Abstract
The neural population spiking activity recorded by intracortical brain-computer interfaces (iBCIs) contain rich structure. Current models of such spiking activity are largely prepared for individual experimental contexts, restricting data volume to that collectable within a single session and limiting the effectiveness of deep neural networks (DNNs). The purported challenge in aggregating neural spiking data is the pervasiveness of context-dependent shifts in the neural data distributions. However, large scale unsupervised pretraining by nature spans heterogeneous data, and has proven to be a fundamental recipe for successful representation learning across deep learning. We thus develop Neural Data Transformer 2 (NDT2), a spatiotemporal Transformer for neural spiking activity, and demonstrate that pretraining can leverage motor BCI datasets that span sessions, subjects, and experimental tasks. NDT2 enables rapid adaptation to novel contexts in downstream decoding tasks and opens the path to deployment of pretrained DNNs for iBCI control. Code: https://github.com/joel99/context_general_bci
1. Introduction
Intracortical neural spiking activity contains rich statistical structure reflecting the processing it subserves. For example, motor cortical activity during reaching is characterized with low-dimensional dynamical models [1, 2], and these models can predict behavior under external perturbation and provides an interpretive lens for motor learning [3–5]. However, new models are currently prepared for each experimental context, meaning separate datasets are collected for each cortical phenomena in each subject, for each session. Meanwhile, spiking activity structure is at least somewhat stable across these contexts; for example, dominant principal components (PCs) of neural activity can remain stable across sessions, subjects, and behavioral tasks [6–9]. This structure persists in spite of turnover in recorded neurons, physiological changes in the subject, or task changes required by the experiment [10, 11]. Conserved neural population structure suggests the opportunity for models that span beyond single experimental contexts, enabling more efficient, potent analysis and application.
In this work we focus on one primary use case: neuroprosthetics powered by intracortical brain computer interfaces (iBCIs). With electrical recordings of just dozens to hundreds of channels of neuronal population spiking activity, today’s iBCIs can relate this observed neural activity to behavioral intent, achieving impressive milestones such as high-speed speech decoding [12] and high degree-of-freedom control of robotic arms [13]. Even so, these iBCIs currently require arduous supervised calibration in which neural activity on that day is mapped to behavioral intent. At best, cutting-edge decoders have included training data from across several days, producing thousands of trials, which is still modest by deep learning standards [12]. Single-session models still dominate the Neural Latents Benchmarks (NLB), a primary representation learning benchmark for spiking activity [14]. Thus, despite the scientifically observed conserved manifold structure, there has been little adoption of neural population models that can productively aggregate data from broader contexts.
One possible path forward is deep learning’s seemingly robust recipe for leveraging heterogeneous data across domains: a generic model backbone (e.g. a Transformer [15]), unsupervised pretraining over broad data, and lightweight adaptation for a target context (e.g. through fine-tuning). The iBCI community has set the stage for this effort, for example with iBCI dataset releases (Section A.1) and Neural Data Transformer (NDT) [16], which shows Transformers, when prepared with masked autoencoding, model single-session spiking activity well. We hereafter refer to NDT as NDT1. Building on this momentum, we report that Transformer pretraining can apply to motor cortical neural spiking activity from iBCIs, and allows productive aggregation of data across contexts.
2. Related Work
Unsupervised neural data pretraining.
Unsupervised pretraining’s broad applicability is useful in neuroscience; little is common across neural datasets except the neural data itself. Remarkably, pretraining approaches across neural data modalities are similar; of the 4 sampled in Table 1, 3 use masked autoencoding as a pretraining objective (EEG uses contrastive learning), and 3 use a Transformer backbone (ECoG uses a CNN). Still, each modality poses different challenges for pretraining; for iBCIs that record spiking activity, the primary challenge is data instability [18]. The high spatial resolution of iBCI microelectrode arrays enables recording of individual neurons and provides the signal needed for high-performance rehabilitation applications, but this fine resolution also causes high sensitivity to shifts in recording conditions [11, 18]. iBCIs typically require recalibration within hours, relative to ECoG-BCIs that may not require recalibration for days [19]. At the macroscopic end, EEG and fMRI can mostly address inter-measurement misalignment through preprocessing (e.g. registration to an atlas).
Table 1. Neural data pretraining.
NDT2, like contemporary neural data models, are intermediate pretraining efforts, comparable to early-modern large language models like BERT [20]. Neural models vary greatly in task quality and data encoding; implanted devices (Microelectrodes, ECoG, SEEG) severely restrict the subject count available (especially with public data). Volume is estimated as full dataset size / model input size. ECoG = Electrocorticography’ SEEG = Stereo electroencephalography; EEG = Electroencephalography; fMRI = Functional magnetic resonance imaging.
| Modality | Task | Estimated Pretraining Volume | Subjects |
|---|---|---|---|
| Microelectrodes (ours) | Motor reaching | 0.25M trials | ~12 |
| ECoG: LFP [21] | Naturalistic behavior | 0.04M trials / 108 days [22] | 12 |
| SEEG: LFP [23] | Movie Viewing | 3.2M trials / 4.5K electrode-hrs | 10 |
| fMRI [24] | Varied (34 datasets) | 1.8M trials (12K scans) | 1.7K |
| EEG [25] | Clinical assessment | 0.5M trials / (26K runs [26]) | 11K |
| BERT [20] | Natural Language | 1M ‘trials’ (3.3B tokens) | - |
Data aggregation for iBCI.
Data aggregation for iBCI has largely been limited to multi-session aggregation, where a session refers to an experimental period lasting up to several hours. These efforts often combine data through a method called stitching [27]. For context, the extracellular spiking signals recorded on microelectrode arrays are sometimes “sorted” by the shape of their electrical waveforms, with different shapes attributed to spikes from distinct putative neurons. Sorted data has inconsistent dimensions across sessions, but as mentioned, activity across sessions has been observed to share consistent subspace structure, as e.g. identified by PCA [7]. Stitching aims to extract this stable subspace (and also resolve neuron count differences) by learning readin and readout layers per session. Stitching has been applied for BCI applications over half a year [28–30, 11]. However, even linear layers can add thousands of parameters, which risks overfitting in clinical iBCI data that comprise only dozens of trials.
Alternatively, many iBCI systems simply forgo spike sorting, which appears to have a minor performance impact [10, 18]. Then, multi-session data have consistent dimensions and can feed directly into a single model [10, 31, 32] (even if the units recorded in those dimensions shift [33]). Note that these referenced models also typically incorporate augmentation strategies centered around channel gain modulation, noising, or dropout, emphasizing robustness as a design goal.
Relative to existing efforts, NDT2 aims to aggregate a larger volume of data by considering other subjects and motor tasks (Fig. 1B). The fundamental tension we empirically study is whether the increased volume of pretraining is helpful despite the decreased relevance of heterogeneous data for any particular target context.
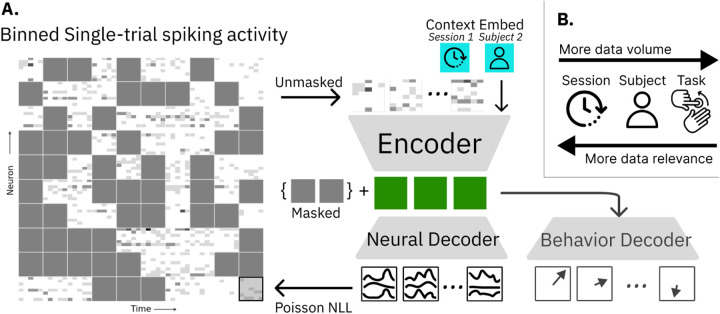
Figure 1.
