacement. J. Appl. Cryst. 41, 641–643 (2008). [Google Scholar]
- 49.P. Emsley, B. Lohkamp, W. G. Scott, K. Cowtan, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D. Liebschner, P. V. Afonine, M. L. Baker, G. Bunkóczi, V. B. Chen, T. I. Croll, B. Hintze, L. W. Hung, S. Jain, A. J. McCoy, N. W. Moriarty, R. D. Oeffner, B. K. Poon, M. G. Prisant, R. J. Read, J. S. Richardson, D. C. Richardson, M. D. Sammito, O. V. Sobolev, D. H. Stockwell, T. C. Terwilliger, A. G. Urzhumtsev, L. L. Videau, C. J. Williams, P. D. Adams, Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.C. J. Williams, J. J. Headd, N. W. Moriarty, M. G. Prisant, L. L. Videau, L. N. Deis, V. Verma, D. A. Keedy, B. J. Hintze, V. B. Chen, S. Jain, S. M. Lewis, W. B. Arendall III, J. Snoeyink, P. D. Adams, S. C. Lovell, J. S. Richardson, D. C. Richardson, MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Y. Han, X. Fan, H. Wang, F. Zhao, C. G. Tully, J. Kong, N. Yao, N. Yan, High-yield monolayer graphene grids for near-atomic resolution cryoelectron microscopy. Proc. Natl. Acad. Sci. U.S.A. 117, 1009–1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.K. Naydenova, M. J. Peet, C. J. Russo, Multifunctional graphene supports for electron cryomicroscopy. Proc. Natl. Acad. Sci. U.S.A. 116, 11718–11724 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.A. Punjani, J. L. Rubinstein, D. J. Fleet, M. A. Brubaker, cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 55.T. Bepler, A. Morin, M. Rapp, J. Brasch, L. Shapiro, A. J. Noble, B. Berger, Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.M. D. Winn, C. C. Ballard, K. D. Cowtan, E. J. Dodson, P. Emsley, P. R. Evans, R. M. Keegan, E. B. Krissinel, A. G. W. Leslie, A. McCoy, S. J. McNicholas, G. N. Murshudov, N. S. Pannu, E. A. Potterton, H. R. Powell, R. J. Read, A. Vagin, K. S. Wilson, Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.F. Long, R. A. Nicholls, P. Emsley, S. Gražulis, A. Merkys, A. Vaitkus, G. N. Murshudov, AceDRG: A stereochemical description generator for ligands. Acta Crystallogr. D Struct. Biol. 73, 112–122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.M. Schapira, M. Totrov, R. Abagyan, Prediction of the binding energy for small molecules, peptides and proteins. J. Mol. Recognit. 12, 177–190 (1999). [DOI] [PubMed] [Google Scholar]
- 59.E. G. Bligh, W. J. Dyer, A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
- 60.Y. Z. Tan, P. R. Baldwin, J. H. Davis, J. R. Williamson, C. S. Potter, B. Carragher, D. Lyumkis, Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Supplementary Text
References