Scott et al. show with ultrastructure expansion microscopy that self-phosphorylation is a major driver of the spatial patterning of PLK4 at the centriole and plays a critical role in selecting a single centriole duplication site.
Abstract
Polo-like kinase 4 (PLK4) is a key regulator of centriole biogenesis, but how PLK4 selects a single site for procentriole assembly remains unclear. Using ultrastructure expansion microscopy, we show that PLK4 localizes to discrete sites along the wall of parent centrioles. While there is variation in the number of sites PLK4 occupies on the parent centriole, most PLK4 localize at a dominant site that directs procentriole assembly. Inhibition of PLK4 activity leads to stable binding of PLK4 to the centriole and increases occupancy to a maximum of nine sites. We show that self-phosphorylation of an unstructured linker promotes the release of active PLK4 from the centriole to drive the selection of a single site for procentriole assembly. Preventing linker phosphorylation blocks PLK4 turnover, leading to supernumerary sites of PLK4 localization and centriole amplification. Therefore, self-phosphorylation is a major driver of the spatial patterning of PLK4 at the centriole and plays a critical role in selecting a single centriole duplication site.
Introduction
Centrioles are small, cylindrical-shaped organelles that recruit a surrounding pericentriolar material (PCM) to form the centrosome, a microtubule-organizing center that arranges the interphase microtubule cytoskeleton in many cell types and forms the poles of the mitotic spindle during cell division (Gönczy, 2012; Nigg and Holland, 2018). Centrioles can also act as basal bodies that template the assembly of cilia, an organelle with important roles in signaling (Breslow and Holland, 2019). Centriole biogenesis is tightly coordinated with cell cycle progression. Centriole duplication begins at G1/S phase when the two parent centrioles form a single new procentriole on their wall. The procentriole elongates as the cell cycle progresses, and in late G2 phase, the two centriole pairs separate and increase PCM recruitment to drive the assembly of the mitotic spindle. At the end of mitosis, both new daughter cells inherit a pair of centrioles that are competent for reduplication in the next cell cycle. This tight control of centriole biogenesis is central to human health, and errors in centriole duplication are linked to multiple human diseases, including growth retardation syndromes, neurodevelopmental disorders, and cancer (Basto et al., 2008; Chavali et al., 2014; Coelho et al., 2015; Levine et al., 2017; Levine and Holland, 2018; Nigg and Raff, 2009; Serçin et al., 2016).
The earliest step in procentriole formation involves the assembly of a cartwheel structure that is catalyzed by the activity of Polo-like kinase 4 (PLK4). PLK4 is recruited to the parent centriole by binding to the receptors CEP152 and/or CEP192 (Cizmecioglu et al., 2010; Hatch et al., 2010; Kim et al., 2013; Park et al., 2014; Sonnen et al., 2013). At the start of the cell cycle, PLK4 encircles the base of the parent centriole. At G1/S phase, PLK4 transitions to a single site that marks the position of cartwheel assembly and procentriole formation (Kim et al., 2013; Ohta et al., 2014). Active PLK4 recruits and phosphorylates its substrate SCL/TAL1 interrupting locus (STIL) in two regions to promote distinct binding interactions with the cartwheel protein SAS6 and the microtubule-binding protein centrosomal P4.1-associated protein (CPAP; Dzhindzhev et al., 2014; Kratz et al., 2015; McLamarrah et al., 2020; Moyer et al., 2015; Moyer and Holland, 2019; Ohta et al., 2014). The restriction of PLK4 to a single site, referred to as the ring-to-dot transition, is thought to be critical to ensure that each parent centriole forms a single procentriole.
A complex pattern of self-phosphorylation regulates multiple aspects of PLK4’s behavior and centriole biogenesis. Dimeric PLK4 trans-phosphorylates a degron to create a binding site for the SCF-βTrCP E3 ubiquitin ligase that targets the kinase for ubiquitination and degradation by the proteasome (Cunha-Ferreira et al., 2009, 2013; Guderian et al., 2010; Holland et al., 2010, 2012; Klebba et al., 2013; Rogers et al., 2009). Consequently, active PLK4 has a short half-life, and inhibition of PLK4 activity leads to increased protein abundance. PLK4 also phosphorylates its T-loop to promote kinase activation (Lopes et al., 2015; Moyer et al., 2015), and self-phosphorylation has also been shown to both promote (Montenegro Gouveia et al., 2018; Park et al., 2019) and oppose (Yamamoto and Kitagawa, 2019) self-association properties of PLK4. How PLK4 self-phosphorylation events act cooperatively to control the ring-to-dot transition and centriole duplication remains unclear.
Here, we examine PLK4 localization at high spatial resolution using a recently developed expansion microscopy technique. We show that inactive PLK4 binds stably on the parent centriole at up to nine sites located on a ring of CEP152. By contrast, active PLK4 turns over at the centriole and concentrates at a dominant site that directs procentriole assembly. Moreover, we show that site selection and PLK4 turnover require self-phosphorylation of an unstructured linker downstream of the PLK4 kinase domain. Together, our data show that PLK4 autophosphorylation controls key principles of PLK4 behavior and is required to restrict centriole assembly to a single site on each parent centriole.
Results
Examining the PLK4 ring-to-dot transition
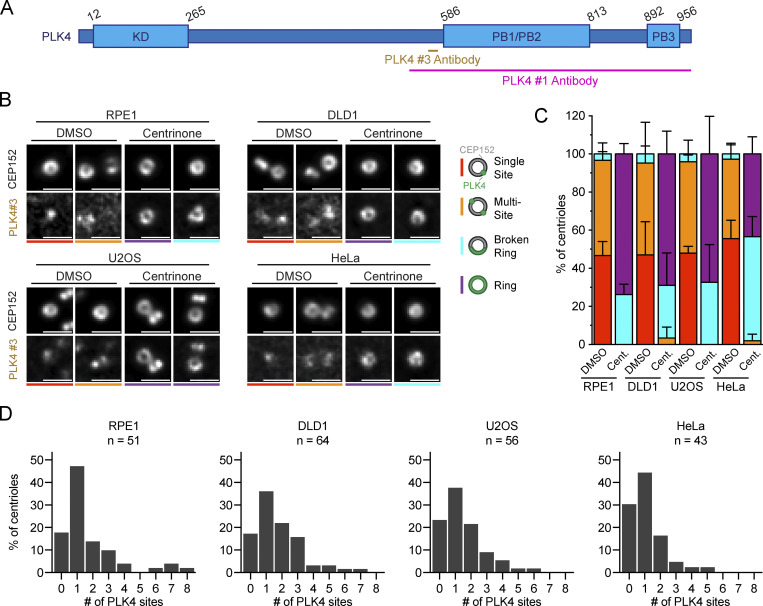
To examine PLK4 localization, we stained non-transformed human telomerase reverse transcriptase RPE1 cells, DLD1 colon cancer cells, U2OS osteosarcoma cells, and HeLa cervical cancer cells with two PLK4 antibodies raised against different epitopes (Fig. S1 A). In untreated cells, homogeneous rings of PLK4 were rarely observed (Fig. 1, A and B; and Fig. S1, B and C). Instead, PLK4 localized as a single spot (Single Site) or at multiple regions (Multi-Site) on the ring of CEP152. Treatment with the PLK4 inhibitor centrinone for 24 h led to the formation of incomplete ring-like structures (Broken Ring) or complete rings (Ring) of PLK4 on all parent centrioles, showing that rings of PLK4 can be detected under these experimental conditions (Fig. 1, A and B; and Fig. S1, B and C; Wong et al., 2015). We conclude that clear rings of PLK4 are only observed in RPE1, DLD1, U2OS, or HeLa cells when PLK4 kinase activity is inhibited.
Figure S1.
Complete rings of PLK4 are not observed in unperturbed cycling cells. (A) Schematic representation of the PLK4 protein. PLK4 #1 and PLK4 #3 antibody epitopes are shown. KD = kinase domain; PB = Polo-box domain. (B) Widefield immunofluorescence of PLK4 localization in DMSO or centrinone-treated RPE1, DLD1, U2OS, and HeLa cells using PLK4 #3 antibody. The total n for each experiment was ≥10 over three biological replicates. (C) Schematic showing the localization of PLK4 (green) on the CEP152 (gray) ring. Quantification of the PLK4 localization pattern in DMSO or centrinone-treated RPE1, DLD1, U2OS, and HeLa cells using the PLK4 #3 antibody. (D) Graphs showing the number of PLK4 sites at parent centrioles in RPE1, DLD1, U2OS, or HeLa cells in DMSO. The n refers to the total number of centrioles analyzed from multiple punches over at least three biological replicas per cell line. Scale bar = 1 µm. Data are represented as mean ± SEM.
Figure 1.
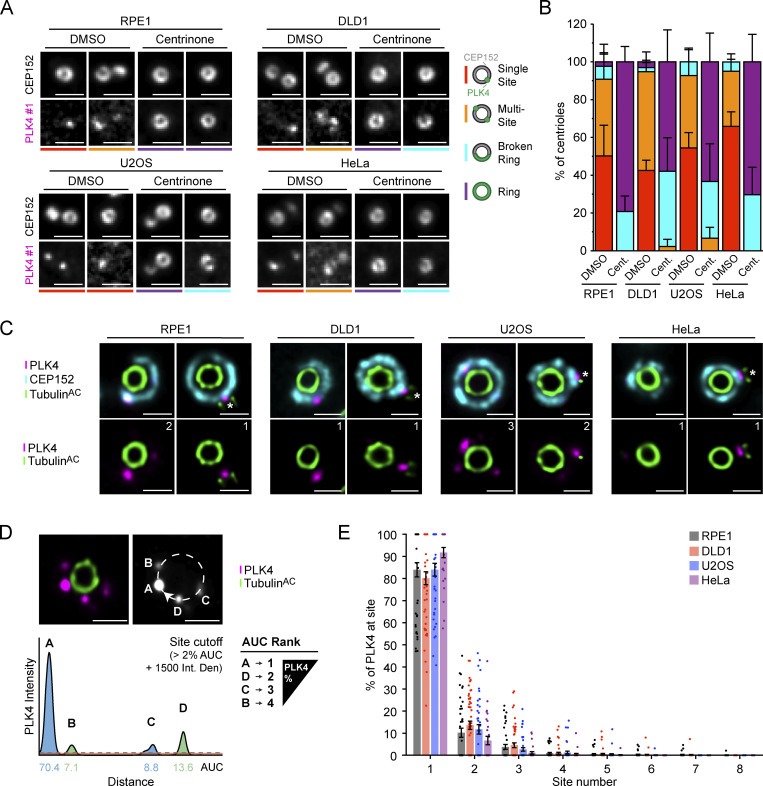
PLK4 localizes at discrete sites on the wall of the parent centriole. (A) Widefield immunofluorescence of PLK4 localization in DMSO or centrinone-treated RPE1, DLD1, U2OS, and HeLa cells using PLK4 #1 antibody. The total n for each experiment was ≥10 over three biological replicates. (B) Schematic showing the localization of PLK4 (green) on the CEP152 (gray) ring. Quantification of the PLK4 localization pattern in DMSO or centrinone-treated RPE1, DLD1, U2OS, and HeLa cells using the PLK4 #1 antibody. (C) Representative U-ExM images of PLK4 (magenta), CEP152 (cyan), and acetylated α-tubulin (green) in unduplicated and duplicated parent centrioles. The number of PLK4 sites observed in each image is indicated. (D) Schematic representation of the analysis pipeline used to quantify the amount of PLK4 localized at individual sites on parent centrioles. An intensity line scan of PLK4 was performed around the wall of the parent centriole and the AUC was calculated for each site. Sites were ordered based on decreasing AUC values and plotted to represent the percent of PLK4 protein occupying each site. Peaks that contained >2% of centriolar PLK4 and had an intensity above 1,500 were considered as a PLK4 site. (E) Graph showing the amount of PLK4 localized at individual sites on parent centrioles. All representative images are scaled independently to best represent phenotypes. The asterisk denotes the procentriole. Data are represented as mean ± SEM. Scale bar = 1 µm for widefield immunofluorescence images and 250 nm for U-ExM images.
PLK4 localizes at distinct sites on a ring of CEP152
To examine the localization of PLK4 at parent centrioles in higher resolution, we used ultrastructure expansion microscopy (U-ExM; Gambarotto et al., 2019, 2021). Expanded punches were stained with antibodies against acetylated α-tubulin to visualize centrioles, CEP152, and PLK4. While CEP152 localized as a ring around the base of the parent centriole, PLK4 was recruited to individual foci on the CEP152 ring (Fig. 1 C). The number of PLK4 sites at parent centrioles ranged from zero to eight and varied among the cell lines analyzed. In RPE1 cells, 61% of parent centrioles had PLK4 localized at one to two sites and no PLK4 was detected on CEP152 in 18% of centrioles (Fig. S1 D). In DLD1 cells, PLK4 localized at one to two sites in 58% of parent centrioles and 17% of centrioles lacked PLK4 (Fig. S1 D). In U2OS cells, PLK4 was localized at one to two sites in 59% of parent centrioles, and no PLK4 was present on CEP152 in 23% of centrioles (Fig. S1 D). Finally, in HeLa cells, PLK4 localized at one to two sites in 60% of parent centrioles, and 30% of centrioles lacked PLK4.