A. NDT2 is a spatiotemporal Transformer encoder-decoder in the style of He et al. [17], operating on binned spiking activity. During masked autoencoding pretraining, a spike rate decoder reconstructs masked spikes from encoder outputs; downstream, additional decoders similarly use encoder outputs for behavior prediction (e.g. cursor movement). The encoder and decoders both receive context embeddings as inputs. These embeddings are learned vectors for each unique type of metadata, such as session or subject ID. B. NDT2 aims to enable pretraining over diverse forms of related neural data. Neural data from other sessions within a single subject are the most relevant but limited in volume, and may be complemented by broader sources of data.
Domain-adaptive vs. domain-robust decoding.
Given little training data, special approaches help enable BCI decoder use in new, shifted contexts. For example, decoders can be designed to be robust to hypothesized variability in recorded populations by promoting invariant representations through model or objective design [32, 34, 35]. Alternatively, decoders can be adapted to a novel context with further data collection, which is reasonable especially if only unsupervised neural data are required. Several approaches align data from novel contexts by learning an input mapping that minimizes distributional distance between novel encodings and pretraining encodings explicitly [11, 36, 37]. NDT2 also allows unsupervised adaptation, simply by fine-tuning without additional objectives.
3. Approach
3.1. Designing Transformers for unsupervised scaling on neural data
Transformers prepared with masked autoencoding (MAE) [20] are a competitive model for representation learning on spiking neural activity in single contexts, as measured by their performance on the NLB [14]. Cross-domain Transformer investment has also produced vital infrastructure for scaling to large datasets. Thus, we retain Transformer MAE [17], as the unsupervised pretraining recipe. This architecture is reviewed in Fig. 1A.
The primary design choice is then data tokenization, i.e. how to decompose the full iBCI spatiotemporal spiking activity into units that NDT2 will compute and predict over. Traditionally, the few hundred neurons in motor populations have been analyzed directly in terms of their population-level temporal dynamics [1]. NDT1 [16] follows this heritage and directly embeds the full population, with one token per timestep. Yet across contexts, the meaning of individual neurons may change, so operations to learn spatial representations may provide benefits. For example, Le and Shlizerman [38] and Liu et al. [35] add spatial attention to NDT’s temporal attention. Yet separate space-time attention can impair performance [39] and requires padding in both space and time when training over heterogeneous data. Still, the other extreme of full neuron-wise spatial attention has prohibitive computational cost. We compromise and group K neurons to a token (padding if needed), akin to pixel patches in ViTs [40]. Neural “patches” are embedded by concatenating projections of the spike counts in the patch. In pilot experiments, we find comparable efficiencies between this patching strategy and factorized attention. We opt for full attention design to easily adopt the asymmetric encoder-decoder proposed in [17]. This architecture first encodes unmasked tokens and uses these encodings to then decode masked tokens. This 2-step approach provides memory savings over the canonical uniform encoder architecture, as e.g. popularized in the language encoder, BERT [20].
We next consider token resolutions. In time, iBCI applications benefit from control rates of 50–100Hz [41]; we adopt 50Hz (20ms bins) and consider temporal context up to 2.5s (i.e. 125 tokens in time). Given a total context budget of 2K tokens (limited by GPU memory), this leaves a few dozen tokens for spatial processing. It is currently common to record from 100–200 electrodes with an interelectrode spacing of 400μm (Blackrock Utah Arrays) [29, 42], so our budget forces 16–32 channels per token. We note that future devices are likely to record from thousands of channels at a time with much higher spatial densities [43], which will warrant new spatial strategies.
NDT2 also receives learned context tokens. These tokens use known metadata to allow cheap specialization, analogous to prompt tuning [44] from language models or environment embeddings [45] from robotics. Specifically, we provide tokens reflecting session, subject, and task IDs.
3.2. Datasets
We pretrain models over an aggregation of datasets; the relative volume of different datasets are compared in Fig. 2A, with details in Section A.1. All datasets contains single-unit (sorted) or multi-unit (unsorted) spiking activity recorded from either monkey or human primary motor cortex (M1) during motor tasks. In particular, we focus evaluation on a publicly available monkey dataset, where the subjects performed self-paced reaching to random targets generated on a 2D screen (Random Target Task, RTT) [46], and unpublished human clinical BCI datasets. RTT data contains both sorted and unsorted activity from two monkeys over 47 sessions (~20K seconds per monkey). RTT is ideal for assessing data scaling as it features long, near-hourly sessions of a challenging task where decoding accuracy increases with data [47]. For comparison, in the Maze NLB task, which uses cued preparation and movement periods, decoding performance saturates by 500 trials [14]. Since RTT is continuous, we split each session into 1s trials in keeping with NLB [14].
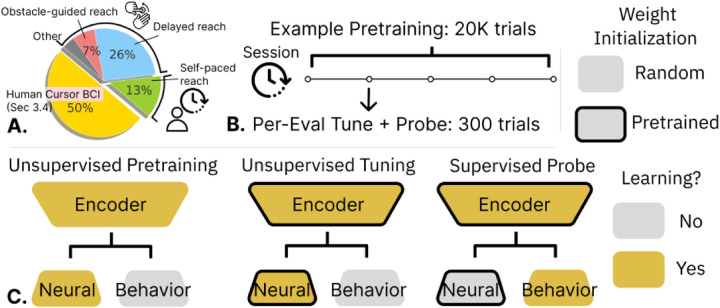
Figure 2. Model Training:
A. We model neural activity from human and monkey reach. In monkey models, evaluation sessions are drawn from a self-paced reaching dataset [46]; multi-session and multi-subject models pretrain with other sessions in these data. The multi-task model pretrains with the other monkey reach data. Human models use a similar volume of data. B. A multi-session model pretrains with data from an evaluation subject, with held-out evaluation sessions. Then, for each evaluation session, we first do unsupervised tuning off the pretrained model, and then train a supervised probe off of this tuned model. C. We show which model components are learned (receive gradients) during pretraining and the two tuning stages. For example, supervised probes use an encoder that received both pretraining and tuning on a target session. All tuning is end to end.
We also study M1 activity in 3 human participants with spinal cord injury (P2-P4). These participants have limited preserved motor function but can modulate neural activity in M1 using attempted movements of their own limbs. This activity can be decoded to control high degree-of-freedom behavior [13]; we restrict our study to 2D cursor control tasks to be most analogous to RTT, which also restricts targets to a 2D workspace. All experiments conducted with humans were performed under an approved Investigational Device Exemption from the FDA and were approved by the Institutional Review Board at the University of Pittsburgh. The clinical trial is registered at clinicaltrials.gov (ID: NCT01894802) and informed consent was obtained before any experimental procedures were conducted. Details on the implants and clinical trial are described in [42, 13], and similar task data are described in [48].
4. Results
We demonstrate the three requirements of a pretrained spiking neural data model for a BCI: a multi-context capable architecture (Section 4.1), beneficial scaled pretraining (Section 4.2) and practical deployment (Section 4.3, Section 4.4). We discuss the impact of context tokens in Section A.4.
Model preparation and evaluation.
Most experiments used a 6-layer encoder (~3M parameters). NDT2 adds a 2-layer decoder (0.7M parameters) over NDT1 [16]; we ran controls to ensure this extra capacity does not confer benefits to comparison models. To ensure that our models were not bottlenecked by compute or capacity in scaling (Section 4.2), models were trained to convergence with early stopping and progressively larger models were trained until no return was observed. We pretrain with causal attention where tokens cannot attend to future timesteps, as would be the case in realtime iBCI use (though bidirectional attention improves modeling). We pretrain with 50% masking and dropout of 0.1. Further hyperparameters were not swept in general experiments; initial settings were manually tuned in pilot experiments and verified to be competitive against hyperparameter sweeps. Further training details are given in Section A.3. We briefly compare against prior reported results, but to our knowledge there is no other work that attempts similar pretraining, so we primarily compare performance within NDT-family design choices.
Model preparation and evaluation follows several steps, as summarized in Fig. 2B and C. We evaluate models on randomly drawn held-out test data from select “target” sessions (selection is later detailed per experiment). Separate models are prepared for each target session, either through fine-tuning or by training from scratch for single-context models. For unsupervised evaluation, we use the Poisson negative log-likelihood (NLL) objective, which measures reconstruction performance of randomly masked bins of test trials (Fig. 2C middle). For supervised evaluation, we report the R2 of decoded kinematics, which for these experiments are 2D velocity of the reaching effector (Fig. 2C right). These supervised models are separately tuned off the per-evaluation session unsupervised model; so that all supervised scores receive the benefit of unsupervised modeling of target data [49].
4.1. NDT2 enables multicontext pretraining
We evaluate on five temporally spaced evaluation sessions of monkey Indy in the RTT dataset, with both sorted and unsorted processing. Both versions are important; sorted datasets contain information about spike identity and are broadly used in neuroscientific analysis, while unsorted datasets are frequently more practical in BCI applications. Velocity decoding is done by tuning all models further with a 2-layer Transformer probe (matching the reconstruction decoder). Here we provide the models with 5 minutes of data (300 training trials) for each evaluation session. This quantity is a good litmus test for transfer as it is sufficient to fit reasonable single-session models but remains practical for calibration. A 10% test split is used in each evaluation session (this small % is due to several sessions not containing much more than 300 trials). We pretrain models using approximately 20K trials of data, either with the remaining non-evaluation sessions of monkey Indy (Multi-Session), the sessions from the other monkey (Multi-Subject), or from non-RTT datasets entirely (Multi-Task, see Fig. 2).
Prior work in multi-session aggregation either use stitching layers or directly train on multi-day data with consistent unit count. Thus, we use NDT1 with stitching as a baseline for sorted data, and with or without stitching for unsorted data. NDT2 pads observed neurons in any dataset to the nearest patch multiple. All models receive identical context tokens.
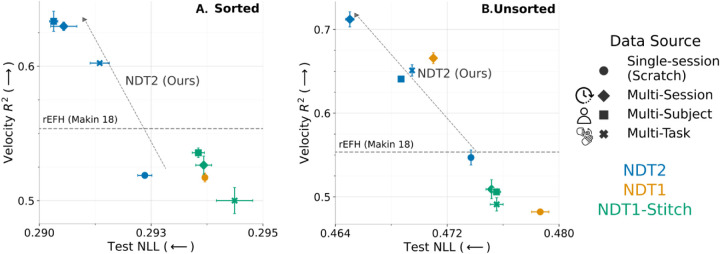
We show the performance of these pretrained models for sorted and unsorted data in Fig. 3. For context, we show single-session performance achieved by NDT1 and NDT2, and the decoding performance of the nonlinear rEFH model released with the dataset [47]. rEFH scores were estimated by linearly interpolating the 16ms and 32ms scores in Makin et al. [47].2 Single session performance for NDT1 and NDT2 is below this baseline. However, consistent with previous findings on the advantage of spatial modeling [38], we find single-session NDT2 provides some NLL gain over NDT1. Underperforming this established baseline is not too unexpected: NDT’s performance can vary widely depending on the extent of tuning (Transformers span a wide performance range on the NLB, see also Section A.3). Part of the value of pretraining is that it greatly simplifies the tuning needed for model preparation [50].
Figure 3.
NDT2 enables pretraining over multi-session, multi-subject, and multi-task data. We show unsupervised and supervised performance (mean of 5 sessions, SEM intervals of 3 seeds) on sorted (A) and unsorted (B) spiking activity. Higher is better for R2, lower is better for negative log-likelihood (NLL). Pretraining data is size-matched at 20Ks, except scratch single-session data. NDT2 improves with pretraining with all data sources, whereas stitching is ineffective. NDT1 aggregation is helpful but does not apply beyond session transfer. A reference well-tuned decoding score from the rEFH model is estimated [47].
However, in sorted data shown in Fig. 3A, all pretrained NDT2 models outperform rEFH and single-session baselines, both in NLL and kinematic decoding. Surprisingly, multi-subject data work as well as multi-session data, and multi-task data provide an appreciable improvement as well. NDT-Stitch performs much worse in all cases, and in fact, cross-task pretraining brings NDT-Stitch below the single-session baseline. We expect that stitching is less useful here than in other works because other works initialize their stitching layers by exploiting stereotyped task structure across settings (see PCR in [28]), and we cannot do this because RTT does not have this structure.
The unsorted data has consistent physical meaning (electrode location) across datasets within a subject, which may particularly aid cross-session transfer. Indeed, unsorted cross-session pretraining achieves the best decoding (> 0.7R2) in these experiments (Fig. 3B, blue diamond). The consistent dimensionality should not affect cross-task and cross-subject decoding, yet they also improve vs their sorted analogs, indicating the unsorted format benefits decoding in this dataset. Given this, we use unsorted formats in subsequent analysis of RTT. Otherwise, relative trends are consistent with the sorted case. Both analyses indicate that different pretraining distributions all provide some benefit for modeling a new target context, but also that not all types of pretraining data are equivalently useful.
4.2. NDT2 scaling across contexts
Naturally, cross-session data are likely the best type of pretraining data for its close relevance, but less relevant data can also occur in much greater volumes. To inform future pretraining data efforts, we perform three analyses to coarsely estimate the data affinity [51] of the three different context classes (cross-session, cross-subject, and cross-task). Previously these relationships have been grounded in shared linear subspaces [6–8]; we now quantify this in the more general generative model encompassed by DNN performance scaling.
Scaling pretraining size.
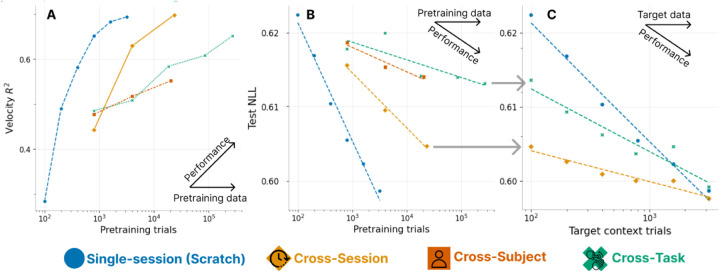
We consider transfer as we scale pretraining size, so as to extrapolate trends that might forecast the utility of progressively larger data. Specifically, we measure performance after tuning varied pretrained models with 100 trials of calibration in a novel context. We do this for both supervised (Velocity R2) and unsupervised (NLL) metrics in Fig. 4A and B respectively. To contextualize performance of cross-context scaling, we measure in-distribution scaling of intra-session data (Scratch). For example, the largest cross-session model tuned with 100 trials achieves a NLL similar to 1K trials of intra-session data (Fig. 4B, Cross-session vs Scratch). This shows that cross-session data can capture a practically long tail of single-session neural variance (experiments rarely exceed 1K trials), with nonsaturated benefits in pretraining dataset size. Alternately, cross-session pretraining allows decoding performance similar to the largest single-session model performance (Fig. 4A, Cross-session vs Scratch), but this scaling is starting to saturate.