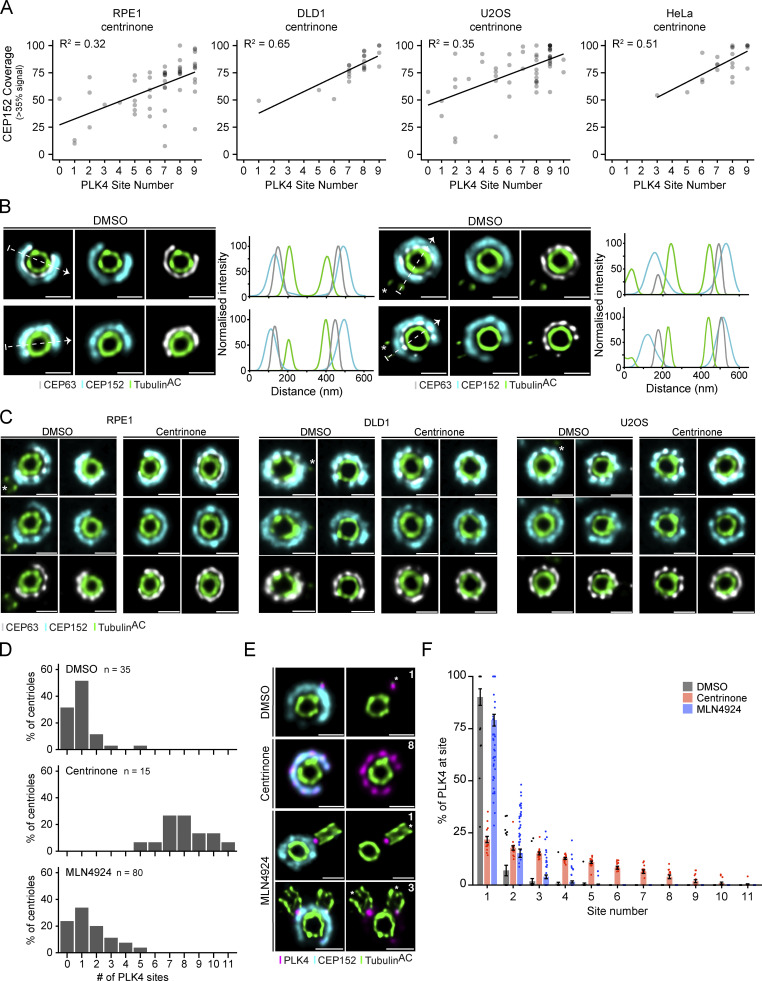
Although PLK4 was frequently observed localizing to more than one site on the CEP152 ring, the intensity of the individual sites on the same centriole often varied. To define the relative abundance of PLK4 at each site, we quantified the intensity of PLK4 around the centriole using a semiautomated image analysis pipeline. A circular profile was drawn around the circumference of the centriole and the area under the curve (AUC) for PLK4 intensity was calculated at each peak (Fig. 1 D). PLK4 sites were reordered based on descending AUC values and plotted to represent the relative occupancy of PLK4 at each site. This revealed that 84%, 80%, 84%, and 92% of centriolar PLK4 exists at the dominant site on parent centrioles in RPE1, DLD1, U2OS, and HeLa cells, respectively (Fig. 1 E).
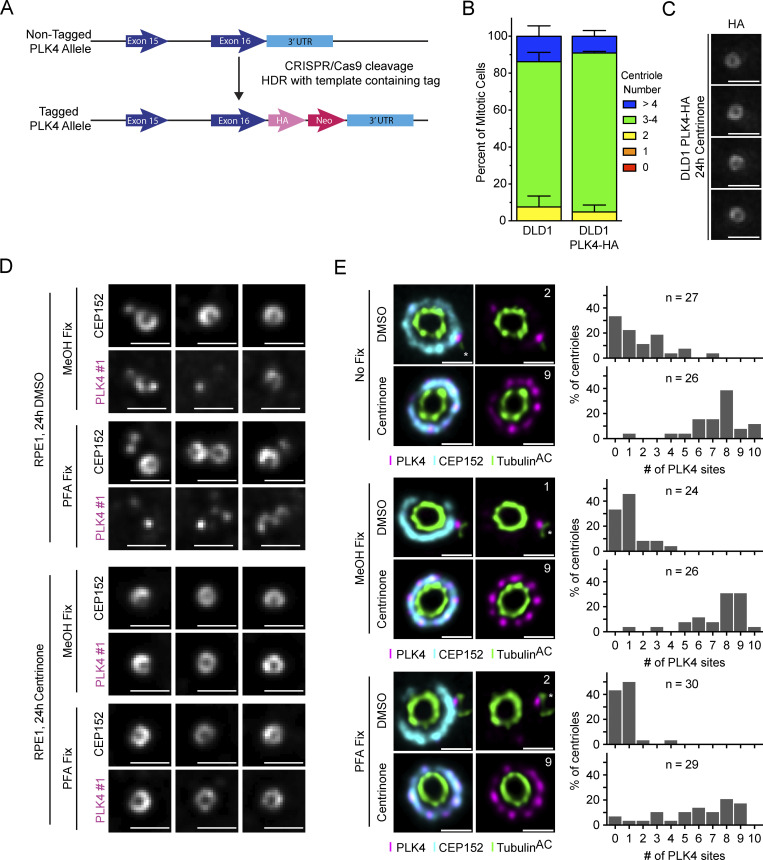
To validate this localization pattern with a different antibody, we used CRISPR/Cas9 to tag the C-terminus of PLK4 with a HA tag in DLD1 cells and examined the localization of PLK4 using a high-affinity, monoclonal anti-HA antibody (Fig. S2 A). C-terminal tagging of PLK4 had no discernable impact on centriole biogenesis (Fig. S2 B). Further, treatment of these PLK4-HA cells with centrinone for 24 h yielded rings of PLK4 similar to that observed in RPE1, DLD1, U2OS, and HeLa cells (Fig. S2 C). Examination by U-ExM revealed a similar distribution of PLK4-HA to that observed in the parental DLD1 cells stained with a PLK4 antibody, with 82% of PLK4-HA at a single site on the parent centriole (Fig. 2, A and B). Taken together, these data show that although there is heterogeneity in the number of sites PLK4 occupies, most PLK4 concentrates at the dominant site on the ring of CEP152.
Figure S2.
Creation of an endogenously tagged PLK4-HA DLD1 cell line. (A) Scheme of the CRISPR/Cas9 tagging of PLK4 with an HA epitope tag in DLD1 cells. (B) Centriole counts for endogenously tagged PLK4-HA and the parental DLD1 cell line. The graph is representative of the average phenotype of ∼150 centrioles over three biological replicates per cell line. (C) Widefield microscopy of centrioles in DLD, PLK4-HA cells after 24 h of centrinone treatment. (D) Widefield microscopy of centrioles from RPE1 cells treated for 24 h with DMSO (left) or centrinone (right). Cells were either fixed via MeOH or PFA. (E) U-ExM of centrioles from RPE1 cells treated for 24 h with DMSO (top) or centrinone (bottom). Cells were either not fixed prior to U-ExM processing or fixed via MeOH or PFA. Graphs showing the number of PLK4 sites at parent centrioles in RPE1 cells treated with DMSO or centrinone. All representative images are scaled independently to best represent phenotypes. The n refers to the total number of centrioles analyzed from multiple punches over at least two biological replicates per fixation condition. Data are represented as mean ± SEM. Scale bar = 1 µm for widefield immunofluorescence images and 250 nm for U-ExM images.
Figure 2.
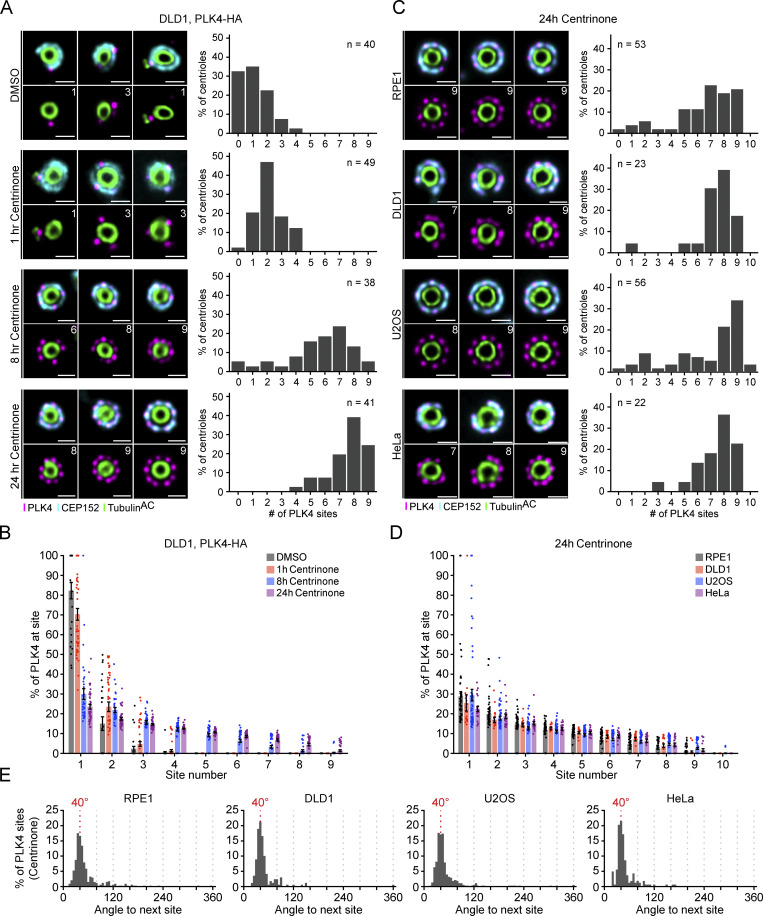
Inactive PLK4 occupies up to nine sites on the CEP152 ring. (A) Representative U-ExM images of PLK4 localization (left) and the number of PLK4 sites (right) at parent centrioles in DLD1, PLK4-HA cells. Cells were fixed after 1, 8, or 24 h of centrinone treatment and processed for U-ExM. The n refers to the total number of centrioles analyzed over several punches within a single biological repeat. (B) Graph showing the amount of PLK4 localized at individual sites on parent centrioles. (C) Representative U-ExM images of PLK4 localization (left) and the number of PLK4 sites (right) at parent centrioles in RPE1, DLD1, U2OS, and HeLa cells upon 24 h treatment with centrinone. All representative images are scaled independently to best show phenotypes. The n refers to the total number of centrioles analyzed from multiple punches over at least three biological replicates per cell line. (D) Graph showing the amount of PLK4 localized at individual sites on parent centrioles. (E) Histograms depicting the angle measured between two adjacent peaks of PLK4. Data are represented as mean ± SEM. Scale bar = 250 nm.
Inactive PLK4 occupies up to nine sites on the CEP152 ring
We next examined how the number of PLK4 foci changed following kinase inhibition. DLD1 PLK4-HA–expressing cells were treated with centrinone for 1, 8, or 24 h, and centrioles were examined by U-ExM (Fig. 2 A). Centrinone treatment caused PLK4-HA to occupy additional distinct sites on the CEP152 ring. After 1 h of centrinone, 70% of centriolar PLK4-HA remained at the dominant site, but this decreased to 30% after 8 h and 24% after 24 h of treatment (Fig. 2 B). The number of PLK4 sites increased with extended centrinone treatment but reached a maximum of nine sites following eight or more hours of kinase inhibition (Fig. 2, A and B). Consequently, the difference in PLK4 abundance across sites was dramatically reduced after prolonged periods of kinase inhibition.
We also examined PLK4 localization in RPE1, DLD1, U2OS, and HeLa cells treated with centrinone for 24 h. Consistent with observations made with the HA antibody, PLK4 predominantly localized across seven to nine sites on parent centrioles in all four centrinone-treated cell lines, and consequently, selection toward the dominant site was substantially diminished (Fig. 2, C and D). We tested the effect of MeOH or PFA fixation prior to expansion on PLK4 staining in untreated or centrinone-treated RPE1 cells. MeOH or PFA fixation did not elicit a clear difference in PLK4 staining, suggesting that the localization of PLK4 to distinct foci is not an artifact of the fixation conditions used (Fig. S2, D and E).
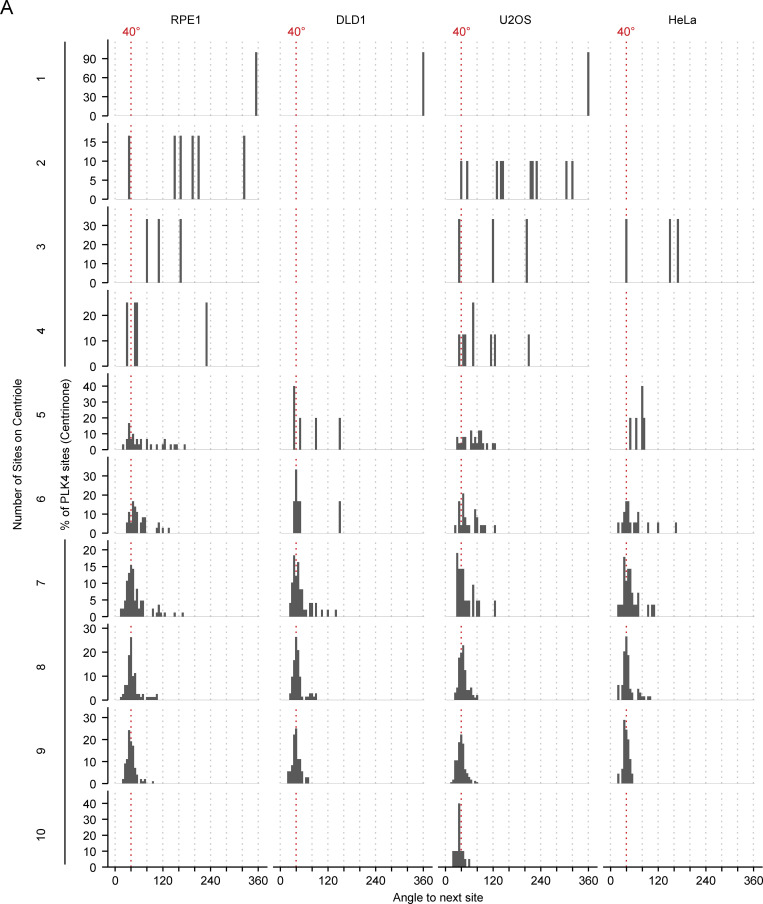
To further examine the localization pattern of PLK4, we measured the angle between adjacent sites of PLK4 foci on the CEP152 ring in centrioles treated with centrinone. We found that the peak of PLK4 foci separation occurred at ∼40° in all cell lines tested, consistent with a ninefold symmetry (Fig. 2 E). We also analyzed the angle between foci in centrioles at each site occupancy (1–10) and found that even when PLK4 was present at seven or eight foci, the average angle between adjacent sites was still ∼40° (Fig. S3). This suggests that the sites of PLK4 are fixed at ∼40° increments rather than free to reorganize when occupancy is less than nine. Taken together, we conclude that inactive PLK4 localizes at up to nine fixed positions around the base of the parent centriole, mirroring the ninefold symmetry of the centriole microtubule wall.