Figure 4. Scaling of transfer on RTT.
We compare supervised R2 (A) and unsupervised NLL scaling (B) as we increase the pretraining dataset size. Each point is a model; non single-session models calibrate to evaluation sessions with 100 trials. All pretraining improves over the leftmost 100-trial single-session from-scratch model, though scaling benefits vary by data sources. C. We seek a convergence point between pretraining and training from scratch, as we increase the number of trials we use in our target context. Models converge by 3K trials.
Shallower slopes for cross-subject and cross-task pretraining on both metrics indicate poorer transfer to evaluation data. In the unsupervised case (Fig. 4B), cross-subject and cross-task transfer never exceed the NLL achieved with 400 single-session trials. Note that our task scaling may be pessimistic as we mix human data (Table 2) with monkey data to prepare the largest model, but the trend before this point is still shallow. However, these limitations do not clearly translate to the supervised metric (note that discrepant scaling metrics are also seen in language models [52]). For example, the decode R2 achieved by the largest task-pretrained model compares with in-session models at 800 trials; the same model’s NLL is most comparable that of a 300 trial single-session model.
Table 2. Human reach intent decoding.
We analyze how pretraining data impacts offline decoding in two people, P2 and P3. Checks (✓) indicate the data used in pretraining beyond task- and subject-specific data; human data totals 100K trials for P2 and 30K trials for P3. Intervals are SEM over 3 fine-tuning seeds. Pretraining transfers across task and somewhat across subject, but there is no benefit from monkey data.
| Neural data (Unsup. pretrain) | Behavior (Sup. pretrain) | Velocity R2 (↑) | ||||
|---|---|---|---|---|---|---|
| Cross-Subject | Cross-Task | +130K-Monkey | +24K RTT (Monkey) | P2 | P3 | |
| ✓ | ✓ | 0.503±0.020 | 0.515±o.oo8 | |||
| 2) | ✓ | 0.487±o.oo7 | 0.509±o.o16 | |||
| 3) | ✓ | 0.444±o.oo7 | 0.493±o.oo2 | |||
| 4) | ✓ | ✓ | ✓ | ✓ | 0.486±o.o12 | 0.472±o.o19 |
| 5) | ✓ | ✓ | ✓ | 0.490±o.oo7 | 0.477±o.oi8 | |
| 6) | ✓ | ✓ | ✓ | 0.474±o.oo9 | 0.491±o.o1o | |
| 7) | ✓ | 0.443±o.oo5 | 0.455±o.o13 | |||
| Smoothed spike ridge regression (OLE) | 0.077 | 0.208 | ||||
Convergence point with from-scratch models.
It remains unclear how rapidly pretraining benefits decrease as we increase target context data. We thus study the returns from pretraining as we vary target context calibration sizes [53] (Fig. 4C). Both models yield returns up to 3K trials, which represents about 50m of data collection in the monkey datasets, and coincidentally is the size of the largest dataset in [46]. Session pretraining provides larger gains, but task pretraining e.g. also respectably halves the data needed to achieve 0.61 NLL. This indicates pretraining is complementary to scaling target session collection efforts. This need not have been the case: even Fig. 4B suggests that task transfer by itself is ineffective at modeling the long tail of neural variance. Note that returns on supervised evaluation are likely similar or better based on Fig. 4A/B; we explore a related idea in Section 4.3.
Overall, the returns on using pretrained BCI models depends on the use case. If we are interested in best explaining neural variance, pretraining alone underperforms a moderately large in-day data collection effort (single-session model achieves lowest NLL in Fig. 4B). However, we do not see interference [53] in our experiments, where pretraining then tuning underperforms a from-scratch model. Thus, so long as we can afford the compute, broad pretraining is advantageous; we show these trends are repeated for two other evaluation sessions in Section A.6. We reiterate that our pretraining effort is modestly scaled; the largest pretraining only has 2 orders more data than the largest intra-context models. These conclusions may further strengthen insofar if we are able to better scale curation of pretraining data over individual experimental sessions.
4.3. Using NDT2 for improved decoding on novel days
RTT Decoding.
A continuously used BCI presents the optimistic setting of having both broad unsupervised data but also multiple sessions worth of supervision for our decoder. To evaluate NDT2 in this case, we follow 1st stage unsupervised pretraining with a 2nd stage of supervised pretraining of a decoder, and finally measure the decoding performance in a novel target session in Fig. 5A. We find that given either supervised or unsupervised tuning in our target session, beyond smoothly improving over the 0-Shot Pretrained model’s performance, achieves decoding performance on par with the best from-scratch models at all data volumes. This is true both in the realistic case where the majority of target-session data are unlabeled (100 Supervised Trials) and in the optimistic case when >1K trials of supervised data are available (Scratch). As expected, though, gains against the Scratch models are largest when target session data are limited. In sum, pretraining and fine-tuning enables practical calibration without explicit domain adaptation designs (as explored e.g. in [11, 36, 37]).
Figure 5. Tuning and evaluating pretrained decoders.
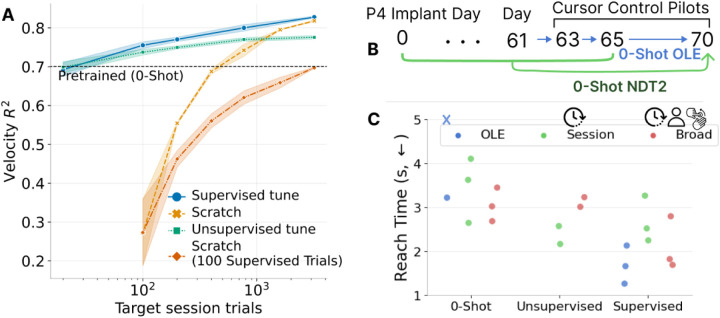
A. In offline experiments, we compare a multisession pretrained model calibrated to a novel day against current approaches of a pretrained model without adaptation (0-Shot) and from-scratch training (yellow, orange). Both supervised and unsupervised outperforms these strategies. Standard error shown over 3 seeds. B. In human control pilot experiments, we evaluated models on 3 test days. “0-Shot” models use no data from test days. C. Average reach times in 40 center-out trials is shown for 3 decoders over 2–3 sessions. This average includes trials where target is not acquired in 10s, though this occurs 1–2 times in 40 trials. Session NDT2 uses <250 trials of data, while Broad NDT2 also includes other human and monkey data. Pretrained models provide consistent 0-Shot control while OLE [23] can sometimes fail (shown by X). Control improves with either unsupervised or supervised tuning. However, OLE appears to overtake NDT2 with supervision.
Human BCI evaluation.
We next evaluate NDT2 in offline decoding of human motor intent, i.e. open loop trials of 2D cursor control. This shift from actual reach in monkeys is challenging: human sessions often contain few trials (e.g. 40) and intent labels are much noisier than movement recordings (intent labeling is described in Section A.1). We now evaluate a temporally contiguous experimental block, but only tune one model over this block, rather than per session, due to the high session count. We also increase test split to 50% to decrease noise from evaluating only a few trials.