Figure S3.
Angle measurements between PLK4 foci per site occupancy. (A) Histograms depicting the angle measured between two adjacent peaks of PLK4 at individual site occupancies in RPE1, DLD1, U2OS, and HeLa cells.
The ring of CEP152 is often discontinuous
CEP152 is thought to localize as a continuous ring that encircles the base of the parent centriole. However, we noted that the ring of CEP152 at parent centrioles was often “broken” and PLK4 did not localize within the areas devoid of CEP152 (Fig. 1 C; Fig. 2, A and C). We used our image analysis pipeline to quantify the fractional coverage of CEP152 around the circumference of the centriole and compared it with the number of PLK4 foci observed. In this analysis, a complete ring of CEP152 corresponds to 100% coverage, while a value below 100% indicates a break or dramatic decrease in intensity for a part of the CEP152 ring (broken ring). We observed a higher fractional CEP152 coverage correlated with more PLK4 foci, with the greatest CEP152 occupancy occurring at centrioles with eight or nine PLK4 foci in cells treated with centrinone (Fig. S4 A). CEP152 forms a complex with CEP63 and the two proteins are dependent on each other for recruitment to the centriole (Brown et al., 2013). CEP63 encircled the base of parent centrioles and was localized inside the ring of CEP152 (Fig. S4 B). As expected, the localization of CEP63 and CEP152 were highly correlated and breaks in the CEP152 ring were matched with similar decreases in CEP63 in the cell lines tested (Fig. S4 C).
Figure S4.
PLK4 site occupancy is dependent on CEP152 availability. (A) Plots showing the correlation between PLK4 site occupancy and CEP152 ring completion (coverage). Breaks in the ring are classified as a region with <35% of maximum CEP152 signal intensity. (B) Representative U-ExM images and corresponding line intensity scans for CEP152 (cyan), CEP63 (gray), and acetylated α-tubulin (green) in RPE1 cells treated with DMSO for 24 h. The arrow shows the direction of the line scan. (C) Representative U-ExM images of the CEP152 and CEP63 rings in RPE1, DLD1, and U2OS cells treated with DMSO or centrinone for 24 h. (D) Graphs showing the number of PLK4 sites at parent centrioles of RPE1 cells treated with DMSO, centrinone, or MLN4924. (E) Representative U-ExM images of RPE1 cells after 24 h of DMSO, centrinone, or MLN4924 treatment. (F) Graph showing the amount of PLK4 localized at individual sites in RPE1 cells after 24 h of DMSO, centrinone, or MLN4924 treatment. The n refers to the total number of centrioles analyzed from multiple punches over at least two biological replicates. All representative images are scaled independently to best represent phenotypes. The asterisk denotes the site of the procentriole. Data are represented as mean ± SEM. Scale bar = 250 nm.
Inhibiting PLK4 activity leads to increased protein abundance. To establish if increasing PLK4 levels is sufficient to increase occupancy to nine sites, we treated RPE1 cells with MLN4924 for 24 h to inhibit PLK4 degradation. MLN4924 is a NEDD8-activating enzyme inhibitor that inactivates all Cullin-Ring Ligases, including the Skp, Cullin, F-box containing complex (SCF; Soucy et al., 2009). While centrinone increased the number of PLK4 sites to a maximum of nine, treatment with MLN4924 only modestly increased the PLK4 site number (Fig. S4, D and E). Further, the amount of PLK4 at the dominant site was similar in DMSO and MLN4924 (90% versus 79%) but declined substantially in centrinone (22%; Fig. S4 F). This suggests that increasing protein abundance is insufficient to drive complete PLK4 occupancy at the centriole. Given that nine sites of PLK4 are only observed after >8 h of kinase inhibition, we conclude that both increased PLK4 levels and kinase inhibition are needed to achieve the full nine-site occupancy for PLK4.
The majority of PLK4 localizes at a dominant site that directs procentriole assembly
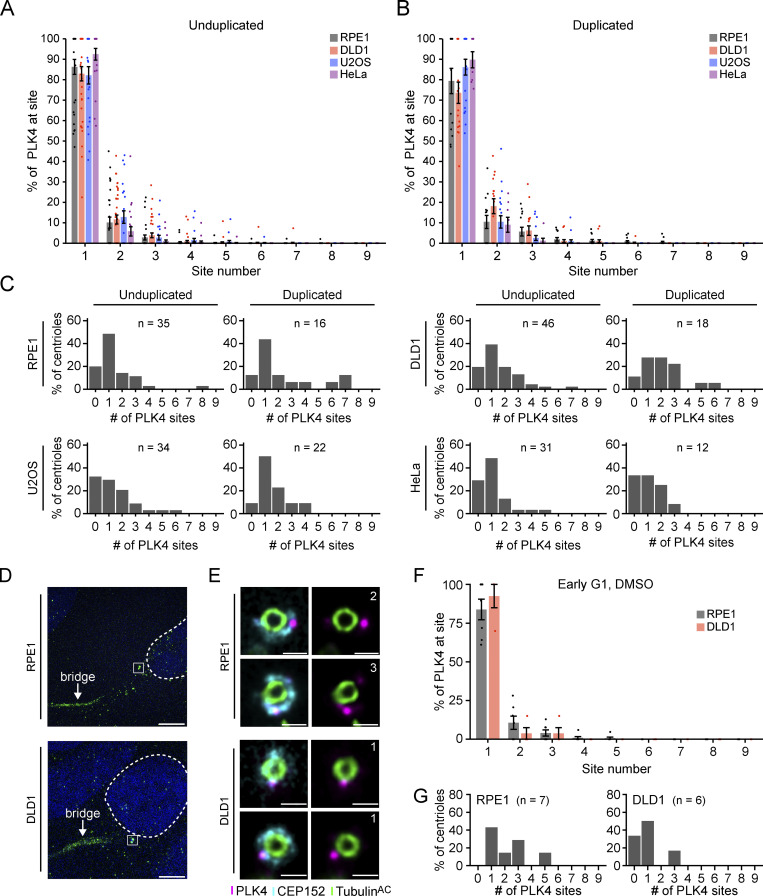
To examine if the distribution of PLK4 changes before and after centriole duplication, we divided our U-ExM images into unduplicated parent centrioles and parent centrioles with an associated procentriole. Surprisingly, we did not observe a clear shift in PLK4 localization following procentriole assembly in RPE1, DLD1, U2OS, or HeLa cells: ∼74–92% of PLK4 localized to the dominant site at parent centrioles with and without procentrioles (Fig. S5, A and B). The number of PLK4 sites also did not exhibit a clear or consistent change between duplicated and unduplicated parent centrioles in the cell lines tested (Fig. S5 C). Therefore, although there is cell-type variation in the number of sites PLK4 occupies at parent centrioles, the dominant PLK4 site is almost always the site of procentriole assembly, and additional minor PLK4 sites do not appear to grossly compromise the numerical fidelity of the centriole duplication. We conclude that most PLK4 concentrates at the dominant site before and after procentriole assembly.
Figure S5.
PLK4 site occupancy at unduplicated and duplicated centrioles. (A and B) Analysis of the amount of PLK4 localized at individual sites on unduplicated (A) and duplicated (B) centrioles. (C) Graphs showing the number of PLK4 sites at unduplicated and duplicated parent centrioles in U2OS, DLD1, RPE1, or HeLa cells. The n refers to the total number of centrioles analyzed from multiple punches over at least three biological replicates. (D) Representative U-ExM images of early G1 RPE1 or DLD1 cells with an α-tubulin cytokinetic bridge. The nucleus is denoted by the dotted line and the centrioles are within the box. (E) Representative U-ExM images of centrioles of early G1 RPE1 or DLD1 cells with an α-tubulin cytokinetic bridge. (F) Graph showing the amount of PLK4 localized at individual sites of early G1 RPE1 or DLD1 cells with an α-tubulin cytokinetic bridge. (G) Graphs showing the number of PLK4 sites in G1 RPE1 or DLD1 cells. All representative images are scaled independently to best represent phenotypes. The n refers to the total number of centrioles analyzed from multiple punches over at least two biological replicates. Data are represented as mean ± SEM. Scale bar = 5 µm for D and 250 nm for E.
It is plausible that our analysis was not sensitive enough to detect a transient increase in the number of PLK4 sites that occurs early in G1 phase. To examine this possibility, we used U-ExM to analyze the centrioles of early G1 phase cells marked by α-tubulin–labeled cytokinetic bridges. In early G1 RPE1 and DLD1 cells, 84% and 88% of PLK4 localized at the dominant site, respectively, and the number of PLK4 sites ranged from one to five (Fig. S5, D–G). Although the number of centrioles analyzed in this experiment was low, our data imply that PLK4 is already selected toward a single dominant site early in the cell cycle, and rings of PLK4 do not stably exist in unperturbed cycling cells.
CEP152 recruits PLK4 to individual sites on the wall of parent centrioles
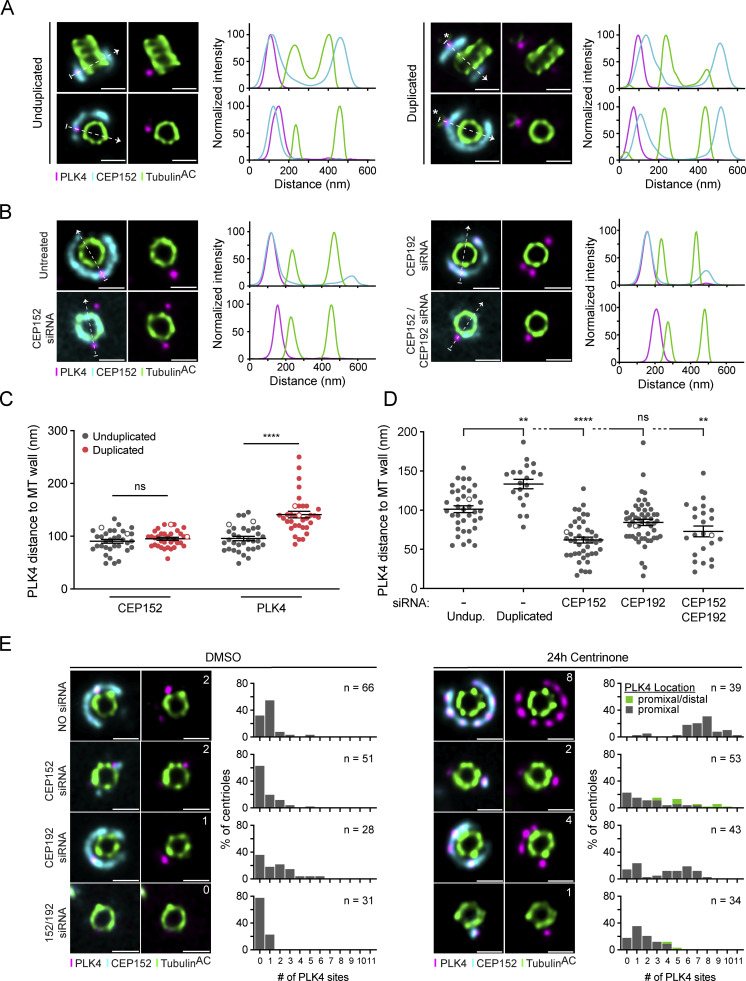
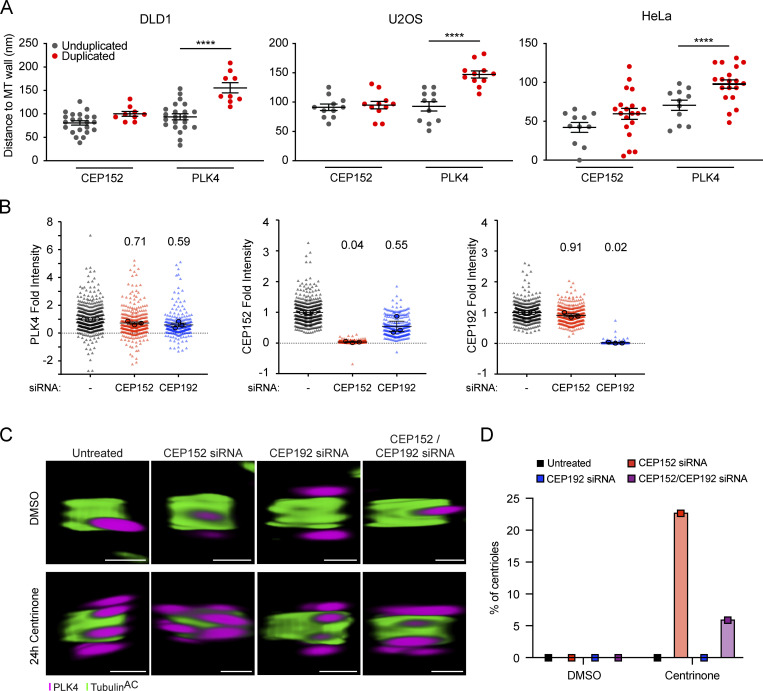
We next examined the positioning of PLK4 relative to CEP152 before and after procentriole assembly. PLK4 localized on top of the ring of CEP152 on unduplicated parent centrioles but transitioned further away from the centriole wall and outside of the CEP152 ring when present at the base of a growing procentriole, possibly as a result of binding to STIL in the cartwheel of the procentriole (Fig. 3, A and C; Fig. S6 A). Previous work using widefield imaging showed that CEP152 and CEP192 cooperate in the recruitment of PLK4 to parent centrioles (Kim et al., 2013; Sonnen et al., 2013). To determine whether CEP152 and/or CEP192 influenced the distance between PLK4 and the wall of parent centrioles, we depleted CEP152 or CEP192 by siRNA in RPE1 cells and examined CEP152 and PLK4 localization by U-ExM (Fig. 3, B and D). Depletion of CEP152 with siRNA decreased the distance of PLK4 to the centriole wall, whereas knockdown of CEP192 had no significant effect. These data are consistent with prior observations that suggested that the depletion of CEP152 leads to the relocalization of PLK4 from an outer ring of CEP152 to an inner ring of CEP192 (Park et al., 2014).
Figure 3.
PLK4 concentrates at a dominant site on parent centrioles. (A) Representative U-ExM images (left) and corresponding line intensity scans (right) for PLK4 (magenta), CEP152 (cyan), and acetylated α-tubulin (green) in unduplicated and duplicated centrioles in RPE1 cells. The arrow shows the direction of the line scan and the * marks the position of the procentriole. (B) Representative U-ExM images (left) and corresponding line intensity scans (right) for PLK4 (magenta), CEP152 (cyan), and acetylated α-tubulin (green) in RPE1 cells either untreated or transfected with CEP152 siRNA, CEP192 siRNA, or both siRNAs for 48 h. The arrow shows the direction of the line scan. (C) Quantification of A; PLK4 and CEP152 distance from the microtubule (MT) wall in unduplicated (gray) or duplicated (red) centrioles from RPE1 cells. Each dot represents one centriole analyzed over three biological replicates per condition. An unfilled dot indicates a centriole shown in the representative images. Statistical significance was determined using two-way ANOVA with post-hoc analysis. (D) Quantification of B; PLK4 distance from the microtubule wall in RPE1 cells transfected with CEP152 siRNA, CEP192 siRNA, or both siRNAs for 48 h. Each dot represents one centriole analyzed over three biological replicates per condition. Statistical significance was determined using two-way ANOVA with post-hoc analysis. Indicated statistical differences relative to unduplicated control. Asterisks indicate the degree of statistical significance between measurements (**P < 0.01, ****P < 0.0001). (E) Representative U-ExM images (left) for PLK4 (magenta), CEP152 (cyan), and acetylated α-tubulin (green), and quantification (right) of PLK4 site number in DMSO (left panel) or centrinone (right panel) in RPE1 cells transfected with CEP152 siRNA, CEP192 siRNA, or both siRNAs for 48 h. All representative images are scaled independently to best represent phenotypes. The n refers to the total number of centrioles analyzed over at least three biological replicas per siRNA condition. Data are represented as mean ± SEM. Scale bar = 250 nm.
Figure S6.
CEP152/CEP63 frequently forms an incomplete ring around the base of parent centrioles. (A) Quantification of PLK4 and CEP152 distance from the microtubule wall in unduplicated (gray) or duplicated (red) parent centrioles in DLD1, U2OS, or HeLa cells. Each dot represents one centriole analyzed over one biological replicate per condition. Statistical significance was determined using two-way ANOVA with post-hoc analysis. Only significant results are indicated. Asterisks indicate the degree of statistical significance between measurements (****P < 0.0001). (B) Average fold change of PLK4, CEP152, and CEP192 in RPE1 cells after 48 h of either CEP152 or CEP192 siRNA. Statistical significance was determined using two-way ANOVA with post-hoc analysis. Only significant results are indicated. (C) Representative 3D U-ExM images of RPE1 cells untreated or treated with the indicated siRNAs showing the proximal and distal PLK4 foci highlighted in Fig. 3 E. (D) Graphs showing the percentage of centrioles with proximal and distal PLK4 foci in RPE1 cells treated with DMSO or centrione for 24 h. Data are represented as mean ± SEM. Scale bar = 250 nm.
To establish whether CEP152 and/or CEP192 were required for the localization of PLK4 to the individual sites on the wall of parent centrioles, we depleted CEP152, CEP192, or both by siRNA in RPE1 cells and examined CEP152 and PLK4 localization. While the knockdown of CEP152 had no impact on CEP192 localization, the depletion of CEP192 reduced CEP152 levels at the centriole by 45% (Fig. S6 B). The number of centrioles with no PLK4 sites increased from 32% in untreated cells to 63% in cells depleted of CEP152 (Fig. 3 E). While depletion of CEP192 did not lead to a significant increase in the number of centrioles with no detectable PLK4 foci, the codepletion of CEP152 and CEP192 increased the number of centrioles with no PLK4 sites to 78% (Fig. 3 E).
We also examined the effect of CEP152 and CEP192 depletion on PLK4 site number in cells treated for 24 h with centrinone. Depletion of either CEP152 or CEP192 resulted in a decrease in the number of sites PLK4 occupied at parent centrioles, while the depletion of both proteins produced a further decline in PLK4 site number. We noted that in cells treated with centrinone, 23% of of CEP152-depleted centrioles and 6% of CEP192/CEP152-depleted centrioles showed both proximal and distal PLK4 foci, which contrasted with the exclusively proximal localization of PLK4 in all other conditions (Fig. 3 E and Fig. S6 C). Both proximal and distal PLK4 foci were counted in our image analysis pipeline, and centrioles represented in this class may overrepresent PLK4 site number and are highlighted separately (Fig. 3 E). Taken together, these data suggest that CEP152 is the major receptor for the recruitment and positioning of PLK4 at individual sites on parent centrioles.
PLK4 activity drives turnover at the centriole
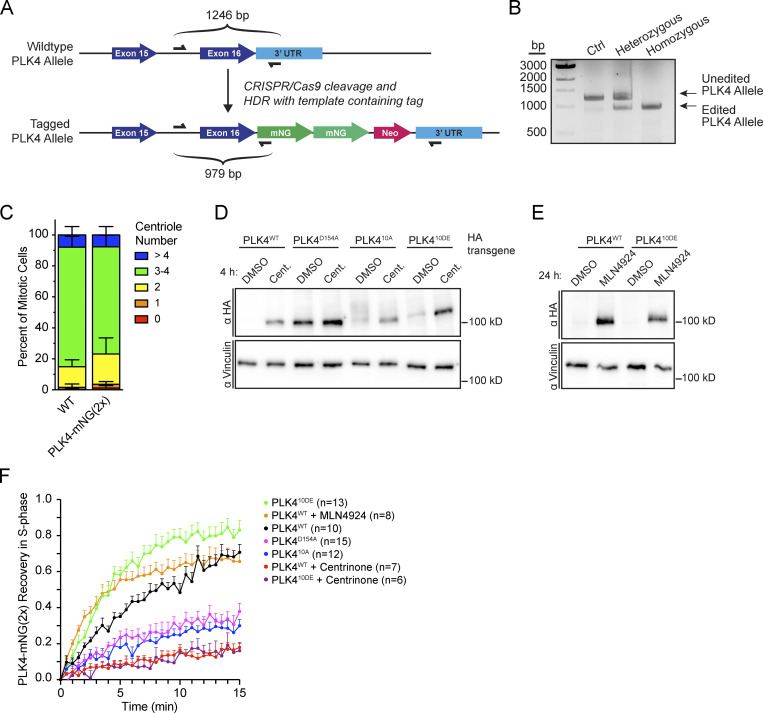
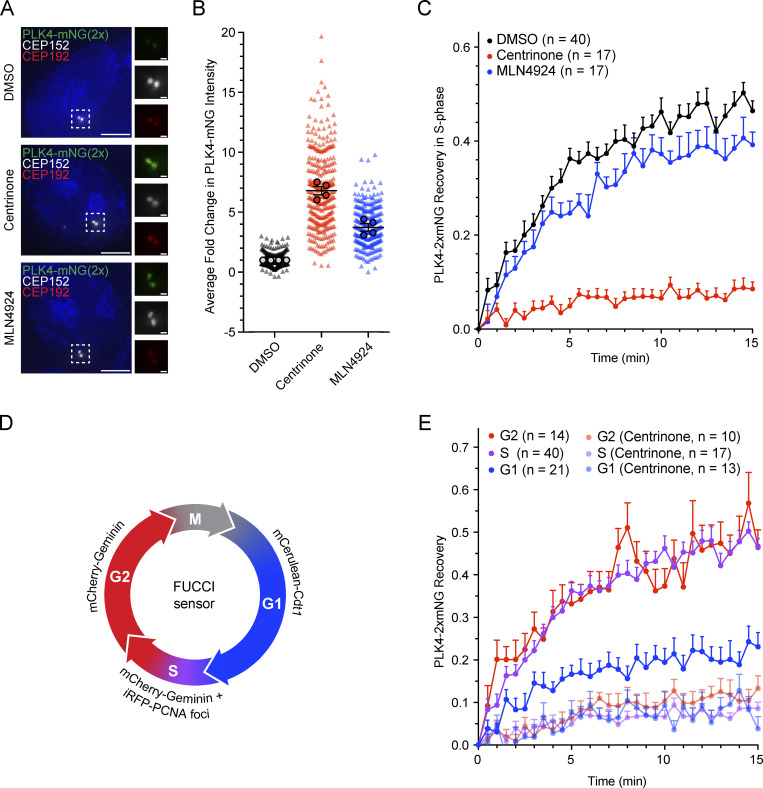
To directly assess how the dynamics of PLK4 contribute to site selection, we tagged the C-terminus of PLK4 with tandem (2×) mNeonGreen in DLD1 cells (Fig. S7 A). A homozygous PLK4-mNeonGreen DLD1 clone was identified (Fig. S7 B) and had a similar centriole content to parental DLD1 cells (Fig. S7 C). Treatment for 4 h with centrinone led to an approximately sevenfold increase in the abundance of PLK4-mNeonGreen at the centriole (Fig. 4, A and B). By contrast, treatment for 4 h with MLN4924 led to an approximately threefold increase in PLK4-mNeonGreen at the centriole (Fig. 4, A and B).
Figure S7.
Dynamics of kinase-active and kinase-inactive PLK4. (A) Scheme for the CRISPR/Cas9 tagging of PLK4 with a 2×-mNeonGreen (PLK4-mNG[2×]) and genotyping strategy in DLD1 cells. (B) Identification of a homozygous PLK4-mNG(2×) tagged clone. (C) Centriole counts for endogenously tagged PLK4-mNG(2×) and the parental DLD1 cell line. The graph is representative of the average phenotype of ∼300 centrioles over six biological replicates per cell line. (D) Western blot of PLK4-HA transgenes showing relative expression after 4 h of DMSO or centrinone. (E) Western blot of PLK4WT-HA or PLK410DE-HA transgenes showing relative expression after 24 h of DMSO or MLN4924 treatment. (F) FRAP analysis of PLK4WT after 24 h MLN4924 treatment (orange). The other data points are replotted from Fig. 5 C. n refers to the total number of cells analyzed. Data are represented as mean ± SEM. Scale bar = 250 nm. Source data are available for this figure: SourceData FS7.
Figure 4.
Inactive PLK4 stably binds to the parent centriole. (A) Immunofluorescence images of endogenously tagged PLK4-mNeonGreen (green), CEP152 (red), and CEP192 (white) in DLD1 cells treated with DMSO (top), centrinone (middle), or MLN4924 (bottom). (B) Quantification of the fold change in centriolar PLK4-mNeonGreen upon treatment with DMSO (black), centrinone (red), or MLN4924 (blue) for 4 h prior to fixation. Fold change was normalized to DMSO. Triangles mark individual cells and circles mark the average for each experiment. (C) FRAP analysis of PLK4-mNeonGreen in S-phase DLD1 cells displaying iRFP-PCNA puncta. Cells were treated with DMSO (black), centrinone (red), or MLN4924 (blue). n refers to the total number of cells analyzed. (D) Schematic of the reporters used to define the cell cycle stage in DLD1 cells expressing endogenously tagged PLK4-mNeonGreen. Drugs were added at least 1 h prior to FRAP. (E) FRAP analysis of PLK4-mNeonGreen in G1 (blue), S (purple), or G2 (red) phase treated with DMSO or centrinone. Recovery in centrinone was also measured. The S-phase FRAP is replotted from C. Drugs were added at least 1 h prior to FRAP. n refers to the total number of cells analyzed. Data are represented as mean ± SEM. Scale bar = 5 µm. Inset scale bar = 500 nm.