We also compare broad pretraining (multi-session, subject, and task) performance (Table 2, row 1) with various data ablations. Ablating cross-subject data only has a minor performance impact (row 1 vs 2), while ablating cross-task data, which here indicates removal of data collected during online control and leaving only open-loop observation trials, hurts performance more (row 3 vs 1). We note that NDT2 fails to train using only single-sessions given extremely low trial count; we use linear decoding performance as a single-session reference linear decoding performance. Overall, the plentiful cross-session data likely occludes further gains from cross-subject and cross-task pretraining, but note that there is no negative transfer.
Next, given reports of monkey to human transfer [54], we assess whether monkey data in either pretraining or decoder preparation improves decoding (Table 2, rows 4–7). We find that monkey data, however incorporated, reduces offline decoding performance (row 4–7 < 1). This cross-species result is the first instance of possible harm from broader pretraining, but warrants further study given the potential of transferring able-bodied monkey decoders to humans.
4.4. NDT2 for human cursor control pilots.
Finally, to assess NDT2’s potential for deployed BCIs, we run realtime, closed-loop cursor control with one person, P4. This person was implanted recently (~ 2 months prior), and has high signal quality but limited historical data. We compare two NDT2 models, one of which pretrains with 10 minutes of participant-specific data, and one broadly pretrained on all human and monkey data, along with a baseline linear decoder (indirect OLE [55]). We evaluate the setting where decoders from recent days are available, and can either be used directly (0-shot) or updated with calibration data on the test day (Fig. 5B). All models can be supervised with test day data, but NDT2 models can also use unsupervised tuning with only neural data. In a standard center-out BCI-reaching task (methods in Section A.2), NDT2 allows consistent 0-shot use, whereas OLE sometimes fails (Fig. 5C, blue X). After either form of tuning, both NDT2 models (multisession, broad) improve. Importantly, NDT2 tunes without any additional distribution-alignment priors, as in [11, 34, 32], showing that pretraining and fine-tuning [20] may be a viable paradigm for closed loop motor BCIs.
Perhaps surprisingly, OLE provides the best control given supervised data. The performance gap and large distribution shift from offline analysis to online control is well known [18], though the specific challenge of DNN control only has basic characterization. For example, NDT2 decodes “pulsar” behavior as in [30]. Costello et al. [56] provides a possible diagnosis: these pulses reflect the ballistic reaches of the open loop training data, whereas OLE, due to its limited expressivity, will always provide continuous (but less stable) control that can be helpful in time-based metrics. Promising approaches to mitigate the pulsar failure mode include further closed-loop tuning [57] or open-loop augmentation [30]; we leave continued study of NDT2 control for future work.
5. Discussion
NDT2 demonstrates that broad pretraining can improve models of neural spiking activity in the motor cortex. With simple changes to the broadly used masked autoencoder Transformer, NDT2 at once spans the different distribution shifts faced in spiking data. We find distinct scaling slopes for different context classes, as opposed to a constant offset in effectiveness [58]. For assistive applications of BCI, NDT2’s simple recipe for multisession aggregation is promising even if the ideal scenario of cross-species transfer remains unclear. More broadly, we conclude that pretraining, even at a modest 10–100K trials, is useful in realistic deployment scenarios with varied levels of supervised data.
Supervising pretrained BCI decoders.
Motor BCIs fundamentally bridge two modalities: brain data and behavior. NDT2 allows neural data pretraining, but leaves open the challenge of decoding of the full range of motor behavior. Without mapping that range, BCIs based on neural data pretraining alone will need continuous supervision. This is still a practical path forward: beyond explicit calibration phases, strategies for supervising BCIs are diverse, and can derive from user feedback [59], neural error signals [60, 61], or task-based estimation of user intent [62, 57, 63]. The ambition of pretrained BCI models, with broad paired coverage of the neural-behavior domain will be challenging given the experimental costs of data collection; the field of robotics suggests that scaled offline or simulated learning are important strategies given this expense. Since we lack convincing closed-loop neural data simulators (though see [64, 62]), understanding how to leverage behavior from existing heterogeneous datasets is an important next step.
Negative NLB result.
NDT2 performance did not exceed current NLB SoTA on motor datasets (RTT, Maze) [65]. We first note the NLB evaluation emphasizes neural reconstruction of held-out neurons, which differs from our primary emphasis on decoding. Second, our unsupervised scaling analysis indicates that modest pretraining (100K trials) provides limited gains for neural reconstruction, especially when the target dataset has many trials (Fig. 4C), as in the NLB RTT dataset, so underperformance does not contradict our results. Revisiting the NLB after further scaling may be fruitful.
Limitations.
NDT2’s design space is under-explored. For example, we do not claim that full space-time attention is necessary over factorization. While NDT2 achieves positive transfer, further gains may come from precise mapping of context relationships [51]. Further, it is difficult to extrapolate the benefits of scaling beyond what was explored here, particularly with gains in unsupervised reconstruction appearing very limited. Our evaluation also has a limited scope, emphasizing reach-like behaviors. While these behaviors are more general than previous demonstrations of context transfer [11, 37, 32, 34], evaluating more complex behavior decoding is a practical priority. Further, NDT2 benefits are modest in human cursor control pilots, reiterating the broadly documented challenge in translating gains in offline analyses to online, human control [30, 18]. Finally, design parameters such as masking ratio may affect scaling trends, which we cannot assess due to compute limits.
Broader Impacts.
Pretrained iBCI models may great improve iBCI usability. However, DNNs may require further safeguards to ensure that decoded behaviors, especially in real-time control scenarios, operate within reasonable safety parameters. Also, pretraining will require data from many different sources, but the landscape around human neural data privacy is still developing. While there are very few humans involved in these experiments, true deidentification remains difficult, requiring, at a minimum, consented data releases.
Supplementary Material
Acknowledgments and Disclosure of Funding
We thank Jeff Weiss, William Hockeimer, Brian Dekleva, and Nicolas Kunigk for technical assistance in trawling human cursor experiments. We thank Cecilia Gallego-Carracedo and Patrick Marino for packaging data for this project (results do not include analysis from PM’s data). We thank Joseph O’Doherty, Felix Pei, and Brianna Karpowicz for early technical advice. JY is supported by the DOE CSGF (i.e. U.S. Department of Energy, Office of Science, Office of Advanced Scientific Computing Research, Department of Energy Computational Science Graduate Fellowship under Award Number DE-SC0023112). RG consults for Blackrock Microsystems and is on the scientific advisory board of Neurowired. This research was supported in part by the University of Pittsburgh Center for Research Computing, RRID:SCR_022735, through the resources provided. Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under Award Numbers R01NS121079 and UH3NS107714. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This report was prepared as an account of work sponsored by an agency of the United States Government. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof.
Footnotes
The rEFH model data splits vary slightly from ours: its data splits are sequential and contiguous in time, whereas we use random draws in keeping with NLB. NDT2 sorted decoding drops from 0.54 to 0.52 when mirroring Makin et al’s data splits.
References
- [1].Vyas Saurabh, Golub Matthew D, Sussillo David, and Shenoy Krishna V. Computation through neural population dynamics. Annual review of neuroscience, 43:249–275, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Altan Ege, Solla Sara A., Miller Lee E., and Perreault Eric J.. Estimating the dimensionality of the manifold underlying multi-electrode neural recordings. PLOS Computational Biology, 17(11):1–23, 11 2021. doi: 10.1371/journal.pcbi.1008591. URL 10.1371/journal.pcbi.1008591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].O’Shea Daniel J, Duncker Lea, Goo Werapong, Sun Xulu, Vyas Saurabh, Trautmann Eric M, Diester Ilka, Ramakrishnan Charu, Deisseroth Karl, Sahani Maneesh, et al. Direct neural perturbations reveal a dynamical mechanism for robust computation. bioRxiv, pages 2022–12, 2022. [Google Scholar]
- [4].Sadtler Patrick T, Quick Kristin M, Golub Matthew D, Chase Steven M, Ryu Stephen I, Tyler-Kabara Elizabeth C, Yu Byron M, and Batista Aaron P. Neural constraints on learning. Nature, 512(7515): 423–426, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vyas Saurabh, Even-Chen Nir, Stavisky Sergey D, Ryu Stephen I, Nuyujukian Paul, and Shenoy Krishna V. Neural population dynamics underlying motor learning transfer. Neuron, 97(5):1177–1186, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gallego Juan A., Perich Matthew G., Naufel Stephanie N., Ethier Christian, Solla Sara A., and Miller Lee E.. Cortical population activity within a preserved neural manifold underlies multiple motor behaviors. Nature Communications, 9(1):4233, Oct 2018. ISSN 2041–1723. doi: 10.1038/s41467-018-06560-z. URL https://www.nature.com/articles/s41467-018-06560-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gallego Juan A., Perich Matthew G., Chowdhury Raeed H., Solla Sara A., and Miller Lee E.. Long-term stability of cortical population dynamics underlying consistent behavior. Nature Neuroscience, 23(2): 260–270, Feb 2020. ISSN 1546–1726. doi: 10.1038/s41593-019-0555-4. URL https://www.nature.com/articles/s41593-019-0555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Safaie Mostafa, Chang Joanna C., Park Junchol, Miller Lee E., Dudman Joshua T., Perich Matthew G., and Gallego Juan A.. Preserved neural population dynamics across animals performing similar behaviour. Sep 2022. doi: 10.1101/2022.09.26.509498. URL https://www.biorxiv.org/content/10.1101/2022.09.26.509498v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dabagia Max, Kording Konrad P., and Dyer Eva L.. Aligning latent representations of neural activity. Nature Biomedical Engineering, 7(4):337–343, Apr 2023. ISSN 2157–846X. doi: 10.1038/s41551-022-00962-7. URL https://www.nature.com/articles/s41551-022-00962-7. [DOI] [PubMed] [Google Scholar]
- [10].Sussillo David, Stavisky Sergey D., Kao Jonathan C., Ryu Stephen I., and Shenoy Krishna V.. Making brain–machine interfaces robust to future neural variability. 7:13749, Dec 2016. ISSN 2041–1723. doi: 10.1038/ncomms13749. URL https://www.nature.com/articles/ncomms13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Karpowicz Brianna M., Ali Yahia H., Wimalasena Lahiru N., Sedler Andrew R., Keshtkaran Mohammad Reza, Bodkin Kevin, Ma Xuan, Miller Lee E., and Pandarinath Chethan. Stabilizing brain-computer interfaces through alignment of latent dynamics. Nov 2022. doi: 10.1101/2022.04.06.487388. URL https://www.biorxiv.org/content/10.1101/2022.04.06.487388v2. [DOI] [Google Scholar]
- [12].Willett Francis, Kunz Erin, Fan Chaofei, Avansino Donald, Wilson Guy, Choi Eun Young, Kamdar Foram, Hochberg Leigh R., Druckmann Shaul, Shenoy Krishna V., and Henderson Jaimie M.. A high-performance speech neuroprosthesis. Jan 2023. doi: 10.1101/2023.01.21.524489. URL https://www.biorxiv.org/content/10.1101/2023.01.21.524489v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].B Wodlinger, Downey J E, Tyler-Kabara E C, Schwartz A B, Boninger M L, and Collinger J L. Ten-dimensional anthropomorphic arm control in a human brain-machine interface: difficulties, solutions, and limitations. Journal of Neural Engineering, 12(1):016011, Feb 2015. ISSN 1741–2560, 1741–2552. doi: 10.1088/1741-2560/12/1/016011. URL https://iopscience.iop.org/article/10.1088/1741-2560/12/1/016011. [DOI] [PubMed] [Google Scholar]
- [14].Pei Felix, Ye Joel, Zoltowski David, Wu Anqi, Chowdhury Raeed H., Sohn Hansem, O’Doherty Joseph E., Shenoy Krishna V., Kaufman Matthew T., Churchland Mark, Jazayeri Mehrdad, Miller Lee E., Pillow Jonathan, Park Memming, Dyer Eva L., and Pandarinath Chethan. Neural latents benchmark ‘21: Evaluating latent variable models of neural population activity, 2022.
- [15].Vaswani Ashish, Shazeer Noam, Parmar Niki, Uszkoreit Jakob, Jones Llion, Gomez Aidan N, Kaiser Ł ukasz, and Polosukhin Illia. Attention is all you need. In Guyon I., Von Luxburg U., Bengio S., Wallach H., Fergus R., Vishwanathan S., and Garnett R., editors, Advances in Neural Information Processing Systems, volume 30. Curran Associates, Inc., 2017. URL https://proceedings.neurips.cc/paper_files/paper/2017/file/3f5ee243547dee91fbd053c1c4a845aa-Paper.pdf. [Google Scholar]
- [16].Ye Joel and Pandarinath Chethan. Representation learning for neural population activity with neural data transformers, 2021. URL 10.51628/001c.27358. [DOI] [Google Scholar]
- [17].He Kaiming, Chen Xinlei, Xie Saining, Li Yanghao, Dollár Piotr, and Girshick Ross. Masked autoencoders are scalable vision learners. Dec 2021. doi: 10.48550/arXiv.2111.06377. URL http://arxiv.org/abs/2111.06377. arXiv:2111.06377 [cs]. [DOI] [Google Scholar]
- [18].Pandarinath Chethan and Bensmaia Sliman J. The science and engineering behind sensitized brain-controlled bionic hands. Physiological Reviews, 102(2):551–604, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Silversmith Daniel B., Abiri Reza, Hardy Nicholas F., Natraj Nikhilesh, Tu-Chan Adelyn, Chang Edward F., and Ganguly Karunesh. Nature Biotechnology, 39(3):326–335, Mar 2021. ISSN 1546–1696. doi: 10.1038/s41587-020-0662-5. URL https://www.nature.com/articles/s41587-020-0662-5. [DOI] [PubMed] [Google Scholar]
- [20].Devlin Jacob, Chang Ming-Wei, Lee Kenton, and Toutanova Kristina. Bert: Pre-training of deep bidirectional transformers for language understanding, 2019. [Google Scholar]
- [21].Talukder Sabera J, Sun Jennifer J., Leonard Matthew K, Brunton Bingni W, and Yue Yisong. Deep neural imputation: A framework for recovering incomplete brain recordings. In NeurIPS 2022 Workshop on Learning from Time Series for Health, 2022. URL https://openreview.net/forum?id=c9qFg8UrIcn. [Google Scholar]
- [22].Peterson Samuel M, Singh Shiva H, Dichter Benjamin, Tan Kelvin, DiBartolomeo Craig, Theogarajan Devapratim, Fisher Peter, and Parvizi Josef. Ajile12: Long-term naturalistic human intracranial neural recordings and pose. Scientific Data, 9(1):184, 2022. ISSN 2052–4463. doi: 10.1038/s41597-022-01280-y. URL 10.1038/s41597-022-01280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang Christopher, Subramaniam Vighnesh, Yaari Adam Uri, Kreiman Gabriel, Katz Boris, Cases Ignacio, and Barbu Andrei. BrainBERT: Self-supervised representation learning for intracranial recordings. In The Eleventh International Conference on Learning Representations, 2023. URL https://openreview.net/forum?id=xmcYx_reUn6. [Google Scholar]
- [24].Thomas Armin W., Ré Christopher, and Russell A. Poldrack. Self-supervised learning of brain dynamics from broad neuroimaging data, 2023.