Given that inhibiting PLK4 led to a greater increase in the levels of centriole-localized PLK4-mNeonGreen than blocking PLK4 degradation, we hypothesized that inactive PLK4 might bind more stably to the centriole. Indeed, a previous study showed that the centriole turnover of overexpressed GFP-PLK4 was reduced following treatment with the PLK4 inhibitor centrinone (Yamamoto and Kitagawa, 2019). To evaluate the impact of kinase activity and proteolysis on the turnover of endogenous PLK4, we performed fluorescence recovery after photobleaching (FRAP) on centriole-localized PLK4-mNeonGreen. To avoid differences due to cell cycle stage, we stably expressed iRFP-PCNA and restricted our analysis to S-phase cells that contained bright iRFP-PCNA puncta. In DMSO-treated cells, 46% of PLK4-mNeonGreen was mobile and turned over at the centriole (Fig. 4 C, black, t½ = 3.2 min). However, only 9% of PLK4-mNeonGreen was turned over in centrinone-treated cells (Fig. 4 C, red). Importantly, the mobile pool of PLK4 was only marginally affected when SCF-directed proteolysis was inhibited with MLN4924 (Fig. 4 C, blue, 39% turnover, t½ = 3.4 min). Together, these observations show that PLK4 turnover at the centriole is largely under the control of PLK4 kinase activity and not proteasomal degradation.
We next examined if the turnover dynamics of centriolar PLK4 changed during the cell cycle. To distinguish cell-cycle phases, tagged PLK4-mNeonGreen cells expressing iRFP-PCNA were transduced with a virus expressing the fluorescence ubiquitination-based cell cycle indicator (FUCCI) reporters mCerulean-Cdt1 and mCherry-Geminin. Cells in G1 express mCerulean-Cdt1, G2 cells express mCherry-Geminin, and mid-S-phase cells display mCherry-Geminin and iRFP-PCNA puncta (Fig. 4 D). Using these markers, we used FRAP to define the turnover dynamics at each cell cycle phase (Fig. 4 E). The mobile fraction of PLK4 turnover was similar in S and G2 phase cells (S: 46% turnover, t½ = 3.2 min; G2: 47% turnover, t½ = 3.3 min), suggesting that PLK4 remains active throughout this time. By contrast, the mobile pool of PLK4 was substantially reduced in G1 phase (23%, t½ = 2.7 min), implying that PLK4 is less active in G1 (Fig. 4 E). As expected, PLK4 turnover was inhibited similarly by centrinone in all cell cycle phases (Fig. 4 E). Together, these data suggest that PLK4 remains active throughout the time procentrioles are assembled during S and G2 phases. This is consistent with work in Drosophila showing that PLK4 activity functions beyond the initiation of procentriole assembly to influence procentriole growth rate (Aydogan et al., 2018).
PLK4 centriole turnover requires Linker 1 (L1) autophosphorylation
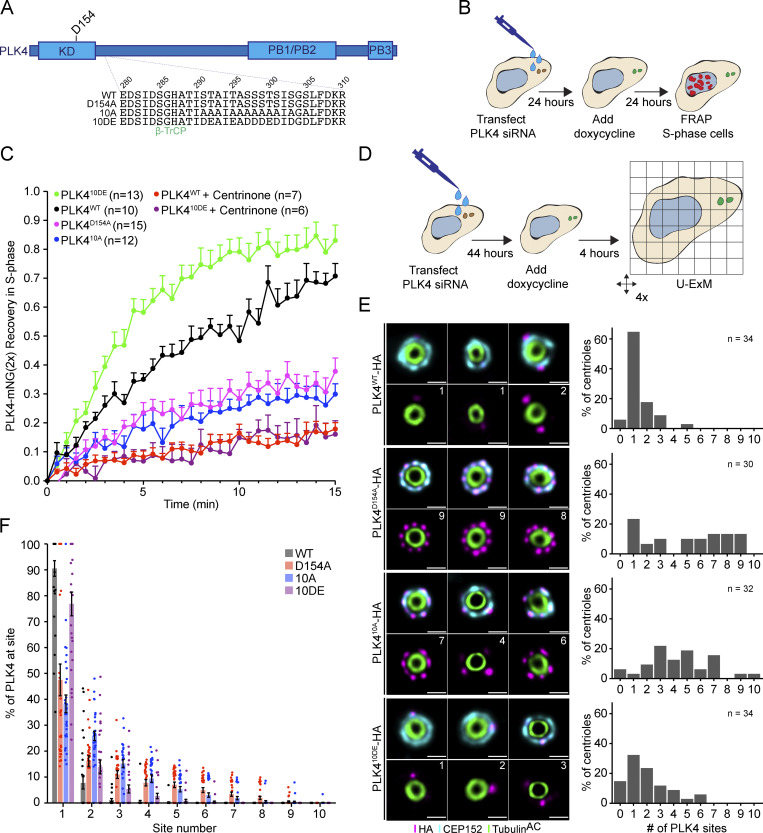
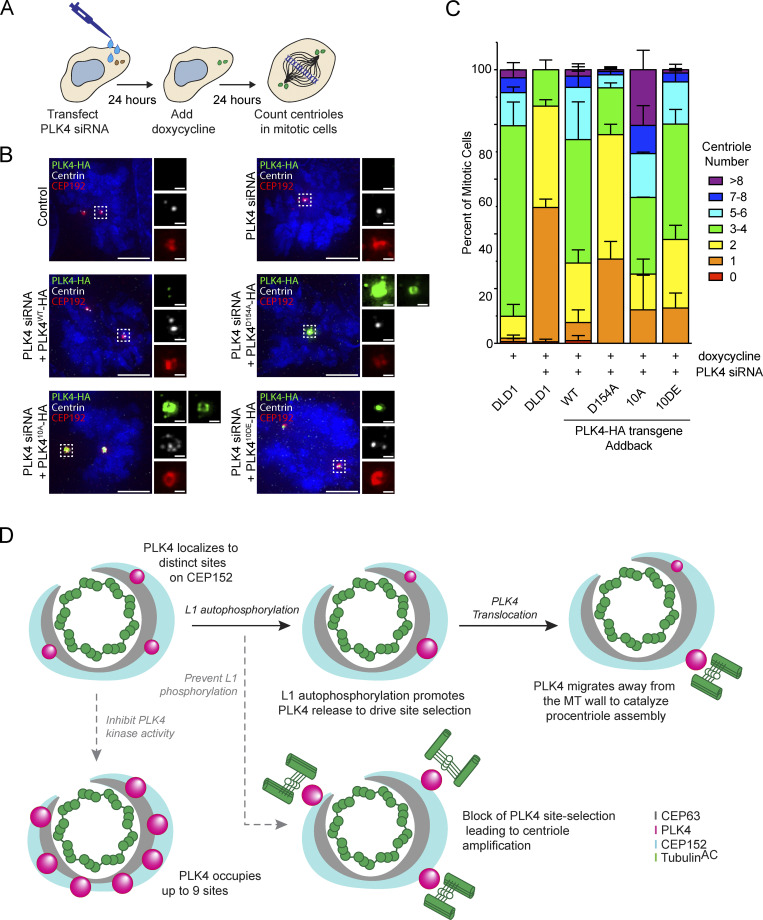
We next set out to investigate how PLK4 kinase activity promotes its turnover at the centriole. PLK4 regulates its stability through self-phosphorylation of a multiphosphodegron located in an unstructured linker following the kinase domain, hereafter the L1 region (Fig. 5 A; Holland et al., 2010). The multiphosphodegron consists of a βTrCP binding motif containing two phosphorylation sites, followed by 10 tightly clustered serine/threonine residues (S291, T292, T295, S297, S298, S299, T300, S301, S303, and S305) that have been reported to impact the centriolar dynamics of GFP-PLK4 (Fig. 5 A; Yamamoto and Kitagawa, 2019). To further study the effects of L1 phosphorylation, we generated isogenic DLD1 Flp-In cell lines carrying single-copy integrations of a doxycycline-inducible wild-type, kinase-dead (D154A), phospho-null (10A), and phospho-mimetic (10DE) PLK4-mNeonGreen transgenes. Endogenous PLK4 was depleted by siRNA, and 24 h later, expression of a siRNA-resistant PLK4-mNeonGreen transgene was induced with doxycycline (Fig. 5 B).
Figure 5.
PLK4 centriole turnover and site selection is driven by linker autophosphorylation. (A) Schematic of PLK4 showing amino acid 280–310 within the L1 linker domain. 10 S/T residues within this region were mutated to alanine (10A) or glutamic/aspartic Acid (10 DE). WT = wild-type, D154A = kinase-dead. KD, kinase domain; PB, Polo box. (B) Experimental design to deplete PLK4 and replace it with a siRNA-resistant transgene prior to analysis by FRAP. (C) FRAP analysis of DLD1 cells expressing the Plk4-mNeonGreen transgenes: WT (black), D154A (magenta), 10A (blue), and 10DE (green). Cells were treated with centrinone at least 1 h before performing FRAP. n refers to the total number of cells analyzed. (D) Experimental design to deplete PLK4 and replace it with a siRNA-resistant transgene prior to analysis by U-ExM. (E) Representative U-ExM images of PLK4 localization (left) and the number of PLK4 sites in DLD1 cells expressing siRNA-resistant transgenes of PLK4 WT, D154A, 10A, or 10DE. The n refers to the total number of centrioles analyzed from multiple punches over at least three biological replicas per cell line. (F) Graph showing the amount of PLK4-HA localized at individual sites on parent centrioles. All representative images are scaled independently to best represent phenotypes. Data are represented as mean ± SEM. Scale bar = 250 nm.
To examine the effect of L1 phosphorylation on PLK4 turnover, we performed FRAP on PLK4-mNeonGreen transgenes in S phase (Fig. 5 C). PLK4WT-mNeonGreen showed a higher mobile fraction (71% turnover, t½ = 7.5 min; Fig. 5 C, black) than kinase-inactive PLK4D154A-mNeonGreen (38% turnover, t½ = 4.1 min; Fig. 5 C, magenta). As expected, centrinone dramatically decreased the mobile fraction of PLK4WT-mNeonGreen (18% turnover, t½ = 8.4 min; Fig. 5 C, red). The recovery of phosphorylation-deficient PLK410A was similar to kinase-dead PLK4 (30% turnover, t½ = 6.1 min; Fig. 5 C, blue), while phosphomimetic PLK410DE-mNeonGreen recovered 83% after bleaching (Fig. 5 C, green). The turnover kinetics of PLK410DE (t½ = 3.0 min) were faster than that of PLK4WT (t½ = 7.5 min; Fig. 5 C). We conclude that phosphorylation of L1 is required for the release of PLK4 from the centriole.
To examine the effect of linker mutations on PLK4 abundance, we generated DLD1 Flp-In cell lines carrying doxycycline-inducible PLK4-HA transgenes and compared total cellular levels of PLK4WT-HA, PLK4D154A, PLK410A-HA, and PLK410DE-HA in cells treated with or without centrinone for 4 h. As expected, the levels of PLK4WT-HA were low but increased following treatment with centrinone, while the PLK4D154A-HA mutant was expressed at higher levels that were unchanged by centrinone addition (Fig. S7 D). PLK410A-HA was more abundant than PLK4WT-HA, and PLK410DE-HA showed a further increase in abundance. Both mutants increased abundance following centrinone treatment indicating that linker phosphorylation does not prevent kinase activity (Fig. S6 D).
To establish if the increased turnover kinetics of PLK410DE was due to an increase in PLK4 abundance, we treated DLD1 PLK4WT-mNeonGreen cells with MLN4924 for 24 h to increase the levels of active PLK4 prior to FRAP (Fig. S7 E). Stabilization of PLK4WT with MLN4924 increased the kinetics of PLK4 turnover to a level similar to PLK410DE (t½ = 2.1 and 3.0 min, respectively; Fig. S7 F). However, both untreated and MLN4924-treated PLK4WT had a similar mobile fraction (71% and 66% turnover respectively) that was less than we observed with PLK410DE (83% turnover) This suggests that the increased abundance of PLK410DE drives more rapid centriole binding while mimicking L1 phosphorylation reduces the binding affinity of PLK4 to the centriole, increasing the mobile pool of PLK4.
If phosphorylation of L1 is the only requirement for PLK4 release from the centriole, then inhibition of kinase activity would not affect PLK410DE-mNeonGreen turnover. However, treatment with centrinone greatly suppressed the turnover of PLK410DE-mNeonGreen (Fig. 5 C, purple, 16% turnover). This suggests that while phosphorylation of L1 is required to release PLK4 from the centriole, it is not sufficient and additional unidentified phosphorylation sites regulate the binding of PLK4 to the centriole.
Self-phosphorylation of PLK4’s unstructured linker drives site selection
To determine the impact of L1 phosphorylation on PLK4 localization, we depleted endogenous PLK4 by siRNA, and PLK4-HA transgenes were expressed for 24 h before processing cells for U-ExM (Fig. 5 D). 91% of PLK4WT-HA localized at the dominant focus on the CEP152 ring, while kinase-inactive PLK4D154A-HA was less concentrated and distributed over more sites on the CEP152 ring (Fig. 5, E and F). The PLK410A-HA and PLK4D154A-HA mutants showed reduced PLK4 focusing with only 38% and 47% of PLK4 localized at the dominant site, respectively. By contrast, 77% of PLK410DE-HA localized at the dominant site (Fig. 5, E and F). These results argue that phosphorylation of sites in the L1 linker is required to concentrate PLK4 at the dominant focus on the CEP152 ring. The increase in the cellular levels of PLK410A-HA and PLK410DE-HA in the presence of centrinone (Fig. S7 D) indicates that both mutants are kinase active, suggesting that the increased site occupancy number of PLK410A-HA is not due to a loss of kinase activity.