- [25].Kostas Demetres, Aroca-Ouellette Stéphane, and Rudzicz Frank. BENDR: Using transformers and a contrastive self-supervised learning task to learn from massive amounts of EEG data. Frontiers in Human Neuroscience, 15, 2021. ISSN 1662–5161. doi: 10.3389/fnhum.2021.653659. URL https://www.frontiersin.org/articles/10.3389/fnhum.2021.653659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Obeid Iyad and Picone Joseph. The temple university hospital EEG data corpus. Frontiers in Neuroscience, 10:196, 2016. ISSN 1662–453X. doi: 10.3389/fnins.2016.00196. URL https://www.frontiersin.org/articles/10.3389/fnins.2016.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Turaga Srini, Buesing Lars, Packer Adam M, Dalgleish Henry, Pettit Noah, Hausser Michael, and Macke Jakob H. Inferring neural population dynamics from multiple partial recordings of the same neural circuit. In Burges C.J., Bottou L., Welling M., Ghahramani Z., and Weinberger K.Q., editors, Advances in Neural Information Processing Systems, volume 26. Curran Associates, Inc., 2013. URL https://proceedings.neurips.cc/paper_files/paper/2013/file/01386bd6d8e091c2ab4c7c7de644d37b-Paper.pdf. [Google Scholar]
- [28].Pandarinath Chethan, O’Shea Daniel J., Collins Jasmine, Jozefowicz Rafal, Stavisky Sergey D., Kao Jonathan C., Trautmann Eric M., Kaufman Matthew T., Ryu Stephen I., Hochberg Leigh R., Henderson Jaimie M., Shenoy Krishna V., Abbott L. F., and Sussillo David. Inferring single-trial neural population dynamics using sequential auto-encoders. Nature Methods, 15(10):805–815, Oct 2018. ISSN 1548–7105. doi: 10.1038/s41592-018-0109-9. URL https://www.nature.com/articles/s41592-018-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Willett Francis R., Avansino Donald T., Hochberg Leigh R., Henderson Jaimie M., and Shenoy Krishna V.. High-performance brain-to-text communication via handwriting. Nature, 593(7858):249–254, May 2021. ISSN 1476–4687. doi: 10.1038/s41586-021-03506-2. URL https://www.nature.com/articles/s41586-021-03506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Deo Darrel R., Willett Francis R., Avansino Donald T., Hochberg Leigh R., Henderson Jaimie M., and Shenoy Krishna V.. Translating deep learning to neuroprosthetic control. Apr 2023. doi: 10.1101/2023.04.21.537581. URL https://www.biorxiv.org/content/10.1101/2023.04.21.537581v1. [DOI] [Google Scholar]
- [31].Ahmadi Nur, Constandinou Timothy G., and Bouganis Christos-Savvas. Robust and accurate decoding of hand kinematics from entire spiking activity using deep learning. Journal of Neural Engineering, 18(2): 026011, Feb 2021. ISSN 1741–2552. doi: 10.1088/1741-2552/abde8a. URL https://iopscience.iop.org/article/10.1088/1741-2552/abde8a/meta. [DOI] [PubMed] [Google Scholar]
- [32].Jude Justin, Perich Matthew G., Miller Lee E., and Hennig Matthias H.. Robust alignment of cross-session recordings of neural population activity by behaviour via unsupervised domain adaptation. Feb 2022. doi: 10.48550/arXiv.2202.06159. URL http://arxiv.org/abs/2202.06159.arXiv:2202.06159 [cs, q-bio]. [DOI] [Google Scholar]
- [33].Downey John E., Schwed Nathaniel, Chase Steven M., Schwartz Andrew B., and Collinger Jennifer L.. Intracortical recording stability in human brain-computer interface users. Journal of Neural Engineering, 15(4):046016, Aug 2018. ISSN 1741–2552. doi: 10.1088/1741-2552/aab7a0. [DOI] [PubMed] [Google Scholar]
- [34].Jude Justin, Perich Matthew G., Miller Lee E., and Hennig Matthias H.. Capturing cross-session neural population variability through self-supervised identification of consistent neuron ensembles. In Proceedings of the 1st NeurIPS Workshop on Symmetry and Geometry in Neural Representations, page 234–257. PMLR, Feb 2023. URL https://proceedings.mlr.press/v197/jude23a.html. [Google Scholar]
- [35].Liu Ran, Azabou Mehdi, Dabagia Max, Xiao Jingyun, and Dyer Eva L.. Seeing the forest and the tree: Building representations of both individual and collective dynamics with transformers. Oct 2022. URL https://openreview.net/forum?id=5aZ8umizItU. [PMC free article] [PubMed] [Google Scholar]
- [36].Farshchian Ali, Gallego Juan A., Cohen Joseph P., Bengio Yoshua, Miller Lee E., and Solla Sara A.. Adversarial domain adaptation for stable brain-machine interfaces. Jan 2019. URL https://openreview.net/forum?id=Hyx6Bi0qYm. [Google Scholar]
- [37].Ma Xuan, Rizzoglio Fabio, Perreault Eric J., Miller Lee E., and Kennedy Ann. Using adversarial networks to extend brain computer interface decoding accuracy over time. Aug 2022. doi: 10.1101/2022.08.26.504777. URL https://www.biorxiv.org/content/10.1101/2022.08.26.504777v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Le Trung and Shlizerman Eli. Stndt: Modeling neural population activity with a spatiotemporal transformer, 2022. [Google Scholar]
- [39].Arnab Anurag, Dehghani Mostafa, Heigold Georg, Sun Chen, Lučić Mario, and Schmid Cordelia. Vivit: A video vision transformer, 2021.