Phosphorylation of PLK4’s linker suppresses centriole amplification
Finally, we examined how the phosphorylation of the L1 linker contributes to centriole duplication. Endogenous PLK4 was depleted by siRNA and RNAi-resistant WT, D154A, 10A, and 10DE PLK4-HA transgenes expressed for 24 h (Fig. 6 A). Parental DLD1 cells had 69% of cells with three to four centrioles in mitosis (Fig. 6, B and C). As expected, depleting PLK4 led to a loss of centrioles in 87% of mitotic cells, and this was largely rescued by the expression of a PLK4WT-HA (45% three to four centrioles) but not kinase-dead PLK4D154A-HA (17% three to four centrioles; Fig. 6, B and C). Addback of the phosphorylation-deficient PLK410A-HA mutant led to dramatic centriole overduplication (28.0% 3–4 centrioles, 47% >5 centrioles, 20% >8 centrioles), indicating a failure of site selection that allowed for the production of multiple daughter centrioles. Expression of phosphorylation-proficient PLK410DE-HA mutant showed similar centriole content to cells expressing the wild-type PLK4 transgene (Fig. 6, B and C). Taken together, these data reveal that L1 phosphorylation is critical to drive assembly of a dominant PLK4 localization site to prevent centriole overduplication.
Figure 6.
Phosphorylation of the PLK4 linker suppresses centriole overduplication. (A) Experimental design to deplete PLK4 and replace it with a siRNA-resistant transgene to determine the role of L1 phosphorylation on centriole duplication. (B) Immunofluorescence images of cells 48 h after PLK4 siRNA treatment. Control DLD1 cells or cells expressing siRNA-resistant PLK4 WT, D154A, 10A, or 10DE transgenes were analyzed. (C) Quantification of centriole number in mitotic cells for conditions described in B. The graph is representative of the average phenotype of ∼150 centrioles over three biological replicates per cell line. (D) Model for PLK4 site selection. PLK4 accumulates at distinct sites on CEP152. Phosphorylation of the L1 region drives PLK4 release and its accumulation at the dominant site. At this site, PLK4 moves away from the microtube (MT) wall to promote procentriole assembly. Inhibition of kinase activity blocks PLK4 turnover at the centriole and leads to the formation of up to nine individual PLK4 sites. Blocking L1 phosphorylation inhibits PLK4 site selection and leads to centriole overduplication. Data are represented as mean ± SEM. Scale bar = 5 µm. Inset scale bar = 500 nm.
Discussion
In this study, we use U-ExM to show that PLK4 accumulates at distinct sites on a ring of CEP152 that encircles the proximal end of the parent centriole. Inactive PLK4 localized at up to nine sites on the CEP152 ring, mirroring the ninefold symmetry of the parent centriole microtubule wall (Fig. 6 D). These data suggest that procentriole assembly can take place at any one of nine sites on a parent centriole. Discrete sites of PLK4 localization were previously observed using stimulated emission depletion (STED) microscopy, but in this case, PLK4 was reported to exhibit a sixfold rotational symmetry around a parent centriole (Takao et al., 2019; Yoshiba et al., 2019). This difference likely arises because the prior measurements were made by averaging the angle between the nearest pairs of PLK4 sites on centrioles with incomplete site occupancy (Takao et al., 2019). An important question is how PLK4 occupies distinct sites on the CEP152 ring. One possibility is that CEP152 and CEP63 have a patterned symmetry that is imparted onto the recruitment of PLK4. Indeed, CEP152 was proposed to have a 12-fold symmetry at parent centrioles (Takao et al., 2019). In addition, the self-association properties of PLK4 could facilitate the coalescence of PLK4 into distinct sites after its initial recruitment (Montenegro Gouveia et al., 2018; Park et al., 2019; Yamamoto and Kitagawa, 2019). Further work will be needed to define how the symmetry of parent centrioles is directed toward the recruitment of PLK4.
PLK4 has been proposed to accumulate as a ring around parent centrioles that resolves into a single dot when STIL is recruited to the procentriole at the start of S phase (Kim et al., 2013; Ohta et al., 2014). In cultured Drosophila cells, PLK4 was briefly detected as a ring during late mitosis and rapidly resolved into a single focus in early G1 (Aydogan et al., 2020; Dzhindzhev et al., 2017). In our hands, complete rings of PLK4 with nine sites were never observed in unperturbed conditions, including unduplicated parent centrioles or early G1 phase cells marked by a cytokinetic bridge. This is somewhat surprising, given that the turnover of PLK4 was reduced in G1 phase cells. However, clear rings of PLK4 with up to nine sites were assembled but only after prolonged periods (>8 h of centrinone treatment) of kinase inhibition. Taken together, our data suggest that both a loss of kinase activity and accumulation of excessive PLK4 protein are both likely to be required for full ring occupancy. While we cannot exclude the transient assembly of PLK4 rings in unperturbed cycling human cells, our data suggest that if complete rings of PLK4 do exist, they are very unstable and rapidly driven toward asymmetry in PLK4 recruitment.
Although PLK4 occupied a variable number of sites on parent centrioles, the majority of PLK4 localized at the dominant site on CEP152/CEP63 both before and after procentriole assembly. This is consistent with recent results that showed PLK4’s localization exhibits complex spatial patterns in U2OS cells (Takao et al., 2019; Yoshiba et al., 2019). In all the cell lines we analyzed, the dominant site of PLK4 marked the location of procentriole assembly. We propose that minor sites of PLK4 remaining on parent centrioles are unable to compete effectively for procentriole assembly factors. Consistently, preventing the phosphorylation of a disordered linker region disrupted PLK4 focusing at the dominant site and resulted in centriole overduplication (Fig. 6 D). Thus, phosphorylation of the PLK4 linker concentrates PLK4 at the dominant site, thereby preserving the numerical fidelity of centriole duplication. Further, as PLK4 remains active through G2 phase, self-phosphorylation may be required to maintain a single site of PLK4 after centriole biogenesis has initiated.
Self-phosphorylation has been shown to control several aspects of PLK4’s behavior, including degradation, activation, and self-association (Cunha-Ferreira et al., 2009, 2013; Guderian et al., 2010; Holland et al., 2010, 2012; Klebba et al., 2013; Lopes et al., 2015; Montenegro Gouveia et al., 2018; Moyer et al., 2015; Park et al., 2019; Rogers et al., 2009; Yamamoto and Kitagawa, 2019). In accordance with prior work, self-phosphorylation of the PLK4 linker is required for the turnover of PLK4 at the centriole. Previous work showed that the phosphorylation of multiple sites in the linker of PLK4 contributes to proteolytic turnover of the kinase (Holland et al., 2010; Klebba et al., 2013), possibly by weakening self-association and allowing access to the βTrCP phosphodegron (Park et al., 2019; Yamamoto and Kitagawa, 2019). In our experiments, alanine or phosphomimicking mutations in the linker had opposite effects on the centriole turnover of PLK4 and centriole amplification. This argues that self-phosphorylation can independently regulate the centriolar turnover and proteolysis of PLK4.
Although a phosphomimicking PLK4 linker mutant turned over rapidly at the centriole, the addition of centrinone blocked this effect. This suggests that phosphorylation of the PLK4 linker is necessary, but not sufficient, to promote centriole turnover. A possible model to explain these data is that phosphorylation of the linker is required to license phosphorylation of a second site (or group of sites) that antagonizes the binding of PLK4 to the centriole. In this case, preventing linker phosphorylation would also prevent phosphorylation of the second site(s), leading to stable binding, while phosphomimetic mutations in the linker would permit phosphorylation of the second site(s), leading to rapid turnover. Treatment with centrinone is expected to prevent phosphorylation of the second site(s), enabling a return to stable binding for the phosphomimicking PLK4 linker mutant. Importantly, the second site(s) of phosphorylation could lie within PLK4 itself or another protein, such as PLK4’s centriole receptor CEP152. Indeed, phosphorylation of Asl/CEP152 has been proposed to reduce its affinity for PLK4 to control the timing of centriole biogenesis in Drosophila embryos (Aydogan et al., 2020).
A surprising finding from our work was that the CEP152/CEP63 “ring” at the base of the parent centriole is frequently discontinuous. It would be interesting to test if this asymmetry in CEP152 distribution can explain the bias in the procentriole assembly position observed when examining the asymmetrically localized centriole protein LRRCC1 (Gaudin et al., 2022). The cause of the incomplete CEP152/CEP63 rings remains unclear. One possibility is that CEP152/CEP63 is locally removed from the wall of the mature parent centriole during centriole disengagement. This could ensure that the previous duplication site on the mature parent centriole is not used for procentriole assembly in the next cell cycle. CEP152/CEP63 would presumably need to be replaced at the site of the break later in the cell cycle to ensure parent centrioles remain competent for repeated rounds of duplication.
We further explored the relative contribution of the two PLK4 receptors, CEP152 and CEP192, to PLK4 recruitment. Several lines of evidence argue that CEP152 plays a major role in the recruitment of PLK4 to distinct sites on the wall of parent centrioles. First, PLK4 colocalized with CEP152 at unduplicated parent centrioles. Second, depletion of CEP152 reduced the number of PLK4 sites in the presence or absence of centrinone and shifted PLK4 localization closer to the wall of the parent centriole, consistent with prior work (Park et al., 2014). Finally, PLK4 was not localized at regions of the centriole where there were breaks in the CEP152 ring. The role of CEP192 in recruiting PLK4 was less clear as depletion of CEP192 also led to a substantial reduction in the centriolar levels of CEP152. Further work will be needed to identify how CEP152 and CEP192 control PLK4 localization and function to ensure faithful control of centriole biogenesis.
Our observations may have utility for mathematical models that have been developed to explain how PLK4 concentrates at a single site to promote procentriole assembly. One model presented by Leda et al. predicts that active PLK4 binds more stably to the centriole. This contrasts with our observations and previously reported FRAP data showing that inhibiting PLK4 leads to more stable binding at the centriole (Leda et al., 2018; Yamamoto and Kitagawa, 2019). A second model proposed by Takao et al. assumed that PLK4 can localize to one of six discrete sites, but we show that PLK4 can bind up to nine potential sites at the parent centriole (Takao et al., 2019). A recent analysis by Wilmott et al. showed that the underlying assumptions from both the Leda et al. (2018) and Takao et al. (2019) models can be described with similar terms as Turing systems (Wilmott et al., 2023 Preprint). Incorporating additional experimental measurements of PLK4 dynamics will strengthen mathematical models to drive a deeper understanding of centriole duplication.
Materials and methods
Cell culture
DLD1 cells (a gift from Stephen Taylor, University of Manchester, Manchester, UK) and HeLa cells (a gift from Carol Greider; University of California, Santa Cruz, Santa Cruz, CA, USA) were grown in Dulbecco’s Modified Eagle Medium (DMEM, Cellgro; Corning) supplemented with 10% FB Essence Serum (FBE; Seradigm), 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM L-glutamine. U2OS cells (a gift from Takanari Inoue; Johns Hopkins, Baltimore, MD, USA) were grown in DMEM supplemented with 10% FBE (Seradigm; VWR),100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM L-glutamine. RPE1 cells (a gift from Prasad Jallepalli; Memorial Sloan Kettering Cancer Center, New York, NY, USA) were grown in DMEM:F12 50:50 medium (Cellgro; Corning) supplemented with 10% FBE, 100 U/ml penicillin, 100 U/ml streptomycin, 2 mM L-glutamine, and 0.35% sodium bicarbonate. All cell lines were maintained at 37°C in 5% CO2 and 21% O2 and routinely checked for mycoplasma contamination. Cell lines were validated via short tandem repeats genotyping.
Generation of stable cell lines
The generation of Flp-In T-REx DLD1 cells (a gift from Stephen Taylor, University of Manchester, Manchester, UK) was performed as previously described (Moyer et al., 2015; Phan et al., 2022). PLK4 constructs were cloned into the pcDNA5 FRT/TO plasmid (Life Technologies). Cell lines were generated by cotransfection of a pcDNA5 FRT/TO plasmid and POG44 Flp-recombinase plasmid into DLD1 Flp-In T-REx cells using X-tremeGene HP (Millipore Sigma). Cells were selected with 400 µg/ml hygromycin B until colony formation (at least 2 wk). Isogeneic colonies were combined and expanded to generate the final cell line. Expression of PLK4 transgenes was induced with 1 µg/ml doxycycline.
To generate iRFP-PCNA expressing Flp-In T-REx DLD1 cells, iRFP and PCNA (#57994; Addgene) were cloned into a FUGW lentiviral vector (Jon Alder, University of Pittsburgh, Pittsburgh, PA, USA). DLD1 cells were transduced with iRFP-PCNA lentivirus and monoclonal cell lines were isolated by limiting dilution. To generate iRFP-PCNA + FUCCI expressing DLD1 cells, iRFP-PCNA expressing DLD1 cells were transduced with a FUCCI lentivirus (pSR1344, a gift of Sergi Regot; Johns Hopkins, Baltimore, MD, USA) and monoclonal cell lines were isolated by limiting dilution.
Lentiviral production
Production and transduction of lentivirus was performed as previously described (Evans et al., 2021). The day prior to transfection, 3 × 106 HEK293FT cells were seeded onto poly-D-lysine–coated 10-cm dishes. The next day, the media was replaced with DMEM + 1% FBS. For each transfection, 600 μl of OptiMEM (Thermo Fisher Scientific) was aliquoted into two 1.5-ml Eppendorf tubes. In the first tube, 4.5 µg of lentiviral vector, 6 µg of psPAX2, and 1.5 µg of pMD2.G were diluted in DMEM. In the second tube, 35 μl polyethylenimine (1 µg/ml; Sigma-Aldrich) was diluted in DMEM. The tubes were incubated for 5 min before the contents were mixed and incubated for 20–30 min. The contents were added dropwise to the media in a 10-cm dish. After 48 h, the media was filtered through a 45-µM syringe filter and flash-frozen until use. 1 d before transduction, 2 × 105 DLD1 cells were seeded into a single well of a 6-well plate. The following day, the media on the cells was replaced with 1 ml of complete media plus 1 ml of virus and 10 µg/ml of polybrene (Sigma-Aldrich).
Drug treatment
Doxycycline was dissolved in H2O and used at a final concentration of 1 µg/ml. Centrinone (Tocris) was dissolved in DMSO and used at a final concentration of 500 nM. MLN4924 (Sigma-Aldrich) was dissolved in DMSO and used at a final concentration of 10 µM.
siRNA experiments
For experiments involving FRAP or U-ExM, siRNA transfection was performed in a 6-well plate 1 d after seeding (∼2.0 × 105 cells/well). For experiments involving widefield microscopy, transfection was performed in a 12-well plate 1 d after seeding (∼0.8 × 105 cells/well). RNAiMAX transfection reagent (Life Technologies) was used to transfect 100 nM siRNA per well, and 24 h later, cells were reseeded and PLK4 transgene expression was induced with 1 µg/ml doxycycline. The inducible PLK4-mNeonGreen transgenes contained silent mutations to render them resistant to PLK4 siRNA (HSS116551; Life Technologies). After 24 h, cells were either imaged or processed for widefield or U-ExM. For knockdown of PLK4 receptors, CEP152 and CEP192, 100 nM siRNA was transfected as described above and incubated for 48 h. Both siRNAs were purchased from Dharmacon: CEP152 sense 5′-GCGGAUCCAACUGGAAAUCUAUU-3′, CEP192 sense 5’-GGAAGACAUUUUCAUCUCUUU-3′.
Generation of tagged PLK4 cell lines using CRISPR/Cas9
To generate the PLK4-2x mNeonGreen tagged cell line, 2 × 105 Flp-In T-REx DLD1, near infrared fluorescent protein–proliferating cell nuclear antigen (iRFP-PCNA) cells were seeded into a well of a 6-well plate. The next day, the cells were transfected with 100 ng of px459 plasmid (#62988; Addgene) containing an sgRNA (5′-AATCAATGAAAATTAGGAGT-3′) targeting close to the STOP codon of the PLK4 gene and 800 ng of pUC19 containing repair template. The repair template consisted of a 432 bp 5′ homology arm, a 2× mNeonGreen-T2A-Neomycin cassette, and a 341 bp 3′ homology arm. Each homology arm recognized the region immediately upstream or downstream of the STOP codon and was synthesized by PCR of DLD1 genomic DNA. At day 5, cells were selected with Neomycin (400 µg/ml) for 2 wk and monoclonal lines were isolated by limiting dilution. The zygosity of each clone was determined using PCR with the following primers: PLK4 Forward: 5′-GTGCTCCAGAAAGTGGAAGG-3′, PLK4 Reverse: 5′-GGTTTTGTCCATGATTCTCAAC-3′, mNeonGreen Reverse: 5′-CCATTGATGCTTCCGAAAAT-3′.
The PLK4-HA line was generated with the same protocol as the mNeonGreen line. 2 × 105 Flp-In T-REx DLD1 cells were seeded into a well of a 6-well plate. The next day, the cells were transfected with 100 ng of px459 plasmid (#62988; Addgene) containing an sgRNA (5′-AATCAATGAAAATTAGGAGT-3′) targeting close to the STOP codon of the PLK4 gene and 800 ng of pUC19 containing repair template with homology arms identical to the ones described above. We did not genotype the monoclonal HA line.
Immunoblotting
Immunoblotting was performed as previously described (Moyer and Holland, 2015). Protein samples were separated by size using SDS-PAGE and transferred onto nitrocellulose membranes with a Trans-Blot Turbo Transfer System (BioRad). The following antibodies were used: HA (#12CA5, 1:1,000; Roche) and vinculin (Clone 7F9, sc73614, 1:1,000; Santa Cruz). Blots were blocked in 1× PBS, 0.1% Triton X-100, and 3% BSA, and washed with 1× PBS and 0.1% Triton X-100. HRP-conjugated secondary antibodies (mouse and rat, 7076S and 7077S; Cell Signaling) were used.
Immunofluorescence staining and imaging
Cells were grown on 18-mm coverslips and fixed in 100% ice-cold methanol at −20°C for 10 min. Cells were quickly washed 3× with PBST (PBS with 0.1% Triton X-100) before blocking. Cells were blocked in blocking buffer (2.5% FBS, 200 mM glycine, 0.1% Triton X-100 in PBS) for 1 h at room temperature or overnight at 4°C. Cells were subsequently incubated in primary antibody diluted in blocking buffer for 1 h followed by three quick washes in PBST. After washing, cells were incubated in secondary antibodies diluted in blocking buffer for 1 h. After 3× quick PBST washes, cells were counterstained with DAPI and mounted in ProLong Gold antifade mountant (Invitrogen). The following primary antibodies were used: PLK4 #01 (aa 510–970, Moyer and Holland, 2015, 1:250), PLK4 #03 (aa 564–579, this study, 1:250), CEP152 (LoMastro et al., 2022, 1:1,000), CEP192 (Moyer and Holland, 2019, 1:1,000), HA (1:1,000; Roche), and Centrin2 (Moyer and Holland, 2019, 1:1,000). Alexa Fluor conjugated secondary antibodies used were donkey anti-rat 488 (#A21208; Thermo Fisher Scientific), donkey anti-mouse 488 (#A21202; Thermo Fisher Scientific), donkey anti-mouse 555 (#A31570; Thermo Fisher Scientific), rat anti-mouse 647 (#406617; BioLegend), donkey anti-mouse 647 (#A32787; Thermo Fisher Scientific), donkey anti-rabbit 488 (#A21206; Thermo Fisher Scientific), donkey anti-rabbit 555 (#A31572; Thermo Fisher Scientific), and donkey anti-rabbit 647 (#A31573; Thermo Fisher Scientific).
Immunofluorescence images were collected with a DeltaVision Elite system (GE Healthcare) controlling a scientific CMOS camera (pco.edge 5.5). Images were acquired at room temperature (25°C) using an Olympus 60×/1.42 NA or an Olympus 100×/1.4 NA oil objective with Applied Precision immersion oil (n = 1.516) in 0.2 µm z-sections. Acquisition parameters were controlled through Soft-WoRx suite (GE Healthcare). For analysis of fold intensity change of PLK4, a semiautomated FIJI macro was utilized to measure background subtracted intensity density.
FRAP
Cells were seeded in a 4-chamber, 35-mm glass-bottom culture dish (Greiner) and maintained at 37°C in a complete medium with 5% CO2 using an environmental control station (Tokai Hit). Centrinone or MLN4924 was added at least 1 h prior. FRAP was performed on a Leica Sp8 inverted microscope using a 63×/1.4 NA objective. Centriolar PLK4-mNeonGreen(2×) was bleached with five pulses of 80% 488-laser intensity (1.050 s exposure). Three z-series of 13 planes (0.5-µm steps) were acquired before bleaching to establish pre-bleach fluorescence. Fluorescence recovery was collected as a 13-plane z-series (0.5-µm steps) every 30 s for 15 min. Fluorescence recovery was measured using ImageJ. The first postbleach intensity measurement was subtracted from all pre- and postbleach values. Values were normalized to reflect the percent recovery of the bleached signal. Kinetic parameters were calculated using GraphPad Prism using a one-phase association nonlinear regression.
U-ExM
Expansion microscopy was performed based on a previously published protocol (Gambarotto et al., 2019). Briefly, cells were seeded on a 25-mm glass coverslip in a single well of a 6-well plate. Cells were rinsed with 1× PBS and 2 ml of the anchor solution (1× PBS formaldehyde/acrylamide mixture at a ratio of 1.4%/2% for RPE1 and U2OS cells and 0.7%/1% for DLD1 and HeLa cells) was added. The plate was wrapped in parafilm and incubated at 37°C for 5 h or overnight. A coverslip was placed on top of a 1-ml drop of precooled monomer solution (23% sodium acrylate [wt/vol], 10% acrylamide, 0.1% N,N’-methylenbisacrylamide, 1× PBS, 0.5% N,N,N′,N′-Tetramethylethylenediamine, and 0.5% ammonium persulfate) sitting on top of parafilm in a humid chamber. The humified chamber was incubated on ice for 15 (DLD1 and HeLa) or 30 min (RPE1 and U2OS) and then moved to 37°C for 1 h. Following gel polymerization, a 4-mm biopsy punch was used to create several punches from each coverslip. Punches were transferred to a 50-ml falcon tube containing denaturation buffer (200 mM SDS, 200 mM NaCL, and 50 mM Tris) and incubated for 15 min at room temperature with gentle agitation. The container was transferred to a water bath at 95°C for 90 min with gentle agitation every 20 min. Punches were washed in water 3× for 20 min each and kept overnight at room temperature with gentle agitation for the first round of gel expansion. Expanded punches were transferred to 1× PBS for 1 h at room temperature and blocked with 2% BSA in PBS at 37°C for 45 min. This step reduced the gel size to ∼2× the original 4-mm width. Primary antibodies were diluted in 2% BSA in PBS and stained for 3 h at 37°C with gentle agitation. After washing the punches 3× for 10 min at room temperature in PBST (0.1% Tween20), secondary antibodies and DAPI were added for 3 h at 37°C. This was followed by another round of washes with PBST (3× for 10 min at room temperature) and three washes with water for 20 min each. Gels were incubated in water overnight at room temperature with gentle agitation for the second round of gel expansion. The expansion factor was determined by measuring the final size of the gel using calipers and dividing by 4 (the initial size of the gel after using the 4-mm biopsy punch). Scale bars were calculated using the expansion factor for each punch.
The following primary antibodies were used: acetylated α-tubulin (6-11 B-1, 1:1,000; Santa Cruz), PLK4 #01 (aa 510–970, Moyer and Holland, 2015, 1:500), PLK4 #03 (aa 564–579, this study, 1:500), CEP152 (LoMastro et al., 2022, 1:500), CEP164 (ABE2621, 1:500; EMD Millipore), CEP63 (16268-1-AP, 1:500; Proteintech), and HA (1:250; Roche). Secondary donkey antibodies conjugated with Alexa Fluor 488, 555, or 650 (all from Thermo Fisher Scientific) were used.
Expanded punches were imaged on poly-L-lysine–coated 35-mm glass-bottom culture dish (Cellvis) using a Leica Sp8 inverted microscope. Punches were imaged using a 63×/1.4 NA oil immersion objective with 0.2 µm z-step size and a pixel size of 22.55 nm. Final images were generated using Lightning processing with the contrast enhancement set to 0.5.
Automated analysis pipeline
ImageJ was used to analyze top-view U-ExM centriole images stained for PLK4, acetylated tubulin, and CEP152. An oval was manually annotated centered on the CEP152 ring using composite images. Integrated density profile traces of both PLK4 and CEP152 signals were measured along the circumference of the oval with a width of 10 pixels. All further analysis of images was conducted in R. In brief, PLK4 sites were determined by identifying local maxima in smoothed PLK4 signal intensity traces. Local minima were used to define PLK4 site boundaries. The cumulative signal for each site (AUC) was then measured for each site from the raw data. Sites containing <2% of total AUC for the centriole or with a maximum intensity <1,500 U were rejected. The angle between sites was then measured for each site across all centrioles. Finally, sites were ranked by AUC in descending order. CEP152 occupancy was determined as the proportion of the CEP152 profile trace >35% of the maximum CEP152 signal for each centriole. Centrioles were excluded from analysis if they shifted and moved through the z-stack, were broken, or rotated to a degree where a clear CEP152 path could not be drawn.
Statistical analysis
All statistics were done using Graphpad Prism. For all experiments, averages and standard error of the mean (SEM) are plotted. For comparisons, we performed two-way ANOVA with post hoc analysis assuming a normal distribution. Normality was not formally tested. Information regarding statistics of an individual experiment is provided in the figure legends.
Online supplemental material
Fig. S1 extends the widefield fluorescence analysis of PLK4 in DMSO and centrinone with a second PLK4 antibody. Fig. S2 details the generation of an endogenously tagged PLK4-HA tool. Fig. S3 contains histograms depicting the angle measured between two adjacent peaks of PLK4 at individual site occupancies rather than the pooled analysis done in Fig. 2 E. Fig. S4 is an extension of the observation that PLK4 occupancy is correlated with CEP152 availability. Fig. S5 is an analysis of PLK4 site occupancy in unduplicated and duplicated centrioles. Fig. S6 is an analysis of CEP63 and CEP152 around the parent centriole in different cell lines. Fig. S7 explores the dynamics between kinase active and inactive PLK4. Data S1, Data S2, Data S3, Data S4, Data S5, and Data S6 contain the source data relevant to Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, and Fig. 6, respectively. Data S7, Data S8, Data S9, Data S10, Data S11, Data S12, and Data S13 contain the source data relevant to Fig. S1, Fig. S2, Fig. S3, Fig. S4, Fig. S5, Fig. S6, and Fig. S7, respectively.