- [40].Dosovitskiy Alexey, Beyer Lucas, Kolesnikov Alexander, Weissenborn Dirk, Zhai Xiaohua, Unterthiner Thomas, Dehghani Mostafa, Minderer Matthias, Heigold Georg, Gelly Sylvain, Uszkoreit Jakob, and Houlsby Neil. An image is worth 16×16 words: Transformers for image recognition at scale. In International Conference on Learning Representations, 2021. URL https://openreview.net/forum?id=YicbFdNTTy. [Google Scholar]
- [41].Shanechi Maryam M., Orsborn Amy L., Moorman Helene G., Gowda Suraj, Dangi Siddharth, and Carmena Jose M.. Rapid control and feedback rates enhance neuroprosthetic control. Nature Communications, 8(1):13825, Jan 2017. ISSN 2041–1723. doi: 10.1038/ncomms13825. URL https://www.nature.com/articles/ncomms13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Collinger Jennifer L, Wodlinger Brian, Downey John E, Wang Wei, Tyler-Kabara Elizabeth C, Weber Douglas J, McMorland Angus JC, Velliste Meel, Boninger Michael L, and Schwartz Andrew B. High-performance neuroprosthetic control by an individual with tetraplegia. The Lancet, 381(9866):557–564, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Trautmann Eric M., Hesse Janis K., Stine Gabriel M., Xia Ruobing, Zhu Shude, O’Shea Daniel J., Karsh Bill, Colonell Jennifer, Lanfranchi Frank F., Vyas Saurabh, Zimnik Andrew, Steinmann Natalie A., Wagenaar Daniel A., Andrei Alexandru, Mora Lopez Carolina, O’Callaghan John, Jan u, Raducanu Bogdan C., Welkenhuysen Marleen, Churchland Mark, Moore Tirin, Shadlen Michael, Shenoy Krishna, Tsao Doris, Dutta Barundeb, and Harris Timothy. Large-scale high-density brain-wide neural recording in nonhuman primates. bioRxiv, 2023. doi: 10.1101/2023.02.01.526664. URL https://www.biorxiv.org/content/early/2023/05/04/2023.02.01.526664. [DOI] [Google Scholar]
- [44].Lester Brian, Al-Rfou Rami, and Constant Noah. The power of scale for parameter-efficient prompt tuning, 2021. [Google Scholar]
- [45].Peng Xue Bin, Coumans Erwin, Zhang Tingnan, Lee Tsang-Wei, Tan Jie, and Levine Sergey. Learning agile robotic locomotion skills by imitating animals. arXiv preprint arXiv:2004.00784, 2020. [Google Scholar]
- [46].O’Doherty Joseph E., Cardoso Mariana M. B., Makin Joseph G., and Sabes Philip N.. Nonhuman primate reaching with multichannel sensorimotor cortex electrophysiology, May 2017. URL 10.5281/zenodo.788569. [DOI] [Google Scholar]
- [47].Makin Joseph G, O’Doherty Joseph E, Cardoso Mariana M B, and Sabes Philip N. Superior arm-movement decoding from cortex with a new, unsupervised-learning algorithm. Journal of Neural Engineering, 15 (2):026010, Apr 2018. ISSN 1741–2560, 1741–2552. doi: 10.1088/1741-2552/aa9e95. URL https://iopscience.iop.org/article/10.1088/1741-2552/aa9e95. [DOI] [PubMed] [Google Scholar]
- [48].Dekleva Brian M, Weiss Jeffrey M, Boninger Michael L, and Collinger Jennifer L. Generalizable cursor click decoding using grasp-related neural transients. Journal of Neural Engineering, 18(4):0460e9, Aug 2021. ISSN 1741–2560, 1741–2552. doi: 10.1088/1741-2552/ac16b2. URL https://iopscience.iop.org/article/10.1088/1741-2552/ac16b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Krishna Kundan, Garg Saurabh, Bigham Jeffrey P., and Lipton Zachary C.. Downstream datasets make surprisingly good pretraining corpora, 2022.
- [50].Kolesnikov Alexander, Beyer Lucas, Zhai Xiaohua, Puigcerver Joan, Yung Jessica, Gelly Sylvain, and Houlsby Neil. Big transfer (bit): General visual representation learning, 2020.
- [51].Zamir Amir, Sax Alexander, Shen William, Guibas Leonidas, Malik Jitendra, and Savarese Silvio. Taskonomy: Disentangling task transfer learning, 2018.
- [52].Vaswani Ashish Teku, Yogatama Dani, Metzler Don, Chung Hyung Won, Rao Jinfeng, Fedus Liam B., Dehghani Mostafa, Abnar Samira, Narang Sharan, and Tay Yi. Scale efficiently: Insights from pre-training and fine-tuning transformers. 2022.
- [53].Hernandez Danny, Kaplan Jared, Henighan Tom, and Sam McCandlish. Scaling laws for transfer, 2021.
- [54].Rizzoglio Fabio, Altan Ege, Ma Xuan, Bodkin Kevin L., Dekleva Brian M., Solla Sara A., Kennedy Ann, and Miller Lee E.. Monkey-to-human transfer of brain-computer interface decoders. bioRxiv, 2022. doi: 10.1101/2022.11.12.515040. URL https://www.biorxiv.org/content/early/2022/11/13/2022.11.12.515040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wang Wei, Chan Sherwin S., Heldman Dustin A., and Moran Daniel W.. Motor cortical representation of position and velocity during reaching. Journal of Neurophysiology, 97(6):4258–4270, Jun 2007. ISSN 0022–3077. doi: 10.1152/jn.01180.2006. [DOI] [PubMed] [Google Scholar]
- [56].Costello Joseph T, Temmar Hisham, Cubillos Luis H, Mender Matthew J, Wallace Dylan M, Willsey Matthew S, Patil Parag G, and Chestek Cynthia A. Balancing memorization and generalization in rnns for high performance brain-machine interfaces. bioRxiv, pages 2023–05, 2023. [Google Scholar]
- [57].Jarosiewicz Beata, Sarma Anish A, Saab Jad, Franco Brian, Cash Sydney S, Eskandar Emad N, and Hochberg Leigh R. Retrospectively supervised click decoder calibration for self-calibrating point-and-click brain–computer interfaces. Journal of Physiology-Paris, 110(4):382–391, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kaplan Jared, McCandlish Sam, Henighan Tom, Brown Tom B., Chess Benjamin, Child Rewon, Gray Scott, Radford Alec, Wu Jeffrey, and Amodei Dario. Scaling laws for neural language models, 2020.
- [59].Gao Jensen, Reddy Siddharth, Berseth Glen, Hardy Nicholas, Natraj Nikhilesh, Ganguly Karunesh, Dragan Anca D, and Levine Sergey. X2t: Training an x-to-text typing interface with online learning from user feedback. arXiv preprint arXiv:2203.02072, 2022. [Google Scholar]
- [60].Even-Chen Nir, Stavisky Sergey D, Kao Jonathan C, Ryu Stephen I, and Shenoy Krishna V. Augmenting intracortical brain-machine interface with neurally driven error detectors. Journal of neural engineering, 14(6):066007, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Iturrate Iñaki, Chavarriaga Ricardo, Montesano Luis, Minguez Javier, and Millán José del R.. Teaching brain-machine interfaces as an alternative paradigm to neuroprosthetics control. Scientific Reports, 5(1): 13893, Sep 2015. ISSN 2045–2322. doi: 10.1038/srep13893. URL https://www.nature.com/articles/srep13893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wilson Guy H., Willett Francis R., Stein Elias A., Kamdar Foram, Avansino Donald T., Hochberg Leigh R., Shenoy Krishna V., Druckmann Shaul, and Henderson Jaimie M.. Long-term unsupervised recalibration of cursor bcis. Feb 2023. doi: 10.1101/2023.02.03.527022. URL https://www.biorxiv.org/content/10.1101/2023.02.03.527022v1. [DOI] [Google Scholar]
- [63].Merel Josh, Carlson David, Paninski Liam, and Cunningham John P. Neuroprosthetic decoder training as imitation learning. PLoS computational biology, 12(5):e1004948, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Willett Francis R., Young Daniel R., Murphy Brian A., Memberg William D., Blabe Christine H., Pandarinath Chethan, Stavisky Sergey D., Rezaii Paymon, Saab Jad, Walter Benjamin L., Sweet Jennifer A., Miller Jonathan P., Henderson Jaimie M., Shenoy Krishna V., Simeral John D., Jarosiewicz Beata, Hochberg Leigh R., Kirsch Robert F., and Ajiboye A. Bolu. Principled bci decoder design and parameter selection using a feedback control model. Scientific Reports, 9(1):8881, Jun 2019. ISSN 2045–2322. doi: 10.1038/s41598-019-44166-7. URL 10.1038/s41598-019-44166-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Unknown. EvalAI leaderboard. https://eval.ai/web/challenges/challenge-page/1256/leaderboard/3184, 2022. Accessed on May 16, 2023.
- [66].Feichtenhofer Christoph, Fan Haoqi, Li Yanghao, and He Kaiming. Masked autoencoders as spatiotemporal learners, 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.