Supplementary Material
contains the source data relevant to Fig. 1.
contains the source data relevant to Fig. 2.
contains the source data relevant to Fig. 3.
contains the source data relevant to Fig. 4.
contains the source data relevant to Fig. 5.
contains the source data relevant to Fig. 6.
contains the source data relevant to Fig. S1.
contains the source data relevant to Fig. S2.
contains the source data relevant to Fig. S3.
contains the source data relevant to Fig. S4.
contains the source data relevant to Fig. S5.
contains the source data relevant to Fig. S6.
contains the source data relevant to Fig. S7.
is the source file for Fig. S7.
Acknowledgments
We thank Cayla Jewett for her helpful comments.
A.J. Holland acknowledges funding by the National Institutes of Health (R01 GM133897, R01 GM114119, and R01 CA266199).
Author contributions: Conceptualization: P. Scott, A. Curinha, and A.J. Holland; Data acquisition: P. Scott, A. Curinha, and C. Gliech; Data analysis: P. Scott, A. Curinha, C. Gliech, and A.J. Holland. Funding acquisition: A.J. Holland; Project administration: A.J. Holland; Supervision: A.J. Holland; Writing—original draft: A.J. Holland and P. Scott; Writing—review & editing: A.J. Holland, P. Scott, A. Curinha, and C. Gliech.
Data availability
Correspondence and requests for data and materials should be addressed to A.J. Holland (aholland@jhmi.edu).
References
- Aydogan, M.G., Steinacker T.L., Mofatteh M., Wilmott Z.M., Zhou F.Y., Gartenmann L., Wainman A., Saurya S., Novak Z.A., Wong S.S., et al. 2020. An autonomous oscillation times and executes centriole biogenesis. Cell. 181:1566–1581 e1527. 10.1016/j.cell.2020.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydogan, M.G., Wainman A., Saurya S., Steinacker T.L., Caballe A., Novak Z.A., Baumbach J., Muschalik N., and Raff J.W.. 2018. A homeostatic clock sets daughter centriole size in flies. J. Cell Biol. 217:1233–1248. 10.1083/jcb.201801014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto, R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., and Raff J.W.. 2008. Centrosome amplification can initiate tumorigenesis in flies. Cell. 133:1032–1042. 10.1016/j.cell.2008.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow, D.K., and Holland A.J.. 2019. Mechanism and regulation of centriole and cilium biogenesis. Annu. Rev. Biochem. 88:691–724. 10.1146/annurev-biochem-013118-111153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, N.J., Marjanović M., Lüders J., Stracker T.H., and Costanzo V.. 2013. Cep63 and cep152 cooperate to ensure centriole duplication. PLoS One. 8:e69986. 10.1371/journal.pone.0069986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali, P.L., Pütz M., and Gergely F.. 2014. Small organelle, big responsibility: The role of centrosomes in development and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130468. 10.1098/rstb.2013.0468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizmecioglu, O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiss U., Antony C., and Hoffmann I.. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191:731–739. 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho, P.A., Bury L., Shahbazi M.N., Liakath-Ali K., Tate P.H., Wormald S., Hindley C.J., Huch M., Archer J., Skarnes W.C., et al. 2015. Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol. 5:150209. 10.1098/rsob.150209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira, I., Bento I., Pimenta-Marques A., Jana S.C., Lince-Faria M., Duarte P., Borrego-Pinto J., Gilberto S., Amado T., Brito D., et al. 2013. Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr. Biol. 23:2245–2254. 10.1016/j.cub.2013.09.037 [DOI] [PubMed] [Google Scholar]
- Cunha-Ferreira, I., Rodrigues-Martins A., Bento I., Riparbelli M., Zhang W., Laue E., Callaini G., Glover D.M., and Bettencourt-Dias M.. 2009. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 19:43–49. 10.1016/j.cub.2008.11.037 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev, N.S., Tzolovsky G., Lipinszki Z., Abdelaziz M., Debski J., Dadlez M., and Glover D.M.. 2017. Two-step phosphorylation of Ana2 by Plk4 is required for the sequential loading of Ana2 and Sas6 to initiate procentriole formation. Open Biol. 7:170247. 10.1098/rsob.170247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev, N.S., Tzolovsky G., Lipinszki Z., Schneider S., Lattao R., Fu J., Debski J., Dadlez M., and Glover D.M.. 2014. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 24:2526–2532. 10.1016/j.cub.2014.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, L.T., Anglen T., Scott P., Lukasik K., Loncarek J., and Holland A.J.. 2021. ANKRD26 recruits PIDD1 to centriolar distal appendages to activate the PIDDosome following centrosome amplification. EMBO J. 40:e105106. 10.15252/embj.2020105106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambarotto, D., Hamel V., and Guichard P.. 2021. Ultrastructure expansion microscopy (U-ExM). Methods Cell Biol. 161:57–81. 10.1016/bs.mcb.2020.05.006 [DOI] [PubMed] [Google Scholar]
- Gambarotto, D., Zwettler F.U., Le Guennec M., Schmidt-Cernohorska M., Fortun D., Borgers S., Heine J., Schloetel J.G., Reuss M., Unser M., et al. 2019. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods. 16:71–74. 10.1038/s41592-018-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin, N., Martin Gil P., Boumendjel M., Ershov D., Pioche-Durieu C., Bouix M., Delobelle Q., Maniscalco L., Phan T.B.N., Heyer V., et al. 2022. Evolutionary conservation of centriole rotational asymmetry in the human centrosome. Elife. 11:e72382. 10.7554/eLife.72382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy, P. 2012. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13:425–435. 10.1038/nrm3373 [DOI] [PubMed] [Google Scholar]
- Guderian, G., Westendorf J., Uldschmid A., and Nigg E.A.. 2010. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J. Cell Sci. 123:2163–2169. 10.1242/jcs.068502 [DOI] [PubMed] [Google Scholar]
- Hatch, E.M., Kulukian A., Holland A.J., Cleveland D.W., and Stearns T.. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191:721–729. 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, A.J., Fachinetti D., Zhu Q., Bauer M., Verma I.M., Nigg E.A., and Cleveland D.W.. 2012. The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev. 26:2684–2689. 10.1101/gad.207027.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, A.J., Lan W., Niessen S., Hoover H., and Cleveland D.W.. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188:191–198. 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.S., Park J.E., Shukla A., Choi S., Murugan R.N., Lee J.H., Ahn M., Rhee K., Bang J.K., Kim B.Y., et al. 2013. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl. Acad. Sci. USA. 110:E4849–E4857. 10.1073/pnas.1319656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba, J.E., Buster D.W., Nguyen A.L., Swatkoski S., Gucek M., Rusan N.M., and Rogers G.C.. 2013. Polo-like kinase 4 autodestructs by generating its Slimb-binding phosphodegron. Curr. Biol. 23:2255–2261. 10.1016/j.cub.2013.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz, A.S., Bärenz F., Richter K.T., and Hoffmann I.. 2015. Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol. Open. 4:370–377. 10.1242/bio.201411023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leda, M., Holland A.J., and Goryachev A.B.. 2018. Autoamplification and competition drive symmetry breaking: Initiation of centriole duplication by the PLK4-STIL network. iScience. 8:222–235. 10.1016/j.isci.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, M.S., Bakker B., Boeckx B., Moyett J., Lu J., Vitre B., Spierings D.C., Lansdorp P.M., Cleveland D.W., Lambrechts D., et al. 2017. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev. Cell. 40:313–322.e5. 10.1016/j.devcel.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, M.S., and Holland A.J.. 2018. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 32:620–638. 10.1101/gad.314351.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoMastro, G.M., Drown C.G., Maryniak A.L., Jewett C.E., Strong M.A., and Holland A.J.. 2022. PLK4 drives centriole amplification and apical surface area expansion in multiciliated cells. ELife. 11:e80643. 10.7554/eLife.80643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, C.A.M., Jana S.C., Cunha-Ferreira I., Zitouni S., Bento I., Duarte P., Gilberto S., Freixo F., Guerrero A., Francia M., et al. 2015. PLK4 trans-autoactivation controls centriole biogenesis in space. Dev. Cell. 35:222–235. 10.1016/j.devcel.2015.09.020 [DOI] [PubMed] [Google Scholar]
- McLamarrah, T.A., Speed S.K., Ryniawec J.M., Buster D.W., Fagerstrom C.J., Galletta B.J., Rusan N.M., and Rogers G.C.. 2020. A molecular mechanism for the procentriole recruitment of Ana2. J. Cell Biol. 219:e201905172. 10.1083/jcb.201905172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro Gouveia, S., Zitouni S., Kong D., Duarte P., Ferreira Gomes B., Sousa A.L., Tranfield E.M., Hyman A., Loncarek J., and Bettencourt-Dias M.. 2018. PLK4 is a microtubule-associated protein that self-assembles promoting de novo MTOC formation. J. Cell Sci. 132:jcs219501. 10.1242/jcs.219501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer, T.C., Clutario K.M., Lambrus B.G., Daggubati V., and Holland A.J.. 2015. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 209:863–878. 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer, T.C., and Holland A.J.. 2015. Generation of a conditional analog-sensitive kinase in human cells using CRISPR/Cas9-mediated genome engineering. Methods Cell Biol. 129:19–36. 10.1016/bs.mcb.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer, T.C., and Holland A.J.. 2019. PLK4 promotes centriole duplication by phosphorylating STIL to link the procentriole cartwheel to the microtubule wall. Elife. 8:e46054. 10.7554/eLife.46054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, E.A., and Holland A.J.. 2018. Once and only once: Mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19:297–312. 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, E.A., and Raff J.W.. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Ohta, M., Ashikawa T., Nozaki Y., Kozuka-Hata H., Goto H., Inagaki M., Oyama M., and Kitagawa D.. 2014. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 5:5267. 10.1038/ncomms6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.E., Zhang L., Bang J.K., Andresson T., DiMaio F., and Lee K.S.. 2019. Phase separation of Polo-like kinase 4 by autoactivation and clustering drives centriole biogenesis. Nat. Commun. 10:4959. 10.1038/s41467-019-12619-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.Y., Park J.E., Kim T.S., Kim J.H., Kwak M.J., Ku B., Tian L., Murugan R.N., Ahn M., Komiya S., et al. 2014. Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis. Nat. Struct. Mol. Biol. 21:696–703. 10.1038/nsmb.2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, T.P., Boatwright C.A., Drown C.G., Skinner M.W., Strong M.A., Jordan P.W., and Holland A.J.. 2022. Upstream open reading frames control PLK4 translation and centriole duplication in primordial germ cells. Genes Dev. 36:718–736. 10.1101/gad.349604.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, G.C., Rusan N.M., Roberts D.M., Peifer M., and Rogers S.L.. 2009. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184:225–239. 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serçin, Ö., Larsimont J.C., Karambelas A.E., Marthiens V., Moers V., Boeckx B., Le Mercier M., Lambrechts D., Basto R., and Blanpain C.. 2016. Transient PLK4 overexpression accelerates tumorigenesis in p53-deficient epidermis. Nat. Cell Biol. 18:100–110. 10.1038/ncb3270 [DOI] [PubMed] [Google Scholar]
- Sonnen, K.F., Gabryjonczyk A.M., Anselm E., Stierhof Y.D., and Nigg E.A.. 2013. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 126:3223–3233. 10.1242/jcs.129502 [DOI] [PubMed] [Google Scholar]
- Soucy, T.A., Smith P.G., Milhollen M.A., Berger A.J., Gavin J.M., Adhikari S., Brownell J.E., Burke K.E., Cardin D.P., Critchley S., et al. 2009. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 458:732–736. 10.1038/nature07884 [DOI] [PubMed] [Google Scholar]
- Takao, D., Yamamoto S., and Kitagawa D.. 2019. A theory of centriole duplication based on self-organized spatial pattern formation. J. Cell Biol. 218:3537–3547. 10.1083/jcb.201904156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmott, Z.M., Goriely A., and Raff J.W.. 2023. A simple Turing reaction-diffusion model can explain how mother centrioles break symmetry to generate a single daughter. bioRxiv. 10.1101/2023.02.02.526828 (Preprint posted February 2 2023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. 2015. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 348:1155–1160. 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S., and Kitagawa D.. 2019. Self-organization of Plk4 regulates symmetry breaking in centriole duplication. Nat. Commun. 10:1810. 10.1038/s41467-019-09847-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba, S., Tsuchiya Y., Ohta M., Gupta A., Shiratsuchi G., Nozaki Y., Ashikawa T., Fujiwara T., Natsume T., Kanemaki M.T., and Kitagawa D.. 2019. HsSAS-6-dependent cartwheel assembly ensures stabilization of centriole intermediates. J. Cell Sci. 132:jcs217521. 10.1242/jcs.217521 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains the source data relevant to Fig. 1.
contains the source data relevant to Fig. 2.
contains the source data relevant to Fig. 3.
contains the source data relevant to Fig. 4.
contains the source data relevant to Fig. 5.
contains the source data relevant to Fig. 6.
contains the source data relevant to Fig. S1.
contains the source data relevant to Fig. S2.
contains the source data relevant to Fig. S3.
contains the source data relevant to Fig. S4.
contains the source data relevant to Fig. S5.
contains the source data relevant to Fig. S6.
contains the source data relevant to Fig. S7.
is the source file for Fig. S7.
Data Availability Statement
Correspondence and requests for data and materials should be addressed to A.J. Holland (aholland@jhmi.edu